SUMMARY
To protect the genome, cells have evolved a diverse set of pathways designed to sense, signal, and repair multiple types of DNA damage. To assess the degree of coordination and crosstalk among these pathways, we systematically mapped changes in the cell's genetic network across a panel of different DNA-damaging agents, resulting in ~1,800,000 differential measurements. Each agent was associated with a distinct interaction pattern, which, unlike single-mutant phenotypes or gene expression data, has high statistical power to pinpoint the specific repair mechanisms at work. The agent-specific networks revealed roles for the histone acetyltranferase Rtt109 in the mutagenic bypass of DNA lesions and the neddylation machinery in cell-cycle regulation and genome stability, while the network induced by multiple agents implicates Irc21, an uncharacterized protein, in checkpoint control and DNA repair. Our multiconditional genetic interaction map provides a unique resource that identifies agent-specific and general DNA damage response pathways.
INTRODUCTION
Failure of cells to respond to DNA damage is associated with genome instability and the onset of diseases such as cancer (Jackson and Bartek, 2009). To combat DNA damage, cells have evolved an intricate system, known as the DNA damage response (DDR), which senses DNA lesions and activates downstream pathways such as chromatin remodeling, cell-cycle checkpoints, and DNA repair (Ciccia and Elledge, 2010). Many studies have sought to use genome-scale technologies to define and map the DDR, including systematic phenotyping of single mutants (Hillenmeyer et al., 2008), RNA interference (RNAi) screening (Paulsen et al., 2009), and gene expression profiling (Caba et al., 2005; Travesa et al., 2012).
While these strategies have met with success in identifying new DDR genes, they have raised a number of questions with regard to how DDR pathways coordinate with one another. For instance, the initial view of DNA damage checkpoints (DDCs) was as a collection of pathways that coordinates cell-cycle progression with DNA repair (Zhou and Elledge, 2000). However, recent studies have implicated checkpoints in other processes, including transcriptional regulation and apoptosis, suggesting that there is extensive crosstalk between such processes during the DDR (Ciccia and Elledge, 2010). Much of this crosstalk may be dependent on the nature of the DNA lesion. For example, the Bloom syndrome helicase interacts with components of the replication checkpoint (e.g., Mrc1/Claspin) when replication forks stall, whereas it cooperates with factors of the DDC (e.g., Rad17) after double-stranded break (DSB) formation (Bjergbaek et al., 2005). An important next step is therefore to understand how functional interconnections between pathways are formed and altered in response to various genotoxic insults.
To address this issue, we turned to a recently developed interaction mapping methodology called differential epistasis mapping, or dE-MAP (Bandyopadhyay et al., 2010). This approach is based on synthetic genetic array technology which enables rapid measurement of genetic interactions (Tong and Boone, 2006), i.e., combinations of two or more mutations which lead to a dramatic departure in growth rate when compared to the product of the individual mutant growth rates (Collins et al., 2006). In the dE-MAP approach, synthetic genetic arrays are used to measure genetic interactions under standard conditions as well as under perturbations of interest and, by comparison of the resulting networks, interactions that are altered in response to perturbation can be quantitatively assessed. These “differential” genetic interactions reveal a unique view of cellular processes and their interconnections under specific stress conditions (Bandyopadhyay et al., 2010).
Here, we systematically map the genetic modules and networks induced by distinct types of DNA damage. Based on this map of both agent-specific and general differential interactions, we validate a number of pathways and factors involved in the DDR. Finally, we demonstrate that differential interaction mapping across a panel of treatments is a powerful and general approach for disentangling a web of distinct but interrelated signaling processes.
RESULTS & DISCUSSION
Mapping Differential Genetic Networks across Distinct Types of DNA Damage
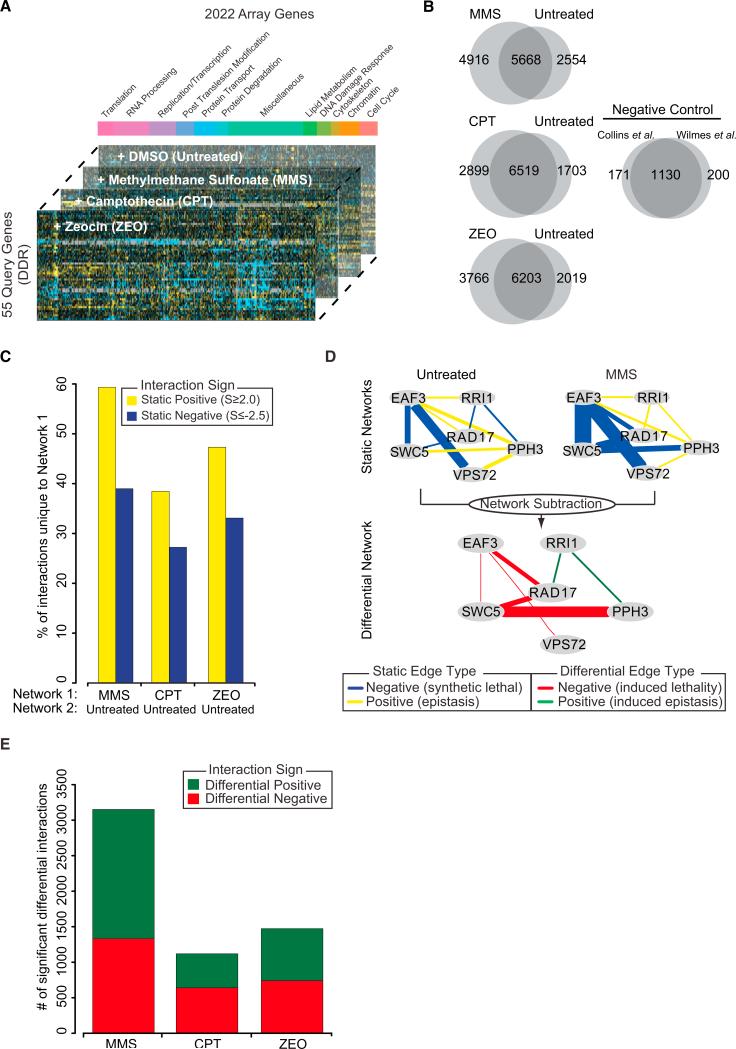
We constructed a dE-MAP in the budding yeast S. cerevisiae centered on the measurement of all possible interactions between a set of 55 query genes and a set of 2,022 array genes (Figure 1A). The 55 queries were chosen to cover pathways that define the DDR including representatives of distinct DNA repair processes (Table S1 available online). The array genes included all of the queries and, to explore crosstalk between DNA repair and other cellular functions, genes involved in cell-cycle regulation, chromatin organization, replication, transcription, and protein transport (Table S1). Double-mutant strains were constructed for each query-array gene pairing (Experimental Procedures) with synthetic gene array technology (Tong and Boone, 2006). In brief, each query strain carrying a gene deletion is mated against an array of strains (in this case 2,022 gene deletion strains), resulting in heterozygous diploids. After sporulation and a series of selection steps, we obtain haploid double-gene deletion strains. Growth rates are determined by measurement of colony sizes after 48 hr in standard conditions (untreated) and in the presence of three chemical agents that induce distinct types of DNA damage: the DNA alkylating agent methyl methanesulfonate (MMS), the topoisomerase I inhibitor camptothecin (CPT), and the DNA intercalating agent zeocin (ZEO). Colony size measurements are normalized and statistically analyzed to assign each double mutant a quantitative S score (Collins et al., 2006), which reports on the extent to which it grew better (positive S score) or worse (negative S score) than expected. Several routine quality control measures were employed to ensure a high-quality data set (Figure S1). In total, the genetic interaction map contains quantitative scores for 97,578 pairs of genes (Table S2).
