Abstract
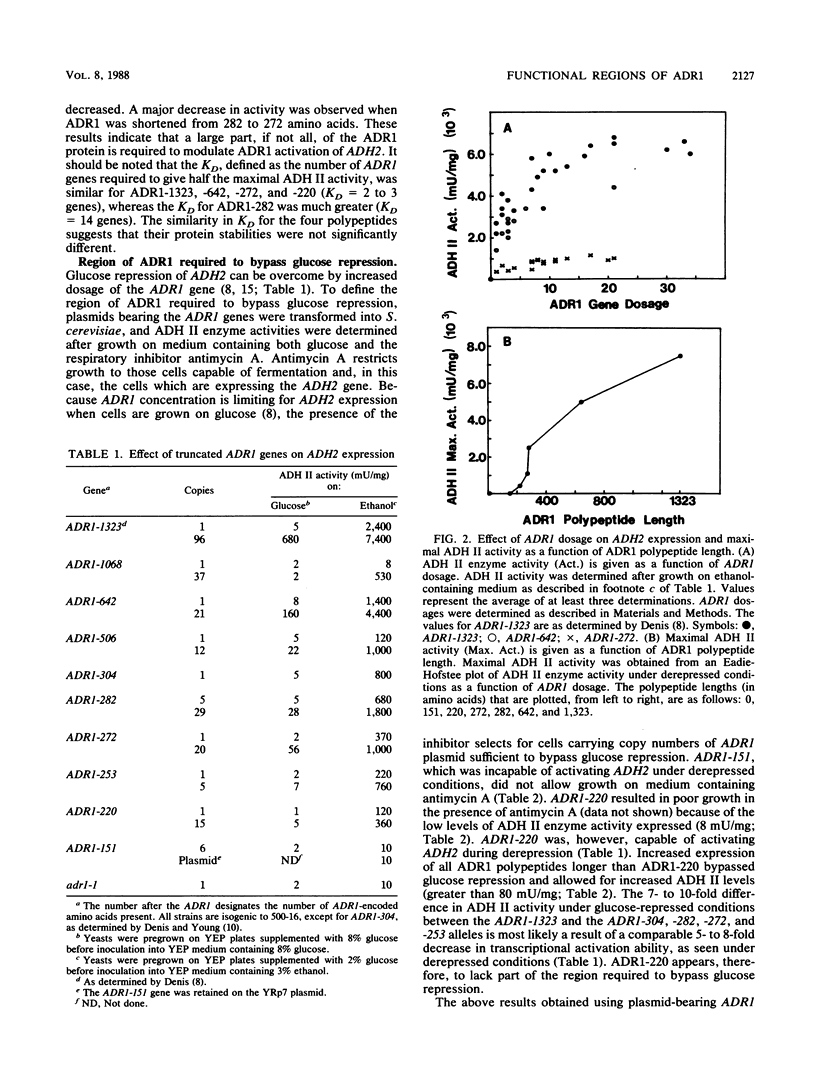
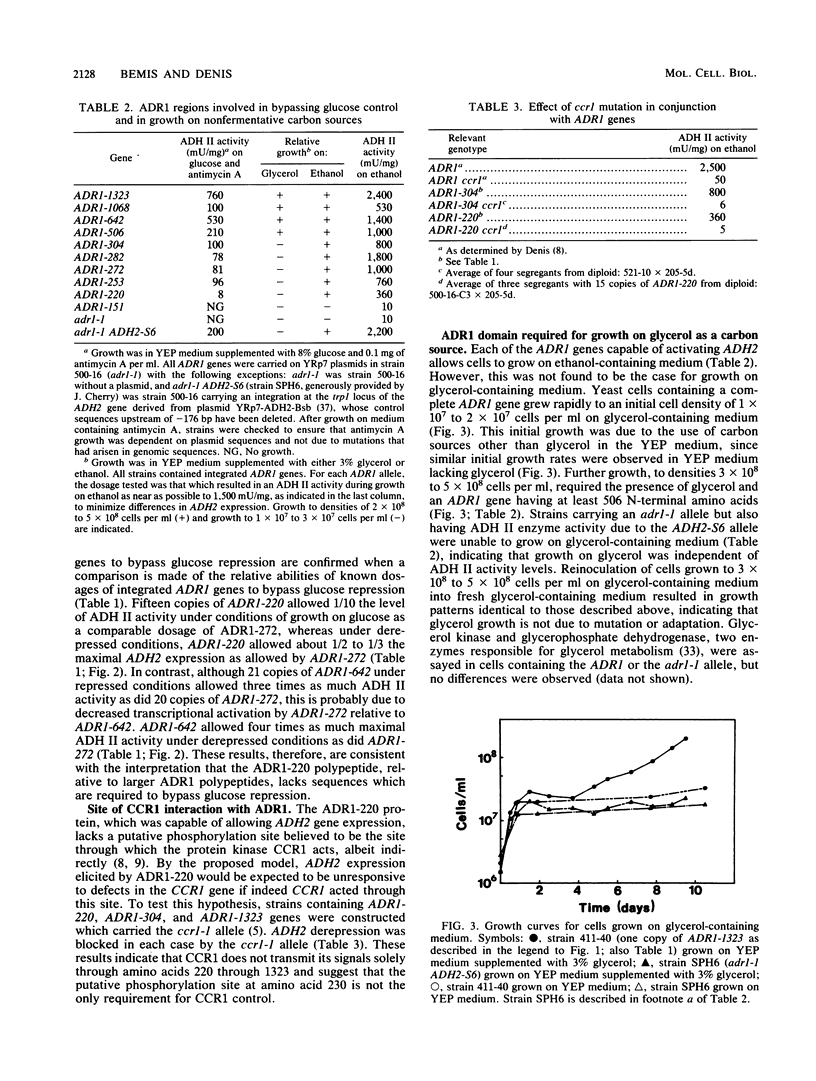
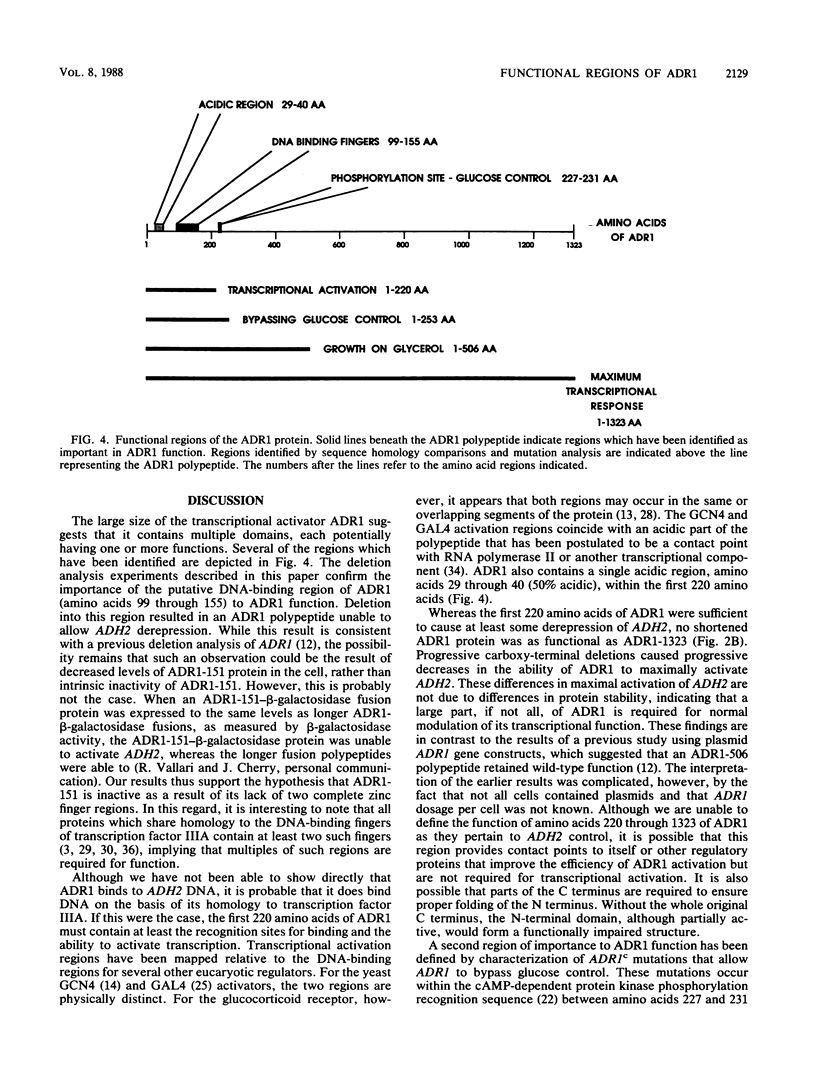
The transcriptional activator ADR1 from Saccharomyces cerevisiae is a postulated DNA-binding protein that controls the expression of the glucose-repressible alcohol dehydrogenase (ADH2). Carboxy-terminal deletions of the ADR1 protein (1,323 amino acids in length) were used to localize its functional regions. The transcriptional activation region was localized to the N-terminal 220 amino acids of ADR1 containing two DNA-binding zinc finger motifs. In addition to the N terminus, a large part of the ADR1 sequence was shown to be essential for complete activation of ADH2. Deletion of the putative phosphorylation region, defined by ADR1c mutations that overcome glucose repression, did not render ADH2 expression insensitive to glucose repression. Instead, this region (amino acids 220 through 253) was found to be required by ADR1 to bypass glucose repression. These results suggest that ADR1c mutations enhance ADR1 function, rather than block an interaction of the putative phosphorylation region with a repressor molecule. Furthermore, the protein kinase CCR1 was shown to affect ADH2 expression when the putative phosphorylation region was removed, indicating that CCR1 does not act solely through this region. A functional ADR1 gene was also found to be necessary for growth on glycerol-containing medium. The N-terminal 506 amino acids of ADR1 were required for this newly identified function, indicating that ADH2 activation and glycerol growth are controlled by separate regions of ADR1.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blumberg H., Eisen A., Sledziewski A., Bader D., Young E. T. Two zinc fingers of a yeast regulatory protein shown by genetic evidence to be essential for its function. 1987 Jul 30-Aug 5Nature. 328(6129):443–445. doi: 10.1038/328443a0. [DOI] [PubMed] [Google Scholar]
- Celenza J. L., Carlson M. A yeast gene that is essential for release from glucose repression encodes a protein kinase. Science. 1986 Sep 12;233(4769):1175–1180. doi: 10.1126/science.3526554. [DOI] [PubMed] [Google Scholar]
- Chowdhury K., Deutsch U., Gruss P. A multigene family encoding several "finger" structures is present and differentially active in mammalian genomes. Cell. 1987 Mar 13;48(5):771–778. doi: 10.1016/0092-8674(87)90074-2. [DOI] [PubMed] [Google Scholar]
