SUMMARY
Negative control in two-component signal transduction results from sensor transmitter phosphatase activity for phospho-receiver dephosphorylation. A hypothetical mechanism for this reaction involves a catalytic residue in the H-box active site region. However, a complete understanding of transmitter phosphatase regulation is hampered by the abundance of kinase-competent, phosphatase-defective missense substitutions (K+ P− phenotype) outside of the active site region. For the Escherichia coli NarX sensor, a model for the HisKA_3 sequence family, DHp domain K+ P− mutants defined two classes. Interaction mutants mapped to the active site-distal base of the DHp helix 1, whereas conformation mutants were affected in the X-box region of helix 2. Thus, different types of perturbations can influence transmitter phosphatase activity indirectly. By comparison, K+ P− substitutions in the HisKA sensors EnvZ and NtrB additionally map to a third region, at the active site-proximal top of the DHp helix 1, independently identified as important for DHp-CA domain interaction in this sensor class. Moreover, the NarX transmitter phosphatase activity was independent of nucleotides, in contrast to the activity for many HisKA family sensors. Therefore, distinctions involving both the DHp and CA domains suggest functional diversity in the regulation of HisKA and HisKA_3 transmitter phosphatase activities.
INTRODUCTION
Two-component systems, which constitute a widespread signaling mechanism in bacteria, operate on the interplay between histidine kinase sensors and their cognate response regulators via protein phosphorylation (Ninfa and Magasanik, 1986, Keener and Kustu, 1988). Signal transduction occurs between the sensor transmitter module and the response regulator receiver (REC) domain, both of which are structurally conserved. Within the transmitter module, the amino-terminal DHp (Dimerization and Histidine phosphotransfer) domain is a dimeric four-helix bundle formed by a pair of helical hairpin subunits (Gao and Stock, 2009). The amino-terminal helix α1 and carboxyl-terminal helix α2 of each monomer harbor the characteristic H- and X-box sequence motifs, respectively (Fig. 1) (Parkinson and Kofoid, 1992, Stock et al., 1995, Hsing et al., 1998). The carboxyl-terminal CA (Catalytic and ATP binding) domain of the conventional transmitter module adopts a monomeric α/β sandwich fold (Parkinson and Kofoid, 1992, Gao and Stock, 2009). The receiver domain, which communicates with the transmitter module, exhibits a highly conserved α/β structure, with a five-stranded β sheet surrounded by five α helices (Bourret, 2010).
Fig. 1.
DHp domain sequences. A. E. coli NarX and B. subtilis DesK, representing the HisKA_3 subfamily. B. E. coli EnvZ and NtrB, representing the HisKA subfamily. Residues at positions of K+ P− substitutions are boxed; for some positions, multiple different substitutions were recovered. The H-box (Parkinson and Kofoid, 1992) and X-box (Hsing et al., 1998) motifs are shown as previously defined. The coiled-coil repeat patterns are heptad for DesK (Albanesi et al., 2009) and hendecad for EnvZ (Ferris et al., 2012). The phospho-accepting His residues are shaded black. The transmitter phosphatase DxxxQ/H and E/DxxT/N motifs are described previously (Huynh et al., 2010); hypothesized catalytic residues are shaded red. Residues shaded in yellow and green were identified as participating directly or indirectly, respectively, in determining specificity for DHp-receiver domain interaction (Weigt et al., 2009).
Despite the overall structural conservation of different transmitter modules, some distinctions exist for their amino acid sequences. Based on DHp domain sequences alone, the majority of sensors belong to the HisKA (pfam 00512) and HisKA_3 (pfam 07730) families (Punta et al., 2012), which also correspond to the HPK1-4 and HPK7 families of transmitter module sequences, respectively (Grebe and Stock, 1999). Well-studied examples include the HisKA sensors EnvZ, NtrB, PhoQ, and HK853, and the HisKA_3 sensors NarX, DesK and UhpB.
Signal transduction events involve both positive and negative control. Many sensors are bifunctional and their transmitter module catalyzes both of these regulatory reactions. The ratio of transmitter positive and negative activities determines the steady state amount of phosphorylated receiver for output control (Gao and Stock, 2009). Positive signal transmission occurs via the transmitter autokinase reaction at an invariant His residue in the H-box region of the DHp domain helix α1, and subsequent phosphoryl transfer to an invariant Asp residue in the receiver domain (Stock et al., 1989).
Negative signaling results from transmitter phosphatase activity, which greatly accelerates the relatively slow receiver autodephosphorylation rate (Huynh and Stewart, 2011). Studies of the Escherichia coli NarX (Huynh et al., 2010) and EnvZ (Dutta et al., 2000) sensors have identified a conserved Gln/Asn/Thr residue, in close proximity to the phospho-accepting His residue, as catalytic for transmitter phosphatase activity but largely uninvolved in the positive reactions. Therefore, the H-box motif contains the transmitter active site region, harboring residues essential for both positive and negative enzymatic activities (Hsing and Silhavy, 1997).
Nevertheless, the understanding of transmitter phosphatase activity is complicated by the abundance of kinase-competent, phosphatase-defective missense mutants (K+ P− phenotype), isolated over the years through both random and site-specific mutagenesis of the E. coli sensors EnvZ (Hall and Silhavy, 1981, Tokishita et al., 1992, Hsing et al., 1998, Zhu and Inouye, 2002, Qin et al., 2003, Capra et al., 2010) and NtrB (Atkinson and Ninfa, 1993, Pioszak and Ninfa, 2003a). These K+ P− mutants harbor substitutions that map through virtually all domains, outside the proposed active site. Most alterations are scattered throughout the DHp domain, with a smaller number located in the CA domain, and in the sensory module. Whereas the sensory module mutants are almost certainly perturbed for signal perception, those associated with the transmitter module are more complicated to account for.
Furthermore, the CA domain appears instrumental in HisKA transmitter phosphatase activity, since it greatly enhances the reaction rates of the NtrB and EnvZ DHp domains (Jiang et al., 2000, Zhu et al., 2000). The involvement of the CA domain is further suggested from the stimulatory effects of nucleotides, such as ATP, ADP, and non-hydrolyzable analogs in several sensors, although they have little or no effect in some others. However, exactly how nucleotide binding regulates the transmitter phosphatase reaction remains unclear (Huynh and Stewart, 2011).
Altogether, these observations emphasize the complexity of transmitter phosphatase activity and its requirement for a coordination of multiple parameters. Here, we address both the broad spectrum of K+ P− mutants and the nucleotide effect in the NarX sensor, representing the HisKA_3 family. Mutants with DHp domain substitutions fall into two distinct classes, being affected either for transmitter-receiver interaction or for conformation. Thus, these non-active-site region alterations affect different aspects of transmitter phosphatase activity. Most CA domain substitutions conferred comparatively weaker phenotypes, and transmitter phosphatase activity was nucleotide independent. Thus, the NarX CA domain appears to serve a relatively minor role in transmitter phosphatase activity. Overall, these results illuminate differences in negative regulation between sensors in the HisKA_3 and HisKA families.
RESULTS
Optimizing the in vivo assay for NarX negative function
The nitrate-responsive two-component NarX-NarL pair is a well-studied prototypical system with a known signal input and rather simple architecture. Nitrate binding at the NarX sensory module triggers transmitter autokinase, resulting in subsequent phosphoryl transfer to NarL. In the absence of nitrate, NarX exhibits transmitter phosphatase activity on phospho-NarL (Stewart, 2003).
We previously have exploited an assay based on the narL505 allele to monitor NarX in vivo negative function on NarL (Williams and Stewart, 1997a, Huynh et al., 2010). This allele encodes the mutant NarL(V88A) protein, that constitutively activates Φ(narG-lacZ) reporter expression even in narX null strains (Egan and Stewart, 1991). Whereas receiver phosphorylation is a prerequisite for DNA binding by NarL(V88A) (Li et al., 1994), its non-cognate phospho-donors are unidentified. Nevertheless, in the absence of nitrate, the narX+ allele substantially reduces Φ(narG-lacZ) expression in narL505 strains, reflecting NarX negative function on phospho-NarL(V88A) (Egan and Stewart, 1991).
In order to optimize this assay, we evaluated the effect of narX gene dosage by comparing pcnB+ and pcnB null strains. Null alleles of the pcnB gene, encoding poly(A) polymerase I (Cao and Sarkar, 1992), reduce the copy number of ColE1-plasmids (Lopilato et al., 1986, Liu and Parkinson, 1989), due to a defect in polyadenylation of short antisense RNAs (Soderbom et al., 1997). In pcnB1 strains, nitrate-responsive Φ(narG-lacZ) activation by plasmid-borne narX+ is indistinguishable from that conferred by the chromosomally-encoded counterpart (Williams and Stewart, 1997b).
