A gene encoding an enzyme involved in abscisic acid biosynthesis is necessary and sufficient to enable high-temperature inhibition of lettuce seed germination. Introgressed and mutant alleles of NCED4 with reduced expression or enzymatic activity will allow breeding of lettuce cultivars with greater temperature tolerance during germination without compromising tolerance to water stress.
Abstract
Thermoinhibition, or failure of seeds to germinate at warm temperatures, is common in lettuce (Lactuca sativa) cultivars. Using a recombinant inbred line population developed from a lettuce cultivar (Salinas) and thermotolerant Lactuca serriola accession UC96US23 (UC), we previously mapped a quantitative trait locus associated with thermoinhibition of germination to a genomic region containing a gene encoding a key regulated enzyme in abscisic acid (ABA) biosynthesis, 9-cis-EPOXYCAROTENOID DIOXYGENASE4 (NCED4). NCED4 from either Salinas or UC complements seeds of the Arabidopsis thaliana nced6-1 nced9-1 double mutant by restoring germination thermosensitivity, indicating that both NCED4 genes encode functional proteins. Transgenic expression of Salinas NCED4 in UC seeds resulted in thermoinhibition, whereas silencing of NCED4 in Salinas seeds led to loss of thermoinhibition. Mutations in NCED4 also alleviated thermoinhibition. NCED4 expression was elevated during late seed development but was not required for seed maturation. Heat but not water stress elevated NCED4 expression in leaves, while NCED2 and NCED3 exhibited the opposite responses. Silencing of NCED4 altered the expression of genes involved in ABA, gibberellin, and ethylene biosynthesis and signaling pathways. Together, these data demonstrate that NCED4 expression is required for thermoinhibition of lettuce seeds and that it may play additional roles in plant responses to elevated temperature.
INTRODUCTION
Seed dormancy and germination are critical phenomena in plant life cycles and reproductive success (Donohue et al., 2010). Germination capacity and dormancy are determined by genetic factors as well as environmental cues, including light, water, and temperature (Bewley et al., 2013). Temperature is one of the most important environmental signals for seeds, which are able to sense and respond to low, high, and alternating temperatures that provide information about the season and the local microenvironment. In crop production, the sensitivity of seed germination to temperature sets the boundaries for planting dates and growing seasons, which in turn directly influence yield (Barnabás et al., 2008; Varshney et al., 2011). Inhibition of germination when seeds are exposed to supraoptimal temperatures, or thermoinhibition, is a common phenomenon in many winter annual or biennial species, preventing germination during summer when water may be inadequate for successful seedling establishment (Allen et al., 2007). Lettuce (Lactuca sativa) originated in the Mediterranean region as a winter annual, and most cultivated lettuce varieties exhibit seed thermoinhibition. Lettuce plantings in warm seasons therefore require seed priming and cooling by sprinkler irrigation to overcome thermoinhibition for crop establishment (Valdes et al., 1985). It would be beneficial to identify the genetic mechanisms underlying lettuce seed thermoinhibition to ameliorate or eliminate it through breeding.
Physiologically, interactions between abscisic acid (ABA) and gibberellin (GA) play key roles in regulating seed germination at different temperatures. Low-temperature (moist chilling) treatment of Arabidopsis thaliana seeds promotes germination by increasing GA content through upregulation of GA biosynthetic enzymes, such as GA-3-OXIDASEs (GA3ox), and downregulation of GA catabolic enzymes, including GA-2-OXIDASEs (GA2ox) (Yamauchi et al., 2004; Rieu et al., 2008). Chilling also induces the expression of ABA 8’-HYDROXYLASE, an ABA catabolic enzyme, lowering ABA content (Penfield and Hall, 2009). On the other hand, high temperature has the opposite effect, upregulating several ABA biosynthetic enzymes, including 9-cis-EPOXYCAROTENOID DIOXYGENASE9 (NCED9), and resulting in thermoinhibition of germination of Arabidopsis seeds (Toh et al., 2008). NCED catalyzes oxidative cleavage of the 9-cis isomer of violaxanthin or neoxanthin to xanthoxin, which is a rate-limiting step in ABA biosynthesis (Nambara and Marion-Poll, 2005). By contrast, seeds of ABA-deficient (aba1) and ABA-insensitive (abi3) mutants in Arabidopsis can germinate at high temperatures, suggesting a major role for ABA in thermoinhibition (Tamura et al., 2006). In lettuce seeds, thermoinhibition of germination was associated with upregulation of ABA biosynthetic and signal transduction genes, such as NCED4 and ABI5, while the combined application of an ABA biosynthesis inhibitor and exogenous GA can alleviate thermoinhibition (Yoshioka et al., 1998; Gonai et al., 2004; Argyris et al., 2008). Ethylene can also increase the upper temperature limit for lettuce seeds, while the expression of genes encoding ethylene biosynthetic enzymes is suppressed at high temperatures (Dutta and Bradford, 1994; Argyris et al., 2008).
