The floral transition is a key decision in the plant life cycle and under complex regulation. Using modeling and experimental approaches, this work determines a core network of hub activities that accounts for many of the properties of the floral switch. This network exhibits switching behavior and suggests the balance of FT and TFL1 is critical for controlling plant architecture.
Abstract
During flowering, primordia on the flanks of the shoot apical meristem are specified to form flowers instead of leaves. Like many plants, Arabidopsis thaliana integrates environmental and endogenous signals to control the timing of reproduction. To study the underlying regulatory logic of the floral transition, we used a combination of modeling and experiments to define a core gene regulatory network. We show that FLOWERING LOCUS T (FT) and TERMINAL FLOWER1 (TFL1) act through FD and FD PARALOG to regulate the transition. The major floral meristem identity gene LEAFY (LFY) directly activates FD, creating a positive feedback loop. This network predicts flowering behavior for different genotypes and displays key properties of the floral transition, such as signal integration and irreversibility. Furthermore, modeling suggests that the control of TFL1 is important to flexibly counterbalance incoming FT signals, allowing a pool of undifferentiated cells to be maintained despite strong differentiation signals in nearby cells. This regulatory system requires TFL1 expression to rise in proportion to the strength of the floral inductive signal. In this network, low initial levels of LFY or TFL1 expression are sufficient to tip the system into either a stable flowering or vegetative state upon floral induction.
INTRODUCTION
Flowering plants are the most diverse and successful group of land plants. The striking diversity of form demonstrated by the angiosperms is evidence for the remarkable flexibility of the fundamental unit of plant development, the phytomer (Leyser, 2003; Kalisz and Kramer, 2008). The appearance of flowers was a crucial event in plant evolution, enabling remarkable adaptation to almost every habitat on Earth. Central to this adaptability has been the adoption of a remarkably wide variety of reproductive strategies and concomitant plant architectures, with life cycles ranging from weeks to hundreds of years. This diversity is dependent on variation in the timing and programming of the transition from vegetative to reproductive growth.
Despite this variety of reproductive strategies, key regulators of the floral transition are widely conserved. In particular, the floral pathway integrator gene FLOWERING LOCUS T (FT) plays a key role in higher plants. FT protein has been shown to be a mobile signal transported from the leaves to the shoot apex where it triggers flowering in diverse species, including tomato (Solanum lycopersicum), rice (Oryza sativa), and Arabidopsis thaliana (Lifschitz and Eshed, 2006; Corbesier et al., 2007; Jaeger and Wigge, 2007; Mathieu et al., 2007; Tamaki et al., 2007). Genetic and modeling studies in the last two decades have converged on a pathway whereby environmental and endogenous signals inductive to flowering lead to increased levels of expression of FT (Kardailsky et al., 1999; Kobayashi et al., 1999; Simpson and Dean, 2002; Salazar et al., 2009; Kumar et al., 2012). FT forms a complex with the bZIP transcription factor FD and a 14-3-3 protein, triggering flowering through the activation of key floral meristem identity genes, such as APETALA1 (AP1) at the shoot apex (Abe et al., 2005; Wigge et al., 2005; Taoka et al., 2011). FD is important for FT signaling, since fd-2 can partially suppress the early flowering phenotypes of FT overexpressing plants (Abe et al., 2005; Wigge et al., 2005). Additionally, the transcription factor LEAFY (LFY) plays a key role in the integration of flowering signals in parallel with FT to activate floral meristem identity genes (Kardailsky et al., 1999; Abe et al., 2005). FT is able to activate LFY expression through the transcription factor SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) (Yoo et al., 2005; Lee et al., 2008). As a result, rising levels of FT expression induce LFY and AP1 in a feedforward circuit. Furthermore, AP1 and LFY are mutual transcriptional activators (Liljegren et al., 1999). As well as floral activators, repressors play important roles. A key floral repressor is the FT-related gene TERMINAL FLOWER1 (TFL1), which maintains the center of the shoot apical meristem (SAM) in a vegetative state. TFL1 acts by repressing LFY and AP1 (Ratcliffe et al., 1998, 1999). AP1 and TFL1 expression is antagonistic, as AP1 represses TFL1, and this is likely to be direct as AP1 directly binds TFL1 regulatory elements (Kaufmann et al., 2010). In the absence of TFL1 activity, all meristem tissues are converted into flowers, causing the architecture of the plant to be converted from an indeterminate to a determinate growth (Shannon and Meeks-Wagner, 1991).
While these major regulators of the floral transition and their interactions have been determined in Arabidopsis and other plant species, the dynamic properties of the floral transition, for example, robustness to varying incoming signals, irreversibility and precise spatial control are less well understood. Complementing genetic studies, valuable insights into the control of the plant life cycles have been obtained through computational modeling. This includes approaches that simulate the network of known regulators of the transition (Welch et al., 2003) as well as mechanistic models describing the detailed molecular interactions controlling the photoperiod pathway (Salazar et al., 2009) and how differences in plant architecture may arise (Prusinkiewicz et al., 2007). Boolean approaches have been employed successfully to capture gene networks that regulate floral organ specification (Mendoza et al., 1999; Espinosa-Soto et al., 2004; Alvarez-Buylla et al., 2008a, 2008b). An approach that distinguishes between direct and indirect genetic interactions, termed molecular regulatory network, has recently been employed to model sepal primordium polarity (La Rota et al., 2011). The application of computational modeling approaches, for example, to determine crop scheduling and to understand the phenology of wild plants, is indicative of their value (Hammer et al., 2006; Wilczek et al., 2009).
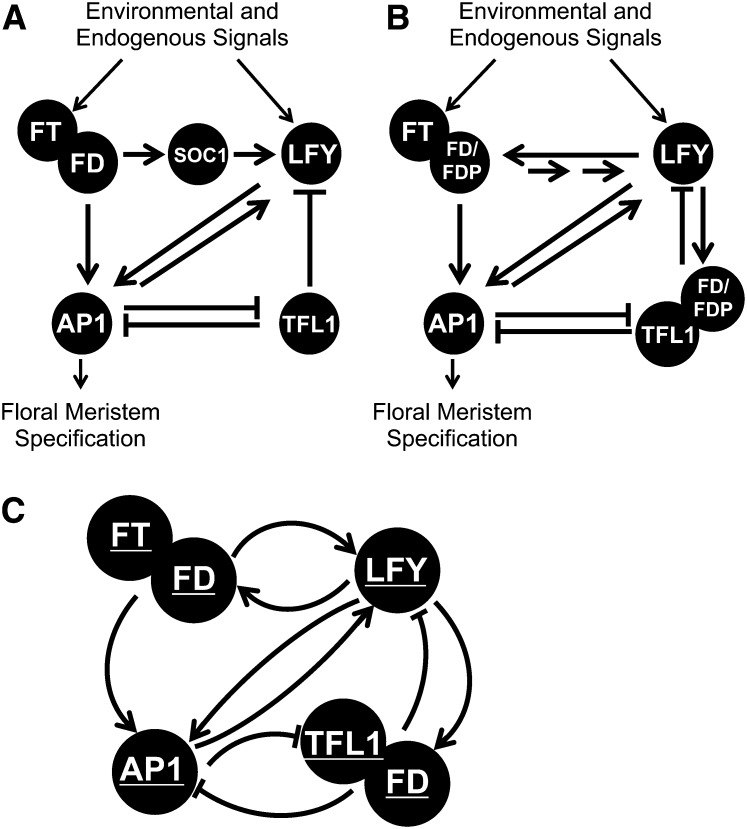
Using an iterative cycle of modeling and experiments, we sought to determine a network that can capture the dynamics of the floral transition. Since many hundreds of genes affect flowering time, our aim was to simplify the network by describing only the major activities of groups of genes, referred to here as regulatory hubs that represent one or more genes and proteins. The known components of the floral transition are well established, and these are summarized in Figure 1A. Hub activities, denoted by underlining, are named after the major known gene activities that contribute to their function (Figure 1C). The activity of each hub is likely to reflect the behavior of many genes in planta. We focus on the decision-making cells on the flanks of the apex. We use the AP1 hub to represent the key integrators that determine whether to flower, or not, and these signals are read out giving rise to three different states: rosette leaves, cauline leaves, or flowers.
Figure 1.
Major Regulatory Networks Governing the Floral Transition.
(A) Environmental and endogenous floral promoting signals stimulate increasing expression of the floral pathway integrator genes FT and LFY. Rising levels of FT and LFY proteins stimulate the expression of floral meristem identity genes, such as AP1, leading to the specification of flowers.
(B) Regulatory interactions in the core network, including activities verified experimentally in this work. FDP acts redundantly with FD to activate flowering. LFY directly binds the FD promoter, accounting for the increase in FD expression upon the floral transition. The TFL1 floral repressive signal is mediated by FD.
(C) A network of five regulatory hubs captures the major properties of the floral transition. Hubs are denoted by underlining, and reflect the likely activity of multiple genes in the plant. For example, the FT hub encompasses the activity of at least FT and TSF, the FD hub FD and FDP, while AP1 includes the biological activity of AP1, CAULIFLOWER, and FRUITFULL.