In the pcnB1 strain grown without nitrate, the narX+ allele reduced basal, uninduced Φ(narG-lacZ) expression by approximately five fold, compared to the narX null strain (Table 1). By contrast, the pcnB+ strain displayed a ten-fold reduction. For a broader spectrum, we also inspected three K− P+ NarX mutants, with the R54K, H399Q and H513Q substitutions. Since the R54K mutant is defective for nitrate binding (Williams and Stewart, 1997a), and the H399Q and H513Q mutants are defective for transmitter autokinase (Cavicchioli et al., 1995, Huynh et al., 2010), all are biased toward the transmitter phosphatase activity. Their strong negative activities observed in the pcnB1 strain were substantially enhanced in the pcnB+ strain, exhibited by the more pronounced reduction of basal expression (Table 1). Since the pcnB+ allele enhanced negative function for both the wild-type and K− P+ mutants, we employed pcnB+ strains to increase the sensitivity for monitoring NarX phosphatase mutant phenotypes.
Table 1.
Effects of pcnB genotype on NarL (V88A)-dependent Φ(narG-lacZ) expression.
| β-Galactosidase a |
||||||
|---|---|---|---|---|---|---|
|
pcnB1
c
|
pcnB
+ d
|
|||||
| Allele b | None | Nitrate | Ratio | None | Nitrate | Ratio |
| pHG165 (vector) | 1,390 | 1,450 | 1.0 | 1,770 | 1,150 | 0.6 |
| narX† | 280 | 1,700 | 6.1 | 180 | 1,100 | 6.1 |
| R54K | 180 | 210 | 1.2 | 59 | 79 | 1.3 |
| H399Q | 120 | 1,650 | 14 | 9 | 27 | 3.0 |
| H513Q | 25 | 1,650 | 66 | 7 | 810 | 116 |
Activity was measured as described in Experimental Procedures, and is expressed in Miller units.
Amino-acyl residue substitutions. narX alleles located on plasmid pVJS1241 or pVJS2474 derivatives.
Strain VJS5720 [Φ(narG-lacZ) narL505 (V88A) narP+ ΔnarX narQ::Tn10 pcnB1].
Strain VJS4033 [Φ(narG-lacZ) narL505 (V88A) narP+ ΔnarX narQ::Tn10 pcnB+].
K+ P− phenotypes of the DHp domain missense mutants
In the in vivo positive function assay, a narX+ allele induces Φ(narG-lacZ) expression by more than 80-fold in response to nitrate, but only weakly in response to nitrite. NarX missense mutants with the nitrite hypersensitive phenotype display increased nitrite response for Φ(narG-lacZ) induction (Williams and Stewart, 1997b, Williams and Stewart, 1997a, Noriega et al., 2010) (Table 2). As previously hypothesized (Williams and Stewart, 1997b), these mutants also are defective for negative function (Noriega et al., 2010) and therefore display the K+ P− phenotype (Table 2). Thus, the NarX K+ P− mutants described here were obtained by localized mutagenesis to the transmitter coding region (Williams and Stewart, 1997b) followed by a random screen for the nitrite-hypersensitive phenotype (Noriega et al., 2010).
Table 2.
Effects of narX missense substitutions on Φ(narG-lacZ) expression.
| β-Galactosidase a |
|||||||
|---|---|---|---|---|---|---|---|
| Negative Function d |
Positive Function e |
||||||
| Allele b | Domain c | None | Nitrate | Ratio | None | Nitrate | Nitrite |
| pHG165 | — | 1,390 | 970 | 0.7 | 35 | 40 | 30 |
| narX† | — | 150 | 1,030 | 6.9 | 26 | 2,060 | 120 |
| M411V | DHp helix αl | 310 | 1,160 | 3.7 | 21 | 2,150 | 310 |
| E586K | CA | 440 | 1,240 | 2.8 | 30 | 2,070 | 390 |
| K531I | CA | 670 | 1,140 | 1.7 | 27 | 2,640 | 460 |
| K481R | CA | 680 | 1,190 | 1.8 | 38 | 1,810 | 440 |
| C415R | DHp helix α1 | 1,320 | 1,110 | 0.8 | 79 | 1,870 | 1,090 |
| K481R+E586K | CA | 1,350 | 1,270 | 0.9 | 55 | 2,350 | 820 |
| Y551F | CA | 1,380 | 1,210 | 0.9 | 44 | 2,250 | 830 |
| S405P | DHp helix α1 | 1,420 | 1,070 | 0.8 | 20 | 1,700 | 720 |
| W442C | DHp helix α2 | 1,420 | 1,230 | 0.9 | 180 | 1,890 | 1,530 |
| F452Y | DHp helix α2 | 1,420 | 1,090 | 0.8 | 45 | 2,090 | 880 |
| W442R | DHp helix α2 | 1,420 | 820 | 0.6 | 30 | 1,820 | 1,500 |
| Q412R+V413M | DHp helix α1 | 1,500 | 1,150 | 0.8 | 24 | 1,800 | 810 |
| K410E | DHp helix α1 | 1,610 | 1,150 | 0.7 | 34 | 1,350 | 1,330 |
| W442G | DHp helix α2 | 1,690 | 1,170 | 0.7 | 120 | 1,670 | 2,380 |
| M411T | DHp helix α1 | 1,840 | 990 | 0.5 | 62 | 1,840 | 1,400 |
| S508C+D558V | CA | 2,280 | 1,030 | 0.5 | 1,950 | 2,010 | 3,120 |
| V580A+F589I | CA | 2,540 | 1,260 | 0.5 | 29 | 940 | 150 |
Activity was measured as described in Materials and Methods, and is expressed in Miller units.
Amino-acyl residue substitutions. narX alleles located on plasmid pVJS1241 or pVJS2474 derivatives.
Location within NarX protein structure; see text for details.
Negative function assayed in strain VJS4033 [Φ(narG-lacZ) narL505 (V88A) narP+ ΔnarX narQ::Tn10 pcnB+].
Positive function assayed in strain VJS5054 [Φ(narG-lacZ) narL+ narP+ ΔnarX narQ::Tn10 pcnB1]; data from reference (Noriega et al., 2010).
Ten K+ P− mutants from this collection have missense substitutions within the DHp domain, outside the H-box active site region on helix α1 and in the X-box on helix α2 (Fig. 1A). The V413M mutant initially was recovered as the Q412R+V413M double substitution. We characterized these mutants for their in vivo NarL negative function phenotypes in Φ(narG-lacZ) narL505 strains as described above.
Among the six mutants in the DHp helix α1 region, five (S405P, K410E, M411T, V413M, and C415R) were severely diminished for in vivo negative function (Tables 2 and S1). By contrast to the M411T mutant, the hydrophobic M411V substitution only conferred a moderate defect. Nevertheless, compared to the wild-type function, the M411V mutant activity was reduced to an appreciable level (Table 2). These substitutions all affect the base of DHp helix α1, distal to the transmitter phosphatase active site residue Gln-404 (Huynh et al., 2010).
All four mutants in the DHp helix α2 X-box region (W442C, W442R, W442G, and F452Y) were also strongly impaired for negative function. Since three substitutions were isolated at the Trp-442 residue, this position appears highly sensitive for negative function disturbance.
Table 2 lists the K+ P− mutants in increasing order for the negative function defect. Overall, the K+ P− phenotypes of these mutants followed a continuum of values and did not assort into distinct categories. Moreover, excluding the exceptional V580A+F589I mutant, there was a good correlation between nitrite hypersensitivity (nitrite response in the narL+ background) and defective negative function (elevated basal Φ(narG-lacZ) expression in the narL505 background) (Fig. S1). This supports the prior conclusion, that the nitrite hypersensitive phenotype reflects loss of negative function, namely transmitter phosphatase activity (Williams and Stewart, 1997b).
Two classes of DHp domain K+ P− mutants dissected by in vitro phosphatase assays
We chose five K+ P− NarX DHp domain mutants for further analysis by direct in vitro assays as described previously (Noriega et al., 2010; (Huynh et al., 2010). We included three of six mutants from the DHp helix α1 region, K410E, M411T and C415R. Of the two Met-411 mutants, M411T had a stronger K+ P− phenotype than M411V, the S405P phenotype probably results indirectly from the structural perturbation of a Pro residue introduced in the α helix, and the V413M mutant was isolated as a double substitution (with Q412R). We also included two of four mutants from the DHp helix α2 region, W442R (representing the three different changes at Trp-442) and F451Y. Finally, these mutants (excepting C415R) were studied previously from a different perspective (Noriega et al., 2010), enabling comparison.