Figure 1. Overview of the Multionditional Differential Network.
(A) Design of the differential genetic interaction screen. The stacked bar plot illustrates the functional breakdown of array genes.
(B) Overlap in static interactions (S ≥ 2.0, S ≤ –2.5) between treated and untreated conditions. The negative control represents the overlap between previously published networks measured in untreated conditions.
(C) The percentage of positive and negative interactions unique to the treated network (Network 1) when compared to the untreated network (Network 2).
(D) Schematic overview of how differential networks are derived by examining the difference between static treated and untreated genetic networks. The thickness of the edge scales with the magnitude of the genetic interaction.
(E) Number of significant positive and negative differential interactions uncovered in each condition.
A comparison of the set of significant positive and negative genetic interactions (S ≥ 2.0 or ≤ –2.5) (Collins et al., 2006) revealed numerous differences between treated and untreated conditions (Figure 1B). On average, 48% of positive interactions and 33% of negative interactions were unique to the treated networks, indicating the presence of DNA damage-induced epistasis and synthetic lethality (Figure 1C). To identify which of these differences were significant, we used a previously published scoring methodology to assess the difference in S score for each gene pair before versus after treatment (Bandyopadhyay et al., 2010). A p value of significance was assigned by comparison of this quantitative difference to a null distribution of differences derived from replicate genetic interaction screens from the same condition. We refer to this network as the “differential” genetic network since it is derived from the difference between two static networks (Figure 1D). At p ≤ 0.002 (FDR ≈12.3%; Supplemental Experimental Procedures), we identified a total of 3,032 significant differential positive and 2,712 differential negative interactions (i.e., interaction becomes either more positive or negative under DNA damage) across all three conditions (Figure 1E).
Differential Interactions Discriminate among Different DNA Damage Responses
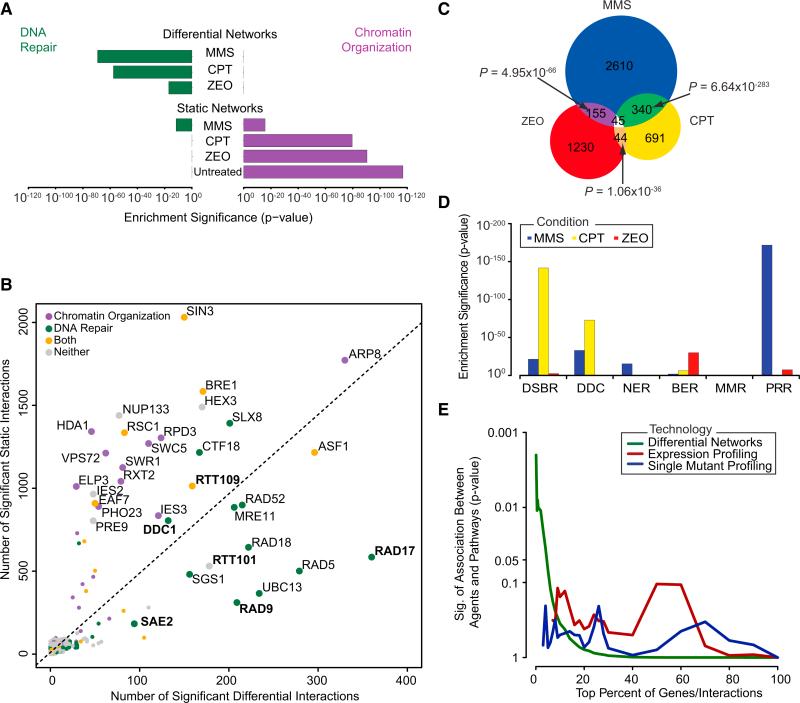
We next examined all networks, differential and static, for their ability to highlight genes that function in the DDR (Supplemental Experimental Procedures). All three differential networks had high enrichment for interactions with known DNA repair genes, while static networks had much less or no enrichment in this regard (Figures 2A and S2A–S2C). Instead, all four static networks showed the strongest enrichment for genes involved in chromatin organization, as had been noted previously (Bandyopadhyay et al., 2010). Moreover, 15 of the top 20 differential interaction “hubs” (genes with the greatest number of interactions) were annotated to DNA repair, whereas in static networks they were largely associated with chromatin organization (Figure 2B). Thus, in contrast to static interactions, differential interactions measured across a shift in conditions tend to highlight gene functions related to that condition.
Figure 2. Differential Networks Reveal Specific Pathways Induced by Different Types of DNA Damage.
(A) The significance of enrichment for interactions with genes that function in DNA repair (green bars) or chromatin organization (purple bars) is plotted for all networks.
(B) The total number of significant static (y axis) and differential (x axis) interactions across all conditions for each gene considered in this study.
(C) Overlap in significant differential interactions induced by each agent.
(D) Enrichment of differential interactions containing genes involved in six major DDR pathways: DSB repair (DSBR), DNA damage checkpoint (DDC), nucleotide excision repair (NER), base excision repair (BER), mismatch repair (MMR), and postreplication repair (PRR).
(E) The significance of association between agents and DDR pathways is computed with differential networks, single-mutant fitness, and differential gene expression across a range of thresholds.
See also Figure S2 and Table S3.
Despite the enrichment for DNA repair genes across all differential networks, we found that these networks were strikingly different from one another. Few interactions (584) were induced by more than one agent, and only 45 interactions were induced by all agents (Figure 2C). In contrast, a control experiment indicated much better agreement between replicate differential networks generated in response to the same agent (Figure S2D). These findings were corroborated in an alternate analysis in which we hierarchically clustered all 55 query genes based on either their static (Figure S2E) or differential (Figure S2F) interaction profiles. Clustering with the differential metric, as opposed to the static interaction scores, lead to a clear separation between the different DNA-damaging agents, underlining the stark differences among the differential genetic interactions induced by these compounds.
To determine whether the distinct interaction patterns were indicative of specific DDR mechanisms, we examined the differential networks for enrichment of interactions with genes involved in six major DDR pathways (Figure 2D and Table S3). The CPT network was highly enriched for DSB repair and DDC functions, consistent with the known mechanism of action of CPT that stabilizes DNA topoisomerase 1-DNA complexes. During S phase, the replication machinery collides with these structures leading to DSB formation (Ulukan and Swaan, 2002). The MMS network displayed only a mild enrichment (p = 0.009) for interactions with components of base excision repair, an unexpected result given that MMS modifies guanine and adenine bases leading to base mispairing and replication fork blocks (Lundin et al., 2005). However, replication-blocking lesions can be bypassed by postreplication repair pathways such as translesion synthesis and DNA damage avoidance or, in case of fork collapse and subsequent chromosome breakage, are counteracted by DSB repair pathways (Chang and Cimprich, 2009). All these pathways showed strong enrichment in the MMS network (Figure 2D). Finally, the ZEO network was enriched for interactions with genes involved in base excision repair and postreplication repair rather than for genes involved in DSB repair (p = 0.002), suggesting that our ZEO treatment generates abasic sites rather than DNA strand breaks, consistent with the mode of action of this compound at lower concentrations (Wu et al., 2008).
