Abstract
Exocytosis involves membrane fusion between secretory vesicles and the plasma membrane. The Soluble N-ethylmaleimide-sensitive factor attachment proteins (SNAPs) and their receptor proteins (SNAREs) interact to fuse vesicles with the membrane and trigger the release of their sialosecretome out of the tick salivary gland cells. In this study, we examined the functional significance of the Vti family of SNARE proteins of blood-feeding Amblyomma maculatum and A. americanum. Vti1A and Vti1B have been implicated in multiple functional roles in vesicle transport. QRT-PCR studies demonstrated that the highest transcriptional expression of vti1a and vti1b genes occurs in unfed salivary glands, suggesting that elevated secretory vesicle formation occurs prior to feeding but continues at low rates after blood feeding commences. Vti1A and Vti1B localize to the secretory vesicles in unfed tick salivary glands in immunofluorescence microscopy studies. Knockdown of vti1a and vti1b by RNA interference resulted in a significant decrease in the engorged tick weight compared to the control during prolonged blood-feeding on the host. RNA interference of vti1a or vti1b impaired oviposition and none of the ticks produced eggs masses. Surprisingly, the double knockdown did not produce a strong phenotype and ticks fed normally on the host and produced egg masses, suggesting a compensatory mechanism exists within the secretory system which may have been activated in the double knockdown. These results suggest an important functional role of the Vti family of SNARE proteins in tick blood feeding and ultimately oviposition. Understanding the basic functions of the Vti family of SNARE proteins in salivary glands may lead to better ways to prevent tick attachment and transmission of tick-borne diseases.
1. Introduction
Ixodid ticks are obligate blood-sucking parasites that feed for a prolonged period of time, varying from a few days to a maximum of 21 days, increasing in mass by up to 100-fold its initial size (Sonenshine, 1991; Teel et al., 2010). Amblyomma tick species have three life stages after hatching from the egg; larvae and nymphs normally feed on small rodents, while adults feed on medium to large mammals (Teel et al., 2010). After finding a suitable site to feed, Amblyomma ticks penetrate the host skin with their mandibles and secrete a glue-like substance that helps to anchor the arthropod to the feeding site. Feeding is accomplished by sucking blood from a hematoma formed in the host skin by mouthpart laceration of the host tissues. This blood pool is maintained in a liquid state by a complex mixture of anti-hemostatic compounds present in tick saliva (Ribeiro 1995, 1989; Chemlar et al., 2012).
A tick's multifunctional salivary glands are vital to its biological success. Saliva secretion has a plethora of bioactive components which facilitate the transmission of tick-borne pathogens, including protein components with anti-complement, anti-inflammatory and antihistaminic activities as well as a variety of other factors that inhibit the host immune response (Sauer et al., 2000; Francischetti et al., 2009; Ribeiro and Francischetti 2003). Most tick salivary proteins are thought to be released from the vesicle contents originating from membrane-bound granular material observed in salivary gland acini II and III (Binnington 1978; Karim et al., 2002) and SNAREs participate in vesicle transport of proteins (Karim et al., 2002; Jaworski 2003; Gong 2007). Intracellular protein trafficking and exocytosis is a complex mechanism and little is known about the secretory pathway in non-neuronal systems like blood-feeding arthropod salivary glands. The events which take place during vesicle fusion and release of saliva proteins are of critical importance with respect to tick attachment and uninterrupted blood-feeding.
SNAREs are the key regulators in all fusion events occurring in the secretory pathway (Jahn et al., 2003). The SNARE complex works as a machine to aid in the fusion of a vesicle with the target membrane (Hong, 2005). SNARE terminology is based upon the requirement of an N-ethylmaleimide-sensitive fusion protein (NSF) and soluble NSF attachment proteins (SNAPs) involvement in the process. Four SNARE proteins associate to form a stable complex needed for vesicle fusion. One of the four proteins is classified as an R-SNARE which provides an arginine residue to the functional component, while the other three proteins are classified as Q-SNAREs which provide glutamine to the structural complex (Fasshauer et al., 1998).
Eukaryotic cells contain several SNARE proteins involved in vesicle transport pathways. So far, 24 members in Saccharomyces cerevisiae, 21 in Aspergillus oryzae, 36 in humans, 68 in Arabidopsis thaliana, and 20 in Drosophila melanogaster have been identified (Jahn and Sudhof 1999; Surpin et a., 2003). Our A. maculatum sialotranscriptome revealed approximately 30 putative SNARE proteins (Karim et al., 2011). The functional roles of some vesicle- and plasma membrane-bound protein receptors (SNAREs) in regulating protein secretion in tick salivary gland cells have previously been examined (Browning and Karim, 2013); Karim et al., 2002; 2004a. 2004, 2005; Gong et al., 2009).
However, the Vti family (vesicle transport through interaction with t-SNAREs) of SNARE proteins has not been studied in ticks (Karim et al., 2011). Vti1A and Vti1B exhibit low sequence similarity with each other, possess different roles, localize to distinct areas of the cell and form distinct SNARE complexes; the former being localized to the Golgi and trans-Golgi network (TGN) and the latter with early and late endosomes as well as the Golgi and TGN (Advani et al., 1998; Antonin et al., 2000; Dressel et al., 2010; Kreykenbohm et al., 2002). Despite these differences, studies suggest that the two can functionally compensate for one another; however, the loss of both Vti1A and Vti1B in murine knockouts resulted in significant developmental impairment and inviability (Atlashkin et al., 2003; Kunwar et al., 2011; Surpin et al., 2003). In the present study, we used a combination of bioinformatic approaches, qRT-PCR, immunoblotting, and confocal microscopy to characterize the Vti family of SNARE proteins in A. maculatum and A. americanum salivary glands. We used RNA interference to demonstrate that the loss of a Vti gene reduces tick engorgement and oviposition in single Vti knockdowns, but not the double knockdown. We then examined the transcriptional expression of other SNARE genes to search for possible activation of a compensatory secretory mechanism.
2. Materials and methods
2.1. Materials
Unless otherwise indicated, general laboratory procedures followed standard protocols (Sambrook et al., 1989). All water used was of 18-megaohm quality, produced by a MilliQ apparatus (Millipore). SNARE antibodies used in this study were rabbit or mouse polyclonal. Anti-Vti1A, anti-Vti1B, and anti-β-actin were from Santa Cruz, BD transduction, and Sigma Aldrich, respectively. Alexa-546 anti-goat, and Alexa-546 anti-mouse, Taq DNA polymerase, reverse transcriptase, and plasmid DNA purification kits were purchased from Invitrogen or Qiagen.
2.2. Bioinformatics
The full length A. maculatum Vti1A and Vti1B sequences were obtained from pyrosequencing an A. maculatum salivary gland cDNA library (Karim et al., 2011). A partial coding sequence for A. americanum Vti1B was obtained from a sequence homology search based on the A. maculatum Vti1B sequence. Amino acid sequences were obtained for comparative analysis to establish phylogenetic relationships aligned using ClustalX2, and graphically presented using Jalview 2.8 (Larkin et al. 2007; Waterhouse et al. 2009). Phylogenetic relationships were inferred using MEGA 5 (Tamura et al. 2011). Protein secretion signals were identified using SignalP 4.0 (Petersen et al., 2011).
2.3. Ticks
Amblyomma maculatum and Amblyomma americanum ticks were raised and obtained from Oklahoma State University's tick rearing facility, and maintained according to the methods of Patrick and Hair (1975). Adult ticks had molted three months before the animals were infested with ticks. Adult A. maculatum and A. americanum were fed on sheep or rabbits, respectively, specifically for this study, and all studies with animals were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee at the University of Southern Mississippi. All unfed ticks were maintained at room temperature with 90% relative humidity under 14 h light/dark photoperiod before being placed on the hosts (Karim et al., 2002). Briefly, six cells are placed on the back of a shaved sheep, and approximately 40 female and 40 male A. maculatum ticks per cell are placed on the animal's back. Alternatively, 40 female and 40 male A. americanum ticks were placed on shaved rabbit ears. For RNAi studies, forty-five ticks are injected with dsRNA, allowed to heal overnight, and forty of the surviving ticks are placed on the animal. Tick survival is determined based on the number of ticks that were placed on an animal. Typically, there is more than 95% attachment success of those ticks that are placed on animals. Experimental groups and controls were placed on the same animal to eliminate inter-animal variation in feeding success. For transcriptional studies, ticks were allowed to feed, and approximately 5 ticks were removed from the host at regular intervals throughout the bloodmeal, and the salivary glands and midguts were dissected and stored in RNAlater. To examine the effect on oviposition, 5-10 ticks were allowed to feed to repletion, and the size of the egg mass was examined. Amblyomma maculatum salivary glands were dissected from the ticks within 4 hrs of being removed from the host (Karim et al., 2002). However, A. americanum were fed on rabbits, and these ticks were immediately dissected following removal. The completely engorged and dropped off ticks were kept under the similar conditions described earlier to oviposit.