- Ciriacy M. Genetics of alcohol dehydrogenase in Saccharomyces cerevisiae. II. Two loci controlling synthesis of the glucose-repressible ADH II. Mol Gen Genet. 1975;138(2):157–164. doi: 10.1007/BF02428119. [DOI] [PubMed] [Google Scholar]
- Ciriacy M. Isolation and characterization of yeast mutants defective in intermediary carbon metabolism and in carbon catabolite derepression. Mol Gen Genet. 1977 Jul 20;154(2):213–220. doi: 10.1007/BF00330840. [DOI] [PubMed] [Google Scholar]
- Danielsen M., Northrop J. P., Ringold G. M. The mouse glucocorticoid receptor: mapping of functional domains by cloning, sequencing and expression of wild-type and mutant receptor proteins. EMBO J. 1986 Oct;5(10):2513–2522. doi: 10.1002/j.1460-2075.1986.tb04529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis C. L., Gallo C. Constitutive RNA synthesis for the yeast activator ADR1 and identification of the ADR1-5c mutation: implications in posttranslational control of ADR1. Mol Cell Biol. 1986 Nov;6(11):4026–4030. doi: 10.1128/mcb.6.11.4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis C. L. Identification of new genes involved in the regulation of yeast alcohol dehydrogenase II. Genetics. 1984 Dec;108(4):833–844. doi: 10.1093/genetics/108.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis C. L. The effects of ADR1 and CCR1 gene dosage on the regulation of the glucose-repressible alcohol dehydrogenase from Saccharomyces cerevisiae. Mol Gen Genet. 1987 Jun;208(1-2):101–106. doi: 10.1007/BF00330429. [DOI] [PubMed] [Google Scholar]
- Denis C. L., Young E. T. Isolation and characterization of the positive regulatory gene ADR1 from Saccharomyces cerevisiae. Mol Cell Biol. 1983 Mar;3(3):360–370. doi: 10.1128/mcb.3.3.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dihanich M., Suda K., Schatz G. A yeast mutant lacking mitochondrial porin is respiratory-deficient, but can recover respiration with simultaneous accumulation of an 86-kd extramitochondrial protein. EMBO J. 1987 Mar;6(3):723–728. doi: 10.1002/j.1460-2075.1987.tb04813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartshorne T. A., Blumberg H., Young E. T. Sequence homology of the yeast regulatory protein ADR1 with Xenopus transcription factor TFIIIA. Nature. 1986 Mar 20;320(6059):283–287. doi: 10.1038/320283a0. [DOI] [PubMed] [Google Scholar]