These functional enrichments suggest that the differential networks help decode the particular combination of DDR pathways underlying the response to each agent. To test this, we measured the statistical association between the three agents and the six major DDR pathways as revealed by differential interactions (Supplemental Experimental Procedures). In contrast to functional enrichment, statistical association measures the extent to which interactions induced by each agent implicate a set of genes that discriminates among the six pathways (i.e., genes which associate with some DNA repair functions but not others). We found that differential interactions were indeed able to elicit a significant association between agents and pathways, especially for the top 5% of interactions (Figure 2E). Moreover, differential interactions performed very favorably at this task in comparison to single-mutant fitness (Hillenmeyer et al., 2008) or differential messenger RNA (mRNA)-expression profiles (Caba et al., 2005; Travesa et al., 2012) gathered for the same or similar agents. Neither of these data types was able to significantly link DNA-damaging agents to particular responses (Figure 2E). The better performance of differential networks may lie in the greater sample size afforded by this technology. Whereas single-mutant fitness and gene expression profiling are limited to measurements of individual genes (181 genes across the six DDR pathways), the differential networks cover interactions between DDR genes and over 30% of the yeast genome (39,973 interactions in total). Thus, while single-mutant and gene expression profiling are adept at defining high-level biological functions (e.g., DNA repair), differential genetic interactions can tease apart a very specific set of (partially overlapping) mechanisms.
Sae2 and Pph3 Cooperate to Promote DNA Repair and Checkpoint Recovery
Hubs identified in static genetic networks have a number of cellular properties. For example, static genetic hubs identified in both S. cerevisiae and S. pombe are more likely to exhibit a severe single-mutant defect and display a greater degree of pleiotropy (Costanzo et al., 2010; Ryan et al., 2012). We found that these features were also present among differential genetic interaction hubs. Genes with a higher number of differential interactions in response to MMS, CPT, or ZEO were more likely to exhibit a single-mutant sensitivity to that particular compound (Figure S3A) and were more likely to be essential for growth in response to numerous drugs, indicating that they may be more pleiotropic (Figure S3B) and help to interconnect the various biological processes required for the DDR.
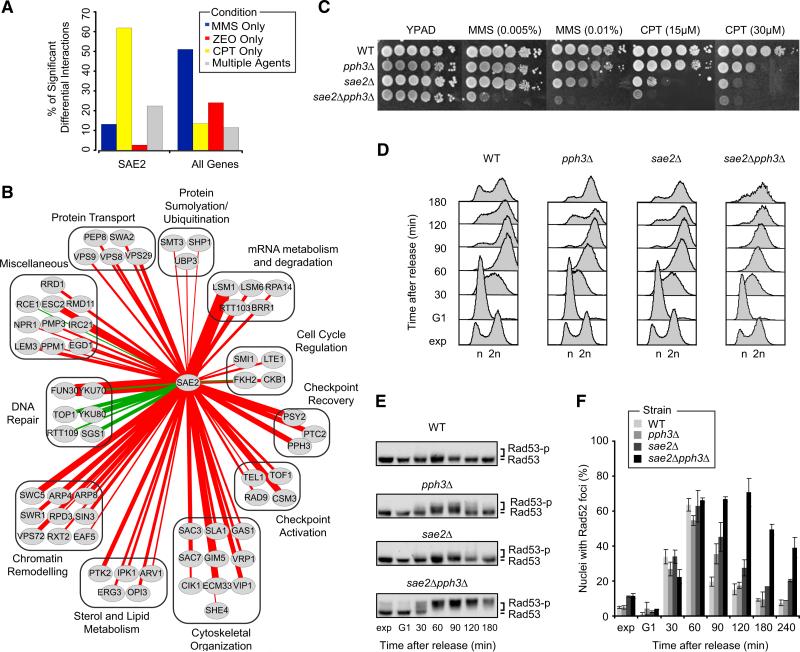
To explore this, we examined one of the hubs identified in our differential network, SAE2, which encodes the homolog of the human endonuclease CtIP, known for its role in the processing of DSBs into 3′ single-stranded tails (Mimitou and Symington, 2008). Consistent with this role, we found that the majority of SAE2's interactions (~60%) were induced specifically by the DSB-inducing agent CPT (Figure 3A). Moreover, SAE2 interacted positively with many DNA repair genes, including SGS1, TOP1, YKU70, and YKU80 (Figure 3B). Surprisingly, we also observed negative interactions between SAE2 and genes encoding components of the PP4 complex (Pph3-Psy2) not only in response to CPT, but also to MMS (Figure 3C). The PP4 complex is required for dephosphorylation of the major checkpoint kinase Rad53 and subsequent recovery from DNA damage-induced cell cycle arrest (O'Neill et al., 2007), suggesting that Sae2 may work in parallel with PP4 in checkpoint deactivation.
Figure 3. Sae2 and Pph3 Cooperate in DNA Repair and Checkpoint Recovery.
(A) Percentage of SAE2's significant differential interactions in response to MMS, CPT, ZEO, or multiple agents. As a control, the average percentage of significant differential interactions in each category across all genes is shown.
(B) Network of all 64 significant positive (green edges) or negative (red edges) differential interactions with SAE2 in response to CPT. Thickness of the edge scales with significance of the interaction.
(C) Serial dilutions (10-fold) of cells were grown on YPAD + MMS or CPT.
(D) Cells were arrested in G1 and transiently exposed to MMS (30 min, 0.02% MMS), then released in fresh medium and analyzed by FACS at the indicated time points.
(E) Western blot analysis of Rad53 phosphorylation in cells from (D).
(F) As in (D), except that cells expressing Rad52-YFP were used. Images were taken at the indicated time points and scored for Rad52-YFP foci. At least 100 nuclei were analyzed per strain and per time point. Data represent the mean ± 1 SD from three independent experiments.
See also Figure S3.
We therefore profiled the pph3Δ and sae2Δ single mutants as well as the sae2Δpph3Δ double mutant for passage through the cell cycle after a transient arrest and MMS exposure in G1. The double mutant displayed a markedly slower progression through S phase, suggesting that cells lacking Sae2 and Pph3 fail to efficiently deactivate the checkpoint (Figure 3D). Indeed, hyperphosphorylation of Rad53 was found to persist for ~3 hr after exposure to MMS in sae2Δpph3Δ cells, whereas either single mutant showed complete dephosphorylation at two hours (Figure 3E).
To test whether the checkpoint hyperactivation in sae2Δ pph3Δ cells is due to a repair defect, we monitored the assembly of the Rad52 repair protein into DNA damage-induced subnuclear foci, which are thought to represent active repair centers (Lisby et al., 2001). While all strains were proficient in assembling the repair machinery (maximum number of Rad52 foci comparable at one hour after treatment; Figure 3F), 4 hr after MMS exposure both wild-type and single mutants had largely completed repair. In contrast, a large proportion of the sae2Δ pph3Δ cells still displayed Rad52 foci, indicating a reduced repair capacity (Figure 3F). Thus, the checkpoint hyperactivation in sae2Δpph3Δ cells could stem from a repair defect. Since exposure to MMS can block replication forks and lead to fork collapse, the data suggest that Sae2 and Pph3 have synergistic roles in promoting checkpoint recovery from MMS-induced replication fork damage. Although further work will be required to resolve how Sae2 and Pph3 cooperate to orchestrate repair and checkpoint recovery, this example illustrates the power of our differential network analysis in identifying connections between different DDR factors.
