Abstract
△Bacterial exopolysaccharide synthesis is a prevalent and indispensible activity in many biological processes, including surface adhesion and biofilm formation. In Caulobacter crescentus, surface attachment and subsequent biofilm growth depend on the ability to synthesize an adhesive polar polysaccharide known as the holdfast. In this work, we show that polar polysaccharide synthesis is a conserved phenomenon among Alphaproteobacterial species closely related to C. crescentus. Among them, mutagenesis of Asticcacaulis biprosthecum showed that disruption of the hfsH gene, which encodes a putative polysaccharide deacetylase, leads to accumulation of holdfast in the culture supernatant. Examination of the hfsH deletion mutant in C. crescentus revealed that this strain synthesizes holdfast; however like the A. biprosthecum hfsH mutant, the holdfasts are shed into the medium and have decreased adhesiveness and cohesiveness. Site-directed mutagenesis at the predicted catalytic site of C. crescentus HfsH phenocopied the ΔhfsH mutant and abolished the esterase activity of HfsH. In contrast, overexpression of HfsH increased cell adherence without increasing holdfast synthesis. We conclude that the polysaccharide deacetylase activity of HfsH is required for the adhesive and cohesive properties of the holdfast, as well as for the anchoring of the holdfast to the cell envelope.
INTRODUCTION
Bacterial adhesion is a complex process influenced by many variables, including cell-wall characteristics, surface properties, and environmental factors (Dunne, 2002). Exopolysaccharides (EPS) are often an indispensible contributor to initial surface attachment and biofilm development (Donlan, 2002). In general, bacterial adhesion to surfaces can be divided into two stages, including (1) reversible attachment, which is typically mediated by extracellular structures such as pili and flagella, and (2) permanent attachment that requires adhesins (Vigeant et al., 2002). In Caulobacter crescentus, the coordinated synthesis of the flagellum, pili, and the holdfast adhesin at the cell pole plays an important role in the transition from reversible to permanent attachment (Bodenmiller et al., 2004, Entcheva-Dimitrov and Spormann, 2004, Levi and Jenal, 2006, Li et al., 2012). Each newborn swarmer cell harbors a flagellum and pili at the same pole. Flagellum-driven motility and adherence mediate the initial reversible adhesion by overcoming surface electrostatic repulsion. Concomitant surface binding by the flagellum and pili causes flagellar rotational arrest and an immediate stimulation of holdfast synthesis at the same pole (Li et al., 2012). For cells that do not contact a surface, the developmental program initiates holdfast synthesis slightly before the loss of motility at the end of the swarmer cell stage (Bodenmiller et al., 2004, Entcheva-Dimitrov and Spormann, 2004, Levi and Jenal, 2006, Li et al., 2012). Permanent attachment requires the holdfast, which has an extremely high adhesive strength in the μN range (Poindexter, 1964, Larson and Pate, 1975, Tsang et al., 2006). Although the detailed composition of the holdfast remains unknown, it is sensitive to lysozyme and chitinase, and can be detected by wheat germ agglutinin (WGA) lectin, indicating that the holdfast contains ß-1,4-N-acetylglucosamine (GlcNAc) polymers (Merker and Smit, 1988, Li et al., 2005).
The holdfast synthesis (hfs) and holdfast anchor (hfa) loci are critical for holdfast polysaccharide biogenesis, export, and anchoring to the cell body (Kurtz Jr and Smith, 1994, Cole et al., 2003, Smith et al., 2003, Toh et al., 2008). The hfs locus is a polysaccharide synthesis gene cluster of the Wzy-dependent type encoding proteins that are predicted to function in holdfast polysaccharide repeat unit synthesis (HfsE and HfsG), modification (HfsH), flipping across the inner membrane (HfsF), polymerization in the periplasmic space (HfsC and HfsI), and export through the outer membrane (HfsD, HfsA, and HfsB) (Smith et al., 2003, Toh et al., 2008, Cuthbertson et al., 2009). Mutation of several genes in this cluster (hfsA, hfsB, hfsD, hfsG, and hfsH) abolishes surface adherence, while mutation of other genes (hfsE and hfsF) leads to a reduction in surface binding (Smith et al., 2003, Toh et al., 2008). The hfa locus, which consists of hfaA, hfaB, and hfaD, encodes proteins required for holdfast anchoring to the cell envelope. Deletion of the hfa genes results in holdfast shedding into the supernatant and on surfaces (Kurtz Jr and Smith, 1994, Cole et al., 2003, Smith et al., 2003, Hardy et al., 2010).
A different type of polysaccharide shedding phenotype has also been reported in Staphylococcus epidermidis and S. aureus mutants of icaB, a polysaccharide deacetylase gene encoded by the icaADBC operon (Heilmann et al., 1996, Vuong et al., 2004, Cerca et al., 2007). Polysaccharide deacetylation is prevalent not only in bacteria, but also in fungi and insects (Itoh et al., 2008, Cantarel et al., 2009). Most polysaccharide deacetylases, which catalyze the N-deacetylation of GlcNAc or the O-deacetylation of O-acetylxylose, belong to the CE4 family, and they include peptidoglycan GlcNAc deacetylases, rhizobial NodB chitooligosaccharide deacetylases, chitin deacetylases, acetyl xylan esterases, and xylanases (Caufrier et al., 2003, Cantarel et al., 2009). icaB mutants synthesize a fully acetylated Polysaccharide Intercellular Adhesin (PIA) polymer that cannot be anchored to the cell surface, which causes defects in biofilm formation (Vuong et al., 2004, Cerca et al., 2007). A polysaccharide deacetylase gene, pgaB, is also present in the pgaABCD operon of E. coli responsible for the synthesis of poly- β -1,6-GlcNAc (PGA), and its deletion leads to a defect in PGA export and its retention in the periplasm, resulting in impaired biofilm formation (Itoh et al., 2008). Both PGA and PIA are linear polymers of β-1,6-GlcNAc, while β-1,4- linked GlcNAc is a component of the holdfast. Notably, the C. crescentus holdfast synthesis protein HfsH shares sequence similarity with IcaB, PgaB and other CE4 family members, suggesting that HfsH may also function as a polysaccharide deacetylase. Although deletion of icaB did not prevent polysaccharide synthesis, previous reports demonstrated that the deletion of hfsH in C. crescentus abolishes surface adhesion (Toh et al., 2008).
In this study, we explored the conservation of holdfast synthesis genes in the Caulobacterales clade of the Alphaproteobacteria, and found that hfsH is broadly conserved in the hfs gene cluster across several bacterial species, consistent with an important function for this protein. To determine if the function of the hfs genes are conserved, we characterized the phenotype of Asticcacaulis biprosthecum transposon mutants with insertions in a number of hfs genes. We discovered that holdfast synthesis continues despite a disruption of A. biprosthecum hfsH. However, the mutant holdfast is shed into the medium and the ability of the shed holdfast to adhere to a surface is impaired. Detailed analysis of the C. crescentus hfsH deletion mutant revealed that it synthesizes defective holdfasts with low adhesiveness and cohesiveness that cannot be anchored to the cell body. In contrast, overexpression of HfsH increased cell adherence. Mutation of the predicted catalytic residue of HfsH phenocopies the ΔhfsH mutant, suggesting that holdfast deacetylation is essential for holdfast anchoring to the cell body and for its surface adherence properties.
RESULTS
The hfs gene cluster is conserved within the Caulobacterales clade of the Alphaproteobacteria
The genes involved in holdfast synthesis were originally characterized in C. crescentus and they are broadly conserved within the Caulobacterales clade of the Alphaproteobacteria (Table S1) (Brown et al., 2008, Chertkov et al., 2011). Notably, all of the genes described as essential for holdfast synthesis in C. crescentus, including hfsD, hfsA, hfsB, hfsG, and hfsH, are conserved within the hfs gene clusters in the Brevundimonas, Asticcacaulis, and Hirschia genera, with the exception of Brevundimonas hfsH, which is found outside of the hfs cluster (Table S1, Fig. 1A). In contrast, the hfsC and hfsF genes, which are dispensable for holdfast synthesis in C. crescentus (Toh et al., 2008), are not conserved in the hfs gene clusters of Hirchia, and hfsC is absent in Brevundimonas. The conservation of genes and the similarity of gene cluster organization is consistent with a conserved function of hfs genes in holdfast synthesis in the Caulobacterales.
Figure 1. Conservation of the holdfast biosynthesis genes and holdfast synthesis in four genera belonging to the Caulobacterales.