Network models can be extended as required by the addition of new hubs and interactions: for example, by adding hubs for key regulators, such as SOC1 (Liu et al., 2008), CO (Samach et al., 2000), and regulatory microRNAs (Wang et al., 2009) as well as other major floral repressor activities, such as SCHLAFMUTZE (Mathieu et al., 2009), to capture further important aspects of the transition. Presently, the activities of these regulators are integrated into the activities of the major hubs, but depending on the question being asked, the model can be suitably expanded. The modeling has enabled us to evaluate likely core regulatory networks suggested by the available genetic data and to account for previously described phenotypes.
RESULTS
FD and FDP Comprise the Major Hub Mediating FT Signaling
Since FT is a major conserved integrator of flowering in higher plants, we focused our modeling to understand how events downstream of FT expression control the flowering transition. While FD is important for FT signaling, fd-2 only partially suppresses 35S:FT (Abe et al., 2005; Wigge et al., 2005). This indicates that factors in addition to FD are required for FT signaling. We therefore investigated a closely related gene, FD PARALOG (FDP). Consistent with its role in flowering, FDP is expressed on the flanks of the shoot apex (Figures 2A and 2B). Significantly, a mutation in the FDP DNA binding domain, fdp-1, causes a slightly later flowering phenotype and enhances the late flowering phenotype of fd-2 (Figure 3A, Table 1). Furthermore, fd-2 fdp-1 double mutants largely suppress FT overexpression (Figure 3B, Table 1), confirming that most FT signaling is mediated by FD/FDP.
Figure 2.
Gene Expression Patterns in the Wild Type the pny pnf Background.
Analysis of the mRNA expression of marker genes in Arabidopsis apices under inductive and noninductive conditions by in situ hybridization. Asterisks denote the SAM.
(A) FDP expression in a wild-type vegetative apex under noninductive conditions. Bar = 50 µm.
(B) FDP expression in a wild-type apex undergoing floral transition.
(C) FDP in pny pnf under inductive conditions.
(D) FD in a wild-type vegetative apex, noninductive conditions.
(E) FD in wild-type apex transitioning, note the increased expression compared with (D).
(F) FD in pny pnf is not upregulated under inductive conditions.
(G) LFY expression in a wild-type vegetative apex under noninductive conditions.
(H) LFY expression is strongly induced during the floral transition.
(I) LFY in pny pnf is not upregulated under inductive conditions.
(J) TFL1 in wild-type plants under noninductive short-day conditions is absent from the apex but note strong expression in the axillary meristem (arrow).
(K) TFL1 is strongly upregulated in the center of the apex in the wild type under inductive conditions.
(L) TFL1 is strongly expressed outside of its normal zone of expression in pny pnf; note the strong expression in the vasculature.
Figure 3.
Genetic Interactions Reveal That FD and FDP Are Necessary for FT to Accelerate Flowering.
According to convention, developmental time is measured by the number of rosette and cauline leaves produced on the main shoot prior to flowering.
(A) A point mutation, fdp-1, was obtained in the DNA binding domain of FDP. fdp-1 (right) enhances the late flowering phenotype of fd-2 (center) such that fdp-1 fd-2 double mutants (left) are even later flowering than fd-2. Plants were grown 39 d.
(B) 35S:FT (right) is suppressed in the fd-2 fdp-1 background (left), indicating that most FT signaling is mediated by FD and FDP, which are partially redundant with each other. Plants were grown 37 d.
(C) The phenotype of tfl1-1 (left) is largely suppressed by fd-2 (tfl1-1 fd-2, center left) compared with the wild type (center right) and fd-2 (right), showing that FD is required for the phenotypic effects of tfl1-1 to be observed. Plants were grown 37 d. Col-0, Columbia-0.
(D) 35S:TFL1 (left) is largely suppressed by fd-2 (right), showing that FD mediates TFL1 signaling. Plants were grown 42 d.
Table 1. Experimental and Model Leaf Number Data.
| Genotype | No. of Plants | Rosette Leaves |
Cauline Leaves |
Model Set | ||
|---|---|---|---|---|---|---|
| Exp. | Model | Exp. | Model | |||
| Wild type (Columbia-0) | 12 | 7.9 | 9.0 | 1.4 | 2.0 | Training |
| 35S:FT | 10 | 4.4 | 3.3 | 1.0 | 1.5 | Training |
| 35S:LFY | 11 | 3.8 | 4.0 | 1.8 | 1.6 | Training |
| 35S:TFL1 | 12 | 27.5 | 27.4 | 15.7 | 15.7 | Training |
| lfy-12 | 9 | 13.0 | 13.5 | 5.3 | 6.4 | Training |
| ft-10 | 10 | 36.4 | 36.4 | 9.3 | 9.0 | Training |
| tfl1-1 | 11 | 7.4 | 7.4 | 0.4 | 1.8 | Training |
| fd-2 | 12 | 18.5 | 18.0 | 5.5 | 4.6 | Training |
| fdp-1 | 10 | 11.2 | 9.7 | 2.0 | 2.2 | Training |
| fd-2 fdp-1 | 10 | 32.9 | 32.8 | 6.3 | 8.1 | Training |
| 35S:TFL1 fd-2 | 12 | 23.8 | 24.4 | 8.2 | 5.4 | Training |
| tfl1-1 fd-2 | 12 | 14.4 | 14.6 | 4.6 | 4.1 | Training |
| 35S:FT fd-2 | 12 | 8.3 | 7.7 | 2.4 | 3.5 | Training |
| tfl1-1 fd-2 fdp-1 | 12 | 24.8 | 30.9 | 6.7 | 7.9 | Prediction |
| 35S:TFL1 fd-2 fdp-1 | 10 | 31.3 | 34.5 | 11.0 | 8.3 | Prediction |
| 35S:FT fd-2 fdp-1 | 12 | 26.0 | 28.6 | 6.0 | 7.9 | Prediction |
For each genotype, the table lists the number of plants, experimental leaf number data, and estimated values for rosette and cauline leaves. The estimated model values were obtained by parameter fitting to the wild type and all single and double mutant data (training set). The triple mutant data are predictions using the fitted parameters.
Floral Activation and Repression Act through Common Mechanisms
The ability of FT to activate LFY and AP1 via FD and FDP represents a regulated feedforward loop, a common motif in circuits involving irreversible transitions (Mangan and Alon, 2003), such as flowering. However, this network alone is unable to account for the regulation of floral signaling, since once activated, it will rapidly convert all competent tissues into a floral fate (determinate growth). By contrast, the SAM of Arabidopsis shows indeterminate growth, which is dependent on the floral repressor TFL1, since tfl1-1 arrests with terminal flowers (Shannon and Meeks-Wagner, 1991) (Figure 3C). Interestingly, the phenotype of tfl1-1 is suppressed in the fd-2 background (Figure 3C) and almost completely in tfl1-1 fd-2 fdp-1 triple mutants (see Supplemental Figure 1A online), indicating that the major role of TFL1 signaling is to counter FT signaling. Consistent with this, tfl1-1 is suppressed under short photoperiods (Shannon and Meeks-Wagner, 1991). As TFL1 is closely related to FT, it has been proposed that it also signals through FD (Ahn et al., 2006). We tested this in 35S:TFL1, which is extremely late flowering and forms lfy-like inflorescences through the repression of LFY and AP1 (Ratcliffe et al., 1998, 1999). Interestingly, 35S:TFL1 can be significantly suppressed in the fd-2 background, indicating that TFL1 signaling is mediated through FD (Figure 3D). These conclusions are supported by the recent demonstration of the ability of TFL1 to act as a transcriptional repressor (Hanano and Goto, 2011). Taken together, these results suggest the simplified network structure shown in Figure 1B, where TFL1 FD counterbalances the floral promoting activity of FT FD.
Feedback in the Floral Transition
FD expression is strongly upregulated during the floral transition (Wigge et al., 2005). Within our regulatory network, two architectures can account for this: FD autoactivating its own transcription or feedback from LFY onto FD. We therefore evaluated these possible networks by fitting to the available leaf number data (see below). We found that both these networks are able to account for the flowering data and therefore sought experimental data to distinguish between these possibilities.
To examine the regulation of FD expression, we created an FD promoter reporter line, FD:GUS (for β-glucuronidase) for in planta studies (Figure 4A). Consistently, we were able to observe strong upregulation of FD expression during floral transition in the SAM (Figure 4B). Genetic dissection of this promoter revealed two regions, containing conserved LEAFY binding sites (LBSs) (Busch et al., 1999), to be essential for FD upregulation (Figures 4A and 4B). To confirm if LFY acts in the upregulation of FD, we used a sensitive reporter line for FD promoter activity, FD:FT. Perturbations in FD promoter upregulation cause a reduction in FT expression and later flowering. In this system, lfy-12 significantly suppresses FD:FT, showing that LFY activates FD (see Supplemental Figure 1B online), consistent with the fact that FT and LFY act in parallel genetic pathways and that 35S:LFY is able to potentiate 35S:FT (Kardailsky et al., 1999; Kobayashi et al., 1999). To determine if LFY activates FD directly, we used chromatin immunoprecipitation (ChIP). This shows robust binding of 3XFLAG:LFY to the promoter of FD at both the predicted LBSs, comparable to LFY binding to the LBS in the AP1 promoter (Figure 4C). Supporting these studies, LFY genome-wide ChIP studies have shown that LFY binds both FD and FDP (Winter et al., 2011).
Figure 4.
Positive Feedback in the Flowering Pathway.