The soluble MBP-NarX protein used in the in vitro tests has maltose binding protein fused to the NarX central GAF domain and transmitter module. All wild-type and mutant MBP-NarX constructs displayed near-normal autokinase and phosphoryl transfer activities in vitro, consistent with their K+ phenotype in vivo, as shown in Fig. S2 and Fig. 3 for the C415R mutant and in reference (Noriega et al., 2010) for the other mutants.
Fig. 3.

Phosphoryl transfer reactions between MBP-NarX227 and His6-NarL. All panels: ●, phospho-His6-NarL. A. □, NarX+. B. ○, C415R. C. ◇, Y551F.
In vitro phosphoryl transfer between phospho-MBP-NarX and His6-NarL provide one assay for transmitter phosphatase activity. MBP-NarX was first autophosphorylated in reactions containing [32P]-ATP and an ATP regeneration system (Noriega et al., 2008), His6-NarL was added, and time points were analyzed for both phospho-MBP-NarX and phospho-His6-NarL (Noriega et al., 2010). In this assay, a transmitter phosphatase defect is revealed by the persistence of phospho-His6-NarL over the time course. Results for the C415R mutant are shown in Fig. 3; results for the others are presented in reference (Noriega et al., 2010). (Note that these latter assays were conducted at low temperature in an attempt to slow the rapid rate of phosphoryl tranfer.) In these assays, all mutants except K410E displayed substantially decreased levels of transmitter phosphatase activity.
A second, more direct assay for transmitter phosphatase activity was performed by adding [32P]-labeled phospho-His6-NarL directly to unphosphorylated MBP-NarX in the absence of ATP regeneration, and analyzing time points for phospho-His6-NarL (Huynh et al., 2010). For these assays, we added a saturating amount of MBP-NarX protein, as described in Experimental Procedures, in order to detect even relatively weak transmitter phosphatase activities. Results are shown in Fig. 2; approximate half-lives (t1/2) for phospho-NarL were estimated from these results. The α1 helix mutant proteins M411T (t1/2 ~ 8 min), C415R (t1/2 ~ 2 min), and K410E (t1/2 ~ 1 min) were progressively more active in that order for the phosphatase reaction with phospho-NarL (Figs. 2A and 2B). By contrast, with the X-box mutant proteins W442R and F452Y, phospho-His6-NarL decayed at the intrinsic autodephosphorylation rate (t1/2 ≥ 20 min) (Fig. 2C).
Fig. 2.
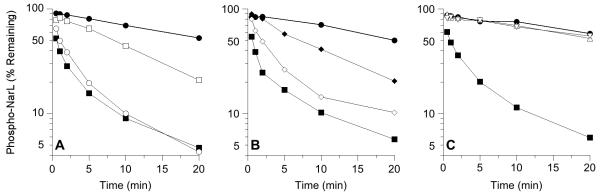
Missense substitutions influence MBP-NarX227 phospho-NarL transmitter phosphatase activity in vitro. All panels: ●, no sensor; ■, NarX+. A. Substitutions in DHp domain helix α1. ○, K410E; □, M411T. B. Substitutions in DHp domain helix α1 and CA domain. ◇, C415R; ◆, Y551F. C. Substitutions in DHp domain helix α2. △, W442R; ▽, F452Y.
Therefore, although these mutants exhibited similar K+ P− phenotypes in vivo (Table 2), the direct transmitter phosphatase assay distinguished two distinct classes of DHp domain mutants. The α1 helix mutants K410E, M411T, and C415R displayed at least detectable activities, whereas the α2 helix mutants W442R and F452Y were devoid of activity even with the excess of added sensor protein.
Two classes of DHp domain K+ P− mutants revealed by bacterial two-hybrid assays
Among other possibilities, substitutions on the DHp domain may impair the transmitter-receiver interaction, since this domain forms the major interface with the receiver domain (Skerker et al., 2008, Casino et al., 2009, Capra et al., 2010). To evaluate the in vivo interaction of NarX K+ P− mutants with the NarL receiver domain, we employed the bacterial adenylate cyclase two-hybrid assay (Karimova et al., 1998). In this assay, the T18 and T25 catalytic fragments of Bortedella pertussis adenylate cyclase reconstitute activity upon interaction of hybrid proteins, thereby complementing an E. coli Δcya strain (Karimova et al., 1998). Unlike the in vitro phosphatase reactions of the soluble, truncated form of NarX, described above, the bacterial two-hybrid assays reflect activity of the native, membrane-bound NarX sensor.
We have used this two-hybrid assay to examine several aspects of the NarX-NarL and NarQ-NarP two-component systems; results will be presented in a separate publication (T. N. Huynh, L.-L. Chen, and V. Stewart, manuscript in preparation). In the course of that work, we evaulated the domain and configuration requirements for optimal detection of NarX-NarL interactions. Accordingly, assays described below used constructs expressing the T25 fragment fused to the carboxyl-terminal end of full-length NarX (NarX-T25), and the T18 fragment fused to the amino-terminal end of the NarL receiver domain (T18-NarLREC). All wild-type and mutant NarX-T25 constructs were active for nitrate-responsive Φ(narG-lacZ) induction (FIg. S3). Thus, these hybrids were stable and displayed normal response to nitrate addition.
In the two-hybrid assays, wild-type NarX exhibited a strong interaction output with the NarL receiver domain (Fig. 4), as expected for a cognate sensor and receiver. Moreover, the active site region mutants H399Q, D400A, and Q404A (Huynh et al., 2010) all displayed the wild-type level of interaction with the NarL receiver, as expected from the catalytic mechanisms for both the phosphoryl transfer and transmitter phosphatase reactions (Stock et al., 1993, Huynh et al., 2010).
Fig. 4.

In vivo interaction between the NarX-T25 and T18-NarLREC hybrid proteins, as determined by bacterial adenylate cyclase two-hybrid assay. LacZ synthesis depends on cAMP resulting from two-hybrid interaction (Karimova et al., 1998). Controls are: Vectors, empty T18 and T25 vectors; ZIP, T18 and T25 vectors with leucine zipper dimerization sequence. The D558V mutant also carries the S508C substitution. Error bars show the standard deviations of the average values determined from three indepenent assays.
The K+ P− mutants described in this study were segregated for interaction output. Whereas the W442R, and F452Y mutants interacted strongly with the NarL receiver domain, the S405P, K410E, M411T, and C415R mutants were defective (Fig. 4). (Again, however, the Pro substitution for Ser-405 also likely disturbed the α helical structure.)
Thus, the two-hybrid assays revealed two distinct classes of DHp domain mutants that were congruent with those revealed by the in vitro phosphatase assay. One class, termed interaction mutants, includes the K410E, M411T, and C415R mutants that also exhibited detectable in vitro phosphatase activities. Note that these mutants interact sufficently well in the phosphoryl transfer conformation to effect normal positive regulation during growth in the presence of nitrate (Table 1) and phosphoryl transfer in vitro (Noriega et al., 2010); (Fig. 3B). The other class, termed conformation mutants (Pioszak and Ninfa, 2003b), includes the W442R and F452Y mutants, which displayed normal interaction but had undetectable transmitter phosphatase activity in vitro.
We conducted these assays for cultures grown in the absence of nitrate, in which the mutant NarX sensors were in the transmitter phosphatase conformation. We hoped to examine interactions also for cultures grown in the presence of nitrate, as yet another test for interactions in the phosphoryl transfer conformation. Unfortunately, despite several attempts with different hybrid configurations and strain backgrounds, we were unable to generate convincingly reliable results for the NarX interaction mutants.
Distinct properties of a CA domain K+ P− mutant
Seven mutants from the K+ P− collection have missense substitutions in the CA domain. Three mutants, with relatively weak P− phenotypes (K481R, K531I, and E586K) have single substitutions, whereas two others with strong P− phenotypes (K481R+E586K and V580A+F589I) have double substitutions. The independent isolation of the K481R and E586K single mutants, with weak P− phenotypes, and the K481R+E586K double mutant, with a stronger P− phenotype, shows the additive effect of these changes.