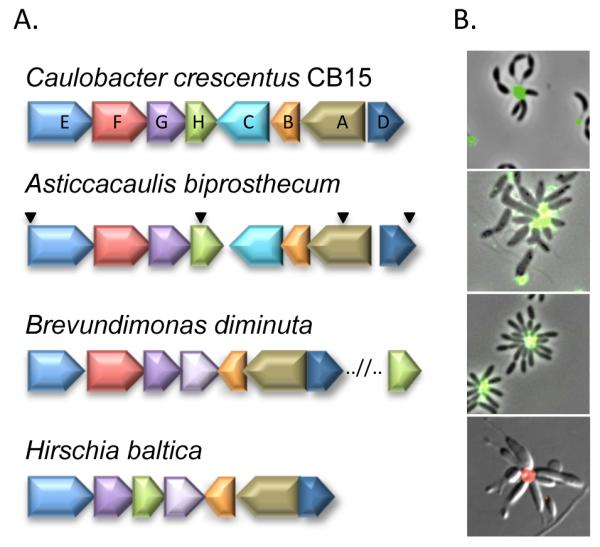
(A) hfs gene names (hfsA – hfsH) are given for C. crescentus, and the homologs from the related organisms are identified by color. Inverted triangles indicate the location of transposon insertions in adhesion deficient mutants of A. biprosthecum. (B) AF488/594-conjugated WGA lectin labeling of the holdfast in C. crescentus, A. biprosthecum, B. diminuta, and H. baltica.
The bioinformatic analysis indicated that holdfast production is a conserved trait in the Caulobacterales. Indeed, production of polar holdfast is readily detected in A. biprosthecum, B. diminuta, and H. baltica using lectin binding assays, where Alexafluor (AF) 488/594 conjugated wheat germ agglutinin (WGA) lectin binds specifically to GlcNAc oligomers in the holdfast (Fig. 1B), suggesting that the holdfast from each bacterium contains GlcNAc. Furthermore, all three species form rosettes (Fig. 1B), clusters of cells attached together with holdfast at the center, suggesting that the holdfasts function as adhesins.
A. biprosthecum hfs mutants are deficient in holdfast production and surface attachment
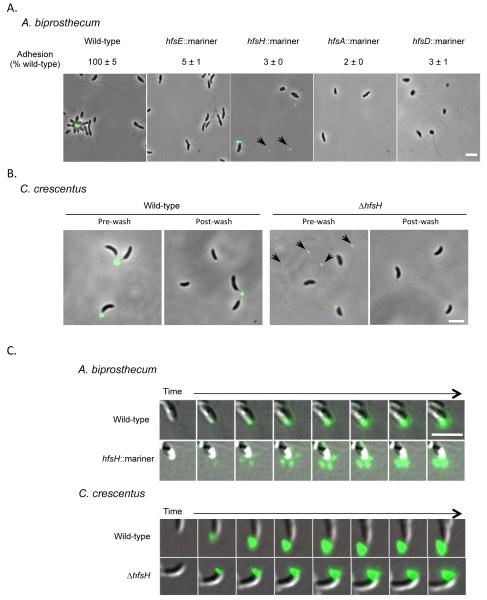
We performed a transposon mutagenesis screen in A. biprosthecum to identify genes required for holdfast biosynthesis and surface adhesion in this species. The site of transposon insertion in four adhesion defective mutants mapped to the A. biprosthecum hfs cluster (Fig. 1A). These four mutants only had 2-5% adherence compared to wild-type in a short term binding assay, which essentially measures single cell attachment to a surface (Fig. 2A). The results of this assay are consistent with the phenotypes of similar C. crescentus hfs mutants (Toh et al., 2008). Since the transposon insertions in hfsD and hfsH do not cause polar effects, we conclude that these genes are essential for surface binding.
Figure 2. Analysis of holdfast synthesis in the hfs mutants of A. biprosthecum and C. cresentus.
(A) Detection and analysis of holdfast synthesis in the hfs insertion mutants of A. biprosthecum by short-term binding assay and AF488-WGA labeling. The short term binding (90 min) data are expressed as a mean percent of wild-type crystal violet staining from two independent experiments with three replicates with the standard error of the mean. (B) Overlay of phase-contrast and fluorescence images showing the detection of holdfasts by AF488-WGA labeling of C. cresentus wild-type and ΔhfsH mutant cells before (left) and after (right) the cells were washed to remove excess lectin. Shed holdfasts are indicated by black arrows in (A) and (B). (C) Overlay of Differential Interference Contrast (DIC) and fluorescence images showing holdfast synthesis/export of the A. biprosthecum wild-type and hfsH mutant and the C. crescentus wild-type and ΔhfsH mutant strains. Images shown were acquired every 10 min. The scale bars represent 2 μm.
Based on the strong adhesion-defective phenotype displayed by the A. biprosthecum hfsA, hfsD, hfsE, and hfsH transposon mutants, we expected that these mutants would fail to synthesize holdfast. We tested for the presence of holdfast in these mutants using lectin binding assays. As expected, the hfsE, hfsA, and hfsD mutants did not synthesize holdfast (Fig. 2A) and did not form rosettes. Rosettes were not observed in A. biprosthecum hfsH mutant cultures, but a low proportion of the hfsH mutant cells had detectable holdfast at the cell pole (Fig. 2A). In addition, cell-free holdfasts were observed in the medium, suggesting that the holdfast is synthesized but not retained at the cell pole (Fig. 2A). This observation differs from the previously described phenotype of a C. crescentus ΔhfsH mutant in which no holdfast was detected on cells or in the medium (Toh et al., 2008).
Deletion of hfsH results in holdfast shedding without affecting holdfast abundance
We hypothesized that the apparent discrepancy in the holdfast synthesis phenotypes of the A. biprosthecum and C. crescentus hfsH mutants was due to the difference in lectin staining conditions or to improvements in the sensitivity of our microscope camera. Previous holdfast staining of the C. crescentus ΔhfsH mutant (Toh et al., 2008) was performed by incubating cells with a relatively high concentration of AF488-WGA lectin, followed by a washing step to remove excess lectin, as described previously (Merker and Smit, 1988, Li et al., 2005). In order to eliminate the variation due to unavoidable small differences in washing conditions in the current study, we labeled holdfasts with a lower concentration of AF488-WGA lectin that robustly labeled wild-type holdfasts without producing a significant background (Fig. 2B), and therefore did not require a subsequent washing step. One possible explanation for the discrepancy in phenotypes was that the washing step in the previous version of the lectin staining assay removed shed holdfasts and holdfasts loosely bound to cells. Indeed, in the absence of a washing step using our improved lectin staining method, we detected a significant amount of shed holdfast and cell-associated holdfasts on C. crescentus ΔhfsH mutant cells (Fig. 2B). In contrast, when a culture of C. crescentus ΔhfsH was labeled with lectin and washed as in the previous method, no holdfasts were detected in the medium or on cells (Fig. 2B), confirming that washing removes shed and cell associated holdfasts and that the different in the washing step caused the discrepancy in phenotypes.
Time-lapse microscopy of holdfast synthesis on agarose pads was performed to assess the dynamics and timing of holdfast production and export (Fig. 2C) in both A. biprosthecum and C. crescentus. In wild-type A. biprosthecum cells, holdfast synthesis occurred in a compact region at the cell pole and the holdfast remained associated with the cell. In contrast, the holdfast of the A. biprosthecum hfsH mutant cells appeared to diffuse from the cell pole upon secretion as shown in early time points (Fig. 2C). This observation is consistent with the observation of shed holdfasts in the culture medium (Fig. 2A). Diffusion of the holdfast material is limited within the agarose, resulting in accumulation of holdfast at the cell pole as seen in later time points (Fig. 2C). Alternatively, the holdfast might be secreted from multiple sites on the cell pole. In C. crescentus, both the wild-type cells and ΔhfsH mutant produced a compact holdfast that remained at the cell pole (Fig. 2C), although occasional holdfasts could be seen detaching from the pole and diffusing away (data not shown). The above results indicate that, while the C. crescentus and A. biprosthecum hfsH mutant cells fail to bind to surfaces, they still synthesize holdfasts, but they are loosely bound to the cell body or shed into the medium.
