ABSTRACT
The question of how HIV-1 interfaces with cellular microRNA (miRNA) biogenesis and effector mechanisms has been highly controversial. Here, we first used deep sequencing of small RNAs present in two different infected cell lines (TZM-bl and C8166) and two types of primary human cells (CD4+ peripheral blood mononuclear cells [PBMCs] and macrophages) to unequivocally demonstrate that HIV-1 does not encode any viral miRNAs. Perhaps surprisingly, we also observed that infection of T cells by HIV-1 has only a modest effect on the expression of cellular miRNAs at early times after infection. Comprehensive analysis of miRNA binding to the HIV-1 genome using the photoactivatable ribonucleoside-induced cross-linking and immunoprecipitation (PAR-CLIP) technique revealed several binding sites for cellular miRNAs, a subset of which were shown to be capable of mediating miRNA-mediated repression of gene expression. However, the main finding from this analysis is that HIV-1 transcripts are largely refractory to miRNA binding, most probably due to extensive viral RNA secondary structure. Together, these data demonstrate that HIV-1 neither encodes viral miRNAs nor strongly influences cellular miRNA expression, at least early after infection, and imply that HIV-1 transcripts have evolved to avoid inhibition by preexisting cellular miRNAs by adopting extensive RNA secondary structures that occlude most potential miRNA binding sites.
IMPORTANCE
MicroRNAs (miRNAs) are a ubiquitous class of small regulatory RNAs that serve as posttranscriptional regulators of gene expression. Previous work has suggested that HIV-1 might subvert the function of the cellular miRNA machinery by expressing viral miRNAs or by dramatically altering the level of cellular miRNA expression. Using very sensitive approaches, we now demonstrate that neither of these ideas is in fact correct. Moreover, HIV-1 transcripts appear to largely avoid regulation by cellular miRNAs by adopting an extensive RNA secondary structure that occludes the ability of cellular miRNAs to interact with viral mRNAs. Together, these data suggest that HIV-1, rather than seeking to control miRNA function in infected cells, has instead evolved a mechanism to become largely invisible to cellular miRNA effector mechanisms.
Introduction
MicroRNAs (miRNAs) are a class of small regulatory RNAs, 22 ± 2 nucleotides (nt) in length, that function by posttranscriptionally inhibiting mRNA function (1). Cellular miRNAs are initially transcribed as part of an ~80-nt stem-loop structure that in turn forms part of a long, capped, and polyadenylated RNA referred to as a primary miRNA (pri-miRNA) transcript (2). This pri-miRNA stem-loop is recognized by the nuclear RNase III enzyme Drosha, which cleaves the ~33-bp stem ~22 bp from the terminal loop to release the ~60-nt-long pre-miRNA hairpin intermediate. After export to the cytoplasm by Exportin 5, the pre-miRNA is cleaved by a second, cytoplasmic RNase III, called Dicer, to liberate the miRNA duplex intermediate. One strand of this duplex is then incorporated into the RNA-induced silencing complex (RISC), consisting minimally of one of the four human Argonaut (Ago) proteins, Ago1 through Ago4, as well as a GW182 protein family member. The miRNA then acts as a guide RNA to target RISC to complementary sites on mRNA molecules, resulting in translational arrest and/or mRNA destabilization (1).
Analysis of mRNA target recognition by miRNA-guided RISCs has shown that the miRNA sequence extending from positions 2 to 7 or 8, the so-called seed region, is particularly important for guiding RISC to target mRNAs, the majority of which show full complementarity to the seed sequence (1). However, noncanonical miRNA binding to mRNAs that show incomplete seed complementarity can contribute up to 40% of miRNA target sites (3). Moreover, many potential target sites that do show full seed complementarity are not functional, and this is often due to the fact that these sites are occluded by mRNA secondary structure (4, 5).
While all mammalian cells express multiple miRNA species, the actual pattern of miRNA expression varies widely between tissues, and miRNAs are thought to play a key role in many aspects of cellular differentiation and organismal development (1). Moreover, cellular miRNAs are not the only miRNAs that have been described, as several viruses are now also known to encode miRNAs (6, 7). In particular, herpesviruses have been shown to encode up to 35 distinct miRNAs that regulate cellular genes involved in cell cycle regulation, apoptosis, and innate immunity as well as viral genes that play a role in regulating viral latency (6, 7). Viral miRNAs have also been detected in polyomavirus family members, as well as in adenoviruses, but so far only one RNA virus, the retrovirus bovine leukemia virus (BLV), has been clearly shown to express high levels of viral miRNAs in infected cells (8). One possible explanation for why RNA viruses, including retroviruses, might not encode miRNAs is that cleavage by Drosha leads to degradation of the pri-miRNA precursor, which in the case of most RNA viruses would likely be the genomic RNA or a viral mRNA (7). The idea that this might be deleterious to efficient viral replication is perhaps supported by the fact that BLV generates its five miRNA species by using RNA polymerase III to transcribe short RNA hairpins that are structurally indistinguishable from pre-miRNA hairpins and that are therefore directly exported to the cytoplasm without cleavage by Drosha (8). Drosha fails to excise these pre-miRNAs from BLV genomic RNAs because their stems are only ~22 bp in length, not ~33 bp as required for recognition by Drosha (8–10). As a result, BLV is able to express viral miRNAs without having to sacrifice viral genomic and/or mRNA species in the process.
Analysis of the miRNA coding potential of HIV-1 has led to considerable controversy. HIV-1 was initially reported to lack any viral miRNAs by conventional sequencing of small RNAs in HIV-1-infected HeLa cells (11), and this report was supported by a subsequent report that failed to detect any HIV-1 miRNAs in chronically HIV-1-infected ACH-2 T cells, again using conventional sequencing (12). In contrast, the first HIV-1 miRNA was reported in 2004, based on Northern analysis of RNA derived from HIV-1-infected cells using probes specific for the viral nef gene (13). This viral miRNA was subsequently proposed to inhibit HIV-1 long terminal repeat (LTR) function at the transcriptional level by targeting the LTR U3 region (14). The second HIV-1 miRNA to be proposed derives from the HIV-1 TAR element, a 59-nt-long RNA stem-loop located at the 5′ end of all HIV-1 miRNAs (15–17). This miRNA was proposed to affect chromatin remodeling and to downregulate proapoptotic cellular genes. More recently, two groups have used moderately deep sequencing approaches to analyze HIV-1-derived small RNAs in infected cells. Yeung et al. (18) obtained 47,773 total reads, of which 125 (0.26%) were of HIV-1 origin, representing a diverse group of small viral RNA sequences that the authors nevertheless proposed likely represented functional viral miRNAs. Similarly, Schopman et al. (19) performed deep sequencing of small RNAs present in HIV-1-infected SupT1 cells and found that HIV-1 contributed ~1% of the total small RNA pool, and several viral small RNA species were recovered at ≤175 total reads each. As cellular miRNAs contributed ~71% of the total of 2,522,374 small RNA reads recovered in this study, this means that even the most prevalent candidate viral miRNAs represented only ~0.01% of the total miRNA pool in these HIV-1-infected T cells, which equates to <5 copies per cell in total (20).
In considering whether small RNA reads of viral origin indeed represent authentic viral miRNAs, the following parameters need to have been satisfied: (i) miRNAs are almost always 22 ± 2 nt in length (1). Therefore, an authentic viral miRNA will be recovered at a discrete size that is close to 22 nt. If viral small RNA reads extend over a wider size range, then this is more consistent with RNA breakdown products. (ii) Because the seed region of the miRNA, nucleotides 2 through 8 from the 5′ end, is the key determinant of mRNA target recognition, authentic viral miRNAs will have a discrete, not a diffuse, 5′ end. (iii) miRNAs tend to regulate >100 mRNA species, due to the small size of the seed recognition sequence, so functional miRNAs are generally highly expressed. Importantly, recent data demonstrate that miRNAs that represent <0.1% of the total viral miRNA pool are unlikely to be functionally relevant (21). (iv) Viral miRNAs should derive from one or a small number of discrete sites in the viral genome that coincide with predicted pri-miRNA stem-loops. If viral small RNA reads are scattered across the genome, they are less likely to represent real miRNA reads (6, 7). (v) Relative to the total small RNA pool, authentic viral miRNAs will be enriched in RNA preparations derived from immunoprecipitated RISCs, while RNA breakdown products will be depleted.
In addition to the question of whether HIV-1 actually encodes miRNAs, it has also been proposed that HIV-1 can modulate cellular miRNA expression to promote its replication (22). Finally, HIV-1 transcripts are, of course, potential targets for binding by RISCs programmed by cellular miRNAs, which could repress virus replication and possibly favor entry into latency (23). Here, we have used deep sequencing as well as techniques that directly recover RISC binding sites to address how HIV-1 interfaces with the miRNA machinery in infected T cells and macrophages. We clearly demonstrate that HIV-1 does not encode any viral miRNAs and also reveal that HIV-1 only minimally affects cellular miRNA expression patterns within 72 h of infection. We also identify a number of cellular miRNA binding sites on the HIV-1 RNA genome and show that some of these can potentially mediate inhibition of viral gene expression. However, we also demonstrate that HIV-1 genomic-length mRNAs represent very poor targets for RISC binding, most probably because large stretches of the HIV-1 genome are occluded by RNA secondary structure.
RESULTS
HIV-1 fails to express any viral miRNAs in infected cells.
To determine the miRNA expression profile in HIV-1-infected cells, we performed deep sequencing of small, ~15- to 30-nt-long RNAs in a range of infected cell types. These included TZM-bl cells, a variant of HeLa cells that express the CD4 receptor and both the CXCR4 and CCR5 coreceptor (24), infected with the CCR5-tropic HIV-1 variant WT/BaL (25); the human CD4+ T-cell line C8166 (26) infected with the CXCR4-tropic HIV-1 laboratory isolate NL4-3 (27); CD4+ peripheral blood mononuclear cells (PBMCs) infected with either the CCR5-tropic BaL isolate (28) or the CXCR4-tropic NL4-3 isolate; and, finally, primary human monocyte-derived macrophages (MDMs) infected with an NL4-3-derived HIV-1 variant, NLHXADA, bearing the CCR5-tropic ADA env gene (29).
TZM-bl cells were directly infected with WT/BaL virus derived from 293T cells transfected with the pWT/BaL proviral clone (25). Uniform infection of the TZM-bl cells was confirmed by staining of a control TZM-bl culture at 72 h postinfection for viral activation of the integrated β-galactosidase (β-Gal) indicator gene (24), which confirmed a >90% infection rate. Quantitation of the level of viral RNA transcripts, by quantitative reverse transcriptase PCR (qRT-PCR) of a region derived from the HIV-1 LTR U3 region that is present in all viral transcripts, indicated that the infected TZM-bl cells contained an average of ~130,367 viral RNA strands per cell (Table 1).