- Hollenberg S. M., Giguere V., Segui P., Evans R. M. Colocalization of DNA-binding and transcriptional activation functions in the human glucocorticoid receptor. Cell. 1987 Apr 10;49(1):39–46. doi: 10.1016/0092-8674(87)90753-7. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986 Sep 12;46(6):885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- Irani M., Taylor W. E., Young E. T. Transcription of the ADH2 gene in Saccharomyces cerevisiae is limited by positive factors that bind competitively to its intact promoter region on multicopy plasmids. Mol Cell Biol. 1987 Mar;7(3):1233–1241. doi: 10.1128/mcb.7.3.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston S. A., Salmeron J. M., Jr, Dincher S. S. Interaction of positive and negative regulatory proteins in the galactose regulon of yeast. Cell. 1987 Jul 3;50(1):143–146. doi: 10.1016/0092-8674(87)90671-4. [DOI] [PubMed] [Google Scholar]
- Kalderon D., Roberts B. L., Richardson W. D., Smith A. E. A short amino acid sequence able to specify nuclear location. Cell. 1984 Dec;39(3 Pt 2):499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Kammerer B., Guyonvarch A., Hubert J. C. Yeast regulatory gene PPR1. I. Nucleotide sequence, restriction map and codon usage. J Mol Biol. 1984 Dec 5;180(2):239–250. doi: 10.1016/s0022-2836(84)80002-9. [DOI] [PubMed] [Google Scholar]
- Keegan L., Gill G., Ptashne M. Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein. Science. 1986 Feb 14;231(4739):699–704. doi: 10.1126/science.3080805. [DOI] [PubMed] [Google Scholar]
- Kemp B. E., Graves D. J., Benjamini E., Krebs E. G. Role of multiple basic residues in determining the substrate specificity of cyclic AMP-dependent protein kinase. J Biol Chem. 1977 Jul 25;252(14):4888–4894. [PubMed] [Google Scholar]
- Kumar V., Green S., Staub A., Chambon P. Localisation of the oestradiol-binding and putative DNA-binding domains of the human oestrogen receptor. EMBO J. 1986 Sep;5(9):2231–2236. doi: 10.1002/j.1460-2075.1986.tb04489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughon A., Gesteland R. F. Primary structure of the Saccharomyces cerevisiae GAL4 gene. Mol Cell Biol. 1984 Feb;4(2):260–267. doi: 10.1128/mcb.4.2.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987 Mar 13;48(5):847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- Ma J., Ptashne M. The carboxy-terminal 30 amino acids of GAL4 are recognized by GAL80. Cell. 1987 Jul 3;50(1):137–142. doi: 10.1016/0092-8674(87)90670-2. [DOI] [PubMed] [Google Scholar]