2.4. Tick salivary glands
A. maculatum and A. americanum salivary glands were dissected in ice-cold 100 mM 3-(N-morpholino)-propanesulfonic acid (MOPS) buffer containing 20 mM ethylene glycol bis-(β-aminoethyl ether)-N,N,N’,N’-tetraacetic acid (EGTA), pH 6.8. After removal, the glands were gently washed in the same ice-cold buffer. The tissues were then immediately stored in RNAlater (Ambion) prior to isolating total RNA or directly kept in lysis buffer. Tissues were either used immediately after dissection, or stored at -80°C in 0.5 M PBS, 0.5 mM EGTA, 1x Complete Mini Protease inhibitor cocktail (Roche), and 40% glycerol for Western blotting. All other manipulations were carried out at 4°C.
2.5. RNA extraction, cDNA synthesis and RT-PCR
Total RNA was extracted from salivary glands dissected from unfed and partially blood-fed adult female ticks using illustra RNAspin Mini RNA isolation kit (GE healthcare) according to the manufacturer's instruction using slight modifications for tick RNA extractions (Karim et al., 2011, 2012). The concentration of extracted total RNA was determined spectrophotometrically using a Nanodrop and samples were stored at -80°C. Reverse transcription of total RNA was done using Moloney murine leukemia virus (M-MLV) reverse transcriptase according to the manufacturer's instructions (Invitrogen). Using tick-specific EST sequences for AmVti1A (Genbank: JO842730), AmVti1B (Genbank: JO843088), and AaVti1B (GenBank: EZ000215), gene-specific primers were designed to amplify gene fragments from salivary glands cDNA of the gulf coast and lone star ticks. All primer sequences used in this study are listed in Table 2. PCR amplification was performed using a program of 94°C for 1 min, 29 cycles of 94°C for 1 min, 49°C for 1 min, and 72°C for 1 min, foll owed by 72°C for 8 min. PCR products were analyzed on a 2% agarose gel stained with eithidium bromide, and visualized using the GelDoc system (Bio-Rad).
Table 2.
Gene-specific primers used in this study
| Gene | Qualitative/Quantitative | Forward primer (5′-3′) | Reverse primer (5′-3′) | Size (bp) |
|---|---|---|---|---|
| AmVti1A | Qualitative | ATGATTGGCATTCTGAAAATGGCG | TCACACATGCCTTCG AACTGTAAAA | 684 |
| AmVti1B | Qualitative | ATGTCCTCGGAAAAGTTTGAGGA | TCACTTCATGATAAACTTCCAGTAGATAA | 684 |
| AmVti1A | Quantitative | TGGAGCTTGAAGTCCGTACGTTGT | TCCTTAATCGTGTGAGTTCCGCCT | 93 |
| AmVtilB | Quantitative | AACTTTCCAAGTCGCCGCTGAAAC | TGTTGGTCATGACGCGCCTATACA | 173 |
| AmStx8 | Quantitative | TGCTAAACAAAGGGCGGGAAATGG | ACCACCTTCACTTACAGCCACACT | 122 |
| AmStx16 | Quantitative | TTGCAAGAGGCTTCAGTCCAGAGA | TGTTGATCTCCTCGTTCCCGCACTT | 182 |
| AmStx17 | Quantitative | TGGGCGCTGGATTCTTAGGTTACA | AACTGAGTGACTCGTCTGGTGGTGCTT | 135 |
| AmStx18 | Quantitative | AAGGCACCTCATGAGACAGAAGGA | ATCACCACATTCTCATCCCAGCCT | 134 |
| AmVAMP1/2 | Quantitative | AGGGTCAATACTGTGAACAGGCCA | ACTGATCCTCTTTGGAGCACGTCA | 182 |
| AmVAMP7 | Quantitative | ACTTCTCGGAGGTCACCGAACAAA | ACTCACTTGGAACCTGCCCTGAAT | 197 |
| AmβActin | Quantitative | TGGCTCCTTCCACCATGAAGATCA | TAGAAGCACTTGCGGTGCACAATG | 177 |
| AaVti1B | Quantitative | TCGAAGAGGACTTTGCGGCACTAA | CTTTGCGCTGCACTTCACGAAGAA | 117 |
| AaCalreticulin | Quantitative | CGTCAAGCACGAGCAGAACATTGA | TGAGGTGGTTCTTGCCCTTGTAGT | 182 |
2.6. Gene expression by qRT-PCR
The transcriptional expression pattern of Vti family of SNARE genes during prolonged tick feeding on the host was performed as described (Browning et al., 2012). Briefly, the cDNA was diluted to approximately 25 ng/μl for qRT-PCR gene expression analysis. First strand cDNA was used to measure mRNA levels through qRT-PCR. For quantification with BIORAD CFX96 Real Time System, the Maxima SYBR Green qPCR Master Mix (Fermentas) was used according to manufacturer's recommendations; approximately 50 ng of cDNA and gene specific primers (150 nM) were used for each reaction mixture. The AmVti1A and AmVti1B amplicons were amplified from A. maculatum cDNA using AmVti1A (qrtpcr) primers, AmVti1B (qrtpcr) primers, respectively, listed in Table 2. From A. americanum cDNA, the AaVti1B amplicon was amplified using AaVti1B gene specific primers (Table 2). Calreticulin and β-actin genes were used as housekeeping genes in QRT-PCR analyses based in A. americanum and A. maculatum tissues, respectively (Table 2). The C1000 Thermal Cycler was used to perform amplification of target gene of interest using following program, 10 minutes at 95 °C, followed by 35 cycles of 15 seconds at 95 °C , 30 seconds at 60 °C, and 30 seconds at 72 °C. All protocols for QRT-PCR experi ments are in line with MIQE guidelines (Bustin et al., 2009).
2.7. In vivo gene knockdown
PCR products of Vti1A or Vti1B were joined to the Block-iT T7 TOPO linker. This TOPO linking reaction was used in two PCRs with gene-specific and T7 PCR primers to produce sense and antisense linear DNA templates. These sense and antisense DNA templates were then used to generate sense and antisense transcripts using the BLOCK-iT RNA TOPO transcription kit (Invitrogen). The sense and antisense ssRNAs were purified and then mixed together, incubated at 100 °C for 5 min, and then left to anneal at room temperature for 4 hr to make Vti1A-dsRNA and dsVti1B-dsRNA, respectively. To make the double knockdown, equal amounts of Vti1A-dsRNA and Vti1B-dsRNA were mixed. For each type of dsRNA, 45 unfed female ticks were injected with 1.5-2.0 μL of 500 ng/μL dsRNA using a 33-gauge needle as described (Karim et al., 2004; Karim and Adamson 2012). After injection, ticks were kept overnight in a 37 °C incubator to confirm tick survival after injection. Forty female ticks injected with irrelevant dsRNA, PBS and Vti-dsRNA were placed in separate cells on a sheep for feeding. An equal number of untreated male ticks were placed to stimulate the female ticks to feed. Several ticks were removed at 120 hr after placement to check the efficiency of transcriptional knockdown using qRT-PCR.
2.8. Immunofluorescence imaging
For immunolocalization, salivary glands from unfed ticks were dissected and fixed in PBS containing 4% formaldehyde in PBS for 30 minutes at room temperature, permeabilized in PBS containing 0.5% Triton X-100 for 30 minutes, and washed in PBS. Next, salivary glands were blocked in PBS containing 3% BSA for 1 hour at room temperature, and then washed thrice for 5 min with PBS. Salivary glands were incubated overnight at 4°C with the primary antibod y (1:100) in PBS containing 3% BSA. Salivary glands were again washed thrice for 15 min in PBS, and then incubated in Alexa-546 anti-goat (Vti1A) and Alexa-546 anti-mouse (Vti1B) secondary antibody (1:100) in PBS containing 3% BSA for 1 hr in the dark. Excess antibody was washed away with three 5 min PBS washes. Salivary glands were mounted on glass slides using VectaShield Mounting Medium with DAPI and viewed under a Leica TCS 4D confocal microscope.