Neddylation Affects Cell Cycle Control and Genome Integrity
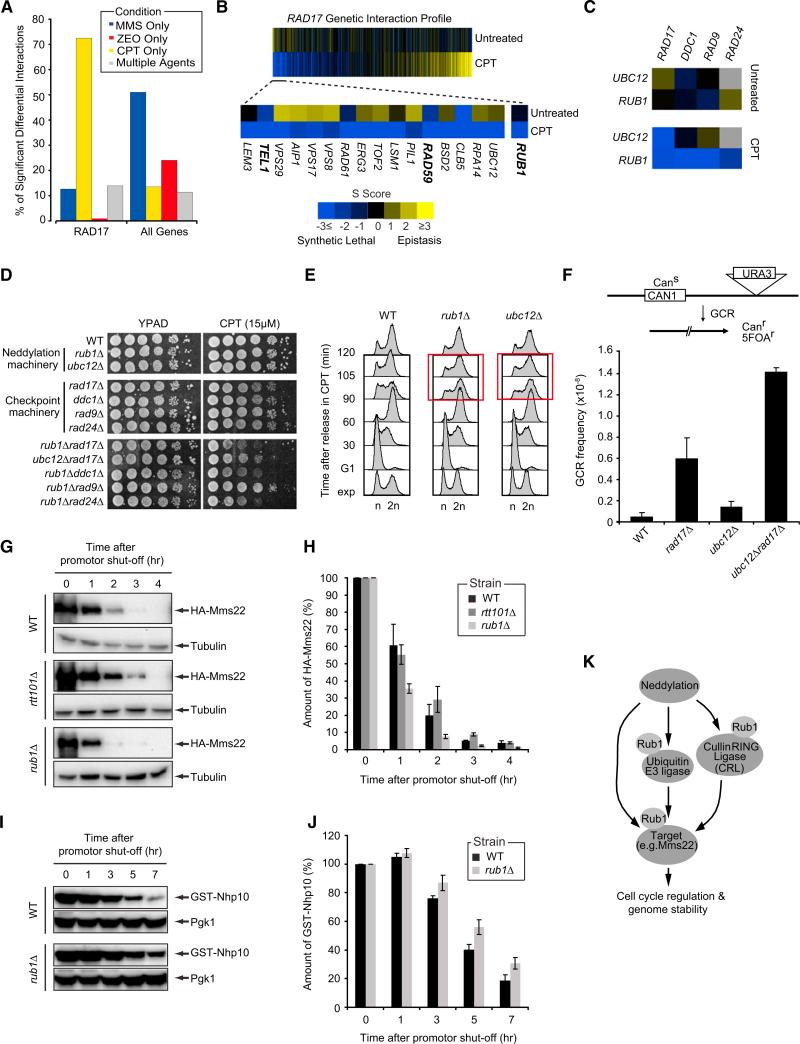
Another major hub in our network was RAD17 (Figure 2B), encoding a component of the 9-1-1 checkpoint complex which is recruited to DSB sites to activate the Mec1 signaling pathway, resulting in cell-cycle arrest and repair (Zhou and Elledge, 2000). Consistent with the role of RAD17 in the DSB response, we found that the majority of its interactions were induced in response to CPT (73%, Figure 4A). Moreover, the CPT-induced genetic interaction profile of RAD17 revealed strong differential negative interactions with DSB repair genes (RAD59) and checkpoint regulators, such as TEL1 (Figure 4B), which is consistent with reports showing that Tel1 functions parallel to Rad17 to regulate checkpoint activation after DSBs (Usui et al., 2001).
Figure 4. Neddylation Regulates Cell-Cycle Progression after DNA Damage and Preserves Genome Integrity.
(A) Percentage of RAD17's significant differential genetic interactions in response to MMS, CPT, ZEO, or multiple agents.
(B) CPT-induced genetic interaction profile for RAD17 sorted (left to right) in order of most differential negative to most differential positive. A subset of the top differential negative interactions is also shown.
(C) Genetic interactions between components of neddylation and DNA damage checkpoint pathways.
(D) Serial dilutions (10-fold) of cells were grown on YPAD + CPT.
(E) Cells were arrested in G1, released in medium + 50 μM CPT and analyzed by FACS at the indicated time points.
(F) GCR frequencies were determined as described in the Experimental Procedures.
(G) GAL1-HA-Mms22 expression was induced for 3 hr in medium + galactose. Cells were released in glucose to shut off expression after which HA-Mms22 levels were monitored by western blot analysis.
(H) Bar plot showing the rate of HA-Mms22 protein degradation in cells from (G). HA-Mms22 protein levels were quantified and normalized to tubulin. The ratio at the start of shutoff was set to 100%.
(I) As in (G), except that GAL1-GST-Nhp10 expression was monitored.
(J) As in (H), except that GST-Nhp10 levels were quantified and normalized to Pgk1.
(K) Schematic illustrating proposed mechanisms by which neddylation regulates cell-cycle progression and genome stability. See the main text for details.
All data represent the mean ± 1 SD deviation of at least three independent experiments. See also Figure S4.
Two additional genes, RUB1 and UBC12, which encode key components of the neddylation machinery, displayed strong differential negative interactions with RAD17 (Figure 4B). Neddylation is a process by which the Rub1 protein (NEDD8 in humans) is conjugated to target proteins in a cascade of reactions involving multiple enzymes, including Ubc12, in a manner analogous to ubiquitylation and SUMOylation (Liakopoulos et al., 1998). In support of a potential link between neddylation and checkpoint pathways, the CPT network revealed a number of additional negative interactions between RUB1/UBC12 and other checkpoint genes, including DDC1, RAD9, and RAD24 (Figure 4C), which we confirmed via spot dilution assays. This suggests that neddylation and DDC cooperate to promote cell survival after exposure to CPT (Figure 4D).
Given the genetic interaction between neddylation and DDCs, we reasoned that neddylation may affect cell-cycle regulation and assessed rub1Δ and ubc12Δ mutants for their progression through the cell cycle in the presence of CPT. After arrest in G1 and release into medium containing CPT, rub1Δ and ubc12Δ cells accumulated in G2/M at 90 min (~60% of cells in G2/M phase), whereas wild-type cells progressed efficiently through G2 and M phase into the next cell cycle (~47% of cells in G2/M phase; Figures 4E and S4A). As this G2/M delay was not observed in the absence of CPT (Figure S4B), we demonstrate that neddylation affects cell-cycle progression in response to DNA damage.
Since defects in cell-cycle regulation can contribute to genome instability (Jackson and Bartek, 2009), we measured the rate of gross chromosomal rearrangements (GCR) in the neddylation mutants. The rate of GCR events in the ubc12Δ mutant was 2.7-fold greater than in wild-type, whereas the rad17Δubc12Δ double mutant showed, respectively, a 7- and 2-fold increase in GCR rates when compared to the ubc12Δ and rad17Δ mutants (Figure 4F), suggesting that neddylation and checkpoint pathways cooperate in promoting genome stability.
We next asked whether the perturbations in cell-cycle progression observed in the neddylation mutants were due to abnormal activation of Rad53- or Chk1-dependent DDCs. However, we failed to detect hyperphosphorylation of Rad53 and Chk1 in both CPT-treated rub1Δ and wild-type cells (Figures S4C–S4D). We reasoned that factors other than DDC proteins are targeted by the neddylation pathway, which in turn may affect cell-cycle progression. The best-studied NEDD8/Rub1 targets are cullin proteins, which are scaffolds for the assembly of multi-subunit cullin-RING ubiquitin ligases (Laplaza et al., 2004; Liakopoulos et al., 1998). These ubiquitin ligases are responsible for the turnover of a vast majority of proteins and consequently play a major role in maintaining cellular homeostasis (Soucy et al., 2010). Strikingly, another interaction hub in the differential network was the cullin Rtt101 (Figure 2B), which has been shown to regulate the progression through G2/M phase by promoting proteasomal degradation of Mms22 (Ben-Aroya et al., 2010). Given the role of neddylation in cullin-RING ubiquitin ligase modification, we examined whether this process affects Mms22 levels. We observed a faster degradation of Mms22 in a rub1Δ strain when compared to wild-type, suggesting that neddylation, in contrast to Rtt101-dependent ubiquitylation (Ben-Aroya et al., 2010), promotes Mms22 stability (Figures 4G–4H).