(A) Analysis of the FD promoter by fusion to the GUS reporter gene and examination of GUS enzyme activity reveals two regions necessary for FD upregulation during the floral transition (+/− denotes promoter upregulation as measured by GUS staining). Each of these regions, shown in gray in the context of the FD promoter sequence, at top, contains an LBS. Promoter constructs, with deletions indicated, are shown below.
(B) Histochemical staining for the GUS reporter gene in Arabidopsis apices under inductive and non-inductive conditions. shows that deletion of the LBS in the FD promoter abolishes FD up-regulation upon the floral transition.
(C) LFY binds directly to the LBS in the FD promoter in vivo as measured by ChIP. 35S:3XFLAG:LFY plants were harvested after 14 d and cross-linked. Enrichment for LBSI and LBSII in the FD promoter was assayed by quantitative PCR in FLAG:LFY as well as control plants (Columbia-0 [Col-0]). The known binding of LFY to LBSI and II in the AP1 promoter was used as a positive control. Error bars are sd for two biological experiments each with three technical replicates.
AP1 has been shown to bind to CaRG boxes in the 3′ region of TFL1 (Kaufmann et al., 2010), and this result is consistent with the observation that constitutive AP1 expression represses TFL1 (Liljegren et al., 1999). We therefore included an inhibitory connection between AP1 and TFL1 in our regulatory network.
Rosette and Cauline Leaf Numbers Can Be Used to Scale the Network
A reliable indicator of developmental time required for initiation of the floral transition is the number of rosette and cauline leaves made on the main stem prior to flowers. In our model, the AP1 hub is the output of the floral induction pathways, and rising levels of AP1 correlate with progress through the floral transition. To directly relate the simulated AP1 curves to key events of the floral transition, we defined two thresholds (Figure 5A). Upon initiation of the floral transition, lateral organs are specified as cauline leaves rather than rosette leaves. Once the transition is complete, flowers are made. The developmental time to reach these states can thus be scaled to the leaf number data. An advantage of this approach is that it provides information on timing of both the initiation of flowering as well as the duration of the transition.
Figure 5.
The Floral Network Model Captures Key Properties of the Floral Transition in Arabidopsis.
(A) The definition of developmental decisions via AP1 hub levels. The AP1 hub determines the output of the floral induction pathway. The introduced thresholds correspond to developmental changes that we map onto and characterize by leaf numbers and allows for leaf number data to be used to fit model parameters. a.u., arbitrary units.
(B) The modeled floral transition is characterized by rising AP1. The initiation of flowering occurs when AP1 crosses the 0.2 threshold, resulting in a switch from rosette to cauline leaf production. Flowering is completed when AP1 exceeds 0.3. The curves show the effect withdrawal of FT production after different lengths of developmental time. When FT production is stopped after the developmental time of 10 leaves (shown in dark blue), flowering is identical to the wild type (WT) where FT is maintained (magenta). Halting FT production after the formation of five leaves (green) in simulations causes a delayed time to flower. Flowering is still accelerated when FT production is withdrawn after the formation of only two leaves (red) compared with a simulated complete lack of FT (ft-10; cyan).
(C) The network is able to integrate simulated circadian oscillations in FT. Strongly modulated FT levels are given as input, but the resulting AP1 from the network is only marginally perturbed.
(D) The network is able to filter simulated noise in FT. Uniform random noise of up to 200% FT was given as input, but the network integrates this out, resulting in an almost smooth AP1 curve.
Genetic networks were represented by ordinary differential equations using Hill-type gene activation and repression, assuming that protein binding is in equilibrium on the time scales of translation (Table 2). With no further constraints, concentrations and binding constants are not independent, so we chose to vary only the binding constants and Hill coefficients in the parameter fitting. Production and degradation rates for the AP1 hub were chosen such that the maximal concentration is unity (AP1max = 1). The two thresholds were chosen to be AP1 = 0.2 for the transition from rosette to cauline and set at AP1 = 0.3 for the change from cauline to flower production. The decision outcome takes on one of these three states at those defined AP1 levels. These threshold values can be chosen arbitrarily as the parameters in the system scaled relatively and will adapt to these settings.
Table 2. Kinetic Model of the Floral Transition.
| Model Equations and Parameters |
| Hub Protein Concentrations |
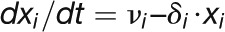 |
| Hub Protein-Protein Binding |
 |
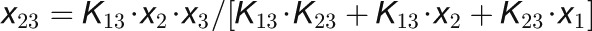 |
| Hub Gene Activation |
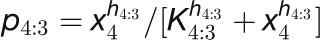 |
 |
 |
 |
 |
 |
 |
| Synthesis Rates |
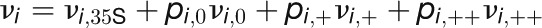 |
 |
 |
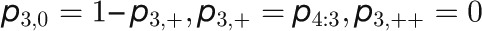 |
 |
 |
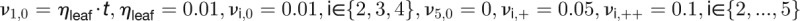 |
| Parameters used in computing leaf number data and figures |
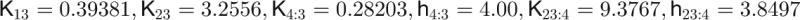 |
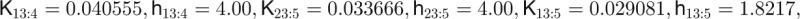 |
 |
The concentrations of the hub activity proteins are denoted by x1 = [FT], x2 = [TFL1], x3 = [FD], x4 = [LFY], and x5 = [AP1]. Kij are effective binding constants between hub activity proteins i and j, Ki:k the effective binding constants between hub activity protein i and a promoter site for the hub activity gene k, Kij:k effective binding constants between complexes of hub activity proteins i and j and a promoter site for gene k, and hi the effective Hill coefficients. pi:k is the fraction of hub activity protein i bound to a promoter site of gene k and pij:k the fraction of the promoter of gene k with the complex i and j . All degradation rates were set to δi = 0.1. We also present the parameters we used to compute the data shown in the figures and tables.
Using a fitness function that measures the fit to several genotypes in the training set (the wild type and single and double mutants), we optimized parameters in 40 independent trials starting from random initial values. The best parameters for the model (Figure 1C) were used to make predictions for the triple mutants. The equations and the optimized parameters are given in Table 2, and their effect on the overall leaf number fit is shown in Supplemental Figures 3 and 4 online. The model is able to capture the flowering time of all genotypes in the training set and predict the leaf numbers for the triple mutants 35S:FT fd-2 fdp-1, 35S:TFL1 fd-2 fdp-1, and tfl1-1 fd-2 fdp-1 (Table 1; see Supplemental Figures 3 to 6 online).
Dynamics of the Floral Transition
An important characteristic of the floral transition is irreversibility. We therefore simulated varying lengths of inductive conditions (i.e., the duration of FT production). Once AP1 has crossed the 0.2 threshold and the apex enters the cauline state, the plant is committed to flowering and halting FT production has little effect on the timing of flowering (Figure 5B). This is consistent with studies showing that short inductions of FT are sufficient to cause floral commitment (Corbesier et al., 1996). Interestingly, although FT is expressed in a circadian fashion in planta (Suárez-López et al., 2001), its output, AP1 expression, does not oscillate (Sundström et al., 2006). To test if our regulatory network also exhibits this ability to integrate and smooth input signals, we examined how oscillating production rates of FT influence the FT and AP1 hubs. The network is able to integrate large oscillations in FT production rate and still generate a smooth increase in AP1 (Figure 5C). FT expression is also strongly influenced by temperature (Balasubramanian et al., 2006); therefore, plants are likely to experience large day-to-day fluctuations in FT levels. We simulated high noise levels in FT production, and even under these conditions, the rise of the AP1 hub is smooth (Figure 5D), showing that our model simulates the ability of this developmental system to integrate noisy environmental signals and make correctly timed decisions. The model therefore captures key properties of the floral transition in Arabidopsis, including irreversibility and the integration of noisy signals.
The Balance of the FT and TFL1 Hubs Is Important for Determining Flowering Behavior
The inflorescence meristem has a distinctive TFL1 expression pattern, with TFL1 mRNA being high in the vegetative center and absent in the floral primordia. Having determined that our regulatory network captures the dynamics of flowering, we sought to see if we could use it to understand aspects of the spatial patterning of gene expression in the inflorescence meristem. To understand how the FT and TFL1 hubs determine flowering behavior, we simulated the dynamics of the transition with varying levels of these control parameters. We find that high TFL1 is able to completely suppress flowering in our model (Figure 6), in contrast with transgenic plants, where 35S:TFL1 delays but does not prevent flowering (Ratcliffe et al., 1998). This observation led us to hypothesize that nonflowering phenotypes may arise through strong ectopic expression of TFL1. Although most single mutants described in the literature eventually flower, the double mutant between two homeodomain proteins, PENNYWISE (PNY) and POUNDFOOLISH (PNF), is notable as it never completes the floral transition (Smith et al., 2004). We therefore examined TFL1 expression in this background. As previously shown, TFL1 is not expressed in the shoot apex under noninductive conditions in the wild type (Conti and Bradley, 2007) and is strongly upregulated upon floral induction (Figures 2J and 2K). Interestingly, we find that TFL1 is ectopically expressed at a high level in the vasculature in pny pnf (Figure 2L). This domain of expression is interesting in the context of the related gene FT, which is also expressed in the vasculature. It is therefore possible that during evolution PNY and PNF have been recruited to TFL1 regulatory regions to prevent vasculature expression and restrict TFL1 expression to the apex. A close relative of TFL1, ATC, is also expressed in the vasculature (Mimida et al., 2001). TFL1 is a mobile molecule and is therefore expected to repress flowering efficiently if expressed in the same domain where the floral inductive signal is transported. We are able to rescue flowering in pny pnf by introducing the tfl1-1 mutation (see Supplemental Figure 1E online), showing that the ectopic expression of TFL1 in pny pnf is responsible for the nonflowering phenotype.