2.9. Statistical analysis
Kruskal-Wallace One Way Analysis of Variance (ANOVA) on Ranks was used to evaluate statistical significance. These analyses were performed using SigmaPlot.
3. Results
3.1. Bioinformatic analysis
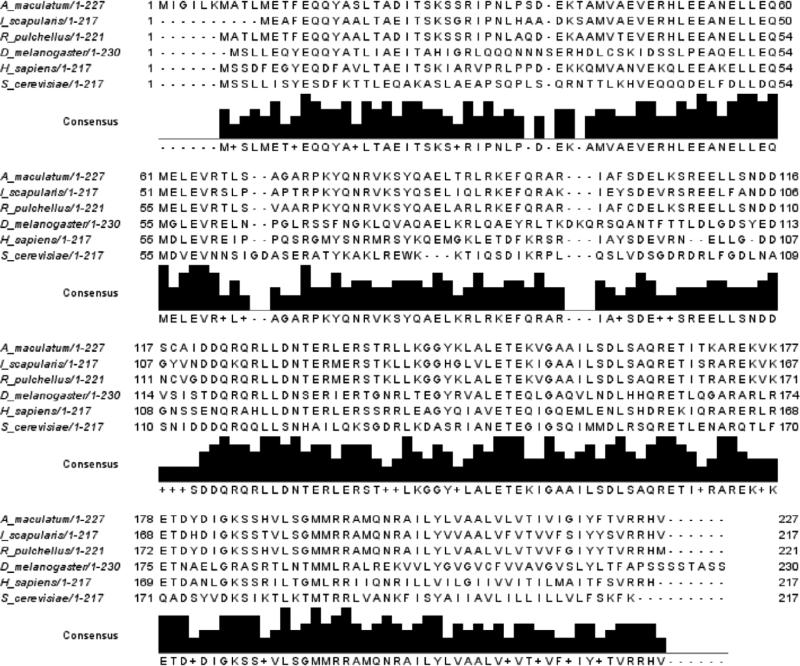
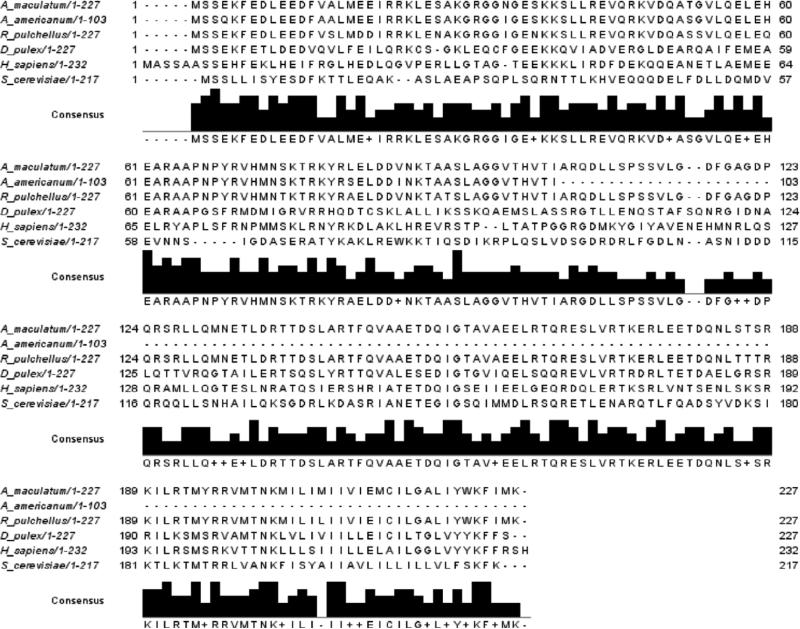
Through pyrosequencing of expressed sequence tags to identify transcripts in A. maculatum salivary glands, two open reading frames were identified that showed significant homology to Vti1A and Vti1B (Karim, 2011). AmVti sequences were aligned with the amino acid sequences of previously identified Vti homologs (Figs. 1-2). AmVti1A exhibited 79% and 91% amino acid identity to tick Vti1A's found in I. scapularis and R. pulchellus, respectively. AmVti1A had 38% identity with the arthropod D. melanogaster Vti1 sequence, and 53% identity to the H. sapiens Vti1A sequence (Fig. 1). AmVti1B had 94% and 92% identity to tick Vti1B sequences found in A. americanum and R. pulchellus, respectively. AmVti1B had 41% identity to Vti1B sequences from the arthropod D. pulex and the vertebrate H. sapiens (Fig. 2). Both AmVti1A and AmVti1B had 26-27% amino acid identity with the distantly related yeast Vti1p sequence (Figs. 1-2). AmVti1A exhibited only 28% amino acid identify with AmVti1B, which is in keeping with previous observations (Advani et al., 1998; Antonin et al., 2000).
Figure 1.
Multiple sequence alignment of Vti1A amino acid sequences from Amblyomma maculatum, Ixodes scapularis, Rhipicephalus pulchellus, Drosophila melanogaster, Homo sapiens, and Saccharomyces cerevisiae.
Figure 2.
Multiple sequence alignment of Vti1B amino acid sequences from Amblyomma maculatum, Amblyomma americanum, Rhipicephalus pulchellus, Daphnia pulex, Homo sapiens, and Saccharomyces cerevisiae.
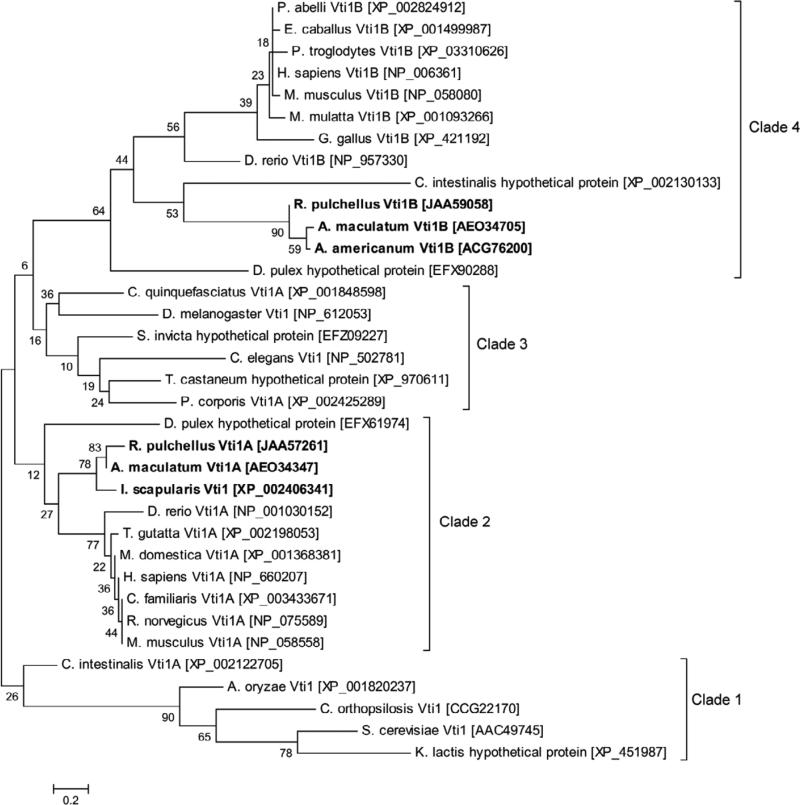
Phylogenetic analysis of the Vti family of SNARE proteins shows the general expected pattern that recapitulates the species phylogeny for eukaryotic organisms, indicating that it behaves like an orthologous family. A single ancestral Vti gene, represented by fungal sequences in Clade 1, has likely diverged into two subgroups, Vti1A (Clades 2-3) and Vti1B (Clade 4) (Fig. 3). The tick Vti sequences group within the arthropods, substantiating their identity as Vti orthologs. It is noteworthy that several arthropod Vti1A sequences (excluding ticks) (Clade 3) appear are more similar to Vti1B sequences than to the vertebrate and tick Vti1A sequences (Clade 2). This could indicate that some arthropod Vti1A sequences arose from an ancestral Vti1B sequence, but bootstrap support for this premise is weak.
Figure 3.
The evolutionary history of the Vti family was inferred using the Maximum Likelihood method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) is shown next to the branches. Clade 1 represents ancestral Vti sequences which have not yet diverged. Clades 2 and 3 represent Vti1A sequences. Clade 4 represents Vti1B sequences. Scale bar represents amino acid substitutions per position.