As another mean of identifying targets of the neddylation machinery, we searched for genes that exhibited strong differential positive interactions with RUB1, as several studies have shown that positive genetic interactions typically occur among genes involved in the same pathway (Fiedler et al., 2009; Sharifpoor et al., 2012). The strongest positive interaction for RUB1 in our CPT network was with NHP10, which encodes a subunit of the INO80 chromatin remodeling complex that participates in the DDR, notably by regulating cell-cycle progression (van Attikum et al., 2007). We examined the levels of Nhp10 and observed a slower degradation in the rub1Δ mutant compared to wild-type (Figures 4I–4J), suggesting that neddylation promotes degradation of Nhp10. Together, the data implicate a role for neddylation in cell-cycle control after DNA damage by regulating the levels of DDR factors such as Mms22 and Nhp10.
While cullin-RING ubiquitin ligases are probably the best-studied Rub1 substrates, many other proteins may be modified by neddylation (Rabut and Peter, 2008). For example, ribosomal proteins and E3 ubiquitin ligases, such as the p53 regulator Mdm2, are substrates for neddylation (Rabut and Peter, 2008). We infer from this that the stability of DDR factors such as Mms22 and Nhp10 may be regulated by direct neddylation, or indirectly by the neddylation of E3 or cullin-RING ubiquitin ligases (Figure 4K). Although further work will be required to resolve the precise mechanisms, the data confirm the power of differential genetic data to identify targets of neddylation.
Irc21 Is a General Response Factor in Checkpoint Control, Repair and Genome Stability
Although the interactions induced by the three agents were largely divergent, the differential data did identify a “conserved” network of 45 interactions that were altered in response to all three agents (Figure S5A). This network contained several known examples of conserved DDR pathways. For example, we observed a strong differential positive interaction between MRE11 and DDC1 (a component of the 9-1-1 complex) in all three conditions. This is consistent with recent work that has shown a nonredundant role for these factors in multiple DDR mechanisms, including the resection of DSBs and subsequent Mec1-dependent activation of the DDC (Nakada et al., 2004).
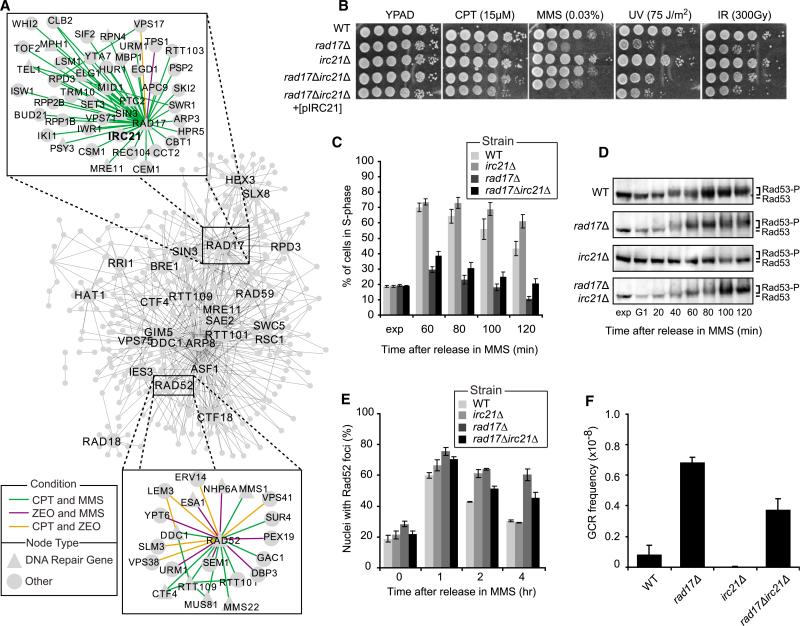
Encouraged by this observation, we turned to the analysis of all 584 differential interactions induced by two or more agents (Figure 5A) to identify general DDR mechanisms. This network again highlighted the damage checkpoint gene RAD17 as a hub not only of the CPT network (see above), but also of conserved interactions across agents (Figure 5A, top inset). These included a positive interaction with IRC21, an as yet un-characterized gene, in response to both CPT (p = 4.7 × 10–7) and MMS (p = 8.3 × 10–7), but not ZEO. We confirmed that Irc21 is expressed in yeast (Figure S5B) and that deletion of IRC21 in a rad17Δ mutant suppresses its sensitivity to CPT and MMS (Figure 5B). Importantly, this suppressive effect was also observed in other checkpoint mutants, including ddc1Δ (another mutant of the 9-1-1 complex) and rad9Δ (Figure S5C). The Irc21 protein contains a cytochrome b5-like domain (Figure S5D), which can be found in proteins involved in cytochrome P450-dependent metabolic processes (Zhang et al., 2005). To rule out that the suppression was due to Irc21 affecting drug metabolism via its cytochrome b5 domain, we exposed cells to ultraviolet light and ionizing radiation and were able to reproduce the suppressive phenotype in both cases (Figure 5B). Ectopic expression of Irc21 in the irc21Δrad17Δ mutant restored the sensitivity to DNA damaging agents to that observed for the rad17Δ mutant (Figure 5B). Thus, Irc21 affects cell survival in response to genotoxic insult by modulating the DDC rather than affecting drug metabolism.
Figure 5. Irc21 Affects Checkpoint Control, DNA Repair, and Genome Stability.
(A) Network of all 584 differential genetic interactions induced by at least two agents. The top 25 hubs in this network are labeled. The subnetworks of interactions involving RAD17 and RAD52 are also shown.
(B) Serial dilutions (10-fold) of cells of the indicated genotypes were grown on YPAD + MMS or CPT, or on YPAD after exposure to UV or IR.
(C) Exponentially (exp) growing cells were arrested in G1, released in medium + 0.02% MMS and 15 μg/ml nocodazole and analyzed by FACS at the indicated time points (see Figure S5E for FACS plots). The bar plot shows the percentage of S phase cells.
(D) Western blot analysis of Rad53 phosphorylation in cells from (C).
(E) Exponentially growing cells expressing Rad52-YFP were exposed to 0.02% MMS for 1 hr and released in fresh medium. Images were taken at the indicated time points and scored for Rad52-YFP foci. At least 100 nuclei were analyzed per strain and per time point.
(F) GCR frequencies were determined as in Figure 4F.
All data represent the mean ± 1 SD from three independent experiments. See also Figure S5.
To explore this further, we monitored the progression of all mutants through the cell cycle in the presence of MMS. While wild-type and irc21Δ strains displayed slow S phase progression and accumulated in G2 2 hr after release from G1, the rad17Δ strain rapidly progressed through S phase and accumulated in G2 within an hour (Figures 5C and S5E). Remarkably, deletion of IRC21 in the rad17Δ strain partially suppressed the checkpoint deficiency as we noted an increased fraction of cells remaining in S phase (Figure 5C). Moreover, we found that while the rad17Δ mutant failed to activate Rad53 kinase (Figure 5D), the irc21-Δrad17Δ double mutant showed a moderate restoration of this phenotype with Rad53 becoming slightly phosphorylated (Figure 5D).
Checkpoint proteins detect DNA lesions, arrest the cell cycle and trigger DNA repair (Zhou and Elledge, 2000). Given that Irc21 modulates the DDC, we examined whether it also affects DNA repair. We monitored the formation of Rad52 foci after exposure to MMS and found that while the rate of assembly of Rad52 foci was similar in all strains (Figure 5E), persistent foci were observed in the rad17Δ mutant for up to 4 hr, indicating abrogation of repair. However, deletion of IRC21 alleviated the repair defect seen in the rad17Δ strain, as indicated by the enhanced dissolution of Rad52 foci in the irc21Δrad17Δ strain compared to that in the rad17Δ strain (Figure 5E).