TABLE 1 .
Overview of the deep sequencing libraries analyzeda
| Cell type | HIV-1 virus isolate used | Envelope gene source | No. of HIV-1 RNA strands/cell | No. of days postinfection |
|---|---|---|---|---|
| TZM-bl | WT/BaL | BaL | 130,367 | 3 |
| C8166 | NL4-3 | NL4-3 | 20,546 | 3 |
| CD4+ PBMC | BaL | BaL | 8,303 | 3 |
| MDM | NLHXADA | ADA | 12,110 | 27 |
| CD4+ PBMC | NL4-3 | NL4-3 | 1,949 | 3 |
This table indicates the cell type used (column 1), the virus used to infect (column 2), and the source of the env gene in that virus (column 3), as well as the total number of HIV-1 transcripts per cell at the time of harvest (column 4) and how many days postinfection RNA was harvested (column 5).
C8166 cells and CD4+ PBMCs were infected with NL4-3 virus cultured in MT4 cells or with BaL virus cultured in CD4+ PBMCs. Cells were initially infected with ~0.7 TZM-bl infectious units per cell and then cultured for 72 h. At this time, infectivity levels were determined by fluorescence-activated cell sorting (FACS) analysis using pooled human anti-HIV-1 IgG followed by fluorescein isothiocyanate (FITC)-conjugated goat anti-human IgG. This analysis (see Fig. S1 in the supplemental material) indicated that ~97% of the C8166 cells, ~78% of the BaL-infected CD4+ PBMCs, and ~94% of the NL4-3-infected CD4+ PBMCs were HIV-1 positive. qRT-PCR analysis of viral transcript levels (Table 1) indicated that these infected cultures contained an average of ~20,546, ~8,303, and ~1,949 viral RNA strands per cell, respectively (Table 1).
Finally, primary MDMs were infected with an HIV-1 virus, NLHXADA, derived from an NL4-3-based proviral clone bearing the CCR5-tropic ADA env gene (29). MDMs were initially infected with a virus stock obtained from pNLHXADA-transfected 293T cells, and supernatant RT levels were then monitored. Once significant RT levels were detected, at 17 days postinfection, the MDMs were cultured for an additional 10 days, for a total of 27 days, prior to lysis and RNA isolation. Previous work has shown that this protocol leads to essentially uniform infection of the MDM culture (30). Analysis indicated an average of ~12,110 HIV-1 RNA strands per cell (Table 1).
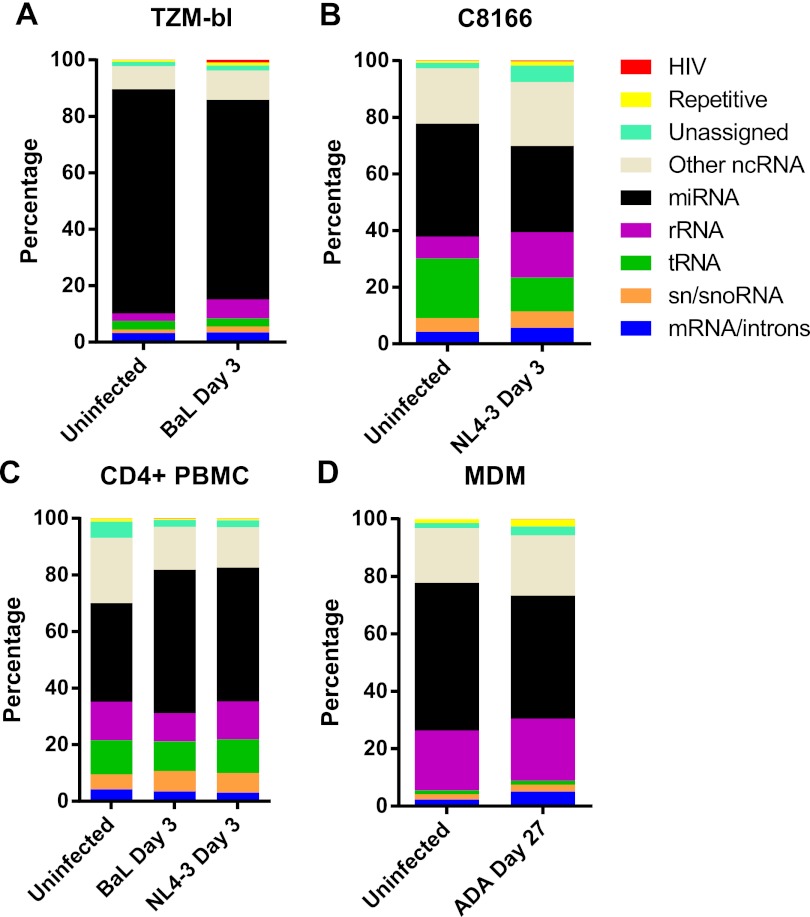
Deep sequencing analysis of the small RNA profile in all five HIV-1-infected cell cultures and in matched uninfected cultures revealed that cellular miRNAs were, as expected, the major RNA variant detected in all 9 deep sequencing libraries (Fig. 1). In addition, we also detected significant levels of reads that could be aligned to human rRNAs, tRNAs, snRNAs, snoRNAs, or mRNAs and likely represent RNA breakdown products (Fig. 1). Very few reads were found to align to the genomes of any of the HIV-1 virus variants in any of the infected cell cultures. This contrasts with previous reports showing that viruses known to encode viral miRNAs, such as Epstein-Barr virus (EBV), Kaposi’s sarcoma-associated herpesvirus (KSHV), and the retrovirus BLV, contribute a substantial share of the small RNAs seen in infected cells (8, 11, 31).
FIG 1 .
Assignment of deep sequencing reads to cellular and viral RNA classes. Small RNA deep sequencing reads were aligned to the human or HIV-1 genome and are shown by their assignment to different subclasses of RNA. Samples were derived from uninfected cells or from cells infected with HIV-1, as indicated. Repetitive sequences are defined as those that map to ≥25 locations in the human genome.
One key characteristic of miRNAs is their size, which ranges from ~20 nt to ~24 nt in length (1). Analysis of the length profile of the total cellular small RNAs recovered in HIV-1-infected TZM-bl cells, C8166 cells, CD4+ PBMCs, and MDMs shows that these indeed peak at ~22 nt in size, as expected (Fig. 2A, C, E, and G). In contrast, the RNA reads that align to the HIV-1 genome are not only rare but also showed a fairly random size distribution, with a tendency for more reads at smaller sizes. Certainly, there was no evidence for high levels of HIV-1-derived small RNAs in the 22 ± 2-nt size range characteristic of miRNAs (Fig. 2B, D, F, and H).
FIG 2 .
HIV-1-specific small RNAs do not cluster at the ~22-nt size predicted for miRNAs. Small RNA reads from 17 to 25 nt in length, derived from HIV-1-infected cells, that align either to the human genome (A, C, E, and G) or the HIV-1 genome (B, D, F, and H) are shown assigned by their size. (A and B) TZM-bl cells infected with the WT/BaL virus. (C and D) C8166 T cells infected with NL4-3. (E and F) CD4+ PBMCs infected with the BaL HIV-1 isolate. (G and H) MDMs infected with the NLHXADA virus.
Analysis of viral small RNAs in cells infected by EBV or KSHV, which are known to encode viral miRNAs, has revealed that virus-derived small RNAs are almost all derived from a small number of locations in the viral genome that coincide with the pri-miRNA RNA stem-loops that are processed to yield viral miRNAs (7). Moreover, these viral miRNAs are characterized by high-level expression (31). Indeed, recent data have shown that miRNAs that contribute <0.1% of the total cellular miRNA pool make no significant functional contribution (21). Cellular and authentic viral miRNAs are also characterized by a discrete 5′ end, consistent with the finding that it is nucleotides 2 through 8 from the 5′ end, the so-called seed region, that primarily determines the mRNA target specificity of the miRNA (1, 11, 31).
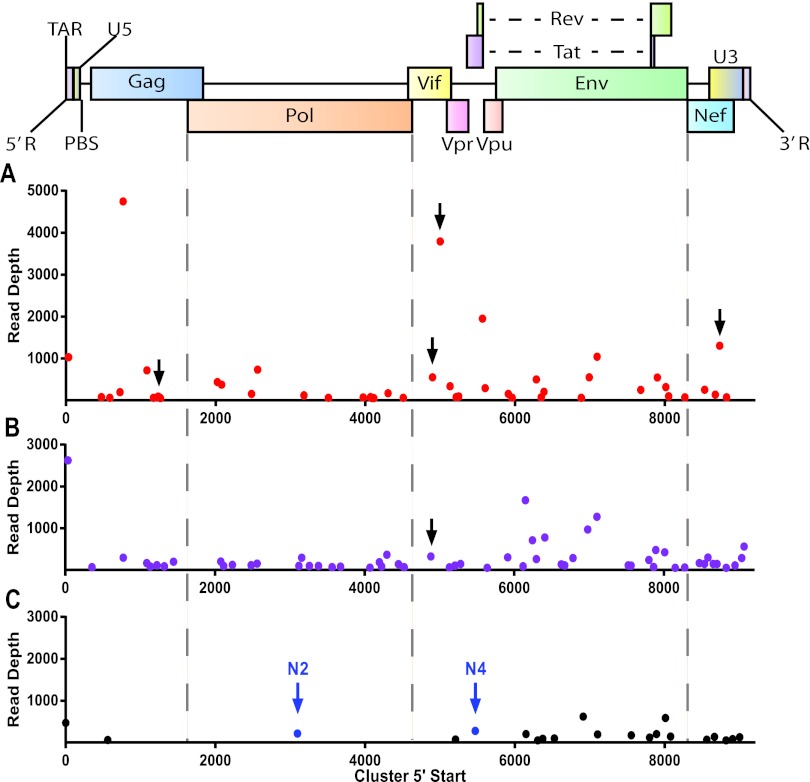
Alignment of the HIV-1-derived small RNAs to the relevant proviral genome, in contrast, shows that these are scattered over the entire proviral sequence (Fig. 3). Some hot spots of small RNA reads were noted, but even these are found to contribute <0.02% of the total miRNA pool in the infected cells (Fig. 3). Moreover, these hot spots were generally not conserved between virus variants and/or cell types. An exception to this generalization arises in the case of small RNA reads that map to the overlap between the HIV-1 Gag and Pol open reading frames, which represent the major reads in the C8166/NL4-3 and CD4+ PBMC/BaL libraries (albeit still contributing only ~0.015% and ~0.0035% of total miRNA reads, respectively) and were also readily detected in TZM-bl/BaL cells (Fig. 3). Analysis of these reads shows that their 5′ ends map to a stretch of 6 U residues found immediately 5′ to an RNA hairpin that facilitates the Gag/Pol frameshift during translation of genome length HIV-1 mRNAs by promoting ribosome stalling at the six-U-residue “slippery site” (see Fig. S2 in the supplemental material) (32). However, this short hairpin does not share any of the characteristics that are typical of a pri-miRNA hairpin structure; in particular, this predicted viral RNA hairpin is only 11 bp long rather than the ~33 bp typical of pri-miRNA hairpins (9, 10). It seems possible that these reads, which almost all begin in the U stretch and then extend into the adjacent hairpin, are actually Gag/Pol mRNA fragments that are protected by the stalled ribosomes that are known to accumulate at this site on the viral mRNA (32).