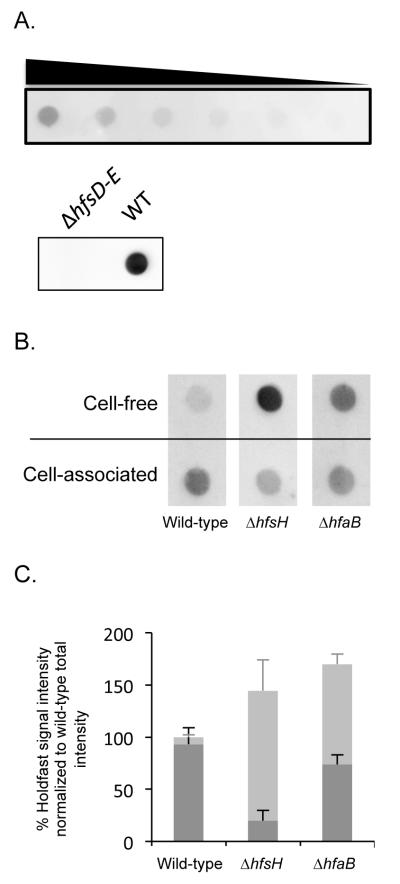
The holdfast shedding phenotype of the hfsH mutants are reminiscent of the observed phenotype of the hfaB mutant of C. crescentus, which also sheds holdfasts into the medium (Cole et al., 2003, Hardy et al., 2010). To compare the levels of holdfast production, secretion, and shedding in different strains, we quantified the amount of holdfast by polysaccharide lectin blotting with horseradish peroxidase (HRP)-conjugated WGA. In this assay, the signal is proportional to quantity of input holdfast, and a deletion mutant that lacks the entire holdfast synthesis gene cluster (ΔhfsD-E) produced no signal (Fig. 3A). Cell-associated holdfasts and cell-free holdfasts in the medium were separated by centrifugation, immobilized onto nitrocellulose membranes, and probed with HRP-WGA (Fig. 3B). In wild-type samples, only a small portion of holdfast signal (6%) was observed in the spent medium (Fig. 3C). In contrast, ΔhfsH and ΔhfaB cells shed significant amounts of holdfast into the medium. Furthermore, a higher fraction of the holdfast material was found in the medium for ΔhfsH cultures (86%) than ΔhfaB cultures (56%). Finally, the C. crecentus ΔhfsH mutant produced a total amount of holdfast polysaccharide similar to that of wild-type and ΔhfaB cells. These results confirm that the ΔhfsH mutant is not defective in the steady-state level of holdfast, and sheds holdfast into the medium. Holdfast shedding prevents efficient attachment of cells to surfaces in the hfaB mutant (Cole et al., 2003, Smith et al., 2003) and explains the impaired surface adhesion of the hfsH mutant.
Figure 3. Detection of holdfast production by hfsH mutants.
(A) Top panel: cell lysates from wild-type cultures were loaded onto a nitrocellulose membrane in a 2-fold serial dilution series from left to right and probed with HRP conjugated WGA. Bottom panel: cell lysates of wild-type and of a ΔhfsD-E mutant were loaded onto a nitrocellulose membrane and probed with HRP conjugated WGA. (B) Dot blots showing holdfast signal from cell-associated and cell-free polysaccharide extracts of C. crescentus wild-type, ΔhfsH mutant, and ΔhfaB mutant cells. All samples were treated with proteinase K and detected on a nitrocellulose membrane using chemiluminescence from an HRP conjugated WGA. (C) Quantification of the holdfast signal from the cell-associated (black bars) and cell-free (grey bars) fractions from each strain. Data are expressed as the percentage of the total holdfast intenstiy (the sum of the cell-associated and cell-free signals) from wild-type, based on dot-blot chemiluminescent signals. Error bars represent the standard error of the mean from three replicates.
A ΔhfsH mutant sheds holdfasts with reduced cohesiveness
The holdfast shedding phenotype of the ΔhfsH mutant is more severe than that of the ΔhfaB mutant (Fig. 3C), suggesting that the phenotype of the ΔhfsH mutant is not simply due to a defect in holdfast anchoring. An alternative explanation for the holdfast shedding phenotype of the ΔhfsH mutant is the modification of the adhesive properties of the holdfast. Indeed, the predicted function of HfsH as a polysaccharide deacetylase suggests that the protein may be responsible for key modifications to the holdfast polysaccharide, thereby disabling permanent surface attachment. The shed holdfasts of the C. crescentus and A. biprosthecum hfsH mutant cells appeared to be smaller than wild-type holdfasts (Fig. 2), and time-lapse microscopy suggested that the holdfast material was more diffuse in the A. biprosthecum hfsH mutant (Fig. 2C). Since cohesiveness involves the ability of components to interact with each other and a decrease in cohesiveness results in dispersal, we hypothesized that the holdfast is an aggregate of adhesin molecules and hfsH mutants may shed holdfast as a consequence of reduced or altered cohesiveness.
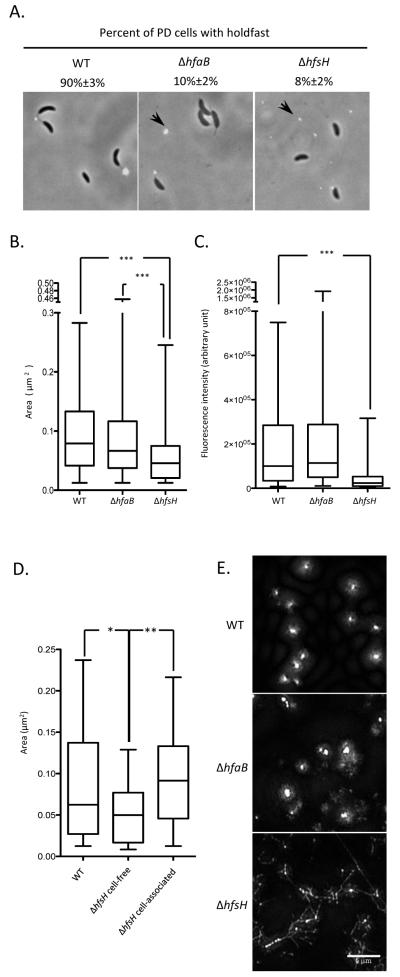
To determine if the C. crescentus ΔhfsH mutant produces smaller, fragmented holdfasts, holdfasts were analyzed by AF488-WGA labeling (Fig. 4A), and the area and fluorescent signal intensity were used as a quantitative measure of the size and amount of holdfast, respectively (Li et al., 2012). The comparison of holdfast size (Fig. 4B) and amount (Fig. 4C) revealed that shed holdfasts of the ΔhfsH mutant are smaller than those of wild-type and the ΔhfaB mutant. Additionally, holdfasts shed by the ΔhfsH mutant were smaller than cell-associated holdfasts (Fig. 4D). Notably, cell-associated holdfasts from ΔhfsH and wild-type cells were not significantly different from one another (Fig. 4D). These results suggest that the hfsH holdfasts are fragmented after secretion, presumably due to low cohesiveness, but are still tight and compact when associated with the cells.
Figure 4. The ΔhfsH mutant sheds small holdfasts.
(A) Overlays of phase-contrast and epifluorescence micrographs. The percentage of AF488-WGA-labeled predivisional (PD) cells with polar holdfasts are expressed as a mean percent from three independent experiments (n=50~75 for each) with the standard error of the mean. The position of some shed holdfasts is indicated by black arrow. The scale bar represents 2 μm. (B) Box plot showing the area of the holdfast (as detected by AF488-WGA signal) for wild-type, ΔhfaB and ΔhfsH holdfasts. (C) Box plot showing the integrated intensity of the holdfast (as detected by AF488-WGA signal) for wild-type, ΔhfaB and ΔhfsH holdfasts. (D) Box plot showing the area of cell-associated holdfasts from wild-type, ΔhfsH, as well as cell-free holdfasts of the ΔhfsH mutant, and cell-associated holdfast from the ΔhfsH mutant. In (B), (C) and (D), data from three experiments (n=50 for each) were used for quantification. In the box-plots, the horizontal bar represents the median, the box represents the 2nd and 3rd quartile, and the whiskers denote the full range of the data. The variance between two groups was analyzed by t-tests. The asterisks denote significant difference between two samples.*** represents P<0.0001. ** represents P<0.01. * represents P<0.05. (E) Surface-attached holdfasts from wild-type, ΔhfsH and ΔhfaB cells imaged by structured illumination microscopy after AF488-WGA labeling. The scale bar represents 5 μm.