Finally we found that, whereas irc21Δ cells showed no alterations in genomic stability, rad17Δ cells displayed an 8.2-fold increase in GCR events compared to the wild-type (Figure 5F). However, irc21Δrad17Δ cells showed only a 4.5-fold increase, suggesting that deletion of IRC21 partially rescues the deleterious impact of Rad17 loss on GCR (Figure 5F). Together, these results suggest that Irc21 not only modulates the DDC, but also promotes efficient DNA repair and contributes to genome stability.
While a previous study had reported only a cytoplasmic localization for Irc21 (Huh et al., 2003), we found that Irc21-GFP localizes to both the cytoplasm and nucleus (Figures S5F–S5G). Irc21-GFP, however, did not accumulate into MMS-induced sub-nuclear foci as observed for Rad52-YFP (Figure S5F), suggesting that it may not operate directly at DNA lesions. Interestingly, we found that irc21Δ strains are hypersensitive to MMS when combined with the TOR inhibitor rapamycin (Figure S5H), a compound that can affect the abundance of proteins including DDR factors (Dyavaiah et al., 2011; Fournier et al., 2010). This may suggest that Irc21 affects the DDR by regulating the steady-state levels of distinct DDR proteins. Although further work is required to work out this intriguing connection, our analysis of the set of commonly perturbed genetic interactions identified Irc21 as a DDR factor that affects cell-cycle progression, DNA repair and genome stability.
An Integrated Module Map Reveals a Role for Rtt109 in Translesion Synthesis
A powerful approach for interpreting genetic interactions is in conjunction with knowledge of physical protein-protein interactions and protein complexes (Kelley and Ideker, 2005). Our previous work has shown that, while static genetic interactions are enriched among components of the same physical complex, differential genetic interactions tend to occur between distinct but functionally related complexes (Bandyopadhyay et al., 2010). Based on this idea, we used a recently described integrative clustering algorithm (Srivas et al., 2011) to transform our differential genetic interaction data for all agents into a map of 179 modules and 452 module-module interactions (Tables S4, Table S5 and the Supplemental Experimental Procedures). Modules group genes with similar patterns of both genetic and physical interactions, many of which were found to coincide with known DNA repair complexes. Module-module interactions represent bundles of differential genetic interactions that span across the genes in the two modules and point to DNA damage-induced cooperativity.
The low overlap among the genetic networks of the three agents (Figure 2C) was reproduced in the module map, as the vast majority (~90%) of module-module interactions were found to occur in response to a single agent. This finding was confirmed in a separate analysis in which we examined differential genetic crosstalk between biological processes (as opposed to protein modules) and observed again very little overlap between conditions (Supplemental Experimental Procedures and Table S6). Indeed, each of the agents highlighted a different module as a central hub of interactions (Figure 6A); these were the 9-1-1 DDC complex (CPT), the Mms2/Ubc13 E2 ubiquitin conjugase complex (MMS), and the Mre11/Rad50/Xrs2 (MRX) DSB repair complex (ZEO). Many of the interactions involving these hub modules recapitulated known DDR mechanisms. For example, the 9-1-1 complex was found to interact with the S phase checkpoint complex Csm3/Tof1, which is consistent with work showing that both complexes are required for the response to CPT (Redon et al., 2006).
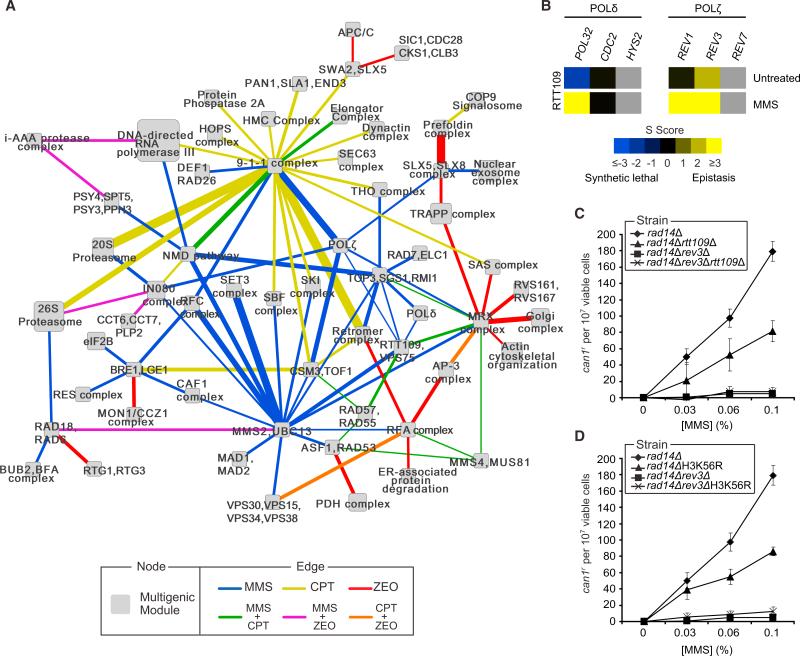
Figure 6. A Global Map of DDR Modules Reveals a Role for RTT109 in Translesion Synthesis.
(A) A map of multiprotein modules connected by bundles of differential genetic interactions. Node size scales with the number of proteins present in the module. Edge size scales with the significance of the enrichment for interactions spanning the two modules. For clarity only a portion of the entire map has been shown. The full list of module-module interactions is provided in Table S5.
(B) Genetic interactions observed between RTT109 and genes encoding members of the Polδ and Polζ complexes.
(C) MMS-induced can1r mutation frequencies in cells of the indicated genotype.
(D) As in (C), except that rad14ΔH3K56R and rad14Δrev3ΔH3K56R cells were used.
All data represent the mean ± 1 SD of three independent experiments. See also Figure S6, Table S4, Table S6, Table S6, and Data sets S1 and S2.
Surprisingly, we also observed a differential positive relationship between Rtt109 and Polδ, as well as the translesion synthesis polymerases Rev1 and Polζ, a complex composed of Rev3 and Rev7 (Figure 6B), which we validated using a spot dilution assay (Figure S6A). Polζ-dependent synthesis enables cells to replicate through DNA lesions, ensuring that such lesions do not lead to replication fork collapse. Moreover, Polζ, in conjunction with Polδ, is responsible for ~85% of the bypass events at abasic sites, with much of this occurring in an error-prone fashion (Andersen et al., 2008).
To validate the link between Rtt109 and translesion synthesis, we utilized a CAN1 forward mutation assay that reports any mutation that disrupts Can1 function, resulting in canavanine-resistance (can1r). Cells with proficient bypass activity accrue mutations at this locus at a much higher rate enabling them to survive selection on canavanine. As expected, deletion of REV3, which impairs Polζ function, produced almost no can1r colonies, indicating an almost complete loss of translesion synthesis activity (Figures 6C and S6B). Interestingly, both rtt109Δ cells and cells expressing H3K56R, a mutant form of histone H3 that cannot be acetylated by Rtt109, showed a 2-fold decrease in the rate of canr colonies when compared to wild-type (Figures 6C, S6D, S6B, and S6C). The rev3Δrtt109Δ and rev3ΔH3K56R mutants on the other hand displayed a reduction in the rate of canr colonies that was comparable to that of the rev3Δ mutant (Figures 6C–D). Together this suggests that Rtt109 affects translesion synthesis through acetylation of H3K56.
Recent work has shown that Rtt109 mediates acetylation of newly synthesized histones that are deposited onto DNA synthesized during DNA replication (Masumoto et al., 2005). In support of this, rtt109Δ or H3K56R mutants have been found to genetically interact with genes involved in DNA replication, including DNA polymerase α and PCNA (Collins et al., 2007). Importantly, the rtt109Δ and H3K56R mutants fail to stabilize Polα and PCNA at stalled replication forks (Han et al., 2007). Since PCNA also serves as a clamp for the loading of translesion synthesis polymerases at these sites (Chang and Cimprich, 2009), we suggest a model in which Rtt109-dependent H3K56 acetylation regulates PCNA-dependent loading of translesion synthesis polymerases at sites of MMS-induced fork stalling.