3.2. Transcriptional gene expression of Vti1A and Vti1B in tick salivary glands
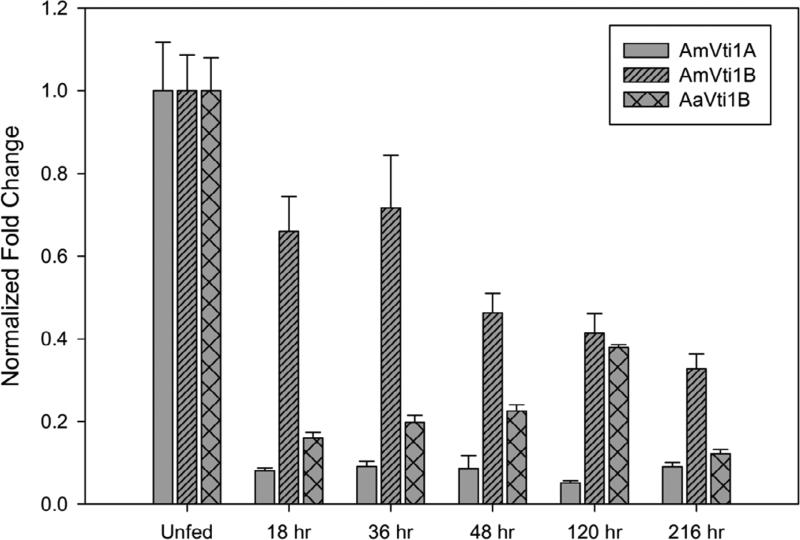
In order to better assess the roles of AmVti1A, AmVti1B and AaVti1B during prolonged blood-feeding, transcriptional gene expression was determined using qRT-PCR. AmVti1A exhibited high expression in the unfed salivary glands, but immediately showed a significant reduction in transcript levels within 18 hrs post-infestation which remained consistent throughout the blood-meal (Fig. 4). Although the expression of AmVti1B was highest in unfed salivary glands and the abundance of AmVti1B transcripts declined slower than that of AmVti1A once feeding began, with the lowest levels observed in ticks 216 hrs after infestation (Fig. 4). Generally, the expression of AmVti1B expression appeared to be higher than that of AmVti1A throughout blood feeding. In A. americanum, the transcriptional expression of Vti1B was also highest in unfed salivary glands but remained at low levels during blood feeding only showing a slight increase 120 hrs after infestation (Fig. 4). If protein abundance mirrors the transcriptional profile, this could suggest that the high expression of Vti family of proteins in unfed tick salivary glands allows for the Vti proteins to be synthesized and assembled onto the surface of secretory vesicles. This expression profile facilitates the formation of abundant secretory vesicles in preparation of blood feeding but continues at low levels after feeding commences. Several attempts to perform semi-quantitative immunoblotting to confirm this premise were unsuccessful, but immunofluorescence microscopy studies do support this supposition (section 3.3). We note that it is also possible that cessation of feeding may have resulted in an immediate decrease in transcription which may not have been observed in QRT-PCR, due to time constraints in tissue dissection.
Figure 4.
Transcriptional expression of AmVti1A, AmVti1B, and AaVti1B in tick salivary glands throughout the bloodmeal. For comparative purposes, transcriptional levels were normalized to levels observed in unfed ticks
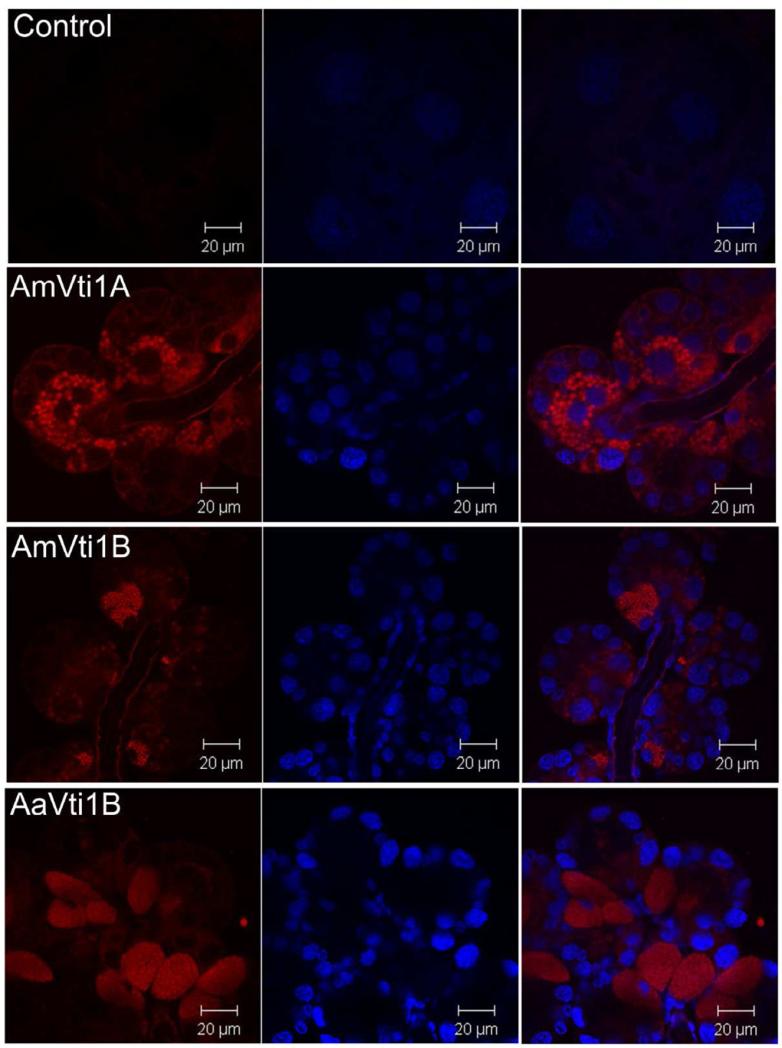
3.3. Immunofluorescence microscopy
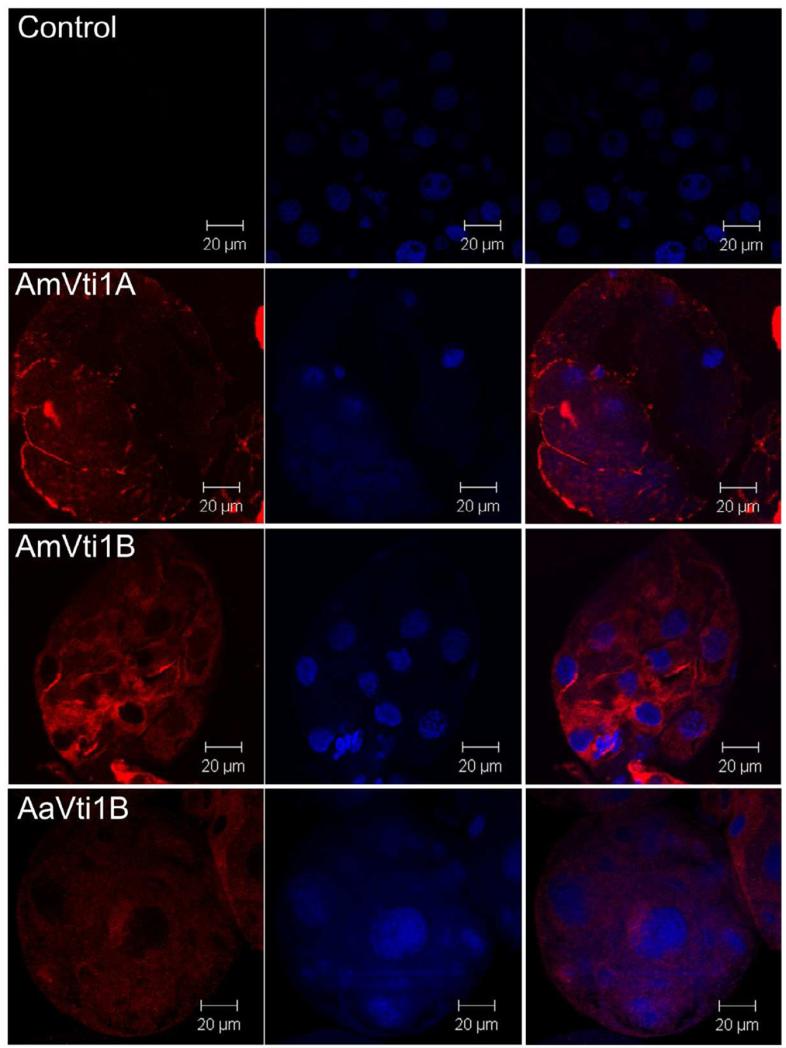
Immunofluorescence microscopy indicates that Vti1A and Vti1B are highly localized to secretory vesicles within salivary gland acini (Fig. 5). However, after partial blood feeding, no distinct localization of Vti proteins was observed (Fig. 6) presumably because the majority of secretory vesicles have fused with the plasma membrane and released their contents into the salivary duct to facilitate feeding. This observation is consistent with the dynamics of Vti transcription, in that the highest expression was observed in unfed ticks (Fig. 4). This observation is surprising, and one might expect the Vti protein family to be expressed only after feeding began. However, we observe few secretory vesicles in partially fed tick salivary glands, relative to the number we observe in unfed tissues, in which secretory cells are literally packed with vesicles. Salivary proteins certainly must be synthesized and secreted through the feeding cycle (5-10 days post-infestation). Indeed, it is common to see dynamic changes in the protein content of tick saliva (Vancova et al., 2007). However, the number of vesicles is greatly reduced in partially fed tick salivary glands (Binnington, 1978) and the low levels of Vti transcription within partially-fed ticks seem to support this observation. Perhaps the low level of secretory vesicle formation is sufficient to support salivary secretions, since vesicle fusion machinery is recycled. Moreover, it is possible that other SNARE proteins (i.e. VAMPS, synaptobrevin, ykt6, syntaxins) may play more important roles in the later stages of blood feeding (Fig. 8).