Figure 6.
Relative Levels of the FT and TFL1 Hubs Determine the Flowering Landscape.
Plot showing the influence of varying levels of the FT and TFL1 hubs on floral transition behavior (as measured by the number of leaves produced before flowering). High levels of the FT hub and moderate levels of the TFL1 hub lead to early flowering, whereas high TFL1 at low to moderate FT levels is able to completely prevent flowering in the model. Levels of the FT and TFL1 hubs are given in arbitrary units.
TFL1 Is Required and Expressed in Proportion to the Strength of the Floral Inductive Signal
An important feature of the Arabidopsis inflorescence meristem is the close juxtaposition of different cell fates. While cells on the flanks of the shoot are irreversibly committed to a floral fate, cells at the center of the apex are maintained in an undifferentiated state. These programming differences must be maintained despite wide variations in signal strength and the fact that most of the key regulators of these processes are mobile signals able to move across many cell layers (Sessions et al., 2000; Wu et al., 2002; Conti and Bradley, 2007; Jaeger and Wigge, 2007). As we could show, the formation of floral meristems appears to be a result of reinforcing positive feedforward signaling, but it is not clear how the meristem is able to maintain a population of undifferentiated cells, which are nonetheless poised to change fate rapidly when they pass to the flanks of the shoot apex. FT expression levels vary strongly during the plant life cycle, yet these changes must be flexibly counterbalanced to avoid conversion of the shoot into a terminal flower. Results from genetic studies suggest this role may be fulfilled by TFL1, which becomes highly expressed during the floral transition in cells that remain permanently vegetative. So, while TFL1 expression is downregulated by AP1 in the floral meristems, the strong upregulation of TFL1 expression upon the floral transition in the vegetative center serves to maintain indeterminate growth.
This suggests that in the absence of floral inductive signals, TFL1 is not required since the upregulation of the floral pathway is absent. Consistent with this, we are able to suppress the architecture and flowering phenotypes of tfl1-1 by growth under noninductive short photoperiods (see Supplemental Figure 1D online). Cells with a vegetative fate will only be able to tolerate a relatively low level of FT in the presence of TFL1, but higher levels of FT will need to be actively counterbalanced to prevent flowering. To test if TFL1 expression rises with FT upon the floral transition, we grew plants under noninductive conditions and then induced the floral transition by shifting to long photoperiods. To minimize the effects of spatial differences, TFL1 expression was analyzed in the entire aerial rosette. Significantly, we observe that TFL1 is expressed in proportion to FT (Figure 7).
Figure 7.
TFL1 Rises with FT Levels.
Quantitative PCR was used to analyze how TFL1 and FT expression varies upon the floral transition in the whole rosette. Plants were grown for 20 d under noninductive 8-h short days and shifted into inductive 16-h long days to trigger flowering. Samples were taken immediately before shifting and every day following the shift for 5 d at dusk. This observation confirms as indicated by in situ hybridization (Figures 2J and 2K) that TFL1 expression rises in proportion to the floral inductive signal (FT).
Proportional Expression of TFL1 in Response to FT Maintains Cells in the Vegetative State
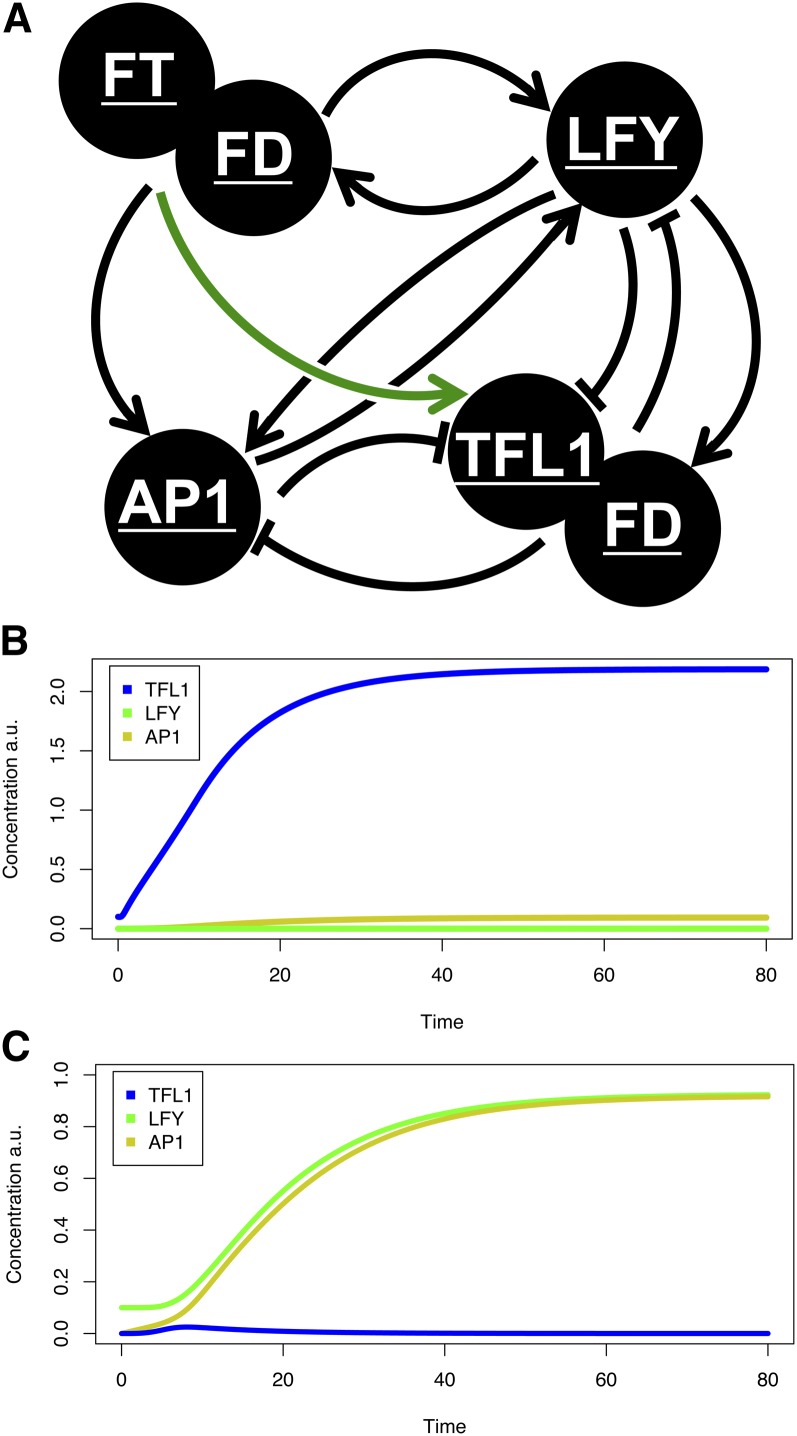
Having established a network architecture that captures the dynamics of the floral transition (Figure 1C), we asked if the model might also help us understand spatial expression patterns of the main floral meristem regulators. The floral transition represents an interesting developmental system, since initially diffuse and variable input signals (FT) gradually increase over time, leading to the expression of floral meristem genes, such as LFY and AP1, on the flanks of the shoot, while the center of the shoot has strong TFL1 expression and remains vegetative. We hypothesized that low initial levels of LFY or TFL1 in our model might be sufficient to determine the stable acquisition of either a flowering (high AP1) or vegetative (high TFL1) state. However, simulations run with this network did not enable a binary outcome between flowering or vegetative fates. Instead, a flowering state was always reached (data not shown).
The failure of our temporal model to capture spatial behavior may be because our initial architecture was optimized to calculate flowering time and thus represented a point on the flanks of the shoot apex. These cells transition to a high AP1 state, but they do not experience upregulation of TFL1, since TFL1 is repressed in floral meristems. By contrast, within the center of the shoot, TFL1 is strongly upregulated upon flowering. Furthermore, we have seen that TFL1 expression correlates across the whole plant with the strength of the floral signal (Figure 7). To account for this behavior in our model, we therefore included a term for the activation of TFL1 by FT FD in the network. While this study was underway, a genome-wide analysis of LFY binding sites showed that LFY binds to TFL1 regulatory elements (Winter et al., 2011). We therefore also included this interaction module in our network architecture (Figure 8A).
Figure 8.
An Extended Network That Captures Cell Fate Determination in the SAM during the Floral Transition.
(A) The initial network used to determine flowering time was unable to produce states with high stable TFL1 hub activity. We found two further interactions were necessary: repression of TFL1 by LFY (Winter et al., 2011) and the proportional upregulation of TFL1 by FT (green arrow).
(B) Priming the system with low initial levels of TFL1 (to simulate the vegetative center of the SAM) results in a stable vegetative state upon the induction of flowering. a.u., arbitrary units.
(C) Reversing the scenario above (B), with TFL1 starting at 0 and LFY at 0.1 (representative of a presumptive floral meristem), gives the opposite behavior with TFL1 becoming stably repressed and AP1 expressed at a high level. The network therefore shows qualitatively flowering or nonflowering behavior depending on the initial conditions. Blue, TFL1; green, LFY; yellow, AP1.