Environmental signals during seed development, particularly temperature and light, also influence seed dormancy. Lettuce seeds that mature at lower temperatures or in far-red light exhibit lower maximum germination temperatures than those that develop at warmer temperatures or in red light (Gray et al., 1988; Contreras et al., 2009). Similarly, Arabidopsis seeds that develop and mature at lower temperature exhibit higher dormancy, while higher temperature during seed development and maturation results in reduced dormancy (Donohue et al., 2005; Chiang et al., 2009). Expression of DELAY OF GERMINATION1 (DOG1), a gene associated with seed dormancy, is higher in seeds matured at cool temperatures (Chiang et al., 2011; Kendall et al., 2011). However, seeds of the ABA-deficient mutant aba1-1 showed no dormancy even when matured at low temperature, indicating that ABA also is involved in the dormancy associated with DOG1 expression (Bentsink et al., 2006; Kendall et al., 2011).
Despite the wealth of empirical information about the role of temperature in regulating seed dormancy and germination (Baskin and Baskin, 1998), relatively little is known about the mechanism(s) by which seeds can sense temperature, especially high temperature, and transduce that signal into gene expression and developmental transitions (Penfield, 2008). An approach to investigate the mechanisms of regulation of seed dormancy and germination by temperature is to identify genetic variation for the trait and isolate temperature-responsive genes associated with germination capacity at high temperatures. In lettuce, we identified an accession of the progenitor species of cultivated lettuce (Lactuca serriola) that exhibited the ability to germinate at temperatures as high as 38°C (Argyris et al., 2005). This accession (UC96US23, hereafter termed UC) was used as a parent with L. sativa cv Salinas (hereafter termed Sal) to develop a recombinant inbred line mapping population that enabled us to identify a major quantitative trait locus (QTL) for this high-temperature germination (HTG) trait. This QTL (Htg6.1) accounted for ∼25% of the total phenotypic variance and was confirmed in near-isogenic lines (Argyris et al., 2011). Fine mapping of Htg6.1 narrowed the QTL interval to ∼1 centimorgan that contained NCED4. In addition, NCED4 expression increased in thermoinhibited Sal seeds but not in thermotolerant UC seeds, and ABA contents were approximately fivefold greater in the former when imbibed at 35°C relative to imbibition at 20°C (Argyris et al., 2008). These results provided strong correlative evidence that NCED4 is associated with the Htg6.1 thermotolerant phenotype.
In this study, we specifically tested whether NCED4 is involved in thermoinhibition of lettuce seed germination and is responsible for the Htg6.1 QTL. Using functional complementation, gene silencing, and mutant alleles, our results confirm that NCED4 expression is necessary and sufficient to inhibit lettuce seed germination at high temperature. Unlike the Sal allele of NCED4, the UC allele is not induced by imbibition at high temperature, reducing ABA biosynthesis and enabling germination. The difference between Sal and UC NCED4 alleles is due to variation in the promoter sequences rather than in the protein coding sequence. We further tested the effect of modifying the expression of NCED4 on seed development and on plant stress tolerance. Our results provide insight into the roles of multiple members of the NCED gene family in seed development, dormancy, and plant stress tolerance.