Figure 5.
Immunolocalization of Vti1A/B in unfed salivary glands. Images were obtained under 63x magnification. The Vti1A/B proteins are stained red (left column), nuclei were counterstained blue (middle column), and then images were merged (right column).
Figure 6.
Immunolocalization of Vti1A/B in salivary glands dissected from partially-fed ticks (5 days post-infestation). Images were obtained under 63x magnification. The Vti1A/B proteins are stained red (left column), nuclei were counterstained blue (middle column), and then images were merged (right column).
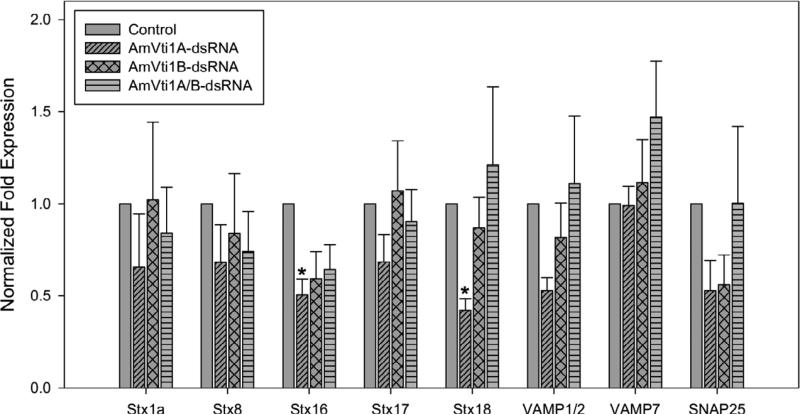
Figure 8.
Transcriptional expression of SNARE genes in the salivary glands of single or double AmVti1A/B knockdowns. Syntaxin 16 and syntaxin 18 have a reduced transcriptional level (p<0.05) in AmVti1A-dsRNA knockdown ticks.
3.4. RNA interference
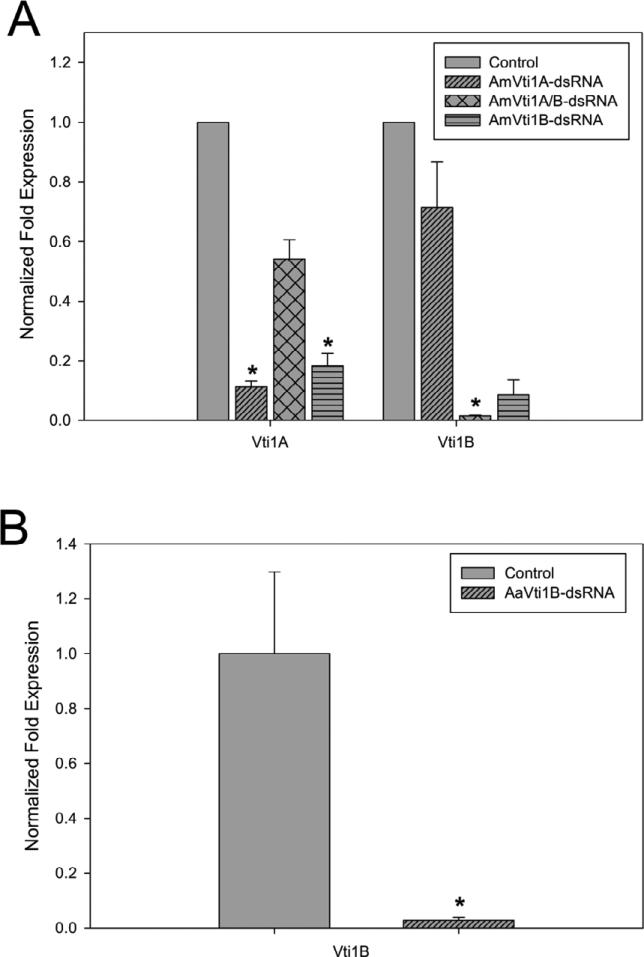
Gene knockdown studies were performed in order to determine the physiological impact on blood feeding in ticks in which the Vti1A and/or Vti1B transcript levels were reduced. Individual knockdowns of AmVti1A and AmVti1B as well as a double AmVti1A/B knockdown consisting of dsRNA of both transcripts were injected to form three separate A. maculatum experimental groups. Only AaVti1b-dsRNA was injected into A. americanum as there were no identified homologs of AmVti1A. Several unsuccessful attempts were made to amplify the Vti1A using A. maculatum Vti1A gene specific primers. A. maculatum injected with AmVti1A-dsRNA showed an 88% decrease in Vti1A transcript levels compared to control (Fig. 7A) while AmVti1B transcript levels remained at 67% expression. In AmVti1B-dsRNA injected ticks, the Vti1B transcript levels were reduced to 99% within the salivary glands, whereas the Vti1A transcript levels remained at 63% (Fig. 4). In the Vti1A/Vti1B knockdown ticks, AmVti1A transcripts were only reduced by 77%, but AmVti1B transcripts were reduced by 98% (Fig. 7A). β-actin levels remained constant in all experiments with A. maculatum. A. americanum injected with AaVti1B-dsRNA showed a 99% reduction in AaVti1B transcript levels compared to control (Fig. 7B).
Figure 7.
Transcriptional knockdown in A. maculatum (A) or A. americanum (B) injected with Vti-dsRNA. Statistical significance is indicated by an asterisk placed above the bar (p<0.05).
Unfed adult female A. maculatum ticks injected with either AmVti1A-dsRNA or AmVti1B-dsRNA showed a large reduction in the mean engorged body mass (83.7mg or 27.2mg, respectively) compared to control ticks (p<0.05) which had a mean engorged body mass of 228.5mg (Table 1). Surprisingly, ticks injected with both AmVti1A/B-dsRNA showed an engorged body mass similar to control ticks (211.7mg vs 228.5mg), that the impact on blood feeding is negligible in the double knock-down. Unfed adult female A. americanum ticks injected with AaVti1b-dsRNA showed a reduced mean engorged body mass compared to control (36.2mg vs 72.9mg; p<0.05).
Table 1.
Data collected from ticks injected with dsRNA.
| Sample | Mean Engorged Body Mass (mg) | Mortality Rate (%) | Mean Egg Mass (mg) |
|---|---|---|---|
| A. maculatum control | 228.5 (n=33) | 17.5 | 459 |
| AmVti1A-dsRNA | 83.7 (n=26) | 40.0 | 0 |
| AmVti1B-dsRNA | 27.2 (n= 12) | 70.0 | 0 |
| AmVti1A/B-dsRNA | 211.7 (n=37) | 7.5 | 308 |
| A. americanum control | 72.9 (n=16) | 21.7 | not determined |
| AaVti1B-dsRNA | 36.2 (n= 22) | 55.0 | 0 |
The mortality rate in knockdown ticks was much higher than control ticks, except for the double-knockdown ticks which had a lower mortality rate than controls (Table 1). Moreover, single-knockdown ticks failed to oviposit, whereas the double-knockdown ticks oviposited normally (Table 1). Taken together, these data strongly indicate a compensatory mechanism exists which may allow the tick to continue to feed in the double-knockdown.