With these two changes to our network, we simulated the vegetative center of the SAM, where TFL1 is initially expressed at moderate levels and LFY is absent. In this scenario, rising levels of FT trigger the further upregulation of TFL1. The negative feedback of TFL1 onto both AP1 and LFY stably prevents their expression (Figure 8B). Under the opposite starting conditions, moderate levels of LFY and no initial TFL1, corresponding to the primordium prior to floral evocation, rising FT activates AP1 and LFY. Since this is a positive feedback loop, high levels of AP1 and LFY are rapidly established, and TFL1 is repressed, leading to a stable floral state (Figure 8C). This simulated outcome corresponds very well to the observed developmental system, where low initial expression levels of either LFY or TFL1 rise sharply upon the transition, leading to stable vegetative or flowering programs being established. This patterning mechanism has parallels with floral induction in tomato, where the floral signal SFT upregulates a repressor of floral meristem fate in lateral meristems adjacent to floral meristems (Thouet et al., 2012).
DISCUSSION
We defined a simplified network that accounts for the major dynamic properties of the floral transition. In order to reduce the model to a minimal set of key activities, we condensed many known genes that have not been modeled explicitly into activity hubs. By its nature, our model is a considerable simplification, but it contains enough detail for us to make and test hypotheses. Furthermore, the modular nature of the hubs means that extra hub activities can be added to the model as required. The proposed network accounts for the flowering behavior of different genotypes, indicating that we have captured the major dynamic properties of the floral transition. A regulated feedforward loop is at the core of the network, and this common motif has been shown to exhibit irreversible behavior and have the ability to filter noise (Mangan and Alon, 2003). Moreover, this model makes interesting predictions, which we are able to confirm experimentally.
Strengths and Limitations of the Model
A challenge to modeling complex biological systems, such as the floral transition, is that many components are involved, and little is known in terms of their physical properties, such as biochemical concentrations, binding affinities for each other, activities, and half-lives within the cell. Our modeling thus involves considerable simplifications, and there are endogenous pathways that stimulate flowering, for example, through the phytohormone gibberellin (Eriksson et al., 2006), the SPL transcription factors (Wang et al., 2009), as well as the floral integrator gene SOC1 (Liu et al., 2008), which we do not explicitly model here but account for with gradually rising levels of the AP1 and LFY hubs. These models were inspired by known biological interactions and the idea of using leaf number data to make the predictions quantitative. Our use of data in the model has similarities with black-box machine learning approaches (reviewed recently in Dalchau, 2012); however, the underlying model is greatly simplified compared with neural networks or similar techniques and is inspired by biological observations. This allows us to train the network to the available data and with the resulting model to suggest experiments that can be related back to biological entities. The resulting model is also simple enough to understand some aspects of it intuitively. This advantage comes at a cost. First, by placing the network in an ordinary differential equation framework, we need to carry out computationally costly parameter space sampling. Second, the reduction to activity hubs means that a direct mapping onto individual genes is complicated. In our case, in Arabidopsis excellent genetic studies have revealed the major components and enabled us to approximate key genes for entire hub activities. For example the strong lfy-12 allele appears to abolish LFY hub activity. Third, the model currently largely neglects important spatial effects. Although we can reproduce the overall behavior of the transition, individual interactions represent spatially averaged behavior, and conclusions from this simplified network about such details must be considered carefully. These issues and limitations of the current model highlight the need to conduct a spatial cell-based modeling approach that accounts for differential expression patterning of these key floral integrators. These limitations notwithstanding, the ability to build modeling frameworks for the regulation of developmental outcomes in this way provides us with a means to test proposed genetic interaction networks. Simplifying the network to key hubs has the advantage of making it potentially easier to identify the critical network interactions that account for the major behaviors of a system. In the era of genome-wide transcription factor binding maps and large-scale data sets, it is particularly timely to develop such approaches.
TFL1 and Plant Lifestyle and Architecture Decisions
The ability of TFL1 to counterbalance FT has been described before, and in particular in tomato it has been shown that the orthologous genes to TFL1 and FT, SELF-PRUNING (SP) and SINGLE FLOWER TRUSS (SFT), have an antagonistic gradient of expression, whereby young leaves have relatively high SP, while this declines as leaves expand, when SFT expression increases (Shalit et al., 2009). By contrast, in Arabidopsis floral induction leads to strong concomitant rises in the levels of both FT and TFL1.
A key property of the floral transition is that it is irreversible in those tissues where flowering is initiated. Despite this, it is vital for most plants that additional meristems remain vegetative, either to prolong the flowering period during a season or, in the case of perennials, to maintain vegetative meristems to allow them to flower in subsequent years. Our work suggests that TFL1 will play a key role in these life cycle choices, and this is supported by studies in apple (Malus domestica), where four TFL1-like genes appear to regulate vegetative and reproductive development (Mimida et al., 2009). Consistent with this, it has been shown that TFL1 in the related species Arabis alpina plays a central role in setting thresholds for flowering and controlling life cycle behavior (Wang et al., 2011).
In addition to the timing of flowering, plant architecture is also influenced by floral signals. Plants forming determinate (cymose) floral structures (e.g., tomato, petunia [Petunia hybrida], and tobacco [Nicotiana tabacum]) do not express TFL1 in the apex after the floral transition. By contrast, indeterminate (racemose) species (e.g., Arabidopsis and Lotus japonicus) express TFL1 in the apex following the floral transition (Bradley et al., 1996; Amaya et al., 1999; Jensen et al., 2001; Ordidge et al., 2005; Guo et al., 2006; Conti and Bradley, 2007; Lifschitz, 2008; Thouet et al., 2008), enabling a pool of undifferentiated cells to be maintained, enabling indeterminate growth. It is therefore likely that axillary meristems are maintained in a vegetative state by high levels of TFL1 expression, while the main apex undergoes the floral transition. Vegetative plants do not express TFL1 in the SAM but show strong TFL1 expression in the axillary meristems (Conti and Bradley, 2007) (Figure 2J); indeed, outgrowth of axillary shoots in tfl1-1 (Shannon and Meeks-Wagner, 1991) or 35S:LFY plants (Weigel and Nilsson, 1995) is precocious and precedes flowering of the SAM.
The Importance of Feedback Regulation in the Floral Transition
A feature of the network architecture we describe is the high degree of interlocked positive and negative feedback loops required for the network to capture temporal and spatial flowering behavior. Positive feedback loops are well described in biology (Freeman, 2000) and indeed have been proposed to play an important role in regulating the floral transition (Wang et al., 2009). They provide a means to amplify initially weak signals and provide an unambiguous output. Interlocked feedback loops as we describe are relevant in a developmental context because they enable the acquisition of a fate outcome that is stable over many cell divisions without requiring changes to the underlying genetic code, for example, white opaque switching in Candida albicans (Zordan et al., 2007). The floral transition is particularly interesting in that it involves indirect signaling across many cells, since the mobile FT signal in combination with FD triggers the upregulation of LFY and AP1, with LFY then feeding back directly to further upregulate FD. AP1 and LFY both positively regulate their own expression, leading to a stable flowering state. This system has parallels with the autoregulation of Ubx during Drosophila melanogaster segmentation (Thüringer and Bienz, 1993). Positive feedback loops provide a means to fine-tune activity as well as irreversibly and stably determine cell fate, but they can also sensitize the system, as illustrated by the role of p53 positive feedback in cancer progression (Harris and Levine, 2005). Integrated positive and negative feedback loops are able to robustly provide pattern formation, for example, in the Drosophila egg (Wasserman and Freeman, 1998). Plants do not appear to rely so much on transmembrane receptors, such as Notch (Wigge and Weigel, 2001), but it appears that the FT binding FD 14-3-3 complex may play an analogous role in providing a developmental readout for a ligand signal. An interesting outcome from the model has been the importance of the balance between FT and TFL1 in determining fate outcomes. Upregulation of TFL1 in proportion to FT is necessary in our model to obtain different cell fate outcomes between the vegetative center of the SAM and floral meristems on the flanks. The ability of TFL1 to flexibly counterbalance strong increases in floral inductive signals is further enhanced by the ability of TFL1 to signal through FD activity, which also rises upon the floral transition. The key role played by TFL1 is highlighted by the dramatic nonflowering phenotype caused by ectopic TFL1 expression in pny pnf. Proportional control, where the output to the control device is modulated in proportion to the deviation from the desired outcome, has been used in engineering for some time (Maxwell, 1868), and it will be interesting to see if the regulation of TFL1 by FT follows the same principles.
METHODS
Computational Methods
The flowering time control network was built iteratively, guided by basic theoretical considerations and constrained by known biological data. For example, as the floral transition is irreversible, we considered network structures known to possess this property. The main objective was to reduce a complex pathway down to its fundamental activities. In order to reduce the number of parameters, the potential networks were simplified by reducing linear pathways to only their end hubs and by grouping similar activities into one representative hub, termed an activity hub. We make the simplifying assumption that in favorable conditions for flowering, FT will be produced in equal amounts by each unit area of a growing leaf. In the simulations, we model this by an incoming FT rate to the apex that increases linearly over time (approximating leaf growth in size and number). The FT production rate therefore increases until the plant flowers. We maintain the production rate at which flowering set in and neglect further leaf growth. The differential equations were solved using the Fortran routine LSODE from ODEPACK (Hindmarsh, 1983). A standard simulated annealing algorithm, SIMANN (Goffe et al., 1994), was employed to fit the parameters of the model. An objective function that measures the fit between leaf numbers obtained by solving the set of ordinary differential equations for sampled model parameters and the experimentally determined values of the training set was used. Further details of derivation of the equations (Table 2) are given in the supplemental material.