RESULTS
Ls-NCED4 Coding Regions from Sal and UC Rescue Arabidopsis NCED Mutants
Genetic mapping and physiological data strongly implicate NCED4 as the causal gene in the Htg6.1 QTL (Argyris et al., 2008, 2011). However, it has not been demonstrated conclusively whether the functionality of this enzyme differs between the two genotypic alleles. We therefore isolated and sequenced the NCED4 protein coding regions from the Sal and UC alleles. Both NCED4 genes contain no intron, and the Sal NCED4 shares 99.3% amino acid sequence identity to UC allele (see Supplemental Figure 1 online). Protein amino acid sequence alignments indicate that NCED4 is more closely homologous to Arabidopsis NCED6 than to NCED9 (see Supplemental Figure 2 and Supplemental Data Set 1 online). However, it was reported that NCED9, but not NCED6, was essential for germination thermoinhibition in Arabidopsis, and seeds of the Arabidopsis mutant nced9-1 displayed loss of thermoinhibition (Toh et al., 2008). We therefore used Arabidopsis nced6-1 nced9-1 double mutants to test the functionality of the lettuce NCED4 alleles.
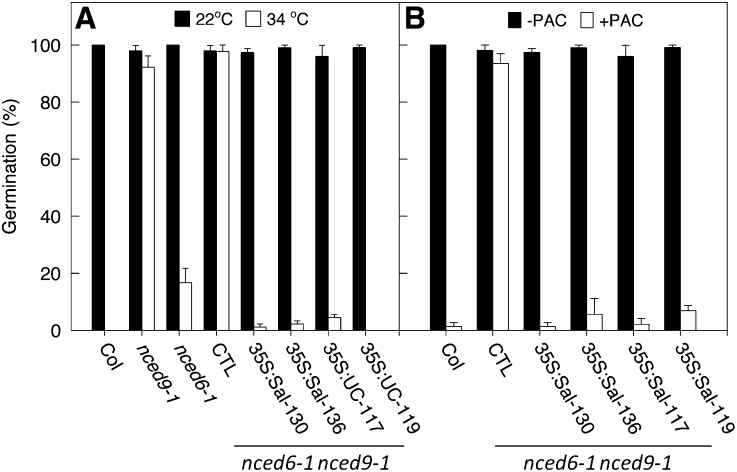
Seeds of both the nced9-1 single mutant and the nced6-1 nced9-1 double mutant exhibited thermotolerance during germination, while only around 20% of seeds of the nced6-1 single mutant germinated at 34°C, indicating that the thermotolerance of the double mutant was largely contributed by loss of function in NCED9 (Figure 1A). To test whether the lettuce NCED4 alleles encoded functional enzymes, we introduced the NCED4 coding and 5′ untranslated regions from Sal and UC driven by the constitutive cauliflower mosaic virus 35S promoter (CaMV35S) into the Arabidopsis nced6-1 nced9-1 double mutant. Germination of seeds of transformed lines containing either 35S:Sal-NCED4 (Sal-130 and Sal-136) or 35S:UC-NCED4 (UC-117 and UC-119) was strongly inhibited at 34°C, indicating that NCED4 coding regions from both Sal and UC could complement the loss of function in At-NCED9. As a further test, freshly harvested seeds of nced6-1 nced9-1 double mutant (empty vector controls) were able to germinate fully at 20°C in the presence of 30 μM paclobutrazol (PAC; a GA biosynthesis inhibitor) (Lefebvre et al., 2006), while seeds of the Columbia wild type and of transformed lines containing 35S:Sal-NCED4 or 35S:UC-NCED4 were inhibited (Figure 1B). We conclude that Ls-NCED4 encodes a NCED enzyme in the ABA biosynthetic pathway and that the proteins encoded by both Sal and UC alleles are equally functional.
Figure 1.
Both Sal and UC Alleles of NCED4 Rescue the Arabidopsis nced6-1 nced9-1 Double Mutant.
(A) Germination at 34°C and 22°C of F4 seeds from Arabidopsis nced6-1 nced9-1 double mutants transformed with 35S:Sal-NCED4 or 35S:UC-NCED4. Data for two independent lines for Sal (35S:Sal-130 and 136) and for UC (35S:UC-117 and 119) are shown, along with nced6-1 nced9-1 seeds containing empty vector (CTL), nced6-1 and nced9-1 single mutants, and Columbia wild type (Col). Final germination percentages (+se) are shown after 3 d incubation in the light at 22 or 34°C.
(B) Germination of F4 seeds from Arabidopsis nced6-1 nced9-1 double mutant transformed with 35S:Sal-NCED4 or 35S:UC-NCED4 in the presence of 30 μM PAC. Two independent lines for Sal (35S:Sal-130 and 136) and for UC (35S:UC-117 and 119) are shown along with the empty vector control (CTL) and the wild type (Col). Seeds were pretreated with PAC for 3 d at 4°C and subsequently incubated in the light at 20°C for 3 d. Error bars denote se (n ≥ 3).