Perspective
Our genetic interaction data and module map (Figure 6A) should provide a major resource for further discovery of DNA damaged-induced functional crosstalk. Furthermore, as DDR pathways are well conserved (Ciccia and Elledge, 2010), this resource is not limited to yeast biology but also informs the DDR in humans and related diseases such as cancer. For example, the dependency we uncovered between neddylation and the DDC (Figure 4C) is echoed in a study in humans, in which an inhibitor targeting the NEDD8 (ortholog of Rub1) activating enzyme (NAE1) lead to sensitivity to ionizing radiation in pancreatic cancer cells expressing mutated p53 (Wei et al., 2012). Differential synthetic lethal interactions may serve as a key resource in the emerging “synthetic-lethal” approach to cancer therapy (Jackson and Bartek, 2009). In this respect, our multiconditional dE-MAP provides a catalog of genes that display differential synthetic-lethal interactions with orthologs of genes implicated in tumorigenesis. Such genes could be targeted (e.g., NAE1) to enhance the killing power of chemotherapeutics in specific cancer types (e.g., p53-deficient cancers).
Finally, this study illustrates that differential network analysis is a powerful approach for annotating gene function that is complementary to existing functional genomics technologies. The resolution of technologies that make measurements at the level of single genes, such as single-mutant or differential mRNA expression profiling, is ultimately limited to the total number of genes in a genome. In contrast, differential interaction mapping taps into a much larger (quadratic) space of gene-gene interactions, which we show enables the dissection of gene function in greater detail (Figure 2D). This power comes at a cost, as screening all gene pairs is presently arduous and expensive even in the model organism S. cerevisiae, requiring us to restrict coverage of the network map to a focused set of query genes. This tradeoff in precision versus coverage is analogous to the two complementary strategies that have been employed in mapping disease-causing mutations: analysis of genotyped pedigrees, involving no more than two to three generations, provides a “coarse” mapping to identify a large candidate region of the genome (Pérez-Enciso, 2003), after which “fine mapping” techniques such as gene association studies are used to pinpoint the location of the causal mutation more precisely (Hästbacka et al., 1994). Here, we have pursued a similar strategy by seeding our differential genetic interaction screen with genes that have been previously annotated to DDR processes. The resulting network highlights dynamic functional connections between numerous pathways and complexes at high resolution, suggesting a paradigm for dissecting the cellular responses to distinct drugs.
EXPERIMENTAL PROCEDURES
Differential Genetic Interaction Screens
Genetic interaction screens were performed as described (Schuldiner et al., 2006), except that the last selection step was performed by replica-plating cells on medium containing 1% DMSO (untreated), 0.01% MMS, 5μg/ml CPT, or 75μM ZEO. Static and differential genetic interaction scores were calculated as described (Bandyopadhyay et al., 2010; Collins et al., 2006).
Spot Dilution Assays
Serial dilutions (10-fold) of mid-log phase cells were spotted on YPAD plates and grown for 2–3 days at 30° C.
Cell-Cycle Checkpoint Assays
Exponentially growing cells were synchronized in G1 with α factor (7.5 μM) and either exposed to MMS in G1 for 30 min and then released in fresh medium, or released in fresh medium containing CPT. FACS analysis was performed using a BD LSRII instrument and WinMDI software. Rad53 phosphorylation analysis was performed as previously described (van Attikum et al., 2007) with anti-Rad53 antibody (Santa Cruz Biotechnologies, SC-6749). Membranes were scanned with a Biorad Universal Hood II instrument and Quantity One software.
GCR and Mutagenesis Assays
Gross chromosomal rearrangement and mutagenesis assays were performed as previously described (Chen and Kolodner, 1999; Johnson et al., 1998).
Analysis of Mms22 and Nhp10 Turnover
Mmm22 and Nhp10 turnover was examined as previously described (Ben-Aroya et al., 2010) with cells expressing GAL1-HA-Mms22 or pGAL1-GST-Nhp10 (Open Biosystems) and anti-HA (Santa Cruz Biotechnology, SC-7392) or anti-GST antibodies (Amersham).
Analysis of Rad52 Foci
Cells expressing pRad52-YFP (Lisby et al., 2001) were grown to midlog phase, exposed to MMS for 1 hr, washed and concentrated in 1% low melting agar (Cambrex). Images were captured with a Leica AF6000 LX microscope at 100-fold magnification with a HCX PL FLUOTAR 1003 1.3 oil objective lens.
Additional experimental methods have been provided in the Supplemental Experimental Procedures.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank G. Hannum and N. de Wind for helpful discussions; J. Shen, M. Tijsterman, and H. Vrieling for critical reading of the manuscript; H. Braberg and A. Roguev for assistance with the genetic interaction screens; M. Lisby, K.J. Myung, S. Ben-Aroya, and P. Hieter for providing reagents. R.S., T.I., and N.J.K., who is a Searle Scholar and a Keck Young Investigator, were generously supported by grants from the U.S. National Institutes of Health (ES014811 and GM084279 to R.S. and T.I.; GM084448, GM084279, GM081879, and GM098101 to N.J.K.). H.v.A. was supported by grants from the Netherlands Organization for Scientific Research (NWO-VIDI) and a CDA grant from Human Frontiers Science Program (HFSP).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, six figures, two data sets, and six tables and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2012.11.023.