The cohesiveness of an adhesive is often influenced by a change in inter-molecular interactions. The alteration of these interactions can modify the aggregation of molecules and disrupt the adhesin structure. To analyze the structure of holdfasts at high resolution, holdfasts from wild-type, ΔhfsH and ΔhfaB cells were attached to surfaces, labeled with AF488-WGA, and imaged by structured illumination super-resolution microscopy (Fig. 4E). Even if holdfasts from both the ΔhfaB and ΔhfsH mutants were less compact than the wild-type holdfasts, ΔhfsH mutant holdfasts appeared drastically different: they had both a punctate and a widespread fiber-like appearance, which was not detectable in the holdfasts of wild-type and the ΔhfaB cells (Fig. 4E). We conclude that the absence of HfsH reduces holdfast cohesiveness and causes its dispersion into small particles and fiber-like structures.
hfsH expression level regulates holdfast adhesiveness
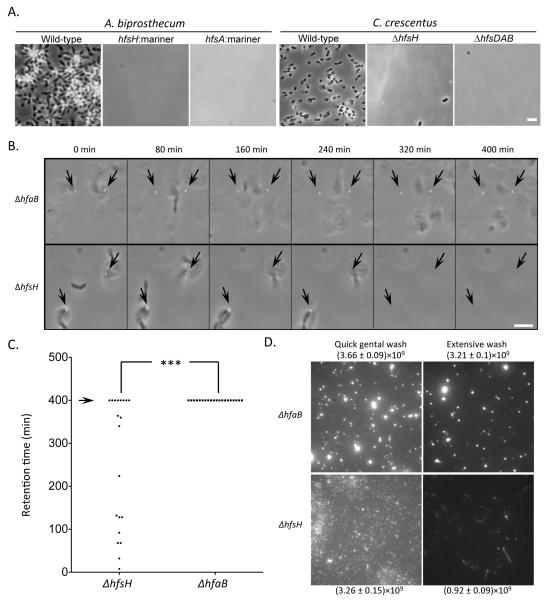
The inability of ΔhfsH cells to attach to surfaces (Toh et al., 2008) suggests that their holdfasts have a decreased adherence. Indeed, while wild-type cells of both C. crescentus and A. biprosthecum tightly adhered to glass coverslips (Fig. 5A), neither hfsH mutant cells nor their shed holdfasts attached to glass coverslips (Fig. 5A), consistent with decreased adhesiveness. To further test the adhesiveness of the mutant holdfasts, we used the ONIX® microfluidics perfusion system to monitor the detachment of surface adhered holdfasts from the ΔhfsH and ΔhfaB mutants under defined flow conditions in real time. The same fluid shear force was applied by a constant flow of medium over attached ΔhfsH and ΔhfaB mutant cells, and the behaviors of holdfasts and cells were recorded (Fig. 5B). The dwell times of 20 attached holdfasts from the ΔhfsH and ΔhfaB mutants were determined over the course of a 400-minute experiment (Fig. 5C). All the ΔhfaB holdfasts remained surface-associated, even after the cell bodies detached from the holdfasts. In contrast, 60% of the ΔhfsH holdfasts detached from the surface (Fig. 5C). Therefore, the ΔhfsH holdfasts are less resistant to shear force, indicating that the ΔhfsH holdfasts have decreased adhesiveness.
Figure 5. Holdfast from the ΔhfsH mutant have reduced adhesiveness.
(A) Coverslip binding assay showing the adhesion deficiency of the A. biprosthecum ΔhfsH mutant and the C. crescentus ΔhfsH mutant. Overlays of phase and fluorescence are shown. (B) Time-lapse images illustrate the effect of fluid shear force on holdfasts of the ΔhfaB and ΔhfsH mutants in ONIX perfusion microfludics channels. The position of holdfasts attached to the surface at the onset of the experiment are indicated by black arrows. (C) Quantification of the retention time (min) of 20 attached holdfasts for each strain after 400 min (arrow) of exposure to 15 μl/min of flow. The asterisks denote significant difference between the ΔhfaB and the ΔhfsH mutants. The variance between two groups was analyzed by t-tests. ***represents P<0.0001. (D) Effect of two different shear forces on attached holdfasts. The amount of holdfast material present after a 10 sec “Quick gentle wash” or a 4 h “extensive wash” was quantified by measuring the sum of the fluorescence intensity from AF488-WGA labeling of a 82 μm× 82 μm area. Fluorescence intensities are expressed as a mean with the standard error of the mean. Data represent the means of eight measurements of three independent experiments.
In order to test the effect of shear stress on an entire population of adhered holdfasts, ΔhfsH and ΔhfaB cells were deposited onto separate wells of a multi-well glass slide under static conditions for 6 h, and different flow regimes were applied to the slide. An extensive wash removed 65% of the ΔhfsH holdfast (Fig. 5D, bottom panels) from the glass surface as compared to 13% for ΔhfaB holdfast (Fig. 5D, top panels). Furthermore, the holdfasts shed by the hfaB mutant retained a punctate appearance, while the holdfasts shed by the hfsH mutant often had a fiber-like appearance after the extensive wash. These results are consistent with the absence of HfsH causing a decrease in both cohesiveness and adhesiveness of the holdfast polysaccharide.
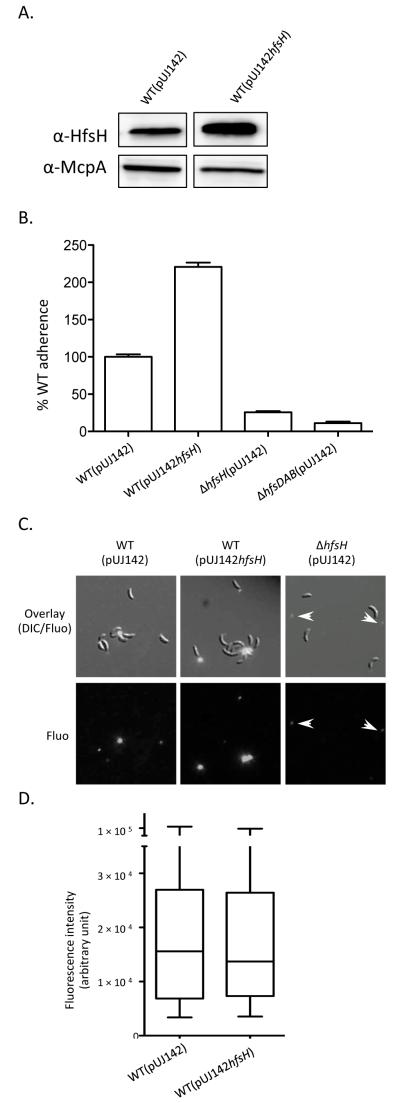
If deletion of hfsH decreases holdfast adhesiveness and cohesiveness because it reduces holdfast deacetylation, then increasing its expression would be expected to increase holdfast deacetylation and cell adhesion. We therefore examined the effect of hfsH overexpression on holdfast adhesiveness by introducing a high-copy replicating plasmid bearing a copy of hfsH (pUJ142hfsH) into the wild-type strain. Overexpression of HfsH (Fig. 6A) for 5 hr more than doubled surface adhesion in a short term adhesion assay that measures single cell adhesion (Fig. 6B). Overexpression of HfsH did not change the fluorescence intensity of the holdfast following fluorescent lectin staining (Fig. 6C and 6D). These results suggest that the elevated surface adherence after HfsH overexpression is due to an increase in holdfast adhesiveness.
Figure 6. The overexpression of hfsH increases surface adherence.
(A) Western blots of whole cell lysates showing the expression level of HfsH in WT(pUJ142hfsH) and WT(pUJ142) after induction with 0.2% w/v xylose for 5 hr. McpA expression level was monitored as a loading control. (B) Quantification of cell adherence to polystyrene following HfsH overexpression for 5hr and short term surface binding (45 min). Data represent the mean of three measurements for three independent experiments. Error bars represent the standard error of the mean. (C) Detection and analysis of holdfast synthesis by AF488-WGA binding under the same conditions as (B). The top panel shows the overlays of DIC and epifluorescence (fluo) micrographs, and the bottom panel shows epifluorescence (fluo) alone. The position of shed holdfasts are indicated by white arrows. (D) Box plot showing the integrated intensity of the holdfast (as detected by AF488-WGA signal). Data from three experiments (n=50 for each) were used for quantification. The horizontal bar represents the median, the box represents the 2nd and 3rd quartile, and the whiskers denote the full range of the data.
A predicted key enzymatic residue of the HfsH polysaccharide deacetylase is required for holdfast adhesive properties
The ability of HfsH to alter adhesive properties of the holdfast may be due to the predicted function of HfsH as a polysaccharide deacetylase of the carbohydrate esterase family 4 (CE4). The CE4 family of polysaccharide deacetylases exhibits enzymatic activities toward a wide range of bacterial polysaccharides (Caufrier et al., 2003). Five catalytic motifs comprise the active site of the deacetylase domain in CE4 family members and the key enzymatic residues in each motif have been identified (Caufrier et al., 2003, Zhao et al., 2010). To investigate the possibility that HfsH modifies holdfast polysaccharides by deacetylation, we first compared the protein sequence of HfsH proteins from C. crescentus and A. biprosthecum with three biochemically characterized CE4 members (Blair and van Aalten, 2004, Blair et al., 2005, Blair et al., 2006) (Fig. S1). Both HfsH proteins contain 4 conserved motifs, including the essential acetate binding residues (blue triangles Fig. S1) and the zinc-binding triad (black box Fig. S1). Thus, HfsH has all the known essential components required to function as polysaccharide deacetylase.