Experimental Methods
Plant Material
The Columbia-0 background was used as the wild type. ft-10, fd-2, 35S:FT, lfy-12, tfl1-1, 35S:LFY, and 35S:TFL1 have been described previously (Weigel et al., 1992; Weigel and Nilsson, 1995; Bradley et al., 1997; Ratcliffe et al., 1998; Kardailsky et al., 1999; Wigge et al., 2005). For FDP analysis, a TILLING line, fdp-1, was obtained that carries a substitution of Arg to Lys at position 181, a change shown to abolish DNA binding in this family of transcription factors (Aukerman et al., 1991). fdp-1 was backcrossed three times in Columbia wild-type background. Plants were grown on soil at 21°C constant temperature under a mixture of Philips Cool White and Osram fluorescent lights, with a fluence rate of 120 µmol m−2 s−1 and a relative humidity of 70%. To simulate long-day conditions, plants were grown under 16-h-light and 8-h-dark cycles or under 16-h-dark and 8-h-light for short-day conditions. Plants used for in situ hybridization and GUS analysis were grown for 21 d under short-day conditions at 21°C and then shifted to long-day conditions at 21°C to induce flowering.
Plant Transformation and Selection
Plants were transformed with the floral dip method (Weigel and Glazebrook, 2002) and selected on germination medium containing 0.5× Murashige and Skoog salt mixture, 0.7% agar, and 50 µg mL−1 kanamycin (Melford), pH 5.7.
In Situ Hybridization
For the paraplast embedding steps, an automated tissue processor was used (TissueTek). Plant material for in situ hybridization was fixed in FAA and embedded overnight (70% ethanol for 1 h, 90% ethanol for 2 × 1 h, 99% ethanol for 2 × 1 h, absolute ethanol for 1 h, xylene for 3 × 1 h, and histowax for 3 × 1 h. In situ antisense probes were synthesized using a DIG RNA labeling kit (Roche). Templates for all genes were the coding regions cloned into pGEM (Promega). Probes greater than 450 bp were hydrolyzed to between 100 and 200 bp by mixing 100 μL of probe with 100 μL of carbonate buffer (80 mM NaHCO3 and 120 mM Na2CO3) and incubated for T minutes (T = (Linitial – Lfinal)/(0.11 × Linitial × Lfinal)), where Linitial and Lfinal are the initial and final lengths of the probe in kilobases. Hydrolysis was stopped with 20 μL of 10% acetic acid. Probes were precipitated overnight with 1 μL glycogen, 1 μL 1 M MgCl2, 600 μL of ethanol at −20°C, and then centrifuged at top speed for 30 min in a microfuge (4°C). Pellets were washed with 80% ethanol, dried, and resuspended in 50 μL water. Probes were diluted 10-fold in hybridization buffer (50% deionized formamide, 10% dextran sulfate, 0.3 M NaCl, 10 mM Tris, pH 8, 10 mM NaPO4, 5 mM EDTA, 0.02% Ficoll 400, 0.02% polyvinylpyrrolidone, 0.02% BSA, and 0.5 mg/mL tRNA) and stored at −20°C. Sections were dewaxed in Histoclear (2 × 10 minutes). Slides were incubated twice with 100% ethanol and rehydrated in the following concentrations of ethanol: 95, 90, 80, and 60%. The last two washes included 150 mM NaCl. Finally, sections were incubated in PBS (130 mM NaCl, 3 mM NaH2PO4, and 70 mM Na2HPO4, pH 7.0) for 2 min. Proteinase K (Roche) digestion was performed at 1 µg/mL for 30 min. The reaction was stopped in 2 mg/mL Gly-PBS and then washed in PBS. Slides were kept in fixative for 5 min (100 mL ethanol, 10 mL acetic acid, and 20 mL formaldehyde to 200 mL with water). Slides were washed twice with PBS, for 5 min each time. Slides were incubated in 130 mM NaCl for 2 min. Slides were then dehydrated in the following incubation series: 30% ethanol (130 mM NaCl), 60% ethanol (130 mM NaCl), 80% ethanol, 90% ethanol, 95% ethanol, and 100% ethanol. Probes were diluted 1:100 in hybridization buffer and denatured at 80°C for 2 min and then placed on ice. One hundred microliter of probe was added to each slide and incubated at 55°C overnight. Slides were incubated in 2× SSC (300 mM NaCl and 30 mM sodium citrate, pH 7.0) at 55°C to remove the cover slips. Slides were washed four times for 2 h total in 0.2× SSC (55°C) (30 mM NaCl and 3 mM sodium citrate, pH 7.0). Slides were incubated in 0.2× SSC at 37°C for 5 min. Slides were washed with 0.2× SSC at room temperature. Slides were stored in 1× PBS. For detection, slides were blocked with 1 to 2 mL of blocking agent (Roche) in TBS (50 mM Tris and 150 mM NaCl, pH 7.5) for 30 min. Repeated with blocking agent in TBS-T (TBS with 0.3× Triton X-100). Anti-DIG solution (Roche) was diluted 1:1250 in 1/10 blocking reagent in TBS-T and applied to each slide. Cover slips were removed in TBS-T and then 1/10 blocking reagent in TBS-T was applied and incubated for 30 min at room temperature. This wash was repeated three times. After removing wash solution, the slides were washed twice with TNM-50 (100 mM Tris, pH 9.5, 100 mM NaCl, and 50 mM MgCl2) for 5 min each time to remove the detergent. The color reaction was performed with NBT-BCIP (Roche) diluted 1:50 in TNM-50, with 150 μL added per slide. Cover slips were applied to each slide and incubated overnight in a dark humidified box overnight at room temperature. When the color reaction was complete, slides were rinsed in TE and mounted in TE with 50% glycerol and sealed. Slides were examined under a Nikon Eclipse 800 microscope and images captured with a Pixera Pro ES600 camera.
Plasmid Construction
All constructs apart from in situ probes were cloned into Gateway system vectors (Invitrogen) and transformed into Arabidopsis thaliana plants by the floral dip method, as described above. Constructs were cloned using standard PCR and restriction enzyme digestion/ligation methods as described (Sambrook and Russell, 2001). For the FDP rescue, a genomic fragment starting 5359 bp upstream of the start ATG and extending 201 bp downstream of the stop codon was introduced in the fd-2 fdp-1 background and plants analyzed in T2. For the promoter analysis of FD, a 3-kb genomic fragment upstream of the start codon was cloned in frame with GUS. Putative LBSs were abolished by overlapping PCR and replaced by random sequence.
Transcript Analysis
Plants were grown under short-day conditions for 3 weeks and then shifted to long days. Three plants were harvested every day at dusk, and the total RNA was extracted using Trizol reagent (Invitrogen). Two micrograms of RNA was treated with DNaseI (Roche) and used for cDNA synthesis (first-strand cDNA synthesis kit; Fermentas). cDNA was diluted 1:10 and 5 µL used for quantitative PCR using a Roche Lightcycler 480 and the corresponding SYBR Green master mix. To detect FT transcript levels, oligos FT_F and FT_R were used and for TFL1, oligos TFL1_F and TFL1_R. Oligos amplifying TUB6 (At5g12250) were used for normalization. Primer sequences are provided in Supplemental Table 1 online. PCR was repeated three times, and at least two biological replicates were used for each time point.