Gain or Loss of Function of NCED4 Can Mediate Lettuce Seed Thermoinhibition
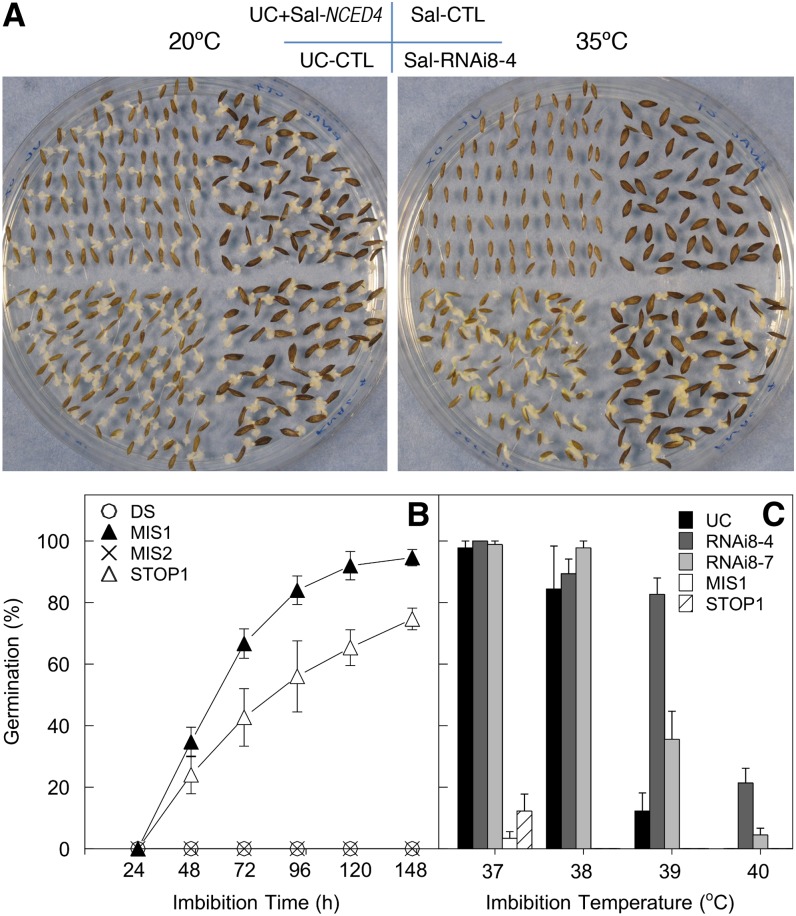
We next tested whether gain of function of Sal-NCED4 can result in thermoinhibition in UC seeds. We transformed UC plants with Sal-NCED4 driven by its native promoter (ProSal:Sal-NCED4). Seeds from T1 transgenic plants were expected to segregate for the transgene. As the UC allele is recessive to the Sal allele (Argyris et al., 2005), seeds that are homozygous or heterozygous for the transgene (75% if there is only one copy) are expected to exhibit thermoinhibition, while seeds that are homozygous for UC NCED4 are expected to germinate at high temperature. Phenotypic screening showed that seeds from seven of 12 transformed UC lines exhibited significant thermoinhibition compared with the controls containing empty vector (see Supplemental Figure 3 online). Genotyping of seeds from three independent lines showed that 96.3% (289/300) of nongerminating seeds contained the Sal-NCED4 transgene, while 92% (87/95) of the germinating seeds genotyped did not contain the transgene. Germination of homozygous transgenic UC seeds derived from one of these T1 lines (UC+Sal-NCED4-3) displayed complete thermoinhibition at 35°C, further supporting that gain of function of the Salinas NCED4 allele with its promoter is sufficient for expression of thermoinhibition (Figure 2A; see Supplemental Figure 3 online). Endogenous ABA content in the dry seeds of this homozygous UC+Sal-NCED4 line was 2.7-fold higher than in control UC seeds and very similar to that in Sal seeds, indicating that the transgenic expression of Sal-NCED4 resulted in greater ABA accumulation (Table 1).
Figure 2.
Gain- or Loss-of-Function Analysis of Sal-NCED4.