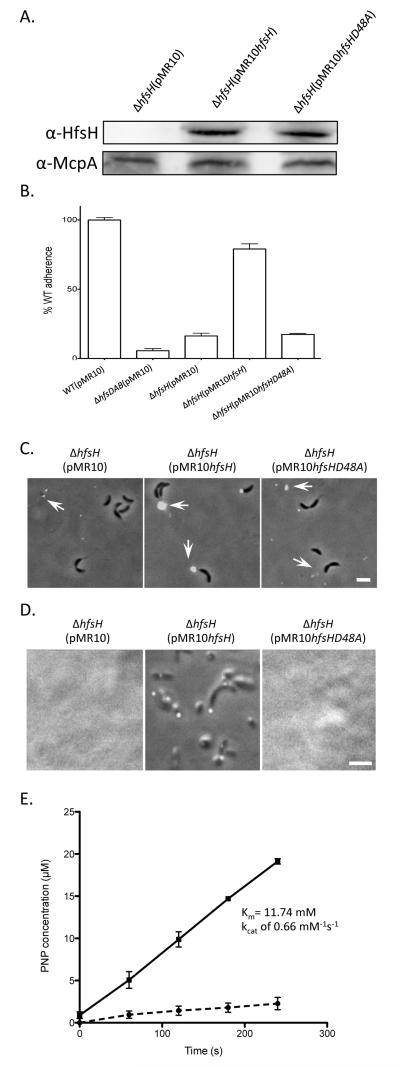
To examine the importance of the deacetylase activity of HfsH in vivo, a point mutation was introduced in C. crescentus hfsH at a predicted key enzymatic residue, resulting in an amino acid change from aspartic acid to alanine at position 48 (D48A) (star in Fig. S1). A low-copy replicating plasmid bearing a copy of either the wild-type (pMR10hfsH) or the mutated hfsH gene (pMR10hfsHD48A) was introduced in the C. crescentus ΔhfsH mutant. Western blot analysis indicated that both the plasmid-borne wild-type and mutant proteins were expressed at similar levels (Fig. 7A). Adhesion assays revealed that wild-type HfsH, but not HfsHD48A, could complement the adhesion defect of ΔhfsH cells (Fig. 7B). Similar to the ΔhfsH mutant, the hfsHD48A mutant was deficient in holdfast anchoring to the cell body and small holdfasts were shed into the medium (Fig. 7C). In contrast, the ΔhfsH mutant complemented with wild-type hfsH produced large, compact, cell-associated holdfasts. Finally, holdfasts produced by the ΔhfsH mutant expressing HfsHD48A could not adhere to surfaces, whereas holdfasts from the ΔhfsH mutant expressing wild-type HfsH readily attached (Fig. 7D). We purified both wild-type HfsH and HfsHD48A from E. coli, and confirmed that they have similar secondary structure based on circular dichroism measurement (Fig. S2). This indicates that the D48A did not affect HfsH structure. Esterase activity assays indicated that HfsH has esterase activity (Km = 11.74 mM, kcat = 0.66 mM−1s−1) that is comparable to that of other characterized polysaccharide deacetylase (Waters et al., 2012). This activity is abolished by the D48A mutation (Fig. 7E). These results indicate that the esterase activity of HfsH is essential for production of a functional cell-associated holdfast with both cohesive and adhesive properties.
Figure 7. HfsHD48A lacks esterase activity and its expression does not complement the ΔhfsH mutant.
(A) Western blots of whole cell lysates showing the expression level of wild-type HfsH and HfsHD48A proteins expressed in a ΔhfsH mutant. (B) Quantification of cell adherence to polystyrene following short term binding (45 min). Data represent the means of three measurements of three independent experiments. Error bars represent the standard error of the mean. (C) Both ΔhfsH (pMR10) and ΔhfsH (pMR10hfsHD48A) produce and shed smaller holdfasts than ΔhfsH (pMR10hfsH) as detected by AF488-WGA binding. The position of holdfasts is indicated by white arrows. (D) Coverslip binding assay showing the adhesion deficiency of CB15 ΔhfsH (pMR10hfsHD48A) in contrast to the binding of CB15 ΔhfsH(pMR10hfsH) cells and holdast. (E) HfsH (black line) and HfsHD48A (black dashed line) esterase activity using PNPA as a substrate. The assay mixture consists of 40 mM phosphate buffer at pH 7.4, 0.2 μM HfsH and 0.5 mM PNPA, in a total volume of 100 μl.
DISCUSSION
The holdfast of C. crescentus forms a small compact structure with elastic properties at the tip of stalks, and tightly adheres to surfaces with an impressive adhesion force in the μN range (Merker and Smit, 1988, Li et al., 2005, Tsang et al., 2006). Disruption of the hfsH polysaccharide deacetylase gene does not abolish holdfast synthesis. However, the mutant holdfasts detach from cells, bind weakly to surfaces, fail to maintain a cohesive structure, and disperse into small fragments that adopt a “fiber-like” appearance. The fiber-like appearance may reflect the structure of the holdfast in solution or may be a footprint of holdfast fragments left on the surface by disintegrating holdfasts (Neu, 1992). Our results revealed that holdfast shedding is due to a disruption of its cohesive properties, leading to a loss of its native compact aggregation state. We also showed that the holdfast anchoring defect of the ΔhfsH mutant is not due to a defect in the ability of the holdfast anchoring machinery to multimerize in the outer membrane or to localize to the cell pole (Fig. S3). Perhaps deacetylation exposes an amine that is required for interaction of the polysaccharide with the anchor. Alternatively, the decreased cohesiveness of the hfsH mutant causes most of the holdfast polysaccharide to be shed with only a small, undetectable, amount still bound to the anchor.
Bacterial exopolysaccharides typically have a regular chemical structure based on two to eight sugar repeat units. These polysaccharides may adopt an ordered helical conformation formed by one to three chains of repeats, which can be detected as strands (Sorlier et al., 2001, Rinaudo, 2004). The adhesive characteristics of exopolysaccharides, strongly depends on chain conformation, and is greatly influenced by substituents that modify interchain and intrachain interactions (Haag, 2006). For example, deacetylation of polysaccharides might facilitate the conformational transition of the polymer strands from random coils to ordered helices so as to promote gel formation, which is mediated by interspersed regions of soluble, hydrated polymer with regions of polymer-polymer interactions (Villain-Simonnet et al., 2000, Rinaudo, 2004). Acetyl groups have been shown to be essential for the stability of bacteria polysaccharides and subsequent biofilm development (Ridout et al., 1997, Villain-Simonnet et al., 2000, Tielen et al., 2005). These findings provide further support for a model in which changes in chemical composition, including the degree of deacetylation, impact the physical properties of the holdfast polysaccharides.
Extensive studies on the chitin-chitosan system also suggest that modifying the acetylation state of polysaccharides alters their chemical-physical properties. Partial deacetylation of chitin, a ubiquitous GlcNAc polymer, leads to the production of chitosan, which contains more exposed amine groups and fewer acetyl groups. During the production of chitosan, the intrinsic pKa (pK0) was found to increase from 6.46 to 7.32 as a function of the degree of deacetylation (Sorlier et al., 2001). The degree of deacetylation influences the physical properties of chitosan by altering electrostatic interactions, hydrogen bonding, and hydrophobic interactions with the surrounding environment (Sorlier et al., 2001). By analogy, our results showing that deletion of hfsH reduces adherence while its overexpressing increases it suggest that proper deacetylation of the holdfast GlcNAc polysaccharide alters holdfast physical properties. We hypothesize that the absence of HfsH prevents proper modification of the holdfast polysaccharide, thereby decreasing holdfast cohesiveness and adhesiveness. In contrast, overexpression of HfsH should introduce a higher level of deacetylation, leading to increased adhesiveness. Unfortunately, the detailed composition of the holdfast is unknown and it is produced in small quantity. These factors have limited our ability to directly test the level of deacetylation of the holdfast polysaccharide in the experiments reported here.
The high sequence similarity of the two HfsH proteins in C. crescentus and A. biprosthecum, and the similar phenotypes resulting from the disruption of hfsH, strongly suggest that HfsH has a conserved role in the regulation of polar holdfast properties. Additionally, the observed shedding of holdfasts from the hfsH mutant cells is consistent with the phenotype caused by the loss of polysaccharide deacetylation in S. epidermidis. S. epidermidis PIA is typically produced as fibrous strands on the cell surface; however, a strain lacking the putative polysaccharide deacetylase enzyme IcaB causes release of PIA from the cell surface (Vuong et al., 2004). This strengthens the argument that deacetylation is a general mechanism for promoting polysaccharide adhesiveness. While IcaB localizes to the cell surface and deaceylates surface-associated PIA at random positions (Vuong et al., 2004), HfsH is a soluble protein lacking a secretion signal sequence, and is therefore predicted to modify the polysaccharide precursors in the cytoplasm (Fig. S4). Cytoplasmic deacetylation of polysaccharide precursors occurs prior to polysaccharide polymerization, and may allow for more precise control of the degree of acetylation in the adhesive polysaccharide.