FIG 3 .
Alignment of small RNA reads to the HIV-1 genome. RNA reads of 19 to 25 nt in length that mapped to the HIV-1 genome were aligned based on their 5′ ends and are shown relative to their genomic position of origin (x axis). The y axis shows the number of reads as a percentage of the total number of cellular miRNA reads in the same RNA sample. (A) TZM-bl infected with WT/BaL; (B) C8166 infected with NL4-3; (C) CD4+ PBMCs infected with BaL; (D) MDMs infected with NLHXADA.
Two groups have reported that HIV-1 encodes miRNAs derived from the TAR RNA stem-loop that is located at the 5′ end of all viral transcripts (15–17). The TAR stem, at ~24 bp, is again much shorter than the optimal ~33-bp stem characteristic of pri-miRNA stem-loops (9, 10), and TAR also differs from authentic pri-miRNAs in that the terminal loop is only 6 nt, not the ≥10 nt characteristic of pri-miRNAs (see Fig. S3 in the supplemental material) (9). Most importantly, Drosha cleavage of pri-miRNAs requires unstructured RNA sequences both 5′ and 3′ to the pri-miRNA stem (10, 33), while TAR is located at the very 5′ end of all HIV-1 RNAs and, moreover, likely has a cap and cap-binding proteins associated with its 5′ end. In fact, we recovered almost no reads that mapped to TAR in all cases except ADA-infected MDMs, where these reads contributed ~0.002% of the viral short RNA reads (Fig. 3). Analysis of these rare TAR-derived RNAs revealed a heterogeneous population of small RNAs that mapped to both sides of the TAR RNA stem-loop and that, in particular, did not have a discrete 5′ end (see Fig. S3).
It could be argued that some of the HIV-1 small RNA reads, despite their low level of expression, are nevertheless functional miRNAs that are efficiently loaded into RISC. To address whether this is the case, we immunoprecipitated RISC using an antibody able to specifically bind Ago1, Ago2, and Ago3, each of which can interchangeably function as a key component of RISC (1), and then compared the profile of small RNAs recovered from the RISC immunoprecipitate with that seen when the total small RNA profile was determined in parallel in WT/BaL-infected TZM-bl cells. As expected, the RNA library derived from the RISC immunoprecipitate indeed showed a strong enrichment in miRNAs, with known cellular miRNAs increasing from ~71% to ~93% of the total RNA library. In contrast, the HIV-1-derived small RNA reads were selectively lost when RISC-associated small RNAs were sequenced (see Fig. S4). In this library, no viral RNA was found to contribute >0.005% of the total human miRNA pool, a level that is far lower than the >0.1% contribution previously reported to be characteristic of functionally relevant miRNAs (21).
Previously, it has been suggested that HIV-1 might generate miRNAs, or even small interfering RNAs (siRNAs), from transcripts derived from the negative-sense RNA strand (19). Even though retroviruses do not generate minus-strand RNAs during their replication cycle, minus-sense strands could in principle arise by transcription from cellular promoters located 3′ to, and in the opposite orientation from, integrated proviruses. However, we failed to detect a significant level of small RNA reads derived from the HIV-1 antisense strand in either infected TZM-bl cells (see Fig. S5A in the supplemental material) or in any other infected cell types (data not shown). While most regions of the HIV-1 minus strand gave read numbers equivalent to ≤0.0002% of the total miRNA population, one region, complementary to the HIV-1 primer binding site, did give rise to a substantial number of reads (see Fig. S5A). These have been previously noted by others (18, 19) and proposed to derive from breakdown products of the lysine tRNA that serves as the primer for HIV-1 reverse transcription. Indeed, reads complementary to the HIV-1 primer binding site were recovered at comparable levels in noninfected cells (data not shown), and analysis of small RNAs bound to RISC in infected TZM-bl cells failed to detect any reads derived from the region complementary to the viral primer binding site, thus strongly suggesting that these are indeed tRNA breakdown products (see Fig. S5B).
HIV-1 fails to significantly affect cellular miRNA expression early after infection.
There are a number of reports documenting cases where cellular miRNAs facilitate aspects of a viral replication cycle and/or where viruses manipulate the cellular miRNA expression profile to enhance some aspect of their replication or pathogenic potential (34–40). This has also been suggested for HIV-1, which has been reported to downregulate the miR-17/92 miRNA cluster, consisting of the six cellular miRNAs miR-17, miR-18a, miR-19a, miR-20a, miR-19b, and miR-92a, to upregulate expression of the p300/CBP-associated factor (PCAF) histone acetyltransferase, which has been proposed to function as a cofactor for the HIV-1 Tat transcription factor (22). To examine whether HIV-1 indeed exerts a significant effect on the cellular miRNA profile, we therefore compared the level of expression of cellular miRNAs in uninfected and matched HIV-1-infected cells, measured either by total small RNA sequencing or, in the case of TZM-bl cells infected with BaL, by deep sequencing RISC-associated small RNAs (Fig. 4). This analysis was performed 72 h after initial infection in all cases. We believe this short time frame is appropriate, given that HIV-1-infected T cells have been reported to have a half-life of only ~24 h in vivo (41).
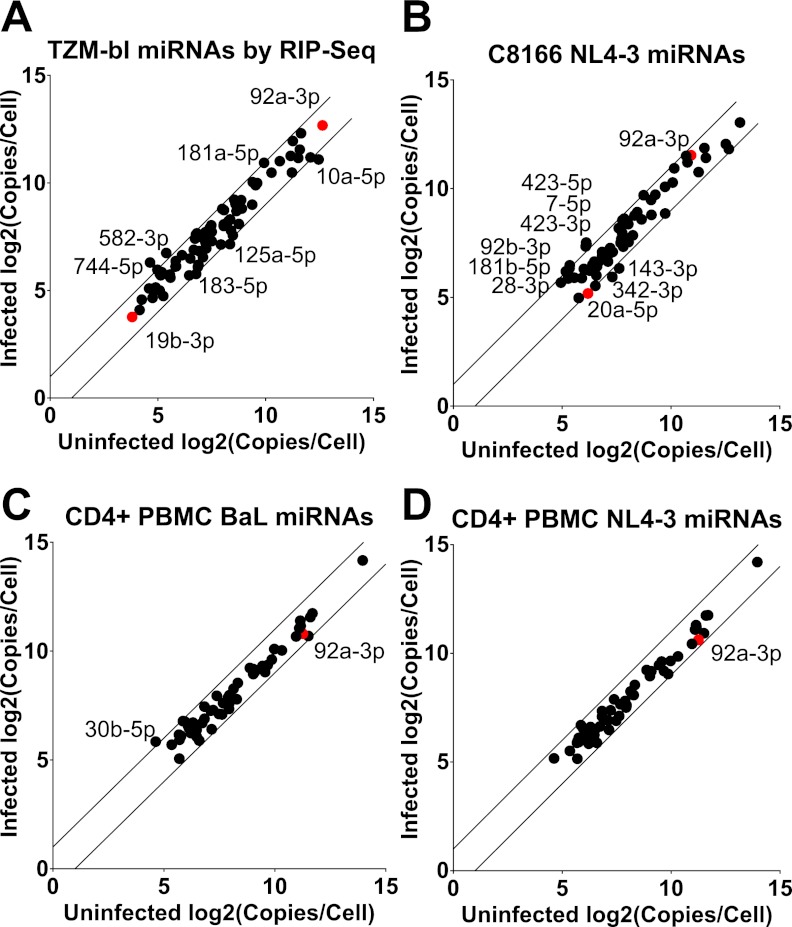
FIG 4 .
Effect of HIV-1 infection on cellular miRNA expression levels. This figure presents a comparison of the level of each cellular miRNA detected in uninfected and HIV-1-infected cells. Only miRNAs that represent ≥0.1% of the total miRNA pool were included. Panel A shows data obtained by comparison of RISC-associated small RNAs, while panels B, C, and D compare total miRNA levels. The lines delineate changes of 2-fold between samples. miRNAs that fall outside this 2-fold limit are indicated by name. Red dots represent miRNAs derived from the miR-17/92 cluster. For illustrative purposes, this figure assumes that cells each express 5 × 104 miRNAs.
As shown in Fig. 4, and presented in detail in Table S1, analysis of the expression level of cellular miRNAs in HIV-1-infected TZM-bl cells, C8166 T cells, and, most importantly, primary CD4+ PBMCs revealed few differences between the infected and uninfected cells despite clear evidence of efficient infection (see Fig. S1 in the supplemental material) and quite high levels of viral transcripts (Table 1). In particular, only one cellular miRNA, miR-30b-5p, increased >2-fold in BaL-infected PBMCs, and no cellular miRNAs increased or decreased >2-fold in NL4-3-infected PBMCs. In both TZM-bl and C8166 cells, several cellular miRNAs changed their expression, in either a positive or negative direction, by slightly over the 2-fold limit indicated in Fig. 4. The functional significance of these changes, if any, is unclear. Perhaps surprisingly, we failed to see any significant (≥2-fold) effect of HIV-1 infection on the expression of members of the miR-17/92 cluster, as previously proposed (22).
Cellular miRNAs bind HIV-1 transcripts inefficiently.
Viruses that infect cells encounter a range of different cellular miRNAs that have the potential to inhibit viral mRNA function, and it is largely unclear how viruses avoid this problem. One virus family, the poxviruses, has been shown to globally degrade cellular miRNAs (42). However, most viruses, including HIV-1, do not block miRNA function in infected cells (43). It is also possible that a virus that is highly tissue tropic, e.g., HIV-1, which infects only CD4+ T cells and macrophages, has simply evolved to avoid mRNA binding sites for the miRNAs specific to these cell types. In that case, however, one would expect HIV-1 to be susceptible to inhibition by the diverse miRNAs found in other, normally nontarget cell types.