3.5. Transcriptional expression of SNARE genes in knockdown tick salivary glands
Since the SNARE hypothesis claims that four SNAREs must work in a complex comprised of four SNAREs working together, we hypothesized that knocking down the expression of Vti1A/B would in turn affect the expression of other proteins involved in SNARE complex assembly. Therefore, we examined the transcriptional expression of other SNAREs previously identified from the pyrosequencing of the cDNA library generated from A. maculatum salivary glands (Karim et al., 2011). The transcriptional expression of syntaxin 16 and syntaxin 18 were reduced in the Vti1A-knockdown, but not in the Vti1B-knockdown or double-knockdown (Fig. 8), No statistically significant differences were observed in the transcriptional expression of syntaxin 1a, syntaxin 8, syntaxin 17, VAMP1/2, VAMP7, or SNAP25 (Fig. 8).
4. Discussion
The knowledge of vesicle transport mechanisms and role of SNARE proteins in non-neuronal secretory systems, such as in tick salivary glands, remains elusive. Our previous work has identified numerous SNARE proteins in salivary glands of A. americanum (Karim et al., 2002). Moreover, we have shown that transcriptional knockdown of these SNARE proteins, via RNA interference, resulted in lower salivary secretions, and blood-feeding abnormalities (Karim et al., 2004, 2004a, 2005). The purpose of this study was to further expand our knowledge on the function of SNARE proteins in tick salivary glands and to specifically determine the effect of Vti knockdown on tick blood-feeding.
Bioinformatics suggests a single ancestral Vti1 protein has functionally diverged to form two proteins in Arabidopsis, Drosophila, higher-order vertebrates, and A. maculatum (Figs. 1-3) (Fisher von Mollard and Stevens 1999; Atlashkin et al., 2003). However, it is apparent that some organisms do not encode both Vti1 sequences. An A. americanum Vti1A sequence has not yet been identified in published databases, although this could be due to limited genomics resources available for A. americanum rather than being absent in the genome, since both Vti1A and Vti1B were present in both the A. maculatum and R. pulchellus sialotranscriptome (Karim et al., 2011; Tan et al., unpublished). However, it is possible that only one, multifunctional, Vti1b-type protein exists in A. americanum, since no homolog of Vti1B was found within the I. scapularis genome. Moreover, there are a number of other vesicle-associated SNARE proteins, such as VAMP1/2, VAMP7, Synaptobrevin, Ykt6, that have been identified within ticks that could potentially operate with some functional redundancy to the Vti-family (Fig. 8) (Karim et al., 2002, 2004, 2005, 2011; Gong et al., 2009),
QRT-PCR data demonstrates the highest levels of transcriptional expression occurred in unfed tick salivary glands, suggesting that Vti1A/Vti1B are synthesized and assembled onto the surface of secretory vesicles prior to tick feeding, and continues at low levels throughout blood feeding (Fig. 4). This is substantiated by the strong immunolocalization of Vti1A/B within secretory vesicles of unfed tick salivary glands (Fig. 5). This observation could reflect a difference between non-neuronal secretory systems, found in tick salivary glands, and neuronal systems, in which most SNARE proteins have been characterized. Adaptations to diapause may require ticks to pre-form numerous secretory vesicles full of the immunosuppressive, antihemostatic, and anti-inflammatory proteins needed to evade the host defense, since ticks’ biological success requires uninhibited feeding. As the tick feeds, the SNARE machinery (including Vti proteins) is likely recycled for subsequent vesicle fusion events. Indeed, it has been shown that NSF and α-/β-SNAP disassemble SNARE complexes, thereby recycling individual SNAREs for subsequent fusion reactions (Burgalossi et al., 2010). Moreover, early electron microscopy work shows abundant secretory vesicles present in unfed tick salivary gland acini (Binnington et al., 1978). As the tick begins to feed, fewer secretory vesicles are observed and a large luminal area forms within the acini, presumably as a result of secretory vesicles fusing with plasma membrane and releasing the contents into the salivary duct (Binnington et al., 1978). These data seem to indicate that Vti's are expressed at much higher levels than needed for maintenance of vesicle fusion.
The disruption of feeding may partially explain the phenotype observed in RNAi knockdowns. The reduction in engorged body mass of Vti1A- or Vti1B-knockdown ticks and higher mortality rate clearly indicates tick feeding was inhibited (Table 1). Higher mortality and feeding inhibition was also observed in previous studies with ticks in which SNARE transcripts have been targeted by RNAi (Karim et al., 2002, 2004, 2004a, 2005), indicating that the secretory vesicle contents are critical to feeding. However, it is difficult to distinguish if the differences in feeding success within the Vti knockdowns stem specifically from functional inhibition in salivary glands or if they result from effects in other tissues. It is still unclear how the SNARE exocytosis machinery functions in the midgut, and we are unable to determine the difference of silencing Vti in the salivary glands versus the midgut, since both were silenced in vivo. Surprisingly, a strong phenotype was not observed in the double-knockdown, which had a similar engorged body mass and mortality rate to the control (Table 1). This data set contrasts with murine studies which show that mice lacking Vti1A or Vti1B do not show an overt phenotype, whereas double knockout mice survive until birth with normal endo-lysosomal trafficking, though massive neurodegeneration was observed (Atlashkin et al., 2003; Kunwar et al., 2011). It is unclear why the Vti1A- or Vti1B-knockdown ticks were unable to oviposit, but it seems more likely that this impact on egg laying arose due to insufficient blood-feeding, rather than any direct role in oviposit. However, it is possible that the failure to oviposit could have resulted from vitellogenin being taken up by the oocytes via micropinocytosis (Sonenshine, 1991). It is likely that that the Vti family of SNARE proteins are involved in the whole process of blood-feeding and oviposition in ticks.
Typically, incomplete reduction in SNARE proteins do not result in discernible phenotypes as these proteins are present in excess quantities beyond minimum requirements (Bethani et al., 2009); therefore, it is notable that knockdown of Vti1A or Vti1B resulted in a reduced engorged tick weight, mortality, and oviposit (Table 1). It is perhaps noteworthy to mention that the knockdown of Vti1A transcript resulted in a ~45% reduction in the transcript levels of Vti1B (Fig. 7). It is unclear whether this is due to biological downregulation of Vti1B or is simply an off-target effect of RNA interference. In either case, this observation suggests the effects of a single-knockdown may be potentiated by the decrease in transcriptional expression of the coordinate Vti (Fig. 7).
At the organismal level, a selective deficit in the release of salivary proteins may not give rise to a lethal phenotype. However, protein release deficit in knockdown ticks may lead to subtle changes in ticks that would require closer examination and in depth analysis of saliva secretions. To further understand the lack of a strong phenotype, particularly in double-Vti knockdowns, we examined the transcriptional levels of A. maculatum SNARE genes (Fig. 8). Transcriptional levels of VAMP1/2 and VAMP7 were slightly elevated in the double-Vti knockdowns, though these were not sufficient to be statistically significant. However, even a slight increase in VAMP proteins levels could accommodate successful vesicle fusion (van den Bogaart et al., 2010). Loss of function studies of key SNAREs involved in fusion in neuronal system suggest that a parallel pathway, possibly involving noncanonical SNAREs typically implicated in constitutive vesicle trafficking, may mediate fusion and recycling of a subset of vesicles (Bronk et al., 2007; Deak et al., 2004; Schoch et al., 2001; Washbourne et al., 2002; Ramirez et al., 2012). Certainly, there was no clear transcriptional activation of any of the SNARE genes that we examined in the knockdowns. The absence of observable SNARE transcriptional activation (Fig. 8) could be attributed to the time when these were sampled. Transcriptional activity of all Vti genes is approximately is 60-90% lower than unfed ticks by 120 hrs post-infestation (Fig. 4), which is the time point at which we examined the efficiency of transcriptional knockdown (Figs. 7). It is uncertain the degree of transcriptional knockdown that occurred at earlier stages of feeding. Any transcriptional coactivation could be occurring earlier in the bloodmeal than we sampled.