In conclusion, we have shown that deacetylation plays an important role in bacterial adhesion by modulating adhesive and cohesive properties of a polysaccharide adhesin. The hfsH mutant may also prove useful in further characterization of the holdfast. Until now, a thorough knowledge of how the the high adhesive force of the holdfast is achieved has been hampered by the fact that the composition of the holdfast remains unknown. Although the physical and chemical properties of the holdfast produced by the ΔhfsH mutant are altered, the resulting holdfast is more soluble and should make it possible to purify sufficient amount of holdfast material for composition and structure analysis. Such an analysis will provide insights into how the holdfast generates such impressive adhesive and cohesive forces, and will have a significant impact on the study of bacterial adhesion.
EXPERIMENTAL PROCEDURES
Bacterial strains, plasmids and growth conditions
Strains and plasmids used in this study are listed in Table S2. Escherichia coli strains were grown at 37°C in Luria-Bertani (LB) broth or on LB agar supplemented with kanamycin (30 μg/ml in broth, 50 μg/ml or 25 μg/ml in agar) as needed. A. biprosthecum strains were grown at 26°C in peptone-yeast extract (PYE) broth or on PYE agar supplemented with kanamycin (5 μg/ml in broth, 20 μg/ml in agar) and nalidixic acid (20 μg/ml in agar) as needed. C. crescentus strains were grown at 30°C in PYE supplemented with kanamycin (5 μg/ml in broth, 20 μg/ml in agar), nalidixic acid (20 μg/ml in agar), or chloramphenicol (1 μg/ml in broth or 0.5 μg in agar) as necessary. Plasmids were introduced into E. coli by transformation and into C. crescentus by conjugation (Ely, 1979, Ely, 1991). B. diminuta was grown at 26°C in PYE broth, and H. baltica was grown at 26°C in Hirschia medium (ATCC medium 1883).
C. crescentus strains carrying pUJ142 derivatives were grown overnight in PYE supplemented with 0.5 μg/ml chloramphenicol and 0.5% glucose (repressing condition), and subcultured to OD600~0.1 in PYE supplemented with 0.5 μg/ml chloramphenicol and 0.2% xylose (inducing condition) and grown to OD600~0.6 before the cells were collected and analyzed as described.
Transposon mutagenesis, adhesion mutant identification, and mutation mapping
Transposon mutants were generated by conjugation. A spontaneous nalidixic acid resistant strain of A. biprosthecum (YB5191) was mated with E. coli cells containing the mariner transposon on a plasmid (Rubin et al., 1999). A. biprosthecum transposon mutants were selected based on kanamycin resistance, and screened for adhesion defects using a short-term binding assay. Genomic DNA from mutants of interest with confirmed phenotypes was isolated using the Bactozol extraction kit, and was subsequently used as a template for Touch-Down PCR (Levano-Garcia et al., 2005) to identify the insertion site of the mariner transposon. The PCR products were purified using the Qiagen QIAquick purification kit and sequenced using amplification from the transposon specific primer at the Institute for Molecular and Cellular Biology, Indiana University, Bloomington. The specific primers are MarLseq (5′ GGGAATCATTTGAAGGTTGGT 3′) and MarRseq (5′ CGGGTATCGCTCTTGAAGG GA 3′); the arbitrary primers are MarTDL2 (5′ GACACGGGCCTCGANGNNNCNTNGG 3′) and MarTDR1 (5′ CAACCGTGGCGGGGNTNCNNGNCNCG 3′).
Short-term binding assays
This assay was performed as previously described with the following modifications (Hardy et al., 2010). Overnight cultures were diluted with fresh PYE to an OD600 of 0.15 and incubated at 30°C until cultures reached an OD600 between 0.35 and 0.60. Cultures were diluted to an OD600 of 0.30 and added to the wells of a 12-well plate (750 μl per well) or a 24-well plate (500 μl per well). Plates were incubated with shaking at room temperature for 15 min for C. crescentus or 90 min for A. biprosthecum. The cell culture was removed, and the wells were washed twice with fresh PYE to remove unattached cells. Each well was stained with 400 μl of 0.1% (wt/vol) crystal violet for 15 min. Wells were washed four times with H2O to remove excess crystal violet. After washing, the crystal violet was eluted with 500 μl of 10% acetic acid. Absorbance was measured at 589 nm.
Lectin binding assays
Alexa Fluor (AF) 488/594-conjugated wheat germ agglutinin (WGA) (Molecular Probes) was added to 100 μl cell cultures to a final concentration of 0.5 μg/ml, and incubated for 10 min before visualization by epifluoresence microscopy.
Epifluorescence microscopy and image analysis
Microscopy was performed on a Nikon Eclipse 90i equipped with Chroma 83000 filter set, a 100X (DIC) oil objective, and a Photometrics Cascade 1K EMCCD camera or a Nikon Eclipse Ti-e equipped with a Chroma 89021 filter set, a 60x plan Apo oil objective (1.5x magnifier), and an Andor iXon EMCCD camera. Images were captured using Metamorph Imaging Software package version 7.5.4. or Nikon NIS Elements advance research version 4.0. The area of individual holdfast was isolated from the background with the image thresholding function in ImageJ software, and area and fluorescence intensity from isolated areas were measured with the particle analysis function in ImageJ (Schneider et al., 2012).
The structured illumination microscopy used a prototype of the commercial model Deltavision|OMX V2.0 (Applied Precision Inc, Issaquah, WA) with a 100x 1.40 NA oil objective (UpanSApo, Olympus, Japan), and 1.516 index immersion oil. Fluorescence emission was collected by the same objective, split by channel, and filtered using an FF01-512/25-25 fluorescence filter (Semrock, Rochester, NY) for the 488 excitation channel. Fluorescence images were captured by a dedicated Cascade II 512 EM-CCD camera (Photometrics, Tuscon AZ). Acquisition was controlled by the DV-OMX controller software (Applied Precision Inc, Issaquah, WA). Reconstructions were made with the OMX specific SoftWorx v4.5.0 software package (Applied Precision Inc., Issaquah, WA).
Time-lapse microscopy
C. crescentus and A. biprosthecum overnight cultures were diluted and allowed to grow to mid-log phase (OD600 ~0.45 to 0.65). PYE medium containing 1% (w/v) agarose and 0.5 μg/ml AF488-WGA was applied to a glass slide to form an agarose pad. Cultures were then diluted to approximately OD600 ~0.2 and spotted on the PYE agarose pads. Cells were imaged at 5-10 min time intervals using either a Nikon Eclipse 90i light microscope equipped with a motorized stage and a software-based autofocusing loop or a Nikon Eclipse Ti with a motorized stage and the hardware autofocusing function (Nikon Perfect Focus®).
Alternatively, cells from exponential phase were diluted with PYE to OD600 ~0.1, and loaded onto the ONIX Microfluidic Platform (CellASIC) with ~100 cells trapped in individual ~10−5 μl chambers (3 μm in depth, and 3.0×1.2 mm in area). The cells were cultured inside the channels for 3 h using PYE containing 0.5μg/ml AF488-WGA at a laminar flow rate of 2.5μl/h to enrich attached cells on the glass bottom. Then the flow rate was increased to 15 μl/h, and the behavior of attached holdfasts and cells was monitored by taking fluorescence and bright field images every 4 min for 400 h using a Nikon Eclipse Ti inverted light microscope equipped with a motorized stage and the hardware autofocus (Nikon Perfect Focus®) function. Image stacks were segmented and processed using NIS elements and ImageJ software.
Polysaccharide lectin blotting
Cells were harvested by centrifugation (30 min at 4,000 × g) from overnight PYE culture, and resuspended in fresh PYE to OD600 ~0.1. The cultures were incubated at 30°C under shaking condition until the desired cell density was reached. The cells and media were separated by centrifugation at 6,000 × g for 10 min. Cells were resuspended in 100 mM Tris pH = 8, and proteinease K was added to a final concentration of 0.4 mg/ml followed by 3-5 h incubation at 70°C. Samples were adsorbed to a nitrocellulose membrane using a Bio-Dot microfiltration apparatus (Bio-Rad Laboratories, Hercules, CA). The membrane was blocked with 3% BSA in TBS for 1 h, then incubated with a 1:20,000 dilution of wheat germ agglutinin (WGA) lectin conjugated to horseradish peroxidase (HRP) (Sigma, St. Louis, MO) for 1 h at room temperature. The blots were developed with SuperSignal West PICO Substrate (Thermo Scientific, Rockford, IL). A Kodak ImageStation 440 CF or 4000mm PRO was used for image processing and chemiluminescent signal quantification for each isolated dot using 96-well grid analysis.