Two CA domain single mutants displayed strong P− phenotypes: Y551F (Table 2), and D558V, segregated away from the second substitution, S508C (Table S1). We performed in vitro tests with the Y551F mutant as a representative of CA domain mutants. The Y551F mutant had normal autokinase (Fig. S2) and phosphoryl transfer (Fig. 3C) activities, but displayed impaired transmitter phosphatase activity in vitro (t1/2 ~ 7 min), comparable to that of the M411T mutant (Fig. 2B). In the two-hybrid assay, the NarX Y551F interacted with the NarL receiver domain to produce output comparable to that of wild-type NarX (Fig. 4). Thus, NarX Y551F corresponds to neither the conformation nor the interaction classes defined by the DHp domain mutants described above. The S508C+D558V mutant was not evaluated in vitro, but also displayed a wild-type interaction output (Fig. 4).
The role of the NarX CA domain is unrelated to nucleotide binding
For sensors such as EnvZ (Igo et al., 1989, Zhu et al., 2000), NtrB (Keener and Kustu, 1988, Kamberov et al., 1994), PhoQ (Sanowar and Le Moual, 2005, Yeo et al., 2012), and WalK (Gutu et al., 2010), transmitter phosphatase activity is stimulated by ATP and/or ADP, and by non-hydrolyzable ATP analogs. By contrast, MBP-NarX transmitter phosphatase activity in vitro was indistinguishable in the presence or absence of these nucleotides (Fig. 5A), demonstrating that the robust NarX transmitter phosphatase function is not stimulated further by nucleotides.
Fig. 5.

Nucleotides (100 μM) do not influence MBP-NarX227 phospho-NarL transmitter phosphatase activity in vitro. A. Effects of nucleotide addition. ●, no sensor; ■, NarX+; ◇, NarX+ plus ATP; △, NarX+ plus ADP; ▽, NarX+ plus AMP-PNP. B. Effect of H513Q substitution. ●, no sensor; ■, NarX+; ◇, H513Q.
In sensors for which the transmitter phosphatase activity is affected by nucleotides, the involvement of the CA domain is implied since it provides the nucleotide binding activity. The NarX CA domain harbors an invariant His-513 residue, analogous to the DesK His-297 residue, which is a critical part of the ATP-binding pocket (Trajtenberg et al., 2010). Thus, the NarX H513Q mutant, which presumably is defective for ATP-binding, was used to test further the role of the CA domain for the transmitter phosphatase reaction. The H513Q mutant is devoid of in vivo positive function as well as in vitro transmitter autokinase activity (Cavicchioli et al., 1995). Nevertheless, it displays wild-type negative function both in vivo and in vitro (Cavicchioli et al., 1995) (Table 1 and Fig. 5B). Thus, at least for NarX, any role of the CA domain for transmitter phosphatase function appears to be independent of nucleotide binding.
DISCUSSION
The transmitter autokinase and phosphoryl transfer reactions universally depend upon an invariant His residue (Stock et al., 1989). By contrast, transmitter phosphatase activity had not been recognized to associate with an analogous invariant residue. We previously hypothesized that this reaction is catalyzed by the amide or hydroxyl functional group of a conserved Gln/Asn/Thr residue within the transmitter active site region (Huynh et al., 2010). Recent work with other sensors provides independent support for a broad application of this hyothesis (Schrecke et al., 2013, Wayne et al., 2012, Willett and Kirby, 2012). Nevertheless, the quest for a common transmitter phosphatase mechanism is hampered by the abundance of missense substitutions conferring the K+ P− phenotype, scattered throughout the transmitter DHp and CA domains. Our analysis of NarX K+ P− mutants suggests how such substitutions likely affect transmitter phosphatase function without directly changing the active site region. Comparisons with EnvZ and NtrB K+ P− mutants also highlight the distinct regulatory strategies employed by HisKA_3 and HisKA DHp domains, as well as the CA domains associated with these families.
Two classes of NarX K+ P− DHp domain mutants altered in interaction or conformation
The Bacillus subtilis DesK DHp domain, the structural model for the HisKA_3 family, is a dimeric four-helix bundle formed by two helical hairpins (Albanesi et al., 2009). The helices α1 and α2 of each monomer exhibit amino acid residues in coiled-coil heptad repeats, conventionally denoted a-b-c-d-e-f-g, in which a and d positions are occupied by hydrophobic residues of the interhelical packing interface (Fig. 1A) (Lupas and Gruber, 2005). The NarX and DesK DHp domain sequences are colinear (Fig. 1A).
Most of the single substitution NarX K+ P− alterations map to the DHp domain, congruent with the catalytic role of this element in transmitter phosphatase activity (Albanesi et al., 2009, Jiang et al., 2000, Zhu et al., 2000). Nevertheless, these substitutions lie outside the active site region. Although they displayed a continuum of in vivo negative function phenotypes (Table 2), they nevertheless could be categorized further into two classes, interaction and conformation mutants.
The interaction mutants were severely defective in the two-hybrid interaction assay with the NarL receiver domain (Fig. 4). Notably, the analogous residues in DesK are all localized to the active site region-distal base of the DHp domain helix α1, immediately downstream of the active site region His-399 and Gln-404 residues (hereafter numbered according to NarX) (Figs. 1A and 5A). In particular, residues Met-411 and Cys-415 are aligned on the same helical face as residue Gln-404. Residue Lys-410 is just offset from this surface. Thus, residues Lys-410, Met-411, and Cys-415 define a transmitter-receiver interaction surface that is critical for the transmitter phosphatase reaction. Mutants with alterations at these positions retained transmitter phosphatase activity in vitro, under sensor-excess assay conditions, implying that the active site geometry is unperturbed.
The analogous interaction surface previously was identified for sensors in the HisKA subfamily (Skerker et al., 2008, Casino et al., 2009, Capra et al., 2010) (Fig. 1). Moreover, work with the HisKA sensor CpxA suggests that positive regulation (phosphoryl transfer) can result from relatively weak sensor-regulator interaction, wheras negative regulation (transmitter phosphatase) requires a stronger interaction (Siryaporn et al., 2010). Properties of the NarX interaction mutants described above are congruent with this hypothesis. These mutants displayed defective interaction in the two-hybrid assay during growth in the absence of nitrate (which favors the phosphatase conformation). Nevertheless, they exhibit near-normal positive regulation in vivo and phosphoryl transfer in vitro (Noriega et al., 2010), indicating that they have sufficiently strong receiver domain interactions when in the phosphoryl transfer conformation.
By contrast to the interaction mutants, the conformation mutants were virtually wild-type for interaction with the receiver (Fig. 4). The corresponding substitutions map to the X-box region of low sequence conservation on helix α2 (Fig. 1A) (Hsing et al., 1998). Residue Trp-442 is at a conserved hydrophobic residue at position e in a heptad repeat, just offset from the coiled-coil packing interface. Nevertheless, based on the DesK DHp domain structure, NarX Trp-442 is placed on the same packing layer and in close proximity to the phosphatase catalytic Gln-404 residue (Fig. 6A). Similarly, residue Phe-452 is a conserved hydrophobic residue at coiled-coil position a in a heptad repeat, immediately adjacent to the strongly conserved NarX Arg-453 residue, of which the analogous DesK Lys-242 residue interacts with the phospho-accepting His residue (Fig. 6A). Since these mutants were also devoid of phosphatase activity in vitro (Fig. 2B), these altered residues in the X-box region may be involved in determining the DHp domain conformation via interhelical interactions, as previously proposed for EnvZ and NtrB X-box mutants (Hsing et al., 1998, Jiang et al., 2000).
Fig. 6.
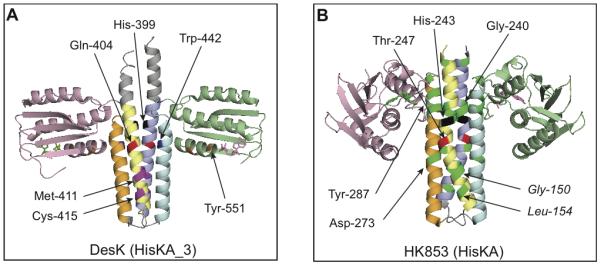
Transmitter module X-ray structures showing positions of K+ P− substitutions and other key residues. A. HisKA_3 family, represented by B. subtilis DesK (V188b version) (PDB 3EHH) (Albanesi et al., 2009). Presumed positions of analogous residues from NarX are indicated by numbers. B. HisKA family, represented by T. maritima HK853 (PDB 3DGE) (Casino et al., 2009). DHp domains are colored blue-cyan and yelow-orange, whereas CA domains are colored rose and pale green. The active site region is delimited by the phospho-accepting His (black) and the phosphatase catalytic Gln/Asn/Thr (red) residues. In DesK, residues in magenta on DHp helix α1 correspond to NarX interaction mutants. Residues in blue on DHp helix α2 correspond to NarX conformation mutants. Residues in orange in the CA domain correspond to NarX Tyr-551 and Asp-558. In HK853, residues in bright green correspond to K+ P− substitutions in EnvZ and NtrB. Residues in roman type are from EnvZ; those in italic are from NtrB.