To address this question, we used the recently described photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation (PAR-CLIP) technique (31, 44) to globally identify all the binding sites for miRNA-programmed RISCs on the HIV-1 genome in infected C8166 T cells or TZM-bl epithelial cells. At 48 h postinfection with the NL4-3 (C8166) or WT/BaL (TZM-bl) virus isolate, the infected cells were incubated with 100 µM 4-thiouridine (4SU) for 16 h prior to cross-linking by irradiation using UV light at 365 nm. mRNAs bound to RISC were then isolated by immunoprecipitation using a monoclonal antibody specific for the human Ago proteins. After RNase treatment, mRNA fragments bound to RISC were gel purified, cDNA cloned, and subjected to Illumina deep sequencing. The resultant reads were then aligned to the human or HIV-1 genome and analyzed using the PARalyzer program, which partitions reads into individual clusters, each of which defines a single RISC binding site (Fig. 5) (45). The PARalyzer program also seeks to use sequencing data defining the cellular miRNAs present in the infected cells, obtained in parallel, to identify the cellular miRNA that is most likely responsible for guiding RISC to that particular viral binding site. This assignment, which assumes perfect complementarity to nucleotides 2 to 7 in the miRNA seed region, is, however, unable to identify “noncanonical” miRNA binding sites, which can contribute a significant percentage of the RISC interaction sites (3).
FIG 5 .
Alignment of PAR-CLIP clusters with the HIV-1 genome. The PAR-CLIP technique was performed on HIV-1-infected cells using a monoclonal antibody specific for RISC. Each HIV-1-specific PAR-CLIP cluster, identified by bioinformatic analysis using the PARalyzer program (45), is aligned to the HIV-1 genome based on the 5′-most nucleotide of the read cluster. Clusters indicated by black arrows were successfully assigned to specific cellular miRNAs and are analyzed in Fig. 6. Clusters indicated by blue arrows derive from the amiRNAs N2 and N4 (5) that were expressed in the TZM-bl ami cells. (A) C8166 cells infected with NL4-3; (B) TZM-bl cells infected with WT/BaL; (C) TZM-bl ami cells infected with WT/BaL.
As shown in Fig. 5, we identified a number of RISC binding clusters on the HIV-1 RNA genome in both C8166 and TZM-bl cells. However, we were surprised by the low level of RISC binding to the viral RNA genome observed. As shown in Table 2, we recovered ~1.7 × 107 assignable PAR-CLIP reads from the HIV-1-infected C8166 cells, of which only 0.21% aligned to the HIV-1 RNA genome. Similarly, we recovered 6.8 × 106 assignable PAR-CLIP reads from the HIV-1-infected TZM-bl cells, of which only 0.3% aligned to the HIV-1 genome. Analysis of the number of HIV-1 RNA strands present in the C8166 cells at the time of cross-linking revealed ~20,707 HIV-1 transcripts, while analysis of the cross-linked TZM-bl cells revealed ~407,661 HIV-1 strands per cell. Previous work has suggested that cells contain between ~105 and ~5 × 105 mRNA molecules per cell, depending on the size of the cell and particularly of the cell cytoplasm (46). As T cells have a small cytoplasm, we can estimate that they likely contain ~200,000 total mRNA molecules per cell, which would mean that HIV-1 contributes ~10% of the total mRNA pool in the infected C8166 cells. In the larger TZM-bl cells, HIV-1 would appear to be responsible for at least 50% of the total mRNA pool, even assuming that the 407,661 HIV-1 strands are largely added to a preexisting mRNA pool of ~5 × 105 molecules. Therefore, HIV-1 contributes 10% of the total mRNA in C8166 cells but is bound only to ~0.21% of the available RISC, an underrepresentation of ~48-fold. This effect is even more extreme in TZM-bl cells, where HIV-1 contributes ~50% of the total mRNA pool but only ~0.31% of the total RISC binding events, an underrepresentation of ~160-fold. It therefore appears that RISC recruitment to HIV-1 mRNAs is inefficient.
TABLE 2 .
Overview of the PAR-CLIP libraries analyzeda
| Library | Total no. of assignable PAR-CLIP reads | % of HIV-1 PAR-CLIP reads | No. of HIV-1 RNA strands per cell |
|---|---|---|---|
| C8166/NL4-3 | 17,347,389 | 0.21 | 20,707 |
| TZM-bl/WT/BaL | 6,817,180 | 0.30 | 407,661 |
| TZM-bl ami/WT/BaL | 11,077,171 | 0.05 | 15,756 |
This table summarizes the characteristics of the HIV-1-infected cell PAR-CLIP libraries, including the total number of assignable (≤1 mismatch) PAR-CLIP reads (column 2), the percentage of PAR-CLIP reads that can be assigned to HIV-1 RNAs (column 3), and the total number of HIV-1 transcripts per cell at the time of cross-linking and RNA harvest (column 4), as determined by qRT-PCR with RNA standards.
While low, RISC binding to HIV-1 is certainly detectable, and we therefore wished to ask if the observed binding sites were functional. In fact, for unclear reasons, relatively few RISC binding sites could be assigned to any of the miRNAs present in the infected cells. However, our analysis did predict that four RISC binding clusters detected in HIV-1-infected C8166 cells, indicated by black arrows in Fig. 5, were likely caused by miR-423 (cluster at position 1233), miR-301a (cluster at position 4898), miR-155 (cluster at position 5001), and miR-29a (cluster at position 8735), respectively. We note that miR-423 represented ~0.63% of total cellular miRNA reads in the C8166 cells used in this experiment, miR-301a represented ~0.10%, miR-155 represented ~5.43%, and miR-29a represented ~3.24% of total miRNA reads, as determined by deep sequencing performed in parallel. We note that the putative miRNA-29a-specific cluster has previously been reported by two other groups, who argued that miR-29a is, in fact, able to inhibit HIV-1 replication (47, 48). In contrast, another group has reported that this proposed miR-29a target site is largely blocked by HIV-1 RNA secondary structure (49). Of these four clusters, only one cluster was also detected in TZM-bl cells, which express miR-301a and miR-423 but not miR-29a or miR-155. The PAR-CLIP reads that contribute to each of these clusters are listed in Fig. S6 in the supplemental material, where they are aligned to the relevant cellular miRNA.
To test whether these miRNAs are indeed able to bind to these regions of the HIV-1 NL4-3 genome, we used PCR to clone ~300-bp segments of the HIV-1 genome centered on each of the PAR-CLIP clusters we had detected. These segments were inserted 3′ to the Renilla luciferase (Rluc) indicator gene present in the psiCHECK2 indicator plasmid, which also contains an internal control firefly luciferase (Fluc) expression cassette in cis. We also constructed derivatives of each of these indicator plasmids in which the predicted seed sequence present in the HIV-1 DNA fragment was mutated, to prevent miRNA binding, as shown in Fig. S6 in the supplemental material. Cotransfection of the indicator plasmids bearing the 1233, 4898, and 5001 clusters into 293T cells, together with either the cognate miRNA expression vector or a control plasmid, revealed specific downregulation in the presence of the relevant cellular miRNA that was lost when the seed target sequence in HIV-1 was mutated (Fig. 6). In contrast, the 8735 cluster did not confer downregulation on the Rluc gene in the presence of an miR-29a expression plasmid, even though we observed strong downregulation of a control Rluc vector bearing two perfect miR-29a target sites (Fig. 6). We therefore conclude that HIV-1 transcripts can indeed bind miR-423, miR-301a, and miR-155 in infected C8166 T cells and that this interaction has the potential to reduce HIV-1 gene expression. In contrast, even though we did detect a RISC binding cluster at a nef miRNA target site previously reported to bind miR-29a, we failed to see downregulation by miR-29a when this cluster was inserted into an Rluc indicator plasmid, a result which agrees with data previously reported by Sun et al. (49). Therefore, it appears that miR-29a binding to this region of the HIV-1 nef gene, while detectable by PAR-CLIP (Fig. 5A), is too inefficient to mediate significant gene repression (Fig. 6B). It remains possible that this interaction is functionally significant in other contexts, for example, in cells expressing very high levels of miR-29a and/or very low levels of HIV-1 transcripts.
FIG 6 .
Identification of specific cellular miRNA binding sites in the HIV-1 genome. We constructed indicator plasmids containing ~300-bp segments of the HIV-1 genome, encompassing the clusters indicated by black arrows in Fig. 5A, inserted 3′ to the Rluc indicator gene. Derivatives in which the predicted seed target for a cellular miRNA were mutated were also generated (see Fig. S6 in the supplemental material), as was a positive-control indicator plasmid containing two perfect target sites for each of the four miRNAs being tested. These were then cotransfected into 293T cells along with the cognate cellular miRNA expression plasmid or a control plasmid. Rluc values are shown normalized to the Fluc internal control and to the level seen in cells lacking the cognate miRNA expression vector, which was set at 1.0. Note that the inhibition in Rluc expression seen with the wild-type (WT) indicator in the presence of the cognate miRNA is lost when the seed target in the HIV-1 sequence is mutated. This figure shows the average of four independent experiments, with the standard error of the mean indicated.
As a further demonstration that miRNAs can indeed guide RISC to the HIV-1 genome, we took advantage of a recent study that identified artificial miRNAs (amiRNAs) that are able to effectively inhibit HIV-1 gene expression and replication by testing over 9,000 different amiRNAs, tiled at 1-nt increments across the HIV-1 RNA genome, for their ability to block HIV-1 replication in culture (5). Interestingly, few of these amiRNAs proved to be effective, thus suggesting that access to the HIV-1 RNA genome was largely occluded for most of the RISCs programmed with these amiRNAs (5). We expressed two amiRNAs that were shown to be effective, termed N2 and N4, by engineering these amiRNAs into retroviral expression vectors and then selecting TZM-bl cells that stably expressed the N2 or N4 amiRNA, as determined by small RNA deep sequencing. The N2- and N4-expressing TZM-bl cells were then infected with HIV-1 strain WT/BaL, and PAR-CLIP was performed as described above. As expected based on previous work (5), we observed that these TZM-bl cells were largely nonpermissive for HIV-1 replication, as the level of HIV-1 transcripts detected in TZM-bl cells expressing N2 and N4, which are fully complementary to the HIV-1RNA genome, was reduced ~25-fold relative to infected wild-type TZM-bl cells (Table 2). Nevertheless, despite the much lower level of HIV-1 RNAs present in these cells, we were able to readily detect PAR-CLIP clusters that precisely aligned with the expected binding sites of RISCs programmed with either the N2 or N4 amiRNA. These interactions were detected despite the potential for Ago2-containing RISCs programmed by these amiRNAs to cleave the viral genome within the binding cluster (Fig. 5) (1). Therefore, these data demonstrate that miRNAs are indeed able to guide RISC to the HIV-1 RNA genome if the target area is accessible.