Western Blot Analysis
Cell lysates were prepared from exponentially growing cultures (OD600 = 0.35 – 0.7). The equivalent of 1.0 ml of culture at OD600 = 1 was centrifuged at 16,000 × g for 2 min at room temperature. The supernatant was removed and the cell pellets were resuspended in 50 μl 10 mM Tris pH 8.0, and 50 uL of 2x SDS sample buffer was added to the cell suspension. Samples were boiled for 5 min before being run on a 12% w/v polyacrylamide gel, and transferred to nitrocellulose membranes. Membranes were blocked for 1 h in 5% w/v nonfat dry milk in TBST (20 mM Tris, pH 8, 0.05% v/v Tween 20), and incubated with antibody overnight at 4°C. HfsH and McpA antibodies were used at a concentration of 1:1000 and 10,000 respectively. Then, a 1:10,000 dilution of secondary antibody, horseradish peroxidase (HRP)-conjugated goat anti-rabbit immunoglobulin, was incubated with membranes at room temperature for 1h. Membranes were developed with SuperSignal West Dura Substrate (Thermo Scientific, Rockford, IL).
Holdfast attachment assay
A 12-well glass multitest slide (MP Biomedicals) or glass coverslips were pre-cleaned by overnight soaking in 100% Micro-90 detergent to clean slides and to maximize holdfast binding (Berne et al., 2010). Cells were harvested by centrifugation (30 min at 4,000 × g) from overnight PYE culture, and resuspended in fresh PYE to OD600 of 0.1. Mulitple 50 μl aliquots of cell culture were spotted on a precleaned 12-well glass multitest slide or coverslip, and incubated for 5 h at 30°C in a humid chamber. After incubation, the slides or coveslips were washed with ddH2O with a squirt bottle or by immersion in a shaking water pool. 10 μl AF 488-WGA (50 μg/ml) was added and incubated in the dark for 20 min at room temperature. Slides were then washed with ddH2O, topped with a large glass coverslip (24 × 60 mm) and sealed with nail polish. All the samples were analyzed by epifluorescence microscopy.
Site-directed mutagenesis of hfsH
The single amino acid substitution was constructed using the Quick Change Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) according to the manufacturer’s instructions, using pMR10::Phfs-hfsH as a template (Toh et al., 2008). The following primer pair was used: 5′ GTG TCG TTC AGC TTC GCG GAC GCC CCC GCC ACC3′ and 5′ GGT GGC GGG GGC GTC CGC GAA GCT GAA CGA CAC3′.
Protein purification and antibody production
The pET28ahfsH or pET28ahfsHD48A construct was transformed into E. coli BL21(DE3). Cultures (10 ml) were grown overnight in LB medium with kanamycin (30 μg/ml) and used to inoculate 1 L of LB medium with kanamycin. The cells were grown at 37°C to OD600= 0.6, expression was induced by the addition of 5 mM isopropyl-d-thiogalactopyranoside. The cells were then cultured for an additional 4 h at 37°C and harvested by centrifugation at 5000 × g for 15 min. For antibody production, recombinant HfsH-His purified by metal-chelate affinity chromatography using Ni2+-NTA-agarose (Qiagen) under denaturing conditions using a standard protocol (Qiagen). A total of 6 mg of purified HfsH protein was used to immunize two rabbits (Josman, LLC) for the production of polyclonal antibodies. Antibodies were affinity purified by treating the crude serum with acetone powders prepared from the ΔhfsH mutant according to standard procedures. (Viollier et al., 2002).
For enzymatic assays, HfsH was purified under native condition using a HisTrap HP column and the A KTA FPLC system (GE Biosciences). Protein was eluted with a linear gradient from 0 to 500 mM imidazole. Fractions containing HfsH were dialyzed twice in 1 L of 40 mM phosphate buffer at pH 7.4, and concentrated to ≈10 mg/mL (as determined by Bradford protein assay) with a 10 kDa cutoff centrifugal filter (Amicon). Samples were kept at −80°C after the addition of 10% v/v glycerol.
Enzymology
An esterase assay was performed as previously described by measuring the degradation of p-nitrophenol acetate (PNPA) into p-nitrophenol at 410 nm with the following modifications (Hassan and Hugouvieux-Cotte-Pattat, 2011). The molar extinction coefficient of p-nitrophenol was determined by plotting A410 versus standard p-nitrophenol solutions with different concentrations. The standard assay mixture consisted of 40 mM phosphate buffer at pH 7.4, 0.5 mM PNPA, and 0.2 μM HfsH in a total volume of 100 μl. The Km and kcat values were determined under the standard conditions at 37°C using substrate concentrations between 0.1 and 4 μM. Under each condition, the spontaneous cleavage of the substrate was tested in parallel by omitting the extract addition. The Data were analyzed by using nonlinear regression analysis with PRISM (GraphPad Software, Inc.) with the default equations for first-order reaction rates and Michaelis-Menten steady-state kinetics.
Bioinformatic analysis
The NCBI BLAST (http://abcis.cbs.cnrs.fr/propsearch/) program was used to identify proteins with amino acid similarity (Camacho et al., 2009). Expresso (http://tcoffee.crg.cat/apps/tcoffee/do:expresso) was used to perform multiple protein sequence alignments using structural information (Notredame et al., 2000).
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health Grants GM102841 to Y.V.B. P.J.B.B. was supported by a postdoctoral National Institutes of Health National Research Service Award number F32AI072992 from the National Institute of Allergy and Infectious Diseases. SIM microscopy was performed in the Indiana University Light Microscopy Imaging Center (LMIC) on a Deltavision OMX microscope, whose purchase was funded by grant S10RR028697 from the National Institutes of Health. We thank members of our laboratory for critical reading of the manuscript.
REFERENCES
- Berne C, Kysela DT, Brun YV. A bacterial extracellular DNA inhibits settling of motile progeny cells within a biofilm. Mol Microbiol. 2010;77:815–829. doi: 10.1111/j.1365-2958.2010.07267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair DE, Hekmat O, Schuttelkopf AW, Shrestha B, Tokuyasu K, Withers SG, van Aalten DM. Structure and mechanism of chitin deacetylase from the fungal pathogen Colletotrichum lindemuthianum. Biochemistry. 2006;45:9416–9426. doi: 10.1021/bi0606694. [DOI] [PubMed] [Google Scholar]
- Blair DE, Schuttelkopf AW, MacRae JI, van Aalten DMF. Structure and metal-dependent mechanism of peptidoglycan deacetylase, a streptococcal virulence factor. Proc Natl Acad Sci U S A. 2005;102:15429–15434. doi: 10.1073/pnas.0504339102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair DE, van Aalten DMF. Structures of Bacillus subtilis PdaA, a family 4 carbohydrate esterase, and a complex with N-acetyl-glucosamine. FEBS Letters. 2004;570:13–19. doi: 10.1016/j.febslet.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Bodenmiller D, Toh E, Brun YV. Development of surface adhesion in Caulobacter crescentus. J Bacteriol. 2004;186:1438–1447. doi: 10.1128/JB.186.5.1438-1447.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PJB, Hardy GG, Trimble MJ, Brun YV. Complex Regulatory Pathways Coordinate Cell-Cycle Progression and Development in Caulobacter crescentus. In: Robert KP, editor. Adv Microb Physiol. Academic Press; 2008. pp. 1–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden T. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic acids research. 2009;37:D233–238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caufrier F, Martinou A, Dupont C, Bouriotis V. Carbohydrate esterase family 4 enzymes: substrate specificity. Carbohydr Res. 2003;338:687–692. doi: 10.1016/s0008-6215(03)00002-8. [DOI] [PubMed] [Google Scholar]
- Cerca N, Jefferson KK, Maira-Litran T, Pier DB, Kelly-Quintos C, Goldmann DA, Azeredo J, Pier GB. Molecular basis for preferential protective efficacy of antibodies directed to the poorly acetylated form of staphylococcal poly-N-acetyl-beta-(1-6)-glucosamine. Infect Immun. 2007;75:3406–3413. doi: 10.1128/IAI.00078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chertkov O, Brown PJ, Kysela DT, de Pedro MA, Lucas S, Copeland A, Lapidus A, Del Rio TG, Tice H, Bruce D, Goodwin L, Pitluck S, Detter JC, Han C, Larimer F, Chang YJ, Jeffries CD, Land M, Hauser L, Kyrpides NC, Ivanova N, Ovchinnikova G, Tindall BJ, Goker M, Klenk HP, Brun YV. Complete genome sequence of Hirschia baltica type strain (IFAM 1418(T)) Standards in genomic sciences. 2011;5:287–297. doi: 10.4056/sigs.2205004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JL, Hardy GG, Bodenmiller D, Toh E, Hinz A, Brun YV. The HfaB and HfaD adhesion proteins of Caulobacter crescentus are localized in the stalk. Mol Microbiol. 2003;49:1671–1683. doi: 10.1046/j.1365-2958.2003.03664.x. [DOI] [PubMed] [Google Scholar]