(A) Germination of homozygous transformed UC seeds containing ProSal:Sal-NCED4 (UC+Sal-NCED4), of homozygous Sal seeds in which NCED4 is silenced (Sal-RNAi8-4), of UC control seeds (UC-CTL) and of Sal control seeds (Sal-CTL). Seeds were imbibed on 1% agarose in the light at 20°C (left) and 35°C (right) for 36 or 48 h, respectively.
(B) Time courses of seed germination of TILLING mutants MIS1, MIS2, STOP1 and the wild type DS at 35°C in the light. Error bars denote se (n = 3).
(C) Seed germination of two TILLING mutants (MIS1 and STOP1), two independent RNAi lines (RNAi8-4 and RNAi8-7) and UC after 120 h imbibition at the indicated temperatures in the light. Error bars denote se (n = 3).
[See online article for color version of this figure.]
Table 1. ABA Content of Lettuce Seeds during Imbibition and Germination at 35°C.
| Genotype | Dry | 35°C for 24 h | 35°C for 48 h |
|---|---|---|---|
| Sal | 248 ± 20 | 99 ± 24 | |
| Sal-RNAi8-4 | 70 ± 11 | 39 ± 3 | 18 ± 10 |
| UC | 106 ± 25 | 32 ± 5 | |
| UC+Sal-NCED4 | 288 ± 20 | 130 ± 14 | 92 ± 11 |
| DS | 216 ± 23 | ||
| MIS1 | 116 ± 29 | ||
| MIS2 | 389 ± 51 | ||
| STOP1 | 173 ± 26 |
Quantification of ABA content (expressed as ng ABA/g dry weight) in dry seeds of Sal, Sal-RNAi8-4, UC, UC+Sal-NCED4, DS, MIS1, MIS2, and STOP1 in the Sal-RNAi8-4 and UC+Sal-NCED4 seeds imbibed at 35°C for 24 h and in the Sal, Sal-RNAi8-4, UC, and UC+Sal-NCED4 seeds imbibed at 35°C for 48 h. Values are means ± se (n = 3). Missing values were not measured.
We also tested whether loss of function of NCED4 due to mutation would result in loss of sensitivity to high temperature imbibition. Three NCED4 mutants were identified by TILLING (Comai and Henikoff, 2006) following chemical mutagenesis of lettuce cultivar Desert Storm (hereafter termed DS). One mutation at nucleotide 1146 of the coding region led to a premature stop codon (W382*, STOP1), and two missense mutations at nucleotides 1186 (MIS1) and 1510 (MIS2) resulted in replacements of Ala by Thr (A396T) and of Gly by Glu (G504E), respectively (see Supplemental Figure 4A online). Homozygous seeds of the MIS1 and STOP1 mutants displayed loss of thermoinhibition with germination percentages of ∼95 and ∼75%, respectively, at 35°C, while seeds of the MIS2 mutant failed to germinate after 148 h of imbibition at 35°C, as did the wild-type DS seeds (Figure 2B). Sequence alignment with VP14 from maize (Zea mays), which shares 51% amino sequence identity with NCED4 and for which a tertiary structure has been published (Messing et al., 2010), showed that both the STOP1 and MIS1 mutations occurred near a His residue (His-412) that is involved in binding an iron atom that is essential to the active site of the enzyme, while the MIS2 mutation is not near the active site (see Supplemental Figures 4A and 4B online). The ABA content of mature MIS1 mutant seeds was about half that of the wild-type DS seeds, while the ABA content of the STOP1 mutant seeds was reduced by ∼20% (Table 1).
We also used RNA interference (RNAi) to silence the NCED4 gene in Sal plants. The RNAi insert, designed to be specific to NCED4, was driven by the Sal-NCED4 promoter. Seeds from six of 10 independent transformed lines exhibited significant loss of thermoinhibition in T1 generation seeds, and since the RNAi insertion would be expected to be dominant, from 50 to 75% of the seeds germinated at 35°C (see Supplemental Figure 5A online). Genotyping showed that all germinated seeds contained RNAi constructs and that 95 of 105 thermoinhibited seeds were nontransgenic. Homozygous seeds from these lines displayed complete loss of thermoinhibition at 35°C (Figure 2A; see Supplemental Figure 5B online). In some lines, silencing of NCED4 resulted in germination at even higher temperatures (39 and 40°C) (Figure 2C) and at a more rapid rate (see Supplemental Figure 5B online) than observed in UC seeds. NCED4 transcripts were essentially undetectable in dry and imbibed RNAi seeds, while the control seeds (nontransgenic lines segregated from the selfed heterozygous RNAi lines) expressed NCED4 in a pattern comparable to previous reports (Figure 3A; Argyris et al., 2008). ABA contents of mature seeds of homozygous RNAi8-4 lines were <30% of that in Sal wild-type seeds and fell to only 20% of Sal seeds after imbibition for 48 h at 35°C (Table 1). These results demonstrate that silencing NCED4 is sufficient to reduce ABA contents at seed maturity and following imbibition at high temperature and to prevent thermoinhibition in lettuce seeds.