DISCUSSION
In this report, we have attempted to comprehensively address how HIV-1 interacts with cellular miRNA biogenesis and effector mechanisms. Our first goal was to unequivocally answer the previously controversial question of whether HIV-1 itself encodes any miRNAs. While two previous studies that used conventional sequencing techniques had failed to detect any HIV-1-encoded miRNAs (11, 12), there are also a substantial number of papers suggesting that HIV-1 does encode miRNAs and/or siRNAs, particularly in the viral nef gene, the viral TAR element, within the HIV-1 Rev response element (RRE), or derived from some form of viral antisense RNA (13–19, 50). Here, we have used both CXCR-4-tropic and CCR5-tropic HIV-1 infection of a range of cell types, combined with deep sequencing of total and RISC-associated small RNAs, to address whether any of these proposed miRNAs or siRNAs in fact exist. The cells used were analyzed at 3 days after infection, with the exception of MDMs, which were analyzed at 27 days after infection. FACS analysis, in the case of CD4+ PBMCs, or analysis of β-gal expression in the case of the indicator cell line TZM-bl, reveals that these cells were efficiently infected (see Fig. S1 in the supplemental material), and we also detected high levels of viral RNA transcripts in all four infected cell types (Table 1). Nevertheless, we failed to detect any HIV-1-specific miRNAs or siRNAs, including from the nef region, TAR, and RRE, even though these putative viral miRNA sequences are all present in one or more of the viruses analyzed (Fig. 3; see also Fig. S5 in the supplemental material). The one region that did give rise to a readily detectable level of viral small RNA reads did not show loading into RISC (see Fig. S4) and likely represents a region of the HIV-1 Gag/Pol mRNA that is protected by stalled ribosomes, as it precisely coincides with the sequence that induces ribosome frameshifting (see Fig. S2) (32). In general, HIV-1-derived small RNAs did not show the predicted 22 ± 2-nt length (Fig. 2), were not loaded into RISC (Fig. S4), did not show the expected discrete 5′ ends (see Fig. S2 and S3), and, perhaps most importantly, never exceeded the 0.1% level of the total cellular miRNA pool recently shown to be critical for miRNA function (21).
In addition to encoding viral miRNAs, viruses can also facilitate their replication by enhancing or repressing the expression of specific cellular miRNAs. HIV-1 infection has in fact been proposed to affect, especially repress, the expression of a number of cellular miRNAs, including the cellular miR-17/92 miRNA cluster (22, 49). The latter effect has been proposed to facilitate HIV-1 replication by derepressing expression of the PCAF acetyltransferase, a proposed cofactor of the HIV-1 Tat transcription factor.
A shared characteristic of these two earlier studies is that they looked at changes in cellular miRNA expression at a substantial time after initial infection with HIV-1. Thus, Triboulet et al. (22) measured cellular miRNA expression at 21 and 42 days postinfection, while Sun et al. (49) saw significant changes in cellular miRNA expression only at 21 days postinfection. However, data obtained in vivo using inhibitors of HIV-1 replication have revealed that HIV-1-infected T cells survive an average of only ~24 h after infection (41). Moreover, long-term culture of HIV-1-infected T cells maximizes the level of cytopathic effect that is observed, and this undoubtedly has the potential to influence cellular gene expression. We therefore felt it was most appropriate to examine the effect of HIV-1 infection at early times after infection, specifically at 72 h postinfection, and our data do not suggest that, at this time point, HIV-1 infection has a strong effect, either positively or negatively, on cellular miRNA expression (Fig. 4). In particular, we saw no significant effect on the expression of members of the miR-17/92 miRNA cluster, including miR-92a itself, which was one of the more highly expressed miRNAs in both TZM-bl cells and T cells (Fig. 4). Nevertheless, we did see some modest but perhaps significant increases or decreases in the expression of some cellular miRNAs (Fig. 4), though these are not the miRNAs previously reported by others. The functional significance of these ≤3-fold changes in a small number of cellular miRNAs is unclear and may not be significant, at least during lytic HIV-1 replication.
While the manuscript was under review, Chang et al. (51) reported an analysis of cellular miRNA expression in the SUP-T1 T-cell line at 5, 12, and 24 h postinfection with HIV-1. They reported that 14 known cellular miRNAs showed significant changes in expression, generally reductions, and they also proposed the existence of a novel cellular miRNA (miR-EPB41L2) that was downregulated by ~10-fold in HIV-1-infected SUP-T1 cells. Analysis of our data in primary CD4+ T cells, C8166 T cells, and TZM-bl cells showed that only one of these 14 miRNAs (miR-21-3p) was expressed at significant (>0.1%) levels in primary T cells, and miR-21-3p expression was not affected by HIV-1 infection (see Table S1C in the supplemental material). Only two of these 14 miRNAs were expressed in C8166 cells at significant levels (miR-21-3p and miR-143-3p), and miR-143-3p indeed showed an ~60% reduction in expression after HIV-1 infection (see Table S1B). Finally, 7 of the 14 miRNAs reported by Chang et al. (51) to be affected by HIV-1 infection were expressed at significant levels in TZM-bl cells, but only one, miR-10a-5p, showed a significant, ~60% reduction in expression level (see Table S1A). The novel miRNA proposed by Chang et al. (51), miR-EPB41L2, was detected at low levels in several of our deep sequencing libraries but showed a variable, small size, ranging from 15 nt to 18 nt, and a highly variable 5′ end (data not shown). We therefore feel that miR-EPB41L2 is unlikely to be an authentic cellular miRNA. Therefore, while we agree with Chang et al. (51) that some cellular miRNAs do show modest changes in expression after HIV-1 infection, these miRNAs vary with the cells being examined, and observed changes are quite limited in the most relevant experimental system, i.e., primary CD4+ T cells (Fig. 4; Table S1C).
Given that HIV-1 neither encodes viral miRNAs nor substantially alters cellular miRNA expression during lytic replication in culture, we are left with the question of how the cellular miRNAs encountered by HIV-1 in infected cells affect HIV-1 replication. Previously, others have proposed that cellular miRNAs might promote HIV-1 latency in resting primary CD4+ T lymphocytes (23) and inhibit productive HIV-1 replication in primary monocytes (52). One particular cellular miRNA, miR-29a, has been suggested to efficiently bind to an RNA target site in the HIV-1 viral nef gene (47, 48), although others have reported that this miR-29a target sequence is actually blocked by viral secondary structure (49).
To comprehensively identify sites on the HIV-1 RNA genome that are actually occupied by miRNA-programmed RISC in infected T cells or epithelial cells, we used the previously described PAR-CLIP technique (31, 44), which uses deep sequencing of short mRNA sequences cross-linked to RISC to identify miRNA binding sites. As shown in Fig. 5, we were able to identify a number of RISC binding sites on the HIV-1 genome. Many of these either did not show seed homology to any of the miRNAs found to be expressed in these cells at the time that PAR-CLIP was performed, as determined by deep sequencing performed in parallel, or did not prove to be inhibited by the cellular miRNAs that were predicted to bind there, when analyzed by indicator assays (data not shown). However, we were able to assign three RISC binding clusters identified in HIV-1-infected C8166 cells, one of which was conserved in TZM-bl cells, to the cellular miRNAs miR-423, miR-301a, and miR-155 and to further show that these targets could indeed confer downregulation in the presence of these miRNAs, when tested in indicator constructs (Fig. 6). Interestingly, we also detected a significant cluster of PAR-CLIP reads in C8166 cells coincident with the previously reported miR-29a target located in the nef/LTR U3 region overlap (Fig. 5A) (47, 48). However, an indicator construct containing this region of the HIV-1 genome was not inhibited by miR-29a when coexpressed in 293T cells, even though we could show that miR-29a was able to inhibit a control indicator construct (Fig. 6). We conclude that miR-29a binding to this site in nef likely occurs but is inefficient, as also previously proposed by others (49).
A possibly surprising result that arose from this PAR-CLIP analysis is that HIV-1 transcripts, despite constituting from 10% to as much as 50% of the total mRNA population in infected cells, only account for ~0.3% of the total assignable PAR-CLIP reads, i.e., HIV-1 transcripts are ~100-fold less likely to bind RISC than the cellular mRNAs present in the same cells. Extensive biochemical analysis has in fact revealed that the HIV-1 RNA genome is highly structured (53), and it has previously been reported that this RNA structure greatly limits the ability of amiRNAs to effectively block HIV-1 replication in culture (5). We have confirmed that two amiRNAs that were reported previously to inhibit HIV-1 replication (5) are indeed able to recruit RISC to the HIV-1 RNA genome (Fig. 5C) and inhibit HIV-1 replication (Table 2), so these data, in total, are most consistent with the hypothesis that miRNA-programmed RISCs can inhibit HIV-1 replication if they can access a target site but that the majority of the HIV-1 genome is occluded, most probably by RNA secondary structure. This may explain why HIV-1 seems able to grow in almost any human cell line that expresses the relevant cell surface receptors, even though these cells can exhibit very different miRNA expression patterns, i.e., HIV-1 is resistant to miRNAs not because it has evolved to selectively exclude binding sites for T-cell- or macrophage-specific miRNAs but because it has evolved a mechanism to render the viral transcripts largely refractory to RISC binding by adopting an extensive secondary structure. While this viral RNA secondary structure may indeed have evolved as a means to reduce inhibition of viral mRNA function by cellular miRNAs, we note that extensive viral RNA secondary structure might also have evolved as a way to reduce inhibition by other cellular innate immune factors, e.g., RNase L, or to facilitate viral RNA packaging into virion particles. Nevertheless, it will be interesting to see whether this is a common mechanism by which viruses, especially RNA viruses, avoid inhibition by the cellular miRNA machinery in infected cells.
MATERIALS AND METHODS
Viral isolates.
The viruses used in this study include the uncloned, CCR5-tropic BaL isolate (28); a CCR5-tropic virus derived from the pWT/BaL proviral clone (25), which contains an SalI/XhoI fragment of BaL, encompassing the HIV-1 tat, rev, vpu, and env genes, cloned into an HXB-3-derived proviral backbone; a CXCR4-tropic virus derived from the widely used pNL4-3 proviral clone (27); and lastly, a CCR5-tropic virus derived from pNLHXADA (29), which contains the env gene of the ADA isolate cloned into a proviral backbone derived primarily from NL4-3 but also partly from HXB-2.
Molecular clones.
The pWT/BaL (25), pNL4-3 (27), and pNLHXADA (29) HIV-1 proviral expression vectors have been previously described. The pMSCV/amiRNA-N2 and -N4 expression plasmids were designed to encode the N2 and N4 amiRNAs previously shown to inhibit HIV-1 replication (5). These retroviral vectors were generated by annealing the amiRNA-encoding oligonucleotides and ligating them into the polylinker present in pMSCV-puro (Clontech).
Fragments of 300 bp of the HIV-1 genome, centered on miRNA target sites detected by PAR-CLIP, were cloned into the 3′ UTR of codon-optimized Rluc via the XhoI and NotI sites of psiCheck2 (Promega), which also contains the Fluc gene in cis as an internal control. Pri-miRNA stem-loops were cloned into the XhoI and XbaI sites of pLCE as ~200-bp DNA fragments generated by PCR of human genomic DNA, as previously described (54).
Cell culture.