- Cuthbertson L, Mainprize IL, Naismith JH, Whitfield C. Pivotal Roles of the Outer Membrane Polysaccharide Export and Polysaccharide Copolymerase Protein Families in Export of Extracellular Polysaccharides in Gram-Negative Bacteria. Microbiol. Mol. Biol. Rev. 2009;73:155–177. doi: 10.1128/MMBR.00024-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan RM. Biofilms: microbial life on surfaces. Emerg Infect Dis. 2002;8:881–890. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunne WM. Bacterial Adhesion: Seen Any Good Biofilms Lately? Clinical Microbiology Reviews. 2002;15:155–166. doi: 10.1128/CMR.15.2.155-166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B. Transfer of drug resistance factors to the dimorphic bacterium Caulobacter crescentus. Genetics. 1979;91:371–380. doi: 10.1093/genetics/91.3.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B. Genetics of Caulobacter crescentus. Methods in enzymology. 1991;204:372–384. doi: 10.1016/0076-6879(91)04019-k. [DOI] [PubMed] [Google Scholar]
- Entcheva-Dimitrov P, Spormann AM. Dynamics and Control of Biofilms of the Oligotrophic Bacterium Caulobacter crescentus. J. Bacteriol. 2004;186:8254–8266. doi: 10.1128/JB.186.24.8254-8266.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag AP. In: Mechanical Properties of Bacterial Exopolymeric Adhesives and their Commercial Development Biological Adhesives. Smith AM, Callow JA, editors. Springer Berlin Heidelberg; 2006. pp. 1–19. [Google Scholar]
- Hardy GG, Allen RC, Toh E, Long M, Brown PJB, Cole-Tobian JL, Brun YV. A localized multimeric anchor attaches the Caulobacter holdfast to the cell pole. Mol Microbiol. 2010;76:409–427. doi: 10.1111/j.1365-2958.2010.07106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan S, Hugouvieux-Cotte-Pattat N. Identification of two feruloyl esterases in Dickeya dadantii 3937 and induction of the major feruloyl esterase and of pectate lyases by ferulic acid. Journal of bacteriology. 2011;193:963–970. doi: 10.1128/JB.01239-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann C, Schweitzer O, Gerke C, Vanittanakom N, Mack D, Gotz F. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol Microbiol. 1996;20:1083–1091. doi: 10.1111/j.1365-2958.1996.tb02548.x. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Rice JD, Goller C, Pannuri A, Taylor J, Meisner J, Beveridge TJ, Preston JF, 3rd, Romeo T. Roles of pgaABCD genes in synthesis, modification, and export of the Escherichia coli biofilm adhesin poly-beta-1,6-N-acetyl-D-glucosamine. J Bacteriol. 2008;190:3670–3680. doi: 10.1128/JB.01920-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz HD, Jr, Smith J. The Caulobacter crescentus holdfast: Identification of holdfast attachment complex genes. FEMS Microbiol Lett. 1994;116:175–182. doi: 10.1111/j.1574-6968.1994.tb06697.x. [DOI] [PubMed] [Google Scholar]
- Larson RJ, Pate JL. Growth and morphology of Asticcacaulis biprosthecum in defined media. Arch Microbiol. 1975;106:147–157. doi: 10.1007/BF00446517. [DOI] [PubMed] [Google Scholar]
- Levano-Garcia J, Verjovski-Almeida S, da Silva AC. Mapping transposon insertion sites by touchdown PCR and hybrid degenerate primers. BioTechniques. 2005;38:225–229. doi: 10.2144/05382ST03. [DOI] [PubMed] [Google Scholar]
- Levi A, Jenal U. Holdfast Formation in Motile Swarmer Cells Optimizes Surface Attachment during Caulobacter crescentus Development. J. Bacteriol. 2006;188:5315–5318. doi: 10.1128/JB.01725-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Brown PJB, Tang JX, Xu J, Quardokus EM, Fuqua C, Brun YV. Surface contact stimulates the just-in-time deployment of bacterial adhesins. Mol Microbiol. 2012;83:41–51. doi: 10.1111/j.1365-2958.2011.07909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Smith CS, Brun YV, Tang JX. The Elastic Properties of the Caulobacter crescentus Adhesive Holdfast Are Dependent on Oligomers of N-Acetylglucosamine. J Bacteriol. 2005;187:257–265. doi: 10.1128/JB.187.1.257-265.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merker RI, Smit J. Characterization of the adhesive holdfast of marine and freshwater caulobacters. Appl Environ Microbiol. 1988;54:2078–2085. doi: 10.1128/aem.54.8.2078-2085.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu TR. Microbial “footprints” and the general ability of microorganisms to label interfaces. Can J Microbiol. 1992;38:1005–1008. [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Poindexter JS. Biological Properties and Classification of the Caulobacter Group. Bacteriological reviews. 1964;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridout MJ, Brownsey GJ, York GM, Walker GC, Morris VJ. Effect of o-acyl substituents on the functional behaviour of Rhizobium meliloti succinoglycan. Int J Biol Macromol. 1997;20:1–7. doi: 10.1016/s0141-8130(96)01140-3. [DOI] [PubMed] [Google Scholar]
- Rinaudo M. Role of Substituents on the Properties of Some Polysaccharides. Biomacromolecules. 2004;5:1155–1165. doi: 10.1021/bm030077q. [DOI] [PubMed] [Google Scholar]
- Rubin EJ, Akerley BJ, Novik VN, Lampe DJ, Husson RN, Mekalanos JJ. In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proc Natl Acad Sci U S A. 1999;96:1645–1650. doi: 10.1073/pnas.96.4.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Meth. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CS, Hinz A, Bodenmiller D, Larson DE, Brun YV. Identification of Genes Required for Synthesis of the Adhesive Holdfast in Caulobacter crescentus. J. Bacteriol. 2003;185:1432–1442. doi: 10.1128/JB.185.4.1432-1442.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlier P, Denuzière A, Viton C, Domard A. Relation between the Degree of Acetylation and the Electrostatic Properties of Chitin and Chitosan. Biomacromolecules. 2001;2:765–772. doi: 10.1021/bm015531+. [DOI] [PubMed] [Google Scholar]
- Tielen P, Strathmann M, Jaeger K-E, Flemming H-C, Wingender J. Alginate acetylation influences initial surface colonization by mucoid Pseudomonas aeruginosa. Microbiol Res. 2005;160:165–176. doi: 10.1016/j.micres.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Toh E, Kurtz HD, Brun YV. Characterization of the Caulobacter crescentus Holdfast Polysaccharide Biosynthesis Pathway Reveals Significant Redundancy in the Initiating Glycosyltransferase and Polymerase Steps. J Bacteriol. 2008;190:7219–7231. doi: 10.1128/JB.01003-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang PH, Li G, Brun YV, Freund LB, Tang JX. Adhesion of single bacterial cells in the micronewton range. Proc Natl Acad Sci U S A. 2006;103:5764–5768. doi: 10.1073/pnas.0601705103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigeant MA, Ford RM, Wagner M, Tamm LK. Reversible and irreversible adhesion of motile Escherichia coli cells analyzed by total internal reflection aqueous fluorescence microscopy. Appl Environ Microbiol. 2002;68:2794–2801. doi: 10.1128/AEM.68.6.2794-2801.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villain-Simonnet A, Milas M, Rinaudo M. A new bacterial polysaccharide (YAS34). I. Characterization of the conformations and conformational transition. Int J Biol Macromol. 2000;27:65–75. doi: 10.1016/s0141-8130(99)00120-8. [DOI] [PubMed] [Google Scholar]
- Viollier PH, Sternheim N, Shapiro L. A dynamically localized histidine kinase controls the asymmetric distribution of polar pili proteins. EMBO J. 2002;21:4420–4428. doi: 10.1093/emboj/cdf454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong C, Kocianova S, Voyich JM, Yao Y, Fischer ER, DeLeo FR, Otto M. A Crucial Role for Exopolysaccharide Modification in Bacterial Biofilm Formation, Immune Evasion, and Virulence. J Biol Chem. 2004;279:54881–54886. doi: 10.1074/jbc.M411374200. [DOI] [PubMed] [Google Scholar]
- Waters DM, Murray PG, Miki Y, Martínez AT, Tuohy MG, Faulds CB. Cloning, Overexpression in Escherichia coli, and Characterization of a Thermostable Fungal Acetylxylan Esterase from Talaromyces emersonii. Appl. Environ. Microbiol. 2012;78:3759–3762. doi: 10.1128/AEM.05659-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Park RD, Muzzarelli RA. Chitin deacetylases: properties and applications. Mar Drugs. 2010;8:24–46. doi: 10.3390/md8010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.