We have recently shown that transcriptional expression of Vti1B and Syntaxin 6 is significantly higher in salivary glands and midguts within Ehrlichia chaffeensis-infected A. americanum compared to uninfected control (Karim et al., 2012). Perhaps the Vti family is utilized by pathogens to aid in survival or dissemination from the tick to the host, Indeed, the Chlamydial IncA protein, and its Legionella homolog, IcmG/DotF, have SNARE-like motifs that inhibit host endocytotic SNARE machinery in order to evade fusion with host lysosomes (Delevoye et al., 2008; Paumet et al., 2009; Wesolowski et al., 2010). Interestingly, Vti1A and Vti1B were found to colocalize with Coxiella burnetii replicative vacuoles, but even in the absence of a functional bacterial secretory system, C. burnetii were able to transit across the endocytic pathway (Campoy et al., 2012). The role of the Vti family in pathogen infection and transmission in A. maculatum and A. americanum is certainly worth further exploration.
It seems possible that pathogens may exploit the ticks’ salivary regulatory signals and mechanism of exocytosis (including vesicle fusion) for transmission to the host. As more detailed information becomes available about transcytotic mechanisms in tick salivary glands, and with an appropriate salivary gland/pathogen model system, it should be possible to test whether regulatory factors controlling the release of salivary proteins also regulate the exocytotic secretion of pathogens. Understanding the role of salivary vesicle transport proteins in secretion is essential to our understanding of tick cellular processes and could result in the discovery of new methods to prevent the transmission of tick-borne diseases.
Our data suggests that AmVti1a and AmVti1B are important members of the endo-lysosomal membrane trafficking. However, they do not seem to be essential, given the lack of a phenotype in the AmVti1A/AmVti1B knockdown. A previous study has similarly shown that Vti1A/B double-knockout mice survive until birth and endo-lysosomal trafficking is largely normal (Kunwar et al. 2011). Murine Vti1A and Vti1B are expendable in maintaining membrane traffic at a basal level and phenotypic defects were only detectable in situations of high demand for endosomal trafficking or specialized endosomal function. This would explain the neuronal defects observed in the double-knockout mice. Taken together these data seem to suggest that a compensatory mechanism exists within the secretory system to maintain endo-lysosomal trafficking at basal levels. Since it is well regarded that various intracellular “rickettisial-type” bacteria hijack the host membrane fusion machinery, it is possible that this process may place an increased burden on membrane trafficking for bacterial replication. It may prove useful to identify SNARE proteins which may function in a compensatory mechanism from the Vti family, particularly since these proteins seem to be recruited by intracellular pathogens. Tick SNAREs (including the Vti family) represent attractive targets in the development of biopharmaceuticals to inhibit tick attachment and prevent the transmission of tick-borne diseases.
Supplementary Material
Highlights.
➢ Functional significance of the VTI family of SNARE proteins, AmVti1a, AmVti1b, and AaVti1b was determined.
➢ The qRT-PCR analysis revealed the highest expression of the VTI family in unfed salivary glands.
➢ The VTI family of SNARE proteins is localized differently in unfed and partially fed salivary glands.
➢ Knockdown of the VTI family of SNARE proteins suggests an important role in tick blood feeding, survival and oviposit.
Acknowledgements
The authors thank Baobin Kang for her expert technical assistance in confocal scanning microscopy. This work was supported by grants from the American Heart Association award (09SDG2280207), NIH NIAID (R15AI099910), and US Department of State award (PGA-P21049) to SK. Support for MSS was provided through the Higher Education Commission of Pakistan's advanced postdoctoral training program. The MS-INBRE Core Facility is supported by the National Institute of General Medical Sciences, National Institute of Health Award (P20RR016476). AMV is deeply indebted to MS-INBRE summer undergraduate research fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests: The authors have declared that no competing interests exist.
References
- Advani RJ, Bae HR, Bock JB, Chao DS, Doung YC, Prekeris R, Yoo JS, Scheller RH. Seven novel mammalian SNARE proteins localize to distinct membrane compartments. J Biol Chem. 1998;273:10317–10324. doi: 10.1074/jbc.273.17.10317. [DOI] [PubMed] [Google Scholar]
- Antonin W, Holroyd C, Fasshauer D, Pabst S, Von Mollard GF, Jahn R. A SNARE complex mediating fusion of late endosomes defines conserved properties of SNARE structure and function. EMBO J. 2000;19:6453–6464. doi: 10.1093/emboj/19.23.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlashkin V, Kreykenbohm V, Eskelinen EL, Wenzel D, Fayyazi A, Fischer von Mollard G. Deletion of the SNARE vti1b in mice results in the loss of a single SNARE partner, syntaxin 8. Mol Cell Biol. 2003;23:5198–5207. doi: 10.1128/MCB.23.15.5198-5207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethani I, Werner A, Kadian C, Geumann U, Jahn R, Rizzoli SO. Endosomal fusion upon SNARE knockdown is maintained by residual SNARE activity and enhanced docking. Traffic. 2009;10:1543–1559. doi: 10.1111/j.1600-0854.2009.00959.x. [DOI] [PubMed] [Google Scholar]
- Binnington KC. Sequential changes in salivary gland structure during attachment and feeding of the cattle tick, Boophilus microplus. Int J Parasitol. 1978;8:97–115. doi: 10.1016/0020-7519(78)90004-8. [DOI] [PubMed] [Google Scholar]
- Bronk P, Deak F, Wilson MC, Liu X, Sudhof TC, Kavalali ET. Differential effects of SNAP-25 deletion on Ca2+ -dependent and Ca2+ -independent neurotransmission. J Neurophysiol. 2007;98:794–806. doi: 10.1152/jn.00226.2007. [DOI] [PubMed] [Google Scholar]
- Browning R, Adamson S, Karim S. Choice of a stable set of reference genes for qRT-PCR analysis in Amblyomma maculatum. J Med Entomol. 2012;49:1339–1346. doi: 10.1603/me12123. [DOI] [PubMed] [Google Scholar]
- Browning R, Karim S. RNA interference mediated depletion of NSF and SNAP-25 results in the inhibition of blood feeding of the Gulf-coast tick, Amblyomma maculatum. Insect Mol Biol. 2013 doi: 10.1111/imb.12017. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgalossi A, Jung S, Meyer G, Jockusch WJ, Jahn O, Taschenberger H, O'Connor VM, Nishiki T, Takahashi M, Brose N, Rhee JS. SNARE protein recycling by αSNAP and βSNAP supports synaptic vesicle priming. Neuron. 2010;68:473–487. doi: 10.1016/j.neuron.2010.09.019. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Campoy EM, Mansilla ME, Colombo MI. Endocytic SNAREs are involved in optimal Coxiella burnetii vacuole development. Cell Microbiol. 2012 doi: 10.1111/cmi.12087. doi: 10.1111/cmi.12087. [DOI] [PubMed] [Google Scholar]
- Chmelar J, Calvo E, Pedra JH, Francischetti IM, Kotsyfakis M. Tick salivary secretion as a source of antihemostatics. J Proteomics. 2012;75:3842–3854. doi: 10.1016/j.jprot.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deak F, Schoch S, Liu X, Sudhof TC, Kavalali ET. Synaptobrevin is essential for fast synaptic-vesicle endocytosis. Nat Cell Biol. 2004;6:1102–1108. doi: 10.1038/ncb1185. [DOI] [PubMed] [Google Scholar]
- Delevoye C, Nilges M, Dehoux P, Paumet F, Perrinet S, Dautry-Varsat A, Subtil A. SNARE protein mimicry by an intracellular bacterium. PLoS Pathogens. 2008;4:e1000022. doi: 10.1371/journal.ppat.1000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressel R, Elsner L, Novota P, Kanwar N, Fischer von Mollard G. The exocytosis of lytic granules is impaired in Vti1b- or Vamp8-deficient CTL leading to a reduced cytotoxic activity following antigen-specific activation. J Immunol. 2010;185:1005–1014. doi: 10.4049/jimmunol.1000770. [DOI] [PubMed] [Google Scholar]