293T and TZM-bl cells (24) were cultured in Dulbecco’s modified Eagle medium (DMEM) (Sigma) supplemented with 10% fetal bovine serum (FBS). The human CD4+ T-cell line C8166 (26) was cultured in RPMI 1640 medium (Sigma) supplemented with 10% FBS. Peripheral blood mononuclear cells (PBMCs) were isolated from total blood by density gradient centrifugation (lymphocyte separation medium; Cellgro number 25-072-CV), and CD4+ T cells were then isolated using the Dynabead CD4 positive isolation kit from Invitrogen (number 1131 D). Cells were activated by incubation in phytohemagglutinin (PHA) and mouse monoclonal antibodies specific for human CD28 and CD49d (BD Biosciences number 347690) for three days. Activation of the CD4+ T cells was confirmed by fluorescence-activated cell sorting (FACS) by using an anti-CD69 antibody, and the cells were then cultured in RPMI supplemented with 10% FBS and interleukin-2 prior to infection. Peripheral blood monocytes were obtained by elutriation and cultured for 4 days in DMEM containing 10% human serum, 1% l-glutamine, and 6 ng/ml monocyte colony-stimulating factor (MCSF) to activate macrophages.
TZM-bl amiRNA cell lines were generated by transfecting 293T cells with 15 µg of a pMSCV amiRNA expression vector, 5 µg of pMLV-gag/pol, and 1 µg pHIT. Forty-eight hours posttransfection, virus-containing supernatants were collected, filtered, and used to transduce TZM-bl cells. Transduced cells were selected for puromycin resistance and then maintained in complete DMEM supplemented with puromycin dihydrochloride (Sigma) at a final concentration of 1 µg/ml. Expression of amiRNAs was confirmed by deep sequencing of small RNAs.
Viral stocks and infection.
NL4-3 virus was prepared in 293T cells via Fugene6 transfection with the pNL4-3 plasmid. Virus-containing supernatants were used to infect MT4 cells maintained in RPMI 1640 supplemented with 10% heat-inactivated FBS. Virus stocks were harvested 4, 5, and 6 days after infection and filtered through a 0.45-µm-pore-size Acrodisc syringe filter. The virus stock with the highest level of infectious virus, as determined by enzyme-linked immunosorbent assay (ELISA) for the p24 Gag protein and by viral titer, was used for subsequent experiments. NL4-3 titers were measured by staining TZM-bl cells (24) with the β-Gal staining kit (mirus Bio) 24 h postinfection and by counting blue foci. The HIV-1BaL isolate stock was maintained in primary CD4+ human T cells, and the titer was determined on TZM-bl as described above. Viral stocks were used to infect C8166 T cells and/or activated CD4+ T cells via spinoculation at 2,000 rpm for 3 h. T cells were infected with HIV-1 using ~0.7 TZM-bl infectious units per cell.
The WT/BaL (as opposed to BaL isolate) virus stock was prepared by transfection of 5 × 106 293T cells with 15 µg of pWT/BaL (25) using FuGENE6 (Roche). Virus-containing supernatants were collected 48 h posttransfection, filtered through a 0.45-µm-pore-size Acrodisc filter, and then used to infect the parental TZM-bl cells or TZM-bl cells engineered to stably express HIV-1-specific amiRNAs.
The NLHXADA virus stock was prepared by transfection of 293T cells with the pNLHXADA proviral clone (29), essentially as described above for pWT/BaL. After four days of culture in medium lacking MCSF, 7 × 105 macrophages were seeded in 24-well plates and directly infected with NLHXADA virus (500 ng p24 total per well) overnight. Supernatants were replaced with fresh medium, and the cells were cultured at 37°C until 10 days after the initial detection of reverse transcriptase activity in the supernatant, a total of 27 days.
Flow cytometry.
A total of 0.3 ml of each infected T-cell culture was resuspended in 1% FBS in phosphate-buffered saline (PBS), stained with pooled human anti-HIV IgG (1:100) (AIDS reagent number 3957) as the primary antiserum and FITC-conjugated goat anti-human IgG (Invitrogen) as the secondary antibody (1:100), and analyzed on a FACSCalibur flow cytometer (BD Biosciences).
HIV-1 strand determinations.
Sections of the NL4-3 and BaL LTR U3 region were cloned into pGEM3 (Promega) using primers 5′ GGGCGAATTCAAGACAAGATATCCTTGATCT 3′ with 5′ GCACTCTAGAAGCTTTATTGAGGCTTAAGC 3′ for NL4-3 and 5′ GGGCGAATTCATCTACCACACACAAGGCTA 3′ with 5′ CTCTCTAGAAGTCCCCAGCGGAAAGTCCCT 3′ for BaL. Vectors were cleaved with XbaI and linear fragments purified by agarose gel electrophoresis. A total of 1 µg of each linear vector was subjected to in vitro transcription followed by DNase treatment with the MEGAscript T7 kit (Invitrogen) and purified with the MEGAclear kit (Invitrogen), both according to the manufacturer’s instructions. Fivefold dilution series beginning at 2 × 1011 RNA strands were diluted into 250 ng of uninfected C8166 RNA and subjected to DNase treatment.
A total of 250 ng of cellular RNA was subjected to RQ1 DNase treatment (Promega) according to the manufacturer’s instructions. One-half of each sample was subjected to reverse transcription using 100 ng of random hexamers (Bioline) and the SuperScript III reverse transcription kit (Invitrogen). Samples were diluted 1:4, and 5 µl of each diluted cDNA was amplified using SYBR green master mix on a StepOnePlus real-time PCR system (Applied Biosystems) in triplicate using 5 pmol of each of the following primers: 5′ TACAAGCTAGTACCAGTTGA 3′ and 5′ GCTGTCAAACCTCCACTCTAAC 3′. Ten 5-fold standard dilutions amplified in parallel were used to quantify total HIV-1 U3 RNA strands in each sample. Primary cells and C8166 cells were estimated to contain ~20 pg of total RNA per cell, and TZMbl cells were estimated to have 40 pg RNA per cell, based upon multiple total RNA extractions by TRIzol.
Deep sequencing of total small RNA.
Total RNA was isolated using TRIzol. The RNA fraction containing ~15- to 30-nt-long RNAs was isolated by polyacrylamide gel electrophoresis (PAGE) purification on 15% Tris-borate-EDTA (TBE)-urea gels (Bio-Rad), electroeluted from the excised gel slice (gel eluter; Hoefer), and cloned as previously described (55). Adapter-ligated small RNAs were reverse transcribed using SuperScript III, amplified using GoTaq green PCR master mix (Promega) with the Tru-Seq 3′ indices (Illumina), and sequenced on an Illumina HiSeq 2000.
TZM-bl deep sequencing and RIP-Seq.
RISC-bound miRNAs from uninfected and WT/BaL-infected TZM-bl cells at 72 h postinfection were isolated by immunoprecipitation using a monoclonal antibody specific for Ago1, Ago2, and Ago3 (Abcam: AB 57113), and proteins were removed by digestion with proteinase K. Recovered small RNAs were then isolated using an miRVana kit (Ambion). The immunoprecipitated RNA (RIP-Seq) cDNA library was constructed essentially as described (56) using an Illumina TruSeq small RNA kit, prior to sequencing using an Illumina HiSeq 2000.
PAR-CLIP.
Forty-eight hours postinfection, cells were labeled with 100 µM 4-thiouridine (Sigma) for 16 h. Cells were washed with PBS and cross-linked using a Stratagene UV Stratalinker 2400 at 365 nm. The samples were then processed as previously described (31, 44). RISC cross-linked RNAs were isolated using anti-Ago2 antibody (Abcam AB57113). (This antibody also recognizes Ago1 and Ago3 [data not shown].) Cloning of isolated small RNAs was performed using the TruSeq small RNA cloning kit (Illumina). TZM-bl cells expressing individual HIV-1-specific amiRNAs were infected independently, but the cell lines were pooled into one sample for PAR-CLIP processing. A sample from each culture untreated with 4-thiouridine was harvested in parallel and deep sequenced in order to determine both the miRNA population present in each PAR-CLIP sample and the number of HIV-1 strands per cell.
Data analysis.
Reads of ≥15 nt were collapsed into FASTA format with the FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html) using the following pipeline: fastq_quality_filter -Q33 | fastq_to_fasta -Q33 | fastx_clipper -a TruSeq-Indexnumber -l 15 –c | fastx_collapser. All reads were then subjected to alignment using Bowtie version 0.12.7 with the following options: –a –best –strata -m 25.
Deep sequencing analysis.
Sequences were sequentially filtered and assigned using the following pipeline (displayed as database: additional bow tie alignment parameters): (i) markers (18- and 24-mer radiolabeled oligos, 3′ and 5′ adapters): -v 0 –noRC; (ii) HIV genome: -v 1; (iii) miRbase version 19 Homo sapiens: -v 1 –noRC (57); (iv) Ensembl ncRNAv70 Homo sapiens: -v 1 –noRC (58); (v) fRNAdb version 3.4 Homo sapiens: -v 1 –noRC (59); (vi) human genome 19: -v 2.
miRNA sequences were given a -5p or -3p designation if they aligned to the 5′ or 3′ stem region, respectively, of the miRNA precursor, as annotated in miRbase version 19. miRNAs that represented ≥0.1% of the total cellular miRNA population were considered significant (21). Reads for sequences aligning to two or more entries in a database were distributed equally between each entry, except that reads aligning to both piwi-interacting RNAs (piRNAs) and rRNAs, tRNAs, snRNAs, Y RNAs, or snoRNAs were assigned to the latter RNA species.
PAR-CLIP analysis.
Alignments to a combined human and HIV-1 genome were performed as previously described (31, 45), allowing 3 mismatches. Alignments containing (+) sense T > C and (−) sense A > G conversions were used to generate clusters with PARalyzer version 1.1 using the following parameters: a minimum of 5 reads for a sequence to be included in a cluster, a minimum cluster read depth of 5, at least 1 conversion event in a read for cluster inclusion, and no nonconversion mismatches allowed for each read determining the cluster. Clusters with a read depth of ≥50 and a conversion-event-to-read-depth ratio of ≥0.75 were considered significant. Significant miRNAs (≥0.1% of the total miRNA pool) expressed in each library were determined as described above, from samples untreated with 4-thiouridine, and were then used by PARalyzer to predict which miRNAs were likely to target a given cluster.
Luciferase indicator assays.
A total of 50 ng of each reporter plasmid was cotransfected with 500 ng of a pLCE-based miRNA expression vector (54), via calcium phosphate coprecipitation, into 24-well plates containing 105 293T cells per well, plated the previous day. At 24 h posttransfection, lysates were harvested and luciferase levels determined using a dual luciferase kit (Promega) according to the manufacturer’s instructions. The ratio between the Rluc and internal control Fluc proteins was determined for each sample and compared to the same reporter plasmid cotransfected with pLCE lacking a miRNA precursor.