HisKA K+ P− substitutions map to additional regions on the transmitter module
Unlike the DesK DHp domain with an essentially continuous α1 helix (Albanesi et al., 2009), HisKA DHp domains are characterized by a helix α1 kink caused by a strongly conserved Pro residue (Marina et al., 2005, Ferris et al., 2012). Consequentially, signal propagation differs in these families. In the DesK DHp domain, the conformational switch between the phosphoryl transfer and transmitter phosphatase states involves a helical phase shift and a coiled-coil formation, in which the imidazole of the phospho-accepting His is buried in the transmitter phosphatase state (Albanesi et al., 2009). In the EnvZ DHp domain, upstream conformational changes are halted at the Pro kink, and the phospho-accepting His residue remains stationary with virtually identical accessibility in both transmitter conformations (Ferris et al., 2012).
In the HisKA sensors EnvZ and NtrB, some substitutions conferring the K+ P− phenotype map to the active site region-distal base of the DHp helices α1 and α2 (Fig. 1B) (Atkinson and Ninfa, 1993, Hsing et al., 1998, Pioszak and Ninfa, 2003a), subsequently identified as the specificity determining region for transmitter-receiver interaction (highlighted in yellow in Fig. 1B) (Skerker et al., 2008, Casino et al., 2009, Yamada et al., 2009, Weigt et al., 2009, Capra et al., 2010). These substitutions are depicted on the HK853 X-ray structure (Fig. 6B), which provides the model for HisKA family transmitters (Casino et al., 2009). Interestingly, although interaction specificity determinants have not yet been determined for HisKA_3 sensors, this region also corresponds to the location of NarX interaction mutants.
However, a larger number of HisKA K+ P− substitutions are found in the active site region-proximal top of the DHp domain helix α1 (Figs. 1B and 5B) (Atkinson and Ninfa, 1993, Hsing and Silhavy, 1997, Hsing et al., 1998, Pioszak and Ninfa, 2003a). This region also harbors residues that are strongly covariant with receiver residues, despite the absence of direct contacts (highlighted in green in Fig. 1B) (Weigt et al., 2009). Furthermore, this region appears to have multiple roles in regulating the DHp domain conformation, including homodimerization (Ashenberg et al., 2011) and DHp-CA domain association in the transmitter phosphatase state (Ferris et al., 2012). By contrast, the corresponding region is not featured in the NarX K+ P− mutant collection. Indeed, the active site region-proximal sequence of the HisKA_3 helix α1 is highly conserved, whereas the corresponding region in HisKA sensors is not (Huynh et al., 2010). This suggests that the DHp helix α1 active site region-proximal top is functionally different in HisKA and HisKA_3 family sensors.
The effects of nucleotides on transmitter phosphatase activity
Stimulation of transmitter phosphatase activity by nucleotides must be related to the CA domain, which provides the nucleotide binding function. In terms of structure, the CA domain associated with HisKA_3 sensors is distinct from its HisKA counterpart. HisKA sensors exhibit the conventional CA domain sequence, featuring the N-, D- (or G1), F-, and G- (or G2) box motifs, in which the F- and G-boxes form a flexible ATP lid (Parkinson and Kofoid, 1992, Stock et al., 1995, Gao and Stock, 2009). By contrast, for HisKA_3 sensors, the CA domain sequences display no apparent F-box region, and the G-box motif has a distinct pattern of conserved Gly residues (Fig. S4; (Grebe and Stock, 1999). Accordingly, the DesK CA domain is comparatively small with a rather short ATP lid and a shallow nucleotide binding pocket, causing the bound nucleotide to be largely exposed to solvent (Trajtenberg et al., 2010).
The NarX transmitter phosphatase activity was not stimulated by ATP, ADP, or the analog AMP-PNP (Fig. 5A). Moreover, both the NarX N509D and H513Q mutants, presumably defective for nucleotide binding, exhibit wild-type transmitter phosphatase activity in vivo and in vitro (Cavicchioli et al., 1995). (We replicated these findings for the H513Q mutant; Table 1 and Fig. 5B.) The NarX N509D mutant is analogous to the EnvZ N347D mutant, which cannot bind ATP in vitro (Tanaka et al., 1998, Zhu and Inouye, 2002). We therefore conclude that the conformational switch between the kinase and phosphatase-states of NarX is not regulated by the CA-bound nucleotides. ATP also has only a very weak, if any, effect on the HisKA_3 UhpB transmitter phosphatase reaction (Wright and Kadner, 2001), extending the generality of our observation for the HisKA_3 family.
By contrast, nucleotides strongly stimulate the transmitter phosphatase activity of the HisKA sensors EnvZ (Igo et al., 1989, Zhu et al., 2000), NtrB (Keener and Kustu, 1988, Kamberov et al., 1994), PhoQ (Sanowar and Le Moual, 2005, Yeo et al., 2012), and WalK (Gutu et al., 2010) among others (Huynh and Stewart, 2011). For the EnvZ N347D mutant, the defect in ATP-binding is coupled with severely perturbed transmitter phosphatase activity that results in reverse phosphoryl transfer rather than hydrolysis (Dutta and Inouye, 1996, Tanaka et al., 1998, Zhu and Inouye, 2002). In addition, for the PhoQ and PmrB sensors, it has been suggested that ADP generated from the autokinase activity stimulates the transmitter phosphatase function by inhibiting ATP binding, thus preventing the kinase conformation (Yeo et al., 2012). Thus, nucleotides have an allosteric effect on the CA domain of these HisKA sensors, unobserved so far for the HisKA_3 family. This difference may in part result from the structural distinctions between these CA domains and/or their different interactions with the DHp domain.
The regulatory role of the CA domain in transmitter phosphatase activity
Although our random screen for the NarX K+ P− phenotype returned substitutions throughout the CA domain, most mapped to residues outside of the conserved sequence motifs (Table 2 and Fig. S4). Two exceptions are substitutions at the conserved G-box residues Tyr-551 and Asp-558. The corresponding positions are respectively occupied by Phe, Tyr or His, and by Glu or Asp, in most HisKA_3 sequences (Grebe and Stock, 1999). Confoundingly, the analogous DesK His-335 and Glu-342 residues are implicated in DHp-CA interaction for the autokinase state but not the phosphatase state (Albanesi et al., 2009, Trajtenberg et al., 2010).
The NarX Y551F mutant exhibited a strong K+ P− phenotype (Table 2), but displayed normal interaction with the NarL receiver domain (Fig. 4). Furthermore, it had detectable transmitter phosphatase activity in vitro (Figs. 2B and 3C). Thus, the Y551F mutant had relatively little if any effect on either transmitter-receiver interaction or active-site conformation. Similarly, the NarX D558V mutant also had a strong K+P− phenotype (Table S1), and Asp-558 did not appear to be in direct contacts with the receiver domain, since the S508C+D558V mutant was wild-type for interaction (Fig. 4). Whereas their exact mechanistic basis is unclear, these CA domain mutants had distinct properties from the DHp domain interaction and conformation mutants described above. Perhaps the NarX CA domain K+ P− substitutions influence the global structure of the CA domain to indirectly influence the DHp domain. Understanding CA domain function in the transmitter phosphatase reaction therefore is in need of further study.
For the EnvZ and NtrB sensors, random K+ P− substitutions affecting the CA domain are also less common than DHp domain mutants (Atkinson and Ninfa, 1993, Hsing et al., 1998, Zhu and Inouye, 2002, Pioszak and Ninfa, 2004), in line with the subsidiary role of the CA domain relative to the DHp domain in transmitter phosphatase activity. Effects of both random and site-specific CA domain mutants are attributed to its function in nucleotide-binding and retention, DHp-CA domain interaction, and interaction with auxiliary regulators (Atkinson and Ninfa, 1993, Hsing et al., 1998, Zhu and Inouye, 2002, Pioszak and Ninfa, 2003a, Yeo et al., 2012).