REFERENCES
- Andersen PL, Xu F, Xiao W. Eukaryotic DNA damage tolerance and translesion synthesis through covalent modifications of PCNA. Cell Res. 2008;18:162–173. doi: 10.1038/cr.2007.114. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S, Mehta M, Kuo D, Sung MK, Chuang R, Jaehnig EJ, Bodenmiller B, Licon K, Copeland W, Shales M, et al. Rewiring of genetic networks in response to DNA damage. Science. 2010;330:1385–1389. doi: 10.1126/science.1195618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Aroya S, Agmon N, Yuen K, Kwok T, McManus K, Kupiec M, Hieter P. Proteasome nuclear activity affects chromosome stability by controlling the turnover of Mms22, a protein important for DNA repair. PLoS Genet. 2010;6:e1000852. doi: 10.1371/journal.pgen.1000852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjergbaek L, Cobb JA, Tsai-Pflugfelder M, Gasser SM. Mechanistically distinct roles for Sgs1p in checkpoint activation and replication fork maintenance. EMBO J. 2005;24:405–417. doi: 10.1038/sj.emboj.7600511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caba E, Dickinson DA, Warnes GR, Aubrecht J. Differentiating mechanisms of toxicity using global gene expression analysis in Saccharomyces cerevisiae. Mutat. Res. 2005;575:34–46. doi: 10.1016/j.mrfmmm.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Chang DJ, Cimprich KA. DNA damage tolerance: when it's OK to make mistakes. Nat. Chem. Biol. 2009;5:82–90. doi: 10.1038/nchembio.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Kolodner RD. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat. Genet. 1999;23:81–85. doi: 10.1038/12687. [DOI] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Schuldiner M, Krogan NJ, Weissman JS. A strategy for extracting and analyzing large-scale quantitative epistatic interaction data. Genome Biol. 2006;7:R63. doi: 10.1186/gb-2006-7-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyavaiah M, Rooney JP, Chittur SV, Lin Q, Begley TJ. Autophagy-dependent regulation of the DNA damage response protein ribonucleotide reductase 1. Mol. Cancer Res. 2011;9:462–475. doi: 10.1158/1541-7786.MCR-10-0473. [DOI] [PubMed] [Google Scholar]
- Fiedler D, Braberg H, Mehta M, Chechik G, Cagney G, Mukherjee P, Silva AC, Shales M, Collins SR, van Wageningen S, et al. Functional organization of the S. cerevisiae phosphorylation network. Cell. 2009;136:952–963. doi: 10.1016/j.cell.2008.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier ML, Paulson A, Pavelka N, Mosley AL, Gaudenz K, Bradford WD, Glynn E, Li H, Sardiu ME, Fleharty B, et al. Delayed correlation of mRNA and protein expression in rapamycin-treated cells and a role for Ggc1 in cellular sensitivity to rapamycin. Mol. Cell. Proteomics. 2010;9:271–284. doi: 10.1074/mcp.M900415-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Zhou H, Li Z, Xu RM, Zhang Z. Acetylation of lysine 56 of histone H3 catalyzed by RTT109 and regulated by ASF1 is required for replisome integrity. J. Biol. Chem. 2007;282:28587–28596. doi: 10.1074/jbc.M702496200. [DOI] [PubMed] [Google Scholar]
- Hästbacka J, de la Chapelle A, Mahtani MM, Clines G, Reeve-Daly MP, Daly M, Hamilton BA, Kusumi K, Trivedi B, Weaver A, et al. The diastrophic dysplasia gene encodes a novel sulfate transporter: positional cloning by fine-structure linkage disequilibrium mapping. Cell. 1994;78:1073–1087. doi: 10.1016/0092-8674(94)90281-x. [DOI] [PubMed] [Google Scholar]
- Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St Onge RP, Tyers M, Koller D, et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Torres-Ramos CA, Izumi T, Mitra S, Prakash S, Prakash L. Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes Dev. 1998;12:3137–3143. doi: 10.1101/gad.12.19.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley R, Ideker T. Systematic interpretation of genetic interactions using protein networks. Nat. Biotechnol. 2005;23:561–566. doi: 10.1038/nbt1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplaza JM, Bostick M, Scholes DT, Curcio MJ, Callis J. Saccharomyces cerevisiae ubiquitin-like protein Rub1 conjugates to cullin proteins Rtt101 and Cul3 in vivo. Biochem. J. 2004;377:459–467. doi: 10.1042/BJ20030755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakopoulos D, Doenges G, Matuschewski K, Jentsch S. A novel protein modification pathway related to the ubiquitin system. EMBO J. 1998;17:2208–2214. doi: 10.1093/emboj/17.8.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Rothstein R, Mortensen UH. Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl. Acad. Sci. USA. 2001;98:8276–8282. doi: 10.1073/pnas.121006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin C, North M, Erixon K, Walters K, Jenssen D, Goldman AS, Helleday T. Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res. 2005;33:3799–3811. doi: 10.1093/nar/gki681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436:294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada D, Hirano Y, Sugimoto K. Requirement of the Mre11 complex and exonuclease 1 for activation of the Mec1 signaling pathway. Mol. Cell. Biol. 2004;24:10016–10025. doi: 10.1128/MCB.24.22.10016-10025.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill BM, Szyjka SJ, Lis ET, Bailey AO, Yates JR, 3rd, Aparicio OM, Romesberg FE. Pph3-Psy2 is a phosphatase complex required for Rad53 dephosphorylation and replication fork restart during recovery from DNA damage. Proc. Natl. Acad. Sci. USA. 2007;104:9290–9295. doi: 10.1073/pnas.0703252104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol. Cell. 2009;35:228–239. doi: 10.1016/j.molcel.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Enciso M. Fine mapping of complex trait genes combining pedigree and linkage disequilibrium information: a Bayesian unified framework. Genetics. 2003;163:1497–1510. doi: 10.1093/genetics/163.4.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut G, Peter M. Function and regulation of protein neddylation. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9:969–976. doi: 10.1038/embor.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon C, Pilch DR, Bonner WM. Genetic analysis of Saccharomyces cerevisiae H2A serine 129 mutant suggests a functional relationship between H2A and the sister-chromatid cohesion partners Csm3-Tof1 for the repair of topoisomerase I-induced DNA damage. Genetics. 2006;172:67–76. doi: 10.1534/genetics.105.046128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan CJ, Roguev A, Patrick K, Xu J, Jahari H, Tong Z, Beltrao P, Shales M, Qu H, Collins SR, et al. Hierarchical modularity and the evolution of genetic interactomes across species. Mol. Cell. 2012;46:691–704. doi: 10.1016/j.molcel.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Weissman JS, Krogan NJ. Quantitative genetic analysis in Saccharomyces cerevisiae using epistatic miniarray profiles (E-MAPs) and its application to chromatin functions. Methods. 2006;40:344–352. doi: 10.1016/j.ymeth.2006.07.034. [DOI] [PubMed] [Google Scholar]
- Sharifpoor S, van Dyk D, Costanzo M, Baryshnikova A, Friesen H, Douglas AC, Youn JY, VanderSluis B, Myers CL, Papp B, et al. Functional wiring of the yeast kinome revealed by global analysis of genetic network motifs. Genome Res. 2012;22:791–801. doi: 10.1101/gr.129213.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucy TA, Dick LR, Smith PG, Milhollen MA, Brownell JE. The NEDD8 Conjugation Pathway and Its Relevance in Cancer Biology and Therapy. Genes Cancer. 2010;1:708–716. doi: 10.1177/1947601910382898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivas R, Hannum G, Ruscheinski J, Ono K, Wang PL, Smoot M, Ideker T. Assembling global maps of cellular function through integrative analysis of physical and genetic networks. Nat. Protoc. 2011;6:1308–1323. doi: 10.1038/nprot.2011.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AH, Boone C. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol. Biol. 2006;313:171–192. doi: 10.1385/1-59259-958-3:171. [DOI] [PubMed] [Google Scholar]
- Travesa A, Kuo D, de Bruin RA, Kalashnikova TI, Guaderrama M, Thai K, Aslanian A, Smolka MB, Yates JR, 3rd, Ideker T, Wittenberg C. DNA replication stress differentially regulates G1/S genes via Rad53-dependent inactivation of Nrm1. EMBO J. 2012;31:1811–1822. doi: 10.1038/emboj.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulukan H, Swaan PW. Camptothecins: a review of their chemotherapeutic potential. Drugs. 2002;62:2039–2057. doi: 10.2165/00003495-200262140-00004. [DOI] [PubMed] [Google Scholar]
- Usui T, Ogawa H, Petrini JH. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell. 2001;7:1255–1266. doi: 10.1016/s1097-2765(01)00270-2. [DOI] [PubMed] [Google Scholar]
- van Attikum H, Fritsch O, Gasser SM. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J. 2007;26:4113–4125. doi: 10.1038/sj.emboj.7601835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D, Li H, Yu J, Sebolt JT, Zhao L, Lawrence TS, Smith PG, Morgan MA, Sun Y. Radiosensitization of human pancreatic cancer cells by MLN4924, an investigational NEDD8-activating enzyme inhibitor. Cancer Res. 2012;72:282–293. doi: 10.1158/0008-5472.CAN-11-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Zhang Z, Che W. Suppression of a DNA base excision repair gene, hOGG1, increases bleomycin sensitivity of human lung cancer cell line. Toxicol. Appl. Pharmacol. 2008;228:395–402. doi: 10.1016/j.taap.2007.12.020. [DOI] [PubMed] [Google Scholar]
- Zhang H, Myshkin E, Waskell L. Role of cytochrome b5 in catalysis by cytochrome P450 2B4. Biochem. Biophys. Res. Commun. 2005;338:499–506. doi: 10.1016/j.bbrc.2005.09.022. [DOI] [PubMed] [Google Scholar]
- Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.