SUPPLEMENTAL MATERIAL
FACS analysis of uninfected and HIV-1-infected C8166 and CD4+ PBMCs. This figure shows a FACS analysis of the C8166 cells and CD4+ PBMCs analyzed in Fig. 1 to 4. Coincident with RNA harvest, HIV-1-infected cells and control cells grown in parallel were stained with pooled human anti-HIV-1 IgG followed by FITC-conjugated goat anti-human IgG and analyzed by FACS, which revealed a substantial and specific increase in the detected level of fluorescence. Red indicates uninfected cells, blue or green indicates HIV-1-infected cells. The approximate percentage of cells infected with HIV-1 in each sample was calculated based on this analysis and is indicated. Download
Alignment of HIV-1 deep sequencing reads with the viral Gag/Pol frameshift signal. The region of the HIV-1 RNA genome that coincides with the Gag/Pol frameshift signal (32) represents the major source of HIV-1-specific small RNA reads in most infected cell types (Fig. 3). This figure collates all the reads, from all four infected cell types shown in Fig. 3, and aligns them to this region of the HIV-1 genome. At the top is shown the HIV-1 sequence, with the loop of the predicted frameshift stem-loop indicated in red. The predicted structure of this stem-loop is shown at the right. Note that most reads initiate 5′ to the actual stem-loop in a U-rich region that also plays a key role in HIV-1 frameshifting and is known to be a site of ribosome pausing. Download
Alignment of HIV-1 small RNA reads to the HIV-1 TAR RNA element. This figure aligns HIV-1-derived small RNA reads, compiled from all four small RNA libraries described in Fig. 3, and aligns them to the HIV-1 TAR RNA stem-loop that forms the 5′ end of all HIV-1 transcripts. The sequence of the TAR region is shown at the top, with the loop indicated in red, and the predicted RNA structure adopted by TAR is shown at the right. Download
Comparison of HIV-1 small RNA reads obtained from a total small RNA and RISC-associated small RNA sample. This figure compares the pattern of total HIV-1 small RNA reads detected in TZM-bl cells infected with the WT/BaL viral clone (A) to the pattern of small RNAs seen in the RISC-associated small RNA fraction, prepared from the same culture in parallel by immunoprecipitation with an Ago-specific monoclonal antibody (B). Data are presented as described for Fig. 3. Download
Alignment of small RNA reads from the HIV-1-infected TZM-bl cells with the minus strand of the HIV-1 genome. This analysis was performed as described for Fig. 3, except that small RNA reads are aligned to the hypothetical antisense strand complementary to the HIV-1 genome. (A) Uses total small RNA reads, obtained from WT/BaL-infected TZM-bl cells; (B) uses reads obtained by analysis of RISC-associated small RNAs. The major peak shown in panel A is located antisense to the HIV-1 primer binding site and likely represents lysine tRNA breakdown products that are not loaded into RISC. Download
PAR-CLIP reads that contribute to the 1233, 4898, 5001, and 8735 clusters indicated in Fig. 6A. The reads that form each of these clusters are listed, including the number of times each read was recovered, and are aligned to the WT HIV-1 sequence (second row) and the likely responsible miRNA, with the seed sequence shown in blue (first row). T to C conversions, resulting from the PAR-CLIP procedure, are indicated in red. The mutant HIV-1 sequences analyzed in Fig. 6 are given at the bottom of each panel, with the mutated bases underlined. Download
These tables show the total number of sequencing reads obtained from all cellular miRNAs that represent ≥0.1% of the total cellular miRNA pool. The total number of reads shown represents the sum of the reads recovered from uninfected and HIV-1-infected cells. For each miRNA, the percentile representation in the uninfected and infected cell contexts is shown, with miRNAs showing ≥2-fold differences indicated in red. (A) TZM-bl cells; (B) C8166 cells; (C) CD4+ PBMCs. These data are presented graphically in Fig. 5.
ACKNOWLEDGMENTS
The research reported in this paper was supported by NIH grants R01-DA030086 to B.R.C., U19-MH081836, P30-AI073961, and U19-AI096109 to M.S., and R01-AI065310 to C.-H.C. A.W.W. and O.F. received support from NIH training grant T32-CA009111. This publication was made possible with help from the Duke University Center for AIDS Research (CFAR), an NIH-funded program (5P30-AI064518).
The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: C8166 cells (number 404) from Robert Gallo; the HIV-1 BaL isolate (number 510) from Suzanne Gartner, Mikulas Popovic, and Robert Gallo; and human anti-HIV-1 immunoglobulin (number 3957) from NABI and NHLBI (Luiz Barbosa).
Footnotes
CitationWhisnant AW, Bogerd HP, Flores O, Ho P, Powers JG, Sharova N, Stevenson M, Chen C-H, Cullen BR. 2013. In-depth analysis of the interaction of HIV-1 with cellular microRNA biogenesis and effector mechanisms. mBio 4(2):e00193-13. doi:10.1128/mBio.00193-13
REFERENCES
- 1. Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136:215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cullen BR. 2004. Transcription and processing of human microRNA precursors. Mol. Cell 16:861–865 [DOI] [PubMed] [Google Scholar]
- 3. Loeb GB, Khan AA, Canner D, Hiatt JB, Shendure J, Darnell RB, Leslie CS, Rudensky AY. 2012. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol. Cell 48:760–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Didiano D, Hobert O. 2006. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat. Struct. Mol. Biol. 13:849–851 [DOI] [PubMed] [Google Scholar]
- 5. Tan X, Lu ZJ, Gao G, Xu Q, Hu L, Fellmann C, Li MZ, Qu H, Lowe SW, Hannon GJ, Elledge SJ. 2012. Tiling genomes of pathogenic viruses identifies potent antiviral shRNAs and reveals a role for secondary structure in shRNA efficacy. Proc. Natl. Acad. Sci. U. S. A. 109:869–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Grundhoff A, Sullivan CS. 2011. Virus-encoded microRNAs. Virology 411:325–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Skalsky RL, Cullen BR. 2010. Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol. 64:123–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kincaid RP, Burke JM, Sullivan CS. 2012. RNA virus microRNA that mimics a B-cell oncomir. Proc. Natl. Acad. Sci. U. S. A. 109:3077–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zeng Y, Yi R, Cullen BR. 2005. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 24:138–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. 2006. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 125:887–901 [DOI] [PubMed] [Google Scholar]
- 11. Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grässer FA, van Dyk LF, Ho CK, Shuman S, Chien M, Russo JJ, Ju J, Randall G, Lindenbach BD, Rice CM, Simon V, Ho DD, Zavolan M, Tuschl T. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2:269–276 [DOI] [PubMed] [Google Scholar]
- 12. Lin J, Cullen BR. 2007. Analysis of the interaction of primate retroviruses with the human RNA interference machinery. J. Virol. 81:12218–12226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Omoto S, Ito M, Tsutsumi Y, Ichikawa Y, Okuyama H, Brisibe EA, Saksena NK, Fujii YR. 2004. HIV-1 nef suppression by virally encoded microRNA. Retrovirology 1:44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Omoto S, Fujii YR. 2005. Regulation of human immunodeficiency virus 1 transcription by nef microRNA. J. Gen. Virol. 86:751–755 [DOI] [PubMed] [Google Scholar]
- 15. Klase Z, Kale P, Winograd R, Gupta MV, Heydarian M, Berro R, McCaffrey T, Kashanchi F. 2007. HIV-1 TAR element is processed by dicer to yield a viral micro-RNA involved in chromatin remodeling of the viral LTR. BMC Mol. Biol. 8:63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Klase Z, Winograd R, Davis J, Carpio L, Hildreth R, Heydarian M, Fu S, McCaffrey T, Meiri E, Ayash-Rashkovsky M, Gilad S, Bentwich Z, Kashanchi F. 2009. HIV-1 TAR miRNA protects against apoptosis by altering cellular gene expression. Retrovirology 6:18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ouellet DL, Plante I, Landry P, Barat C, Janelle ME, Flamand L, Tremblay MJ, Provost P. 2008. Identification of functional microRNAs released through asymmetrical processing of HIV-1 TAR element. Nucleic Acids Res. 36:2353–2365 DOI: 10.1093/nar/gkn076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yeung ML, Bennasser Y, Watashi K, Le SY, Houzet L, Jeang KT. 2009. Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res. 37:6575–6586 DOI: 10.1093/nar/gkp707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schopman NC, Willemsen M, Liu YP, Bradley T, van Kampen A, Baas F, Berkhout B, Haasnoot J. 2012. Deep sequencing of virus-infected cells reveals HIV-encoded small RNAs. Nucleic Acids Res. 40:414–427 DOI: 10.1093/nar/gkr719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Neilson JR, Zheng GX, Burge CB, Sharp PA. 2007. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev. 21:578–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mullokandov G, Baccarini A, Ruzo A, Jayaprakash AD, Tung N, Israelow B, Evans MJ, Sachidanandam R, Brown BD. 2012. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat. Methods 9:840–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Triboulet R, Mari B, Lin YL, Chable-Bessia C, Bennasser Y, Lebrigand K, Cardinaud B, Maurin T, Barbry P, Baillat V, Reynes J, Corbeau P, Jeang KT, Benkirane M. 2007. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science 315:1579–1582 [DOI] [PubMed] [Google Scholar]
- 23. Huang J, Wang F, Argyris E, Chen K, Liang Z, Tian H, Huang W, Squires K, Verlinghieri G, Zhang H. 2007. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat. Med. 13:1241–1247 [DOI] [PubMed] [Google Scholar]
- 24. Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hwang SS, Boyle TJ, Lyerly HK, Cullen BR. 1991. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science 253:71–74 [DOI] [PubMed] [Google Scholar]
- 26. Salahuddin SZ, Markham PD, Wong-Staal F, Franchini G, Kalyanaraman VS, Gallo RC. 1983. Restricted expression of human T-cell leukemia—lymphoma virus (HTLV) in transformed human umbilical cord blood lymphocytes. Virology 129:51–64 [DOI] [PubMed] [Google Scholar]
- 27. Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gartner S, Markovits P, Markovitz DM, Kaplan MH, Gallo RC, Popovic M. 1986. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science 233:215–219 [DOI] [PubMed] [Google Scholar]