Overall, the CA domain of HisKA sensors clearly has a more active role in transmitter phosphatase function compared to that of the HisKA_3 family. First, the isolated NtrB and EnvZ DHp domain phosphatase rate is substantially slower than that of the intact transmitter (Jiang et al., 2000, Zhu et al., 2000), whereas the DesK DHp domain exhibits a comparable activity to the transmitter module (Albanesi et al., 2009). Second, a structural analysis of EnvZ suggests that partial sequestration of the CA domain via DHp-CA domain association is required for the transmitter phosphatase state (Ferris et al., 2012), in contrast to the DesK interdomain interaction that is modulated by DHp coiled-coil alteration in response to a conformational switch (Albanesi et al., 2009). Finally, as described above, nucleotides stimulate the HisKA activity, but have virtually no effect on HisKA_3 negative function. Indeed, the transmitter X-ray structures for the presumed transmitter phosphatase conformations of DesK (HisKA_3) and HK853 (HisKA) exhibit strikingly different DHp-CA domain associations (Fig. 6) (Albanesi et al., 2009, Casino et al., 2009).
Concluding remarks
Results described here, together with work from other laboratories, highlight three major issues concerning negative regulation by two-component sensors. First, negative control seems to require more precise interaction between the DHp and receiver domains than does positive control (Siryaporn et al., 2010). Second, the role(s) for the CA domain remain obscure considering the differing requirements for adenine nucleotide allosteric effectors. Finally, two major DHp domain sequence subclasses, despite sharing similarity in overall structure and function, nevertheless display substantial differences in detail with respect to both mechanism and regulation of negative control. These differences undoubtedly reflect the widespread nature and diversity of two-component signaling.
EXPERIMENTAL PROCEDURES
Cultures
Strains and plasmids are listed in Table S2. The narX† and narX‡ alleles in plasmids pVJS1241 and pVJS2474 encode wild-type NarX, but contain silent restriction endonuclease sites to facilitate recloning (Williams and Stewart, 1997b). These plasmids are based on the vector pHG165, which has the moderate copy number of its parent, pBR322 (Stewart et al., 1986).
Cultures were grown in a 50:50 v/v mixture of defined and complex media as described previously (Appleman and Stewart, 2003). Defined medium was buffered with 3-N-morpholino propanesulfonic acid (MOPS) and supplemented with 80 mM glucose. Complex medium was TY broth containing tryptone (0.8 gm L−1), yeast extract (0.5 gm L−1), and NaCl (0.5 gm L−1). Where indicated, nitrate as NaNO3 was added at 40 mM. Cultures to assay Nar functions were grown anaerobically at 37°C and harvested at early exponential phase. Cultures for two-hybrid assays were grown anaerobically at 30°C with 0.5 mM IPTG, and harvested at stationary phase (Karimova et al., 1998).
For two-hybrid assays, DNA fragments encoding narX+ and site-specific mutant alleles were cloned N-terminally to the T25-encoding fragment in the pKNT25 vector, and the DNA fragment encoding the NarL receiver domain, encompassing the first 142 amino acid residues, was cloned C-terminally to the T18-encoding fragment in the pUT18C vector. These vectors were purchased from Euromedex (Souffelweyersheim, France)
Enzyme assay
β-Galactosidase activities were measured in CHCl3-sodium dodecyl sulfate-permeabilized cells. Specific activities are expressed in arbitrary (Miller) units (Miller, 1972). Assays were performed at room temperature, approximately 21°C. Reported values are averaged from at least two independent cultures, grown on different days, for which the measured values differed by 10% or less. Each culture was assayed in duplicate, with one assay reaction mixture containing twice as much cell extract as the other.
Protein purification
MBP-NarX227 and His6-NarL proteins were expressed and purified as described previously (Noriega et al., 2010). For MBP-NarX-His6 and MBP-NarX(M411T)-His6 proteins, a hexahistidine tag was PCR-incorporated at the C-terminal coding region of the malE-narX DNA-fragment, encoding MBP-NarX227. Protein preparations were evaluated by electrophoresis through Laemmli denaturing slab gels, which were prepared, stained and visualized as described previously (Noriega et al., 2008). All protein preparations were highly enriched, with only trace amounts of contaminating species. Proteins were stored at −80°C as frozen aliquots from dry ice-ethanol exposure. Final protein concentrations (as monomers) were determined by measuring absorbance at 280 nm. Chemicals and reagents were from Sigma-Aldrich Corp. (St. Louis, MO) except as noted. [γ-32P]-ATP (10 uCi/ml, 3000 Ci/mmol) was from Perkin Elmer, Waltham, MA.
Phospho-His6-NarL preparation
Phospho-His6-NarL was generated in a phosphoryl transfer reaction mix with His6-NarL and phospho-MBP-NarX(M411T)-His6, which is severely defective for the transmitter phosphatase activity. The response regulator protein was subsequently recovered from the reaction matrix by metal ion affinity chromatography. Details are as follows.
First, approximately 200 μl of phospho-MBP-NarX (M411T)-His6 was prepared from [32P]-ATP as described previously (Noriega et al., 2008) and loaded onto Micro Bio-Spin chromatography columns (BIO-RAD, Hercules, CA) containing 200 μl amylose resin (New England Biolabs, Ipswich, MA) equilibrated with reaction buffer (100 mM HEPES pH 7.0, 50 mM KCl, 12.5 mM MgCl2, 12.5 mM MnCl2, 2.5 mM DTT and 10% (w/v) glycerol). The amylose resin was used as an immobilizing matrix for separating the sensor from the response regulator, since maltose is tightly bound to MBP. Nucleotides were removed by repeated washing and centrifugation with 500 μl reaction buffer. All centrifugation described hereafter was performed at 100 × g for 30 sec.
Next, phosphoryl transfer was performed in an ice water bath for 5 min by resuspending the amylose resin in 100 μl of reaction buffer containing the response regulator (10-12 μM monomer final concentration when including resin volume to a total of 300 μl). The suspension was centrifuged and the eluate was reloaded only the amylose column for a second incubation of 5 min.
In the recovery step, the suspension was again centrifuged to collect the first fraction of isolated phospho-response regulator (approximately 100 μl). Remaining phospho-His6-NarL was collected from the column as three 50 μl fractions by repeated loading and centrifugation with reaction buffer. Approximately 250 μl of 0.4-0.6 μM phospho-His6-NarL was collected in a total of 10-12 μM protein. To minimize the receiver autophosphatase activity, all tubes and columns were kept in an ice water bath when possible. Removal of MBP-NarX-His6 and nucleotides was confirmed by Laemmli gel electrophoresis and thin-layer chromatography (Noriega et al., 2008), respectively. The yield of [32P]-labeled response regulator was estimated to be at least 0.5 μM in preparations containing up to 8 μM total protein (monomers).
In vitro assays for transmitter phosphatase activity
Time course reactions to monitor phosphoryl transfer and subsequent transmitter phosphatase activities were performed as described previously (Noriega et al., 2010) except that reactions were done at 19°C.
For direct phosphatase reactions, phospho-His6-NarL was pre-incubated for 2 min at 19°C. Receiver autophosphatase reaction was initiated by transferring aliquots into tubes containing reaction buffer that had been pre-incubated for 30 min at 19°C. Transmitter phosphatase reactions were initiated by transferring aliquots into tubes containing MBP-NarX (final concentration 0.5 μM dimers) that had been pre-incubated for 30 min at 19°C. Time point samples were added to stop solution (20 mM HEPES, pH 7.0, 100 mM EDTA, 0.01 gm L−1 SDS). The remaining phospho-His6-NarL was quantified by filter binding as described previously (Noriega et al., 2008) and plotted as the percentage of the initial value. The reaction rate depended on the concentration of added MBP-NarX227 (Fig. S5). We used a saturating concentration, 5 μM MBP-NarX227 dimers, for all assays reported.
In these assays, NarX-catalyzed NarL dephosphorylation was exponential over a relatively short time period, whereupon the rates decelerated. In other experiments, to be presented in a separate publication, we observed exponential dephosphorylation of the paralogous NarP response regulator over a much longer time period (C. E. Noriega and V. Stewart, manuscript in preparation). Thus, the substrate (response regulator) determines the shape of the transmitter phosphatase curve. We hypothesize that the curves for NarL result from aggregation of a subpopulation of phospho-NarL into a phosphatase-inaccessible state.
Reproducibility of results
Independent phosphatase reactions were initiated with different amounts of phosphorylated response regulator, due to variability in fresh batches made on different days with different protein preparations and lots of radiolabel. Thus, meaningful standard errors cannot be calculated for a series of replicate experiments. Overall results are supported qualitatively by dozens of experiments, conducted over several years by different investigators using different protein preparations and reaction conditions. Results summarized in Figs. 2 and 4 are representative of at least duplicate assays.