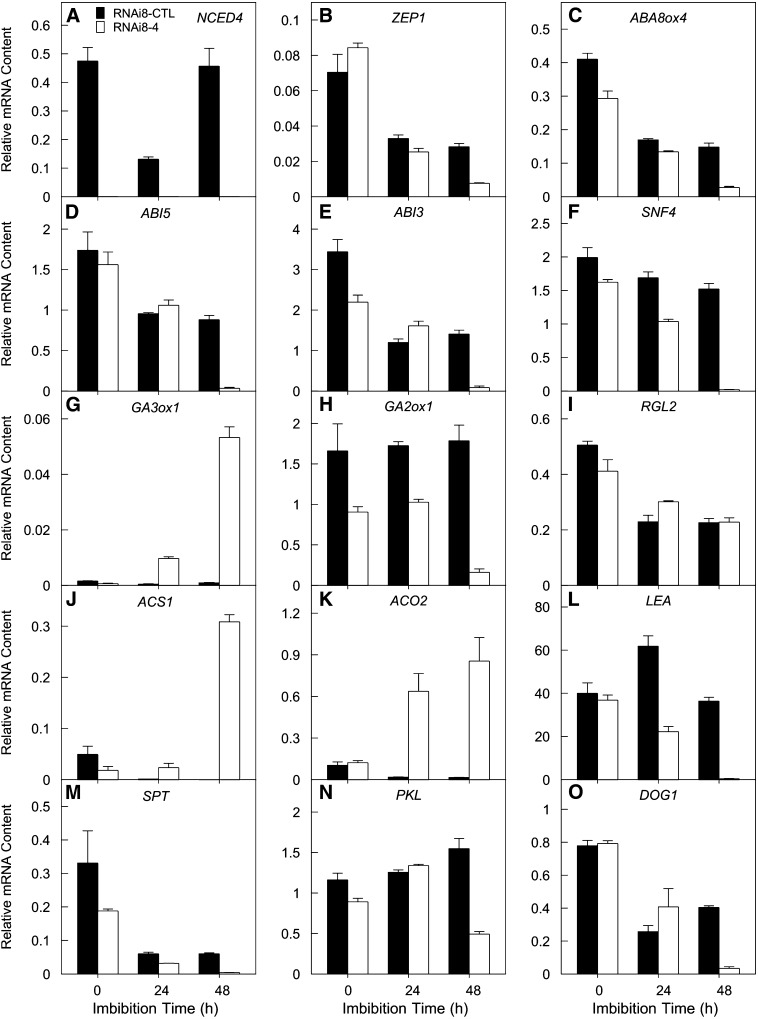
Figure 3.
Silencing of NCED4 Influences the Expression of Seed Dormancy-Related Genes.
Normalized mRNA abundances in homozygous RNAi (RNAi8-4) and control (RNAi8-CTL) seeds imbibed at 35°C in the light for genes associated with ABA biosynthesis and signal transduction ([A] to [F]), GA biosynthesis and signal transduction ([G] to [I]), ethylene biosynthesis ([J] and [K]), seed desiccation tolerance (L), temperature perception (M), chromatin remodeling (N), and dormancy maintenance (O). Error bars denote se of three biological replicates.