- Fasshauer D, Sutton RB, Brunger AT, Jahn R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc Natl Acad Sci USA. 1998;95:15781–15786. doi: 10.1073/pnas.95.26.15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer von Mollard G, Stevens TH. The Saccharomyces cerevisiae v-SNARE Vti1p is required for multiple membrane transport pathways to the vacuole. Mol Biol Cell. 1999;10:1719–1732. doi: 10.1091/mbc.10.6.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francischetti IM, Sa-Nunes A, Mans BJ, Santos IM, Ribeiro JM. The role of saliva in tick feeding. Front Biosci. 2009;14:2051–2088. doi: 10.2741/3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong H, Umemiya R, Zhou J, Liao M, Zhang H, Jia H, Nishikawa Y, Xuan X, Fujisaki K. Blocking the secretion of saliva by silencing the HlYkt6 gene in the tick Haemaphysalis longicornis. Insect Biochem Mol Biol. 2009;39:372–381. doi: 10.1016/j.ibmb.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Gong H, Zhou J, Liao M, Hatta T, Harnnoi T, Umemiya R, Inoue N, Xuan X, Fujisaki K. Characterization of a carboxypeptidase inhibitor from the tick Haemaphysalis longicornis. J Insect Physiol. 2007;53:1079–1087. doi: 10.1016/j.jinsphys.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Hong W. SNAREs and traffic. Biochim Biophys Acta. 2005;1744:493–517. [PubMed] [Google Scholar]
- Jahn R, Lang T, Sudhof TC. Membrane fusion. Cell. 2003;112:519–533. doi: 10.1016/s0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- Jahn R, Sudhof TC. Membrane fusion and exocytosis. Annu Rev Biochem. 1999;68:863–911. doi: 10.1146/annurev.biochem.68.1.863. [DOI] [PubMed] [Google Scholar]
- Jaworski DC. Tick “talk”: protein release by tick salivary cells. Trends Parasitol. 2003;19:427–429. doi: 10.1016/j.pt.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Karim S, Adamson S. RNA interference in ticks: a functional genomics tool for the study of physiology. Adv in Insect Phys. 2012;42:119–146. [Google Scholar]
- Karim S, Browning R, Ali L, Truhett R. Laboratory-infected Ehrlichia chaffeensis female adult Amblyomma americanum salivary glands reveal differential gene expression. J Med Entomol. 2012;49:547–554. doi: 10.1603/me11214. [DOI] [PubMed] [Google Scholar]
- Karim S, Essenberg RC, Dillwith JW, Tucker JS, Bowman AS, Sauer JR. Identification of SNARE and cell trafficking regulatory proteins in the salivary glands of the lone star tick, Amblyomma americanum (L.). Insect Biochem Mol Biol. 2002;32:1711–1721. doi: 10.1016/s0965-1748(02)00111-x. [DOI] [PubMed] [Google Scholar]
- Karim S, Miller NJ, Valenzuela J, Sauer JR, Mather TN. RNAi-mediated gene silencing to assess the role of synaptobrevin and cystatin in tick blood feeding. Biochem Biophys Res Commun. 2005;334:1336–1342. doi: 10.1016/j.bbrc.2005.07.036. [DOI] [PubMed] [Google Scholar]
- Karim S, Ramakrishnan VG, Tucker JS, Essenberg RC, Sauer JR. Amblyomma americanum salivary gland homolog of nSec1 is essential for saliva protein secretion. Biochem Biophys Res Commun. 2004;324:1256–1263. doi: 10.1016/j.bbrc.2004.09.189. [DOI] [PubMed] [Google Scholar]
- Karim S, Ramakrishnan VG, Tucker JS, Essenberg RC, Sauer JR. Amblyomma americanum salivary glands: double-stranded RNA-mediated gene silencing of synaptobrevin homologue and inhibition of PGE2 stimulated protein secretion. Insect Biochem Mol Biol. 2004;34:407–413. doi: 10.1016/j.ibmb.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Karim S, Singh P, Ribeiro JM. A deep insight into the sialotranscriptome of the gulf coast tick, Amblyomma maculatum. PLoS One. 2011;6:e28525. doi: 10.1371/journal.pone.0028525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreykenbohm V, Wenzel D, Antonin W, Atlachkine V, von Mollard GF. The SNAREs vti1a and vti1b have distinct localization and SNARE complex partners. Eur J Cell Biol. 2002;81:273–280. doi: 10.1078/0171-9335-00247. [DOI] [PubMed] [Google Scholar]
- Kunwar AJ, Rickmann M, Backofen B, Browski SM, Rosenbusch J, Schoning S, Fleischmann T, Krieglstein K, Fischer von Mollard G. Lack of the endosomal SNAREs vti1a and vti1b led to significant impairments in neuronal development. Proc Natl Acad Sci USA. 2011;108:2575–2580. doi: 10.1073/pnas.1013891108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Patrick CD, Hair JA. Laboratory rearing procedures and equipment for multi-host ticks (Acarina: Ixodidae). J Med Entomol. 1975;12:389–390. doi: 10.1093/jmedent/12.3.389. [DOI] [PubMed] [Google Scholar]
- Paumet F, Wesolowski J, Garcia-Diaz A, Delevoye C, Aulner N, Shuman HA, Subtil A, Rothman JE. Intracellular bacteria encode inhibitory SNARE-like proteins. PLoS One. 2009;4:e7375. doi: 10.1371/journal.pone.0007375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Ramirez DM, Kavalali ET. The role of non-canonical SNAREs in synaptic vesicle recycling. Cell Logist. 2012;2:20–27. doi: 10.4161/cl.20114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JM. Role of saliva in tick/host interactions. Exp Appl Acarol. 1989;7:15–20. doi: 10.1007/BF01200449. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM. Blood-feeding arthropods: live syringes or invertebrate pharmacologists? Infect Agents Dis. 1995;4:143–152. [PubMed] [Google Scholar]
- Ribeiro JM, Francischetti IM. Role of arthropod saliva in blood feeding: sialome and post-sialome perspectives. Annu Rev Entomol. 2003;48:73–88. doi: 10.1146/annurev.ento.48.060402.102812. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritish EF, Maniatis T. Molecular Cloning, A Laboratory Manual. Cold Spring Harbor Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Sauer JR, Essenberg RC, Bowman AS. Salivary glands in ixodid ticks: control and mechanism of secretion. J Insect Physiol. 2000;46:1069–1078. doi: 10.1016/s0022-1910(99)00210-3. [DOI] [PubMed] [Google Scholar]
- Schoch S, Deak F, Konigstorfer A, Mozhayeva M, Sara Y, Sudhof TC, Kavalali ET. SNARE function analyzed in synaptobrevin/VAMP knockout mice. Science. 2001;294:1117–1122. doi: 10.1126/science.1064335. [DOI] [PubMed] [Google Scholar]
- Sonenshine DE. Biology of Ticks Oxford University Press; New York: 1991. [Google Scholar]
- Surpin M, Zheng H, Morita MT, Saito C, Avila E, Blakeslee JJ, Bandyopadhyay A, Kovaleva V, Carter D, Murphy A, Tasaka M, Raikhel N. The VTI family of SNARE proteins is necessary for plant viability and mediates different protein transport pathways. Plant Cell. 2003;15:2885–2899. doi: 10.1105/tpc.016121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teel PD, Ketchum HR, Mock DE, Wright RE, Strey OF. The Gulf Coast tick: a review of the life history, ecology, distribution, and emergence as an arthropod of medical and veterinary importance. J Med Entomol. 2010;47:707–722. doi: 10.1603/me10029. [DOI] [PubMed] [Google Scholar]
- Vancova I, Slovak M, Hajnicka V, Labuda M, Simo L, Peterkova K, Hails RS, Nuttall PA. Differential anti-chemokine activity of Amblyomma variegatum adult ticks during blood-feeding. Parasite Immunol. 2007;29:169–177. doi: 10.1111/j.1365-3024.2006.00931.x. [DOI] [PubMed] [Google Scholar]
- van den Bogaart G, Holt MG, Bunt G, Riedel D, Wouters FS, Jahn R. One SNARE complex is sufficient for membrane fusion. Nat Struc Mol Biol. 2010;17:358–364. doi: 10.1038/nsmb.1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washbourne P, Thompson PM, Carta M, Costa ET, Mathews JR, Lopez-Bendito G, Molnar Z, Becher MW, Valenzuela CF, Partridge LD, Wilson MC. Genetic ablation of the t-SNARE SNAP-25 distinguishes mechanisms of neuroexocytosis. Nat Neurosci. 2002;5:19–26. doi: 10.1038/nn783. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesolowski J, Paumet F. SNARE motif: A common motif used by pathogens to manipulate membrane fusion. Virulence. 2010;1:319–324. doi: 10.4161/viru.1.4.12195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.