Supplementary Material
Acknowledgements
We are indebted to Li-Ling Chen for her expert assistance throughout this study. We thank Chet Price for his very helpful critiques of two draft versions. This study was supported by U. S. Public Health Service Grant GM036877 from the National Institute of General Medical Sciences. The authors declare no conflicts of interest.
REFERENCES
- Albanesi D, Martin M, Trajtenberg F, Mansilla MC, Haouz A, Alzari PM, de Mendoza D, Buschiazzo A. Structural plasticity and catalysis regulation of a thermosensor histidine kinase. Proc Natl Acad Sci USA. 2009;106:16185–16190. doi: 10.1073/pnas.0906699106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleman JA, Stewart V. Mutational analysis of a conserved signal-transducing element: the HAMP linker of the Escherichia coli nitrate sensor NarX. J Bacteriol. 2003;185:89–97. doi: 10.1128/JB.185.1.89-97.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashenberg O, Rozen-Gagnon K, Laub MT, Keating AE. Determinants of homodimerization specificity in histidine kinases. J Mol Biol. 2011;413:222–235. doi: 10.1016/j.jmb.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson MR, Ninfa AJ. Mutational analysis of the bacterial signal-transducing protein kinase/phosphatase nitrogen regulator II (NRII or NtrB) J Bacteriol. 1993;175:7016–7023. doi: 10.1128/jb.175.21.7016-7023.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourret RB. Receiver domain structure and function in response regulator proteins. Curr Opin Microbiol. 2010;13:142–149. doi: 10.1016/j.mib.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao GJ, Sarkar N. Identification of the gene for an Escherichia coli poly(A) polymerase. Proc Natl Acad Sci USA. 1992;89:10380–10384. doi: 10.1073/pnas.89.21.10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra EJ, Perchuk BS, Lubin EA, Ashenberg O, Skerker JM, Laub MT. Systematic dissection and trajectory-scanning mutagenesis of the molecular interface that ensures specificity of two-component signaling pathways. PLoS Genet. 2010;6:e1001220. doi: 10.1371/journal.pgen.1001220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casino P, Rubio V, Marina A. Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell. 2009;139:325–336. doi: 10.1016/j.cell.2009.08.032. [DOI] [PubMed] [Google Scholar]
- Cavicchioli R, Schroder I, Constanti M, Gunsalus RP. The NarX and NarQ sensor-transmitter proteins of Escherichia coli each require two conserved histidines for nitrate-dependent signal transduction to NarL. J Bacteriol. 1995;177:2416–2424. doi: 10.1128/jb.177.9.2416-2424.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta R, Inouye M. Reverse phosphotransfer from OmpR to EnvZ in a kinase−/phosphatase+ mutant of EnvZ (EnvZ·N347D), a bifunctional signal transducer of Escherichia coli. J Biol Chem. 1996;271:1424–1429. doi: 10.1074/jbc.271.3.1424. [DOI] [PubMed] [Google Scholar]
- Dutta R, Yoshida T, Inouye M. The critical role of the conserved Thr247 residue in the functioning of the osmosensor EnvZ, a histidine Kinase/Phosphatase, in Escherichia coli. J Biol Chem. 2000;275:38645–38653. doi: 10.1074/jbc.M005872200. [DOI] [PubMed] [Google Scholar]
- Egan SM, Stewart V. Mutational analysis of nitrate regulatory gene narL in Escherichia coli K-12. J Bacteriol. 1991;173:4424–4432. doi: 10.1128/jb.173.14.4424-4432.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris HU, Dunin-Horkawicz S, Hornig N, Hulko M, Martin J, Schultz JE, Zeth K, Lupas AN, Coles M. Mechanism of regulation of receptor histidine kinases. Structure. 2012;20:56–66. doi: 10.1016/j.str.2011.11.014. [DOI] [PubMed] [Google Scholar]
- Gao R, Stock AM. Biological insights from structures of two-component proteins. Annu Rev Microbiol. 2009;63:133–154. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebe TW, Stock JB. The histidine protein kinase superfamily. Adv Microb Physiol. 1999;41:139–227. doi: 10.1016/s0065-2911(08)60167-8. [DOI] [PubMed] [Google Scholar]
- Gutu AD, Wayne KJ, Sham LT, Winkler ME. Kinetic characterization of the WalRKSpn (VicRK) two-component system of Streptococcus pneumoniae: dependence of WalKSpn (VicK) phosphatase activity on its PAS domain. J Bacteriol. 2010;192:2346–2358. doi: 10.1128/JB.01690-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MN, Silhavy TJ. The ompB locus and the regulation of the major outer membrane porin proteins of Escherichia coli K12. J Mol Biol. 1981;146:23–43. doi: 10.1016/0022-2836(81)90364-8. [DOI] [PubMed] [Google Scholar]
- Hsing W, Russo FD, Bernd KK, Silhavy TJ. Mutations that alter the kinase and phosphatase activities of the two-component sensor EnvZ. J Bacteriol. 1998;180:4538–4546. doi: 10.1128/jb.180.17.4538-4546.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsing W, Silhavy TJ. Function of conserved histidine-243 in phosphatase activity of EnvZ, the sensor for porin osmoregulation in Escherichia coli. J Bacteriol. 1997;179:3729–3735. doi: 10.1128/jb.179.11.3729-3735.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh TN, Noriega CE, Stewart V. Conserved mechanism for sensor phosphatase control of two-component signaling revealed in the nitrate sensor NarX. Proc Natl Acad Sci USA. 2010;107:21140–21145. doi: 10.1073/pnas.1013081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh TN, Stewart V. Negative control in two-component signal transduction by transmitter phosphatase activity. Mol Microbiol. 2011;82:275–286. doi: 10.1111/j.1365-2958.2011.07829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo MM, Ninfa AJ, Stock JB, Silhavy TJ. Phosphorylation and dephosphorylation of a bacterial transcriptional activator by a transmembrane receptor. Genes Dev. 1989;3:1725–1734. doi: 10.1101/gad.3.11.1725. [DOI] [PubMed] [Google Scholar]
- Jiang P, Atkinson MR, Srisawat C, Sun Q, Ninfa AJ. Functional dissection of the dimerization and enzymatic activities of Escherichia coli nitrogen regulator II and their regulation by the PII protein. Biochemistry. 2000;39:13433–13449. doi: 10.1021/bi000794u. [DOI] [PubMed] [Google Scholar]
- Kamberov ES, Atkinson MR, Chandran P, Ninfa AJ. Effect of mutations in Escherichia coli glnL (ntrB), encoding nitrogen regulator II (NRII or NtrB), on the phosphatase activity involved in bacterial nitrogen regulation. J Biol Chem. 1994;269:28294–28299. [PubMed] [Google Scholar]
- Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci USA. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener J, Kustu S. Protein kinase and phosphoprotein phosphatase activities of nitrogen regulatory proteins NTRB and NTRC of enteric bacteria: roles of the conserved amino-terminal domain of NTRC. Proc Natl Acad Sci USA. 1988;85:4976–4980. doi: 10.1073/pnas.85.14.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Kustu S, Stewart V. In vitro interaction of nitrate-responsive regulatory protein NarL with DNA target sequences in the fdnG, narG, narK and frdA operon control regions of Escherichia coli K-12. J Mol Biol. 1994;241:150–165. doi: 10.1006/jmbi.1994.1485. [DOI] [PubMed] [Google Scholar]
- Liu JD, Parkinson JS. Genetics and sequence analysis of the pcnB locus, an Escherichia coli gene involved in plasmid copy number control. J Bacteriol. 1989;171:1254–1261. doi: 10.1128/jb.171.3.1254-1261.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopilato J, Bortner S, Beckwith J. Mutations in a new chromosomal gene of Escherichia coli K-12, pcnB, reduce plasmid copy number of pBR322 and its derivatives. Mol Gen Genet. 1986;205:285–290. doi: 10.1007/BF00430440. [DOI] [PubMed] [Google Scholar]
- Lupas AN, Gruber M. The structure of alpha-helical coiled coils. Adv Protein Chem. 2005;70:37–78. doi: 10.1016/S0065-3233(05)70003-6. [DOI] [PubMed] [Google Scholar]
- Marina A, Waldburger CD, Hendrickson WA. Structure of the entire cytoplasmic portion of a sensor histidine-kinase protein. EMBO J. 2005;24:4247–4259. doi: 10.1038/sj.emboj.7600886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, N. Y.: 1972. [Google Scholar]