ChIP
35S:LFY:FLAG seedlings were grown on half-strength Murashige and Skoog plates for 14 d before harvesting. Plant tissue (2.5 g) was fixed in 30 mL of fixation buffer (1× PBS [130 mM NaCl, 3 mM NaH2PO4, and 70 mM Na2HPO4, pH 7.0] and 1% formaldehyde) under vacuum for 10 min. Fixation was stopped with 2.5 mL of 2 M Gly for 5 min under vacuum. Seedlings were rinsed with 1× PBS and blotted dry before flash freezing in liquid nitrogen. Nuclei from fixed material were purified as follows. Tissue was ground on dry ice and resuspended in nuclei isolation buffer (10 mM MES, 200 mM Suc, and 0.01% Triton X-100 with protease inhibitors). Supernatant was filtered through Miracloth and made up to 0.3% Triton X-100 and mixed by swirling every 2 min for a total time of 10 min. Nuclei were harvested by centrifuging for 15 min at 3600 rpm at 4°C in a Sorvall centrifuge. The pellet was resuspended in 1 mL of nuclear lysis buffer (50 mM Tris-HCl, pH 8, 10 mM EDTA, 0.5% SDS, and protease inhibitors) and sonicated in a Diagenode Bioruptor 3 × 5 min on the low setting with 10-min intervals. This gave DNA sheared within the range of 300 to 1000 bp. One-tenth volume of 10% Triton X-100 was added to the fragmented chromatin and centrifuged at 13,000 rpm for 10 min at 4°C in an Eppendorf benchtop centrifuge. The supernatant was recovered and used for immunopurification. Anti-FLAG M2 agarose beads (Sigma-Aldrich) were washed three times with ChIP dilution buffer, and aliquots of fragmented chromatin were incubated in ChIP dilution buffer with M2 agarose beads rotating at 4°C for 3 to 4 h. The beads were recovered by brief centrifugation (2000 rpm) and washed as follows: low salt wash buffer (150 mM NaCl, 0.1% SDS, 1% Triton X-100, 2 mM EDTA, and 20 mM Tris-HCl, pH 8.0), two washes, 5 min each; high salt wash buffer (500 mM NaCl, 0.1% SDS, 1% Triton X-100, 2 mM EDTA, and 20 mM Tris-HCl, pH 8.0), two washes, 5 min each; TE (10 mM Tris-HCl, pH 8.0, and 1 mM EDTA), two washes, 5 min each. Chromatin was eluted by resuspending beads in 100 μL TE containing 100 ng/μL 3× FLAG peptide (Sigma-Aldrich) and incubating for 30 min at 4°C. Cross-links were removed by bringing input and ChIP samples to 200 mM NaCl and adding 1 μL of Proteinase K to each sample and incubating at 65°C for 4 h. Ten microliters 0.5 M EDTA, 20 μL Tris HCl, pH 6.5, and 1.5 μL of 14 mg/mL Proteinase K was added to each 500 μL sample and incubated for 1 h at 45°C to elute chromatin. DNA was extracted using phenol/chloroform and precipitated in sodium acetate. The pellet was washed with ethanol and resuspended in TE after drying. DNA was diluted 1:5 and 5 µL used for quantitative PCR using a Roche Lightcycler 480 and the corresponding SYBR Green master mix. Oligonucleotides for detecting LFY binding to the identified LBS were used as described in Supplemental Table 1 online. HSP101 was used as a negative control. The PCR was done with biological duplicates for the 35S:LFY:FLAG line and one Columbia control. PCR was done as triplicates with the standard deviation for each PCR shown on the graph.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: FD, AT4G35900; FDP, AT2G17770; FT, AT1G65480; TFL1, AT5G03840; AP1, AT1G69120; LFY, AT5G61850; HSP101, AT1G74310; PNY, AT5G02030; and PNF, AT2G27990.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Flowering Phenotypes.
Supplemental Figure 2. Hub Dynamics.
Supplemental Figure 3. Overall Fit Changes as a Function of Each Parameter.
Supplemental Figure 4. Overall Fit Changes as a Function of Each Parameter Zoomed in.
Supplemental Figure 5. Fit between Experimental and Modeled Leaf Numbers for Wild Type and Single Mutants.
Supplemental Figure 6. Fit between Experimental and Modeled Leaf Numbers for Double and Triple Mutants.
Supplemental Table 1. Primer Sequences.
Supplementary Material
Acknowledgments
We thank Detlef Weigel who first hypothesized that 35S:TFL1 might be suppressed by fd-2 and provided support and mentoring to P.A.W. as a junior group leader in his department. We thank Takashi Araki and Markus Schmid for sharing unpublished data. We thank Des Bradley, Detlef Weigel, Vava Grbic, Mary Byrne, and Harley Smith for seeds. We thank Caroline Dean, Enrico Coen, Martin Howard, Verônica Grieneisen, Stan Maree, and Melinda Mayer for critical reading of the article and helpful suggestions. We thank three anonymous reviewers for their thoughtful comments and constructive critique. This work was funded by start-up funds from the John Innes Centre to the P.A.W. and R.J.M. groups and a Biotechnology and Biological Science Research Council Responsive mode grant to P.A.W. (BB/D010047/1). The P.A.W. group is supported by a core grant from the Gatsby Foundation.
AUTHOR CONTRIBUTIONS
K.E.J. performed the biological experiments. N.P., S.L., and R.J.M. built the computational models, and N.P. and R.J.M. carried out the numerical analyses. K.E.J., N.P., R.J.M., and P.A.W. designed the research and wrote the article.
Glossary
- SAM
shoot apical meristem
- GUS
β-glucuronidase
- LBS
LEAFY binding site
- ChIP
chromatin immunoprecipitation
References
- Abe M., Kobayashi Y., Yamamoto S., Daimon Y., Yamaguchi A., Ikeda Y., Ichinoki H., Notaguchi M., Goto K., Araki T. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 [DOI] [PubMed] [Google Scholar]
- Ahn J.H., Miller D., Winter V.J., Banfield M.J., Lee J.H., Yoo S.Y., Henz S.R., Brady R.L., Weigel D. (2006). A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J. 25: 605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Buylla E.R., Balleza E., Benítez M., Espinosa-Soto C., Padilla-Longoria P. (2008a). Gene regulatory network models: A dynamic and integrative approach to development. SEB Exp. Biol. Ser. 61: 113–139 [PubMed] [Google Scholar]
- Alvarez-Buylla E.R., Chaos A., Aldana M., Benítez M., Cortes-Poza Y., Espinosa-Soto C., Hartasánchez D.A., Lotto R.B., Malkin D., Escalera Santos G.J., Padilla-Longoria P. (2008b). Floral morphogenesis: Stochastic explorations of a gene network epigenetic landscape. PLoS ONE 3: e3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaya I., Ratcliffe O.J., Bradley D.J. (1999). Expression of CENTRORADIALIS (CEN) and CEN-like genes in tobacco reveals a conserved mechanism controlling phase change in diverse species. Plant Cell 11: 1405–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S., Sureshkumar S., Lempe J., Weigel D. (2006). Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet. 2: e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley D., Vincent C., Carpenter R., Coen E. (1996). Pathways for inflorescence and floral induction in Antirrhinum. Development 122: 1535–1544 [DOI] [PubMed] [Google Scholar]
- Busch M.A., Bomblies K., Weigel D. (1999). Activation of a floral homeotic gene in Arabidopsis. Science 285: 585–587 [DOI] [PubMed] [Google Scholar]
- Conti, L., and Bradley, D. (2007). TERMINAL FLOWER1 is a mobile signal controlling Arabidopsis architecture. Plant Cell 19: 767–778. [DOI] [PMC free article] [PubMed]
- Corbesier L., Gadisseur I., Silvestre G., Jacqmard A., Bernier G. (1996). Design in Arabidopsis thaliana of a synchronous system of floral induction by one long day. Plant J. 9: 947–952 [DOI] [PubMed] [Google Scholar]
- Corbesier L., Vincent C., Jang S., Fornara F., Fan Q., Searle I., Giakountis A., Farrona S., Gissot L., Turnbull C., Coupland G. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Dalchau N. (2012). Understanding biological timing using mechanistic and black-box models. New Phytol. 193: 852–858 [DOI] [PubMed] [Google Scholar]
- Eriksson S., Böhlenius H., Moritz T., Nilsson O. (2006). GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell 18: 2172–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa-Soto C., Padilla-Longoria P., Alvarez-Buylla E.R. (2004). A gene regulatory network model for cell-fate determination during Arabidopsis thaliana flower development that is robust and recovers experimental gene expression profiles. Plant Cell 16: 2923–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M. (2000). Feedback control of intercellular signalling in development. Nature 408: 313–319 [DOI] [PubMed] [Google Scholar]
- Goffe W.L., Ferrier G.D., Rogers J. (1994). Global optimization of statistical functions with simulated annealing. J. Econom. 60: 65–100 [Google Scholar]
- Guo X., Zhao Z., Chen J., Hu X., Luo D. (2006). A putative CENTRORADIALIS/TERMINAL FLOWER 1-like gene, Ljcen1, plays a role in phase transition in Lotus japonicus. J. Plant Physiol. 163: 436–444 [DOI] [PubMed] [Google Scholar]
- Hammer G., Cooper M., Tardieu F., Welch S., Walsh B., van Eeuwijk F., Chapman S., Podlich D. (2006). Models for navigating biological complexity in breeding improved crop plants. Trends Plant Sci. 11: 587–593 [DOI] [PubMed] [Google Scholar]
- Hanano, S., and Goto, K. (2011). Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. Plant Cell 23: 3172–3184. [DOI] [PMC free article] [PubMed]
- Harris S.L., Levine A.J. (2005). The p53 pathway: Positive and negative feedback loops. Oncogene 24: 2899–2908 [DOI] [PubMed] [Google Scholar]
- Hindmarsh, A.C. (1983). ODEPACK, A Systematized Collection of ODE Solvers. (Amsterdam: North-Holland). [Google Scholar]
- Jaeger K.E., Wigge P.A. (2007). FT protein acts as a long-range signal in Arabidopsis. Curr. Biol. 17: 1050–1054 [DOI] [PubMed] [Google Scholar]
- Jensen C.S., Salchert K., Nielsen K.K. (2001). A TERMINAL FLOWER1-like gene from perennial ryegrass involved in floral transition and axillary meristem identity. Plant Physiol. 125: 1517–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalisz S., Kramer E.M. (2008). Variation and constraint in plant evolution and development. Heredity (Edinb) 100: 171–177 [DOI] [PubMed] [Google Scholar]