- Messenguy F., Dubois E., Descamps F. Nucleotide sequence of the ARGRII regulatory gene and amino acid sequence homologies between ARGRII PPRI and GAL4 regulatory proteins. Eur J Biochem. 1986 May 15;157(1):77–81. doi: 10.1111/j.1432-1033.1986.tb09640.x. [DOI] [PubMed] [Google Scholar]
- Miesfeld R., Godowski P. J., Maler B. A., Yamamoto K. R. Glucocorticoid receptor mutants that define a small region sufficient for enhancer activation. Science. 1987 Apr 24;236(4800):423–427. doi: 10.1126/science.3563519. [DOI] [PubMed] [Google Scholar]
- Miller J., McLachlan A. D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster J., Yu J., Cox D., Chan R. V., Smith M., Young E. ADR1-mediated regulation of ADH2 requires an inverted repeat sequence. Mol Cell Biol. 1986 Jun;6(6):1894–1902. doi: 10.1128/mcb.6.6.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons D. T. DNA-binding region of the simian virus 40 tumor antigen. J Virol. 1986 Mar;57(3):776–785. doi: 10.1128/jvi.57.3.776-785.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague G. F., Cronan J. E. Isolation and characterization of Saccharomyces cerevisiae mutants defective in glycerol catabolism. J Bacteriol. 1977 Mar;129(3):1335–1342. doi: 10.1128/jb.129.3.1335-1342.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Promoters, activator proteins, and the mechanism of transcriptional initiation in yeast. Cell. 1987 May 8;49(3):295–297. doi: 10.1016/0092-8674(87)90277-7. [DOI] [PubMed] [Google Scholar]
- Van Heuverswyn H., Van de Voorde A., Van Herreweghe J., Volckaert G., De Winne P., Fiers W. Nucleotide sequence of simian virus 40 DNA: structure of the middle segment of the HindII + III restriction fragment B (sixth part of the T antigen gene) and codon usage. Eur J Biochem. 1980 May;106(1):199–209. doi: 10.1111/j.1432-1033.1980.tb06011.x. [DOI] [PubMed] [Google Scholar]
- Vincent A., Colot H. V., Rosbash M. Sequence and structure of the serendipity locus of Drosophila melanogaster. A densely transcribed region including a blastoderm-specific gene. J Mol Biol. 1985 Nov 5;186(1):149–166. doi: 10.1016/0022-2836(85)90265-7. [DOI] [PubMed] [Google Scholar]
- Williamson V. M., Young E. T., Ciriacy M. Transposable elements associated with constitutive expression of yeast alcohol dehydrogenase II. Cell. 1981 Feb;23(2):605–614. doi: 10.1016/0092-8674(81)90156-2. [DOI] [PubMed] [Google Scholar]