- Ninfa AJ, Magasanik B. Covalent modification of the glnG product, NRI, by the glnL product, NRII, regulates the transcription of the glnALG operon in Escherichia coli. Proc Natl Acad Sci USA. 1986;83:5909–5913. doi: 10.1073/pnas.83.16.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noriega CE, Lin H-Y, Chen L-L, Williams SB, Stewart V. Asymmetric cross-regulation between the nitrate-responsive NarX-NarL and NarQ-NarP two-component regulatory systems from Escherichia coli K-12. Mol Microbiol. 2010;75:394–412. doi: 10.1111/j.1365-2958.2009.06987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noriega CE, Schmidt R, Gray MJ, Chen L-L, Stewart V. Autophosphorylation and dephosphorylation by soluble forms of the nitrate-responsive sensors NarX and NarQ from Escherichia coli K-12. J Bacteriol. 2008;190:3869–3876. doi: 10.1128/JB.00092-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson JS, Kofoid EC. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- Pioszak AA, Ninfa AJ. Genetic and biochemical analysis of phosphatase activity of Escherichia coli NRII (NtrB) and its regulation by the PII signal transduction protein. J Bacteriol. 2003a;185:1299–1315. doi: 10.1128/JB.185.4.1299-1315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pioszak AA, Ninfa AJ. Mechanism of the PII-activated phosphatase activity of Escherichia coli NRII (NtrB): how the different domains of NRII collaborate to act as a phosphatase. Biochemistry. 2003b;42:8885–8899. doi: 10.1021/bi030065p. [DOI] [PubMed] [Google Scholar]
- Pioszak AA, Ninfa AJ. Mutations altering the N-terminal receiver domain of NRI (NtrC) That prevent dephosphorylation by the NRII-PII complex in Escherichia coli. J Bacteriol. 2004;186:5730–5740. doi: 10.1128/JB.186.17.5730-5740.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L, Cai S, Zhu Y, Inouye M. Cysteine-scanning analysis of the dimerization domain of EnvZ, an osmosensing histidine kinase. J Bacteriol. 2003;185:3429–3435. doi: 10.1128/JB.185.11.3429-3435.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabin RS, Stewart V. Dual response regulators (NarL and NarP) interact with dual sensors (NarX and NarQ) to control nitrate- and nitrite-regulated gene expression in Escherichia coli K-12. J Bacteriol. 1993;175:3259–3268. doi: 10.1128/jb.175.11.3259-3268.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanowar S, Le Moual H. Functional reconstitution of the Salmonella typhimurium PhoQ histidine kinase sensor in proteoliposomes. Biochem J. 2005;390:769–776. doi: 10.1042/BJ20050060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrecke K, Jordan S, Mascher T. Stoichiometry and perturbation studies of the LiaFSR system of Bacillus subtilis. Mol Microbiol. 2013;87:769–788. doi: 10.1111/mmi.12130. [DOI] [PubMed] [Google Scholar]
- Siryaporn A, Perchuk BS, Laub MT, Goulian M. Evolving a robust signal transduction pathway from weak cross-talk. Mol Syst Biol. 2010;6:452. doi: 10.1038/msb.2010.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerker JM, Perchuk BS, Siryaporn A, Lubin EA, Ashenberg O, Goulian M, Laub MT. Rewiring the specificity of two-component signal transduction systems. Cell. 2008;133:1043–1054. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderbom F, Binnie U, Masters M, Wagner EG. Regulation of plasmid R1 replication: PcnB and RNase E expedite the decay of the antisense RNA, CopA. Mol Microbiol. 1997;26:493–504. doi: 10.1046/j.1365-2958.1997.5871953.x. [DOI] [PubMed] [Google Scholar]
- Stewart GS, Lubinsky-Mink S, Jackson CG, Cassel A, Kuhn J. pHG165: a pBR322 copy number derivative of pUC8 for cloning and expression. Plasmid. 1986;15:172–181. doi: 10.1016/0147-619x(86)90035-1. [DOI] [PubMed] [Google Scholar]
- Stewart V. Biochemical Society Special Lecture. Nitrate- and nitrite-responsive sensors NarX and NarQ of proteobacteria. Biochem Soc Trans. 2003;31:1–10. doi: 10.1042/bst0310001. [DOI] [PubMed] [Google Scholar]
- Stock AM, Martinez-Hackert E, Rasmussen BF, West AH, Stock JB, Ringe D, Petsko GA. Structure of the Mg2+-bound form of CheY and mechanism of phosphoryl transfer in bacterial chemotaxis. Biochemistry. 1993;32:13375–13380. doi: 10.1021/bi00212a001. [DOI] [PubMed] [Google Scholar]
- Stock JB, Ninfa AJ, Stock AM. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock JB, Surette MG, Levit M, Park P. Two-component signal transduction systems: structure-function relationships and mechanisms of catalysis. In: Hoch JA, Silhavy TJ, editors. Two-component signal transduction. ASM Press; Washington D. C.: 1995. pp. 25–51. [Google Scholar]
- Tanaka T, Saha SK, Tomomori C, Ishima R, Liu D, Tong KI, Park H, Dutta R, Qin L, Swindells MB, Yamazaki T, Ono AM, Kainosho M, Inouye M, Ikura M. NMR structure of the histidine kinase domain of the E. coli osmosensor EnvZ. Nature. 1998;396:88–92. doi: 10.1038/23968. [DOI] [PubMed] [Google Scholar]
- Tokishita S, Kojima A, Mizuno T. Transmembrane signal transduction and osmoregulation in Escherichia coli: functional importance of the transmembrane regions of membrane-located protein kinase, EnvZ. J Biochem. 1992;111:707–713. doi: 10.1093/oxfordjournals.jbchem.a123823. [DOI] [PubMed] [Google Scholar]
- Trajtenberg F, Grana M, Ruetalo N, Botti H, Buschiazzo A. Structural and enzymatic insights into the ATP binding and autophosphorylation mechanism of a sensor histidine kinase. J Biol Chem. 2010;285:24892–24903. doi: 10.1074/jbc.M110.147843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne KJ, Li S, Kazmierczak KM, Tsui HC, Winkler ME. Involvement of WalK (VicK) phosphatase activity in setting WalR (VicR) response regulator phosphorylation level and limiting cross-talk in Streptococcus pneumoniae D39 cells. Mol Microbiol. 2012;86:645–660. doi: 10.1111/mmi.12006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigt M, White RA, Szurmant H, Hoch JA, Hwa T. Identification of direct residue contacts in protein-protein interaction by message passing. Proc Natl Acad Sci USA. 2009;106:67–72. doi: 10.1073/pnas.0805923106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett JW, Kirby JR. Genetic and biochemical dissection of a HisKA domain identifies residues required exclusively for kinase and phosphatase activities. PLoS Genet. 2012;8:e1003084. doi: 10.1371/journal.pgen.1003084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SB, Stewart V. Discrimination between structurally related ligands nitrate and nitrite controls autokinase activity of the NarX transmembrane signal transducer of Escherichia coli K-12. Mol Microbiol. 1997a;26:911–925. doi: 10.1046/j.1365-2958.1997.6262002.x. [DOI] [PubMed] [Google Scholar]
- Williams SB, Stewart V. Nitrate- and nitrite-sensing protein NarX of Escherichia coli K-12: mutational analysis of the amino-terminal tail and first transmembrane segment. J Bacteriol. 1997b;179:721–729. doi: 10.1128/jb.179.3.721-729.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JS, 3rd, Kadner RJ. The phosphoryl transfer domain of UhpB interacts with the response regulator UhpA. J Bacteriol. 2001;183:3149–3159. doi: 10.1128/JB.183.10.3149-3159.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S, Sugimoto H, Kobayashi M, Ohno A, Nakamura H, Shiro Y. Structure of PAS-linked histidine kinase and the response regulator complex. Structure. 2009;17:1333–1344. doi: 10.1016/j.str.2009.07.016. [DOI] [PubMed] [Google Scholar]
- Yeo WS, Zwir I, Huang HV, Shin D, Kato A, Groisman EA. Intrinsic negative feedback governs activation surge in two-component regulatory systems. Mol Cell. 2012;45:409–421. doi: 10.1016/j.molcel.2011.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Inouye M. The role of the G2 box, a conserved motif in the histidine kinase superfamily, in modulating the function of EnvZ. Mol Microbiol. 2002;45:653–663. doi: 10.1046/j.1365-2958.2002.03061.x. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Qin L, Yoshida T, Inouye M. Phosphatase activity of histidine kinase EnvZ without kinase catalytic domain. Proc Natl Acad Sci USA. 2000;97:7808–7813. doi: 10.1073/pnas.97.14.7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