- Kardailsky I., Shukla V.K., Ahn J.H., Dagenais N., Christensen S.K., Nguyen J.T., Chory J., Harrison M.J., Weigel D. (1999). Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Kaufmann K., Wellmer F., Muiño J.M., Ferrier T., Wuest S.E., Kumar V., Serrano-Mislata A., Madueño F., Krajewski P., Meyerowitz E.M., Angenent G.C., Riechmann J.L. (2010). Orchestration of floral initiation by APETALA1. Science 328: 85–89 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Kaya H., Goto K., Iwabuchi M., Araki T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962 [DOI] [PubMed] [Google Scholar]
- Kumar S.V., Lucyshyn D., Jaeger K.E., Alós E., Alvey E., Harberd N.P., Wigge P.A. (2012). Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484: 242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rota C., Chopard J., Das P., Paindavoine S., Rozier F., Farcot E., Godin C., Traas J., Monéger F. (2011). A data-driven integrative model of sepal primordium polarity in Arabidopsis. Plant Cell 23: 4318–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Oh M., Park H., Lee I. (2008). SOC1 translocated to the nucleus by interaction with AGL24 directly regulates leafy. Plant J. 55: 832–843 [DOI] [PubMed] [Google Scholar]
- Leyser O. (2003). Regulation of shoot branching by auxin. Trends Plant Sci. 8: 541–545 [DOI] [PubMed] [Google Scholar]
- Lifschitz E. (2008). Multiple regulatory roles for SELF-PRUNING in the shoot system of tomato. Plant Physiol. 148: 1737–1738, author reply 1738–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschitz E., Eshed Y. (2006). Universal florigenic signals triggered by FT homologues regulate growth and flowering cycles in perennial day-neutral tomato. J. Exp. Bot. 57: 3405–3414 [DOI] [PubMed] [Google Scholar]
- Liljegren S.J., Gustafson-Brown C., Pinyopich A., Ditta G.S., Yanofsky M.F. (1999). Interactions among APETALA1, LEAFY, and TERMINAL FLOWER1 specify meristem fate. Plant Cell 11: 1007–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Chen H., Er H.L., Soo H.M., Kumar P.P., Han J.H., Liou Y.C., Yu H. (2008). Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 135: 1481–1491 [DOI] [PubMed] [Google Scholar]
- Mangan S., Alon U. (2003). Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. USA 100: 11980–11985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J., Warthmann N., Küttner F., Schmid M. (2007). Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr. Biol. 17: 1055–1060 [DOI] [PubMed] [Google Scholar]
- Mathieu J., Yant L.J., Mürdter F., Küttner F., Schmid M. (2009). Repression of flowering by the miR172 target SMZ. PLoS Biol. 7: e1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell J.C. (1868). On governors. Proceedings of the Royal Society 16: 270–283 [Google Scholar]
- Mendoza L., Thieffry D., Alvarez-Buylla E.R. (1999). Genetic control of flower morphogenesis in Arabidopsis thaliana: A logical analysis. Bioinformatics 15: 593–606 [DOI] [PubMed] [Google Scholar]
- Mimida N., Goto K., Kobayashi Y., Araki T., Ahn J.H., Weigel D., Murata M., Motoyoshi F., Sakamoto W. (2001). Functional divergence of the TFL1-like gene family in Arabidopsis revealed by characterization of a novel homologue. Genes Cells 6: 327–336 [DOI] [PubMed] [Google Scholar]
- Mimida N., Kotoda N., Ueda T., Igarashi M., Hatsuyama Y., Iwanami H., Moriya S., Abe K. (2009). Four TFL1/CEN-like genes on distinct linkage groups show different expression patterns to regulate vegetative and reproductive development in apple (Malus x domestica Borkh.). Plant Cell Physiol. 50: 394–412 [DOI] [PubMed] [Google Scholar]
- Ordidge M., Chiurugwi T., Tooke F., Battey N.H. (2005). LEAFY, TERMINAL FLOWER1 and AGAMOUS are functionally conserved but do not regulate terminal flowering and floral determinacy in Impatiens balsamina. Plant J. 44: 985–1000 [DOI] [PubMed] [Google Scholar]
- Prusinkiewicz P., Erasmus Y., Lane B., Harder L.D., Coen E. (2007). Evolution and development of inflorescence architectures. Science 316: 1452–1456 [DOI] [PubMed] [Google Scholar]
- Ratcliffe O.J., Amaya I., Vincent C.A., Rothstein S., Carpenter R., Coen E.S., Bradley D.J. (1998). A common mechanism controls the life cycle and architecture of plants. Development 125: 1609–1615 [DOI] [PubMed] [Google Scholar]
- Ratcliffe O.J., Bradley D.J., Coen E.S. (1999). Separation of shoot and floral identity in Arabidopsis. Development 126: 1109–1120 [DOI] [PubMed] [Google Scholar]
- Salazar J.D., Saithong T., Brown P.E., Foreman J., Locke J.C., Halliday K.J., Carré I.A., Rand D.A., Millar A.J. (2009). Prediction of photoperiodic regulators from quantitative gene circuit models. Cell 139: 1170–1179 [DOI] [PubMed] [Google Scholar]
- Samach A., Onouchi H., Gold S.E., Ditta G.S., Schwarz-Sommer Z., Yanofsky M.F., Coupland G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and Russell, D.W. (2001). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press). [Google Scholar]
- Sessions A., Yanofsky M.F., Weigel D. (2000). Cell-cell signaling and movement by the floral transcription factors LEAFY and APETALA1. Science 289: 779–782 [DOI] [PubMed] [Google Scholar]
- Shalit A., Rozman A., Goldshmidt A., Alvarez J.P., Bowman J.L., Eshed Y., Lifschitz E. (2009). The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proc. Natl. Acad. Sci. USA 106: 8392–8397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon S., Meeks-Wagner D.R. (1991). A mutation in the Arabidopsis TFL1 gene affects inflorescence meristem development. Plant Cell 3: 877–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson G.G., Dean C. (2002). Arabidopsis, the Rosetta stone of flowering time? Science 296: 285–289 [DOI] [PubMed] [Google Scholar]
- Smith H.M., Campbell B.C., Hake S. (2004). Competence to respond to floral inductive signals requires the homeobox genes PENNYWISE and POUND-FOOLISH. Curr. Biol. 14: 812–817 [DOI] [PubMed] [Google Scholar]
- Suárez-López P., Wheatley K., Robson F., Onouchi H., Valverde F., Coupland G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120 [DOI] [PubMed] [Google Scholar]
- Sundström J.F., Nakayama N., Glimelius K., Irish V.F. (2006). Direct regulation of the floral homeotic APETALA1 gene by APETALA3 and PISTILLATA in Arabidopsis. Plant J. 46: 593–600 [DOI] [PubMed] [Google Scholar]
- Tamaki S., Matsuo S., Wong H.L., Yokoi S., Shimamoto K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036 [DOI] [PubMed] [Google Scholar]
- Taoka K., et al. (2011). 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476: 332–335 [DOI] [PubMed] [Google Scholar]
- Thouet J., Quinet M., Lutts S., Kinet J.M., Périlleux C. (2012). Repression of floral meristem fate is crucial in shaping tomato inflorescence. PLoS ONE 7: e31096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thouet J., Quinet M., Ormenese S., Kinet J.M., Périlleux C. (2008). Revisiting the involvement of SELF-PRUNING in the sympodial growth of tomato. Plant Physiol. 148: 61–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thüringer F., Bienz M. (1993). Indirect autoregulation of a homeotic Drosophila gene mediated by extracellular signaling. Proc. Natl. Acad. Sci. USA 90: 3899–3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.W., Czech B., Weigel D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749 [DOI] [PubMed] [Google Scholar]
- Wang R., Albani M.C., Vincent C., Bergonzi S., Luan M., Bai Y., Kiefer C., Castillo R., Coupland G. (2011). Aa TFL1 confers an age-dependent response to vernalization in perennial Arabis alpina. Plant Cell 23: 1307–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman J.D., Freeman M. (1998). An autoregulatory cascade of EGF receptor signaling patterns the Drosophila egg. Cell 95: 355–364 [DOI] [PubMed] [Google Scholar]
- Weigel, D., and Glazebrook, J. (2002). Arabidopsis: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press). [Google Scholar]
- Weigel D., Nilsson O. (1995). A developmental switch sufficient for flower initiation in diverse plants. Nature 377: 495–500 [DOI] [PubMed] [Google Scholar]
- Welch S.M., Roe J.L., Dong Z.H. (2003). A genetic neural network model of flowering time control in Arabidopsis thaliana. Agron. J. 95: 71–81 [Google Scholar]
- Wigge P.A., Kim M.C., Jaeger K.E., Busch W., Schmid M., Lohmann J.U., Weigel D. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059 [DOI] [PubMed] [Google Scholar]
- Wigge P.A., Weigel D. (2001). Arabidopsis genome: Life without notch. Curr. Biol. 11: R112–R114 [DOI] [PubMed] [Google Scholar]
- Wilczek A.M., et al. (2009). Effects of genetic perturbation on seasonal life history plasticity. Science 323: 930–934 [DOI] [PubMed] [Google Scholar]
- Winter C.M., et al. (2011). LEAFY target genes reveal floral regulatory logic, cis motifs, and a link to biotic stimulus response. Dev. Cell 20: 430–443 [DOI] [PubMed] [Google Scholar]
- Wu X., Weigel D., Wigge P.A. (2002). Signaling in plants by intercellular RNA and protein movement. Genes Dev. 16: 151–158 [DOI] [PubMed] [Google Scholar]
- Yoo S.K., Chung K.S., Kim J., Lee J.H., Hong S.M., Yoo S.J., Yoo S.Y., Lee J.S., Ahn J.H. (2005). CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol. 139: 770–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zordan R.E., Miller M.G., Galgoczy D.J., Tuch B.B., Johnson A.D. (2007). Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS Biol. 5: e256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.