- 29. Westervelt P, Gendelman HE, Ratner L. 1991. Identification of a determinant within the human immunodeficiency virus 1 surface envelope glycoprotein critical for productive infection of primary monocytes. Proc. Natl. Acad. Sci. U. S. A. 88:3097–3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sharova N, Wu Y, Zhu X, Stranska R, Kaushik R, Sharkey M, Stevenson M. 2008. Primate lentiviral Vpx commandeers DDB1 to counteract a macrophage restriction. PLoS Pathog. 4:e1000057 http://dx.doi.org/10.1371/journal.ppat.1000057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gottwein E, Corcoran DL, Mukherjee N, Skalsky RL, Hafner M, Nusbaum JD, Shamulailatpam P, Love CL, Dave SS, Tuschl T, Ohler U, Cullen BR. 2011. Viral microRNA targetome of KSHV-infected primary effusion lymphoma cell lines. Cell Host Microbe 10:515–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Giedroc DP, Cornish PV. 2009. Frameshifting RNA pseudoknots: structure and mechanism. Virus Res. 139:193–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zeng Y, Cullen BR. 2005. Efficient processing of primary microRNA hairpins by Drosha requires flanking nonstructured RNA sequences. J. Biol. Chem. 280:27595–27603 [DOI] [PubMed] [Google Scholar]
- 34. Linnstaedt SD, Gottwein E, Skalsky RL, Luftig MA, Cullen BR. 2010. Virally induced cellular microRNA miR-155 plays a key role in B-cell immortalization by Epstein-Barr virus. J. Virol. 84:11670–11678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Libri V, Helwak A, Miesen P, Santhakumar D, Borger JG, Kudla G, Grey F, Tollervey D, Buck AH. 2012. Murine cytomegalovirus encodes a miR-27 inhibitor disguised as a target. Proc. Natl. Acad. Sci. U. S. A. 109:279–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cazalla D, Yario T, Steitz JA, Steitz J. 2010. Down-regulation of a host microRNA by a herpesvirus Saimiri noncoding RNA. Science 328:1563–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309:1577–1581 [DOI] [PubMed] [Google Scholar]
- 38. Ho BC, Yu SL, Chen JJ, Chang SY, Yan BS, Hong QS, Singh S, Kao CL, Chen HY, Su KY, Li KC, Cheng CL, Cheng HW, Lee JY, Lee CN, Yang PC. 2011. Enterovirus-induced miR-141 contributes to shutoff of host protein translation by targeting the translation initiation factor eIF4E. Cell Host Microbe 9:58–69 [DOI] [PubMed] [Google Scholar]
- 39. Marcinowski L, Tanguy M, Krmpotic A, Rädle B, Lisnic VJ, Tuddenham L, Chane-Woon-Ming B, Ruzsics Z, Erhard F, Benkartek C, Babic M, Zimmer R, Trgovcich J, Koszinowski UH, Jonjic S, Pfeffer S, Dölken L. 2012. Degradation of cellular mir-27 by a novel, highly abundant viral transcript is important for efficient virus replication in vivo. PLoS Pathog. 8:e1002510 http://dx.doi.org/10.1371/journal.ppat.1002510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Melar-New M, Laimins LA. 2010. Human papillomaviruses modulate expression of microRNA 203 upon epithelial differentiation to control levels of p63 proteins. J. Virol. 84:5212–5221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Simon V, Ho DD. 2003. HIV-1 dynamics in vivo: implications for therapy. Nat. Rev. Microbiol. 1:181–190 [DOI] [PubMed] [Google Scholar]
- 42. Backes S, Shapiro JS, Sabin LR, Pham AM, Reyes I, Moss B, Cherry S, tenOever BR. 2012. Degradation of host microRNAs by poxvirus poly(A) polymerase reveals terminal RNA methylation as a protective antiviral mechanism. Cell Host Microbe 12:200–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sanghvi VR, Steel LF. 2011. A re-examination of global suppression of RNA interference by HIV-1. PLoS One 6:e17246 http://dx.doi.org/10.1371/journal.pone.0017246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141:129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Corcoran DL, Georgiev S, Mukherjee N, Gottwein E, Skalsky RL, Keene JD, Ohler U. 2011. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol. 12:R79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Islam S, Kjällquist U, Moliner A, Zajac P, Fan JB, Lönnerberg P, Linnarsson S. 2011. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 21:1160–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nathans R, Chu CY, Serquina AK, Lu CC, Cao H, Rana TM. 2009. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol. Cell 34:696–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ahluwalia JK, Khan SZ, Soni K, Rawat P, Gupta A, Hariharan M, Scaria V, Lalwani M, Pillai B, Mitra D, Brahmachari SK. 2008. Human cellular microRNA hsa-miR-29a interferes with viral nef protein expression and HIV-1 replication. Retrovirology 5:117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sun G, Li H, Wu X, Covarrubias M, Scherer L, Meinking K, Luk B, Chomchan P, Alluin J, Gombart AF, Rossi JJ. 2012. Interplay between HIV-1 infection and host microRNAs. Nucleic Acids Res. 40:2181–2196 DOI: 10.1093/nar/gkr961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bennasser Y, Le SY, Benkirane M, Jeang KT. 2005. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity 22:607–619 [DOI] [PubMed] [Google Scholar]
- 51. Chang ST, Thomas MJ, Sova P, Green RR, Palermo RE, Katze MG. 2013. Next-generation sequencing of small RNAs from HIV-infected cells identifies phased microRNA expression patterns and candidate novel microRNAs differentially expressed upon infection. mBio 4(1):e00549-00512 http://dx.doi.org/10.1128/mBio.00549-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang X, Ye L, Hou W, Zhou Y, Wang YJ, Metzger DS, Ho WZ. 2009. Cellular microRNA expression correlates with susceptibility of monocytes/macrophages to HIV-1 infection. Blood 113:671–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Jr, Swanstrom R, Burch CL, Weeks KM. 2009. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460:711–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gottwein E, Cullen BR. 2010. A human herpesvirus microRNA inhibits p21 expression and attenuates p21-mediated cell cycle arrest. J. Virol. 84:5229–5237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Umbach JL, Cullen BR. 2010. In-depth analysis of Kaposi’s sarcoma-associated herpesvirus microRNA expression provides insights into the mammalian microRNA-processing machinery. J. Virol. 84:695–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Keene JD, Komisarow JM, Friedersdorf MB. 2006. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat. Protoc. 1:302–307 [DOI] [PubMed] [Google Scholar]
- 57. Kozomara A, Griffiths-Jones S. 2011. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39:D152–D157 DOI: 10.1093/nar/gkr817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, Gil L, Gordon L, Hendrix M, Hourlier T, Johnson N, Kähäri AK, Keefe D, Keenan S, Kinsella R, Komorowska M, Koscielny G, Kulesha E, Larsson P, Longden I, McLaren W, Muffato M, Overduin B, Pignatelli M, Pritchard B, Riat HS, Ritchie GR, Ruffier M, Schuster M, Sobral D, Tang YA, Taylor K, Trevanion S, Vandrovcova J, White S, Wilson M, Wilder SP, Aken BL, Birney E, Cunningham F, Dunham I, Durbin R, Fernandez-Suarez XM, Harrow J, Herrero J, Hubbard TJ, Parker A, Proctor G, Spudich G, Vogel J, Yates A, Zadissa A, Searle SM. 2012. Ensembl 2012. Nucleic Acids Res. 40:D84–D90 DOI: 10.1093/nar/gks210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mituyama T, Yamada K, Hattori E, Okida H, Ono Y, Terai G, Yoshizawa A, Komori T, Asai K. 2009. The functional RNA. Database 30: databases to support mining and annotation of functional RNAs. Nucleic Acids Res. 37:D89-D92 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FACS analysis of uninfected and HIV-1-infected C8166 and CD4+ PBMCs. This figure shows a FACS analysis of the C8166 cells and CD4+ PBMCs analyzed in Fig. 1 to 4. Coincident with RNA harvest, HIV-1-infected cells and control cells grown in parallel were stained with pooled human anti-HIV-1 IgG followed by FITC-conjugated goat anti-human IgG and analyzed by FACS, which revealed a substantial and specific increase in the detected level of fluorescence. Red indicates uninfected cells, blue or green indicates HIV-1-infected cells. The approximate percentage of cells infected with HIV-1 in each sample was calculated based on this analysis and is indicated. Download
Alignment of HIV-1 deep sequencing reads with the viral Gag/Pol frameshift signal. The region of the HIV-1 RNA genome that coincides with the Gag/Pol frameshift signal (32) represents the major source of HIV-1-specific small RNA reads in most infected cell types (Fig. 3). This figure collates all the reads, from all four infected cell types shown in Fig. 3, and aligns them to this region of the HIV-1 genome. At the top is shown the HIV-1 sequence, with the loop of the predicted frameshift stem-loop indicated in red. The predicted structure of this stem-loop is shown at the right. Note that most reads initiate 5′ to the actual stem-loop in a U-rich region that also plays a key role in HIV-1 frameshifting and is known to be a site of ribosome pausing. Download
Alignment of HIV-1 small RNA reads to the HIV-1 TAR RNA element. This figure aligns HIV-1-derived small RNA reads, compiled from all four small RNA libraries described in Fig. 3, and aligns them to the HIV-1 TAR RNA stem-loop that forms the 5′ end of all HIV-1 transcripts. The sequence of the TAR region is shown at the top, with the loop indicated in red, and the predicted RNA structure adopted by TAR is shown at the right. Download
Comparison of HIV-1 small RNA reads obtained from a total small RNA and RISC-associated small RNA sample. This figure compares the pattern of total HIV-1 small RNA reads detected in TZM-bl cells infected with the WT/BaL viral clone (A) to the pattern of small RNAs seen in the RISC-associated small RNA fraction, prepared from the same culture in parallel by immunoprecipitation with an Ago-specific monoclonal antibody (B). Data are presented as described for Fig. 3. Download
Alignment of small RNA reads from the HIV-1-infected TZM-bl cells with the minus strand of the HIV-1 genome. This analysis was performed as described for Fig. 3, except that small RNA reads are aligned to the hypothetical antisense strand complementary to the HIV-1 genome. (A) Uses total small RNA reads, obtained from WT/BaL-infected TZM-bl cells; (B) uses reads obtained by analysis of RISC-associated small RNAs. The major peak shown in panel A is located antisense to the HIV-1 primer binding site and likely represents lysine tRNA breakdown products that are not loaded into RISC. Download
PAR-CLIP reads that contribute to the 1233, 4898, 5001, and 8735 clusters indicated in Fig. 6A. The reads that form each of these clusters are listed, including the number of times each read was recovered, and are aligned to the WT HIV-1 sequence (second row) and the likely responsible miRNA, with the seed sequence shown in blue (first row). T to C conversions, resulting from the PAR-CLIP procedure, are indicated in red. The mutant HIV-1 sequences analyzed in Fig. 6 are given at the bottom of each panel, with the mutated bases underlined. Download
These tables show the total number of sequencing reads obtained from all cellular miRNAs that represent ≥0.1% of the total cellular miRNA pool. The total number of reads shown represents the sum of the reads recovered from uninfected and HIV-1-infected cells. For each miRNA, the percentile representation in the uninfected and infected cell contexts is shown, with miRNAs showing ≥2-fold differences indicated in red. (A) TZM-bl cells; (B) C8166 cells; (C) CD4+ PBMCs. These data are presented graphically in Fig. 5.