Arabidopsis mutants deficient in ABA biosynthesis have been reported to exhibit increased GA biosynthesis (Seo et al., 2006; Toh et al., 2008). The high temperature–resistant germination phenotype of Arabidopsis aba2-1 mutant seeds was almost completely suppressed by the GA biosynthesis inhibitor PAC (Toh et al., 2008). We therefore tested whether PAC would suppress the germination thermotolerance of RNAi-NCED4 lettuce seeds. Although control seeds (null segregants from RNAi transformation) could germinate completely at 31°C (near their maximum temperature limit), their germination was reduced to only 40% by 200 μM PAC, while PAC did not block the germination of RNAi seeds (see Supplemental Figure 5C online). On the other hand, the high-temperature germination phenotype of UC seeds was completely suppressed by the PAC treatment (see Supplemental Figure 5C online). This is consistent with the requirement of UC seeds for light (which stimulates GA biosynthesis) for germination at any temperature, while Sal seeds can germinate at low temperatures even in the dark (Argyris et al., 2008). Thus, a reduction in GA biosynthesis could be expected to have a greater effect on germination in UC than in Sal seeds. When ABA biosynthesis is suppressed by silencing of NCED4, Sal seeds apparently contain sufficient GA to permit germination even in the presence of PAC, as did Arabidopsis nced mutants (Figure 1B).
Gene Expression in NCED4-RNAi Lettuce Seeds Imbibed at High Temperature
To gain further insight into the consequences of NCED4 action in thermoinhibition of germination, we tested whether loss of thermoinhibition in RNAi-NCED4 lettuce seeds is associated with altered expression of other genes involved in related regulatory pathways. We previously reported on the expression of a large number of genes in lettuce seeds in relation to thermoinhibition (Argyris et al., 2008); here, we compare the expression patterns of some of these genes in control and RNAi seeds imbibed at 35°C. In control seeds in which germination was inhibited, mRNA abundance of a number of genes either declined somewhat or was constant following imbibition, but remained elevated after 48 h at 35°C. Among these were Ls-ZEP1 (Figure 3B), a homolog of the Arabidopsis ABA1 gene encoding an ABA biosynthetic enzyme (Seo and Koshiba, 2002); Ls-ABA8ox4 (Figure 3C), a homolog of the Arabidopsis CYP707A2 gene encoding an ABA 8’-hydroxlase that initiates ABA catabolism (Okamoto et al., 2006; Sawada et al., 2008); transcription factors involved in the ABA signal transduction pathway regulating germination (e.g., Ls-ABI5 and Ls-ABI3; Figures 3D and 3E) (Finkelstein et al., 2008); Ls-SNF4 (Figure 3F), an ABA-induced gene encoding a regulatory subunit of the SnRK1 kinase family involved in metabolic regulation in seeds (Bradford et al., 2003; Radchuk et al., 2006; Rosnoblet et al., 2007); Ls-GA2ox1 (Figure 3H), encoding a GA catabolic enzyme (Toh et al., 2008); Ls-LEA (Figure 3L), encoding a late embryogenesis abundant protein (Delseny et al., 2001); SPATULA (Figure 3M), a temperature-dependent germination repressor (Penfield et al., 2005); and Ls-DOG1 (Figure 3O), a homolog of Arabidopsis DOG1, which is involved in the induction and maintenance of seed dormancy (Bentsink et al., 2006; Chiang et al., 2011; Footitt et al., 2011; Kendall et al., 2011). All of these genes exhibited a tendency for a more rapid decrease in transcript abundance in the RNAi seeds following imbibition, and their mRNAs were very low or absent by 48 h when germination had been completed.
Another group of genes exhibited the opposite pattern, in which expression was very low in the control seeds but increased over imbibition time in the RNAi-NCED4 seeds. Among these genes were Ls-GA3ox1 (Figure 3G), encoding an enzyme forming active GA (Toh et al., 2008); Ls-ACS1 (Figure 3J), encoding 1-aminocyclopropane-1-carboxylic acid (ACC) synthase; and Ls-ACO2 (Figure 3K), encoding an ACC oxidase. Together, ACS1 and ACO2 constitute the ethylene biosynthetic pathway. Increases in transcripts of both GA- and ethylene-related genes are associated with alleviation of thermoinhibition and promotion of germination in lettuce seeds (Argyris et al., 2008). Somewhat in contrast with this was the expression pattern for a lettuce homolog of RGL2, a DELLA gene that encodes a GA signaling repressor that may act in part through stimulating ABA biosynthesis and ABI5 activity (Piskurewicz et al., 2008). Although transcript abundance declined by ∼50% during imbibition at 35°C, this was not affected by silencing of NCED4 (Figure 3I). Expression of PICKLE (Figure 3N), encoding a SWI/SNF-class chromatin-remodeling factor that is proposed to repress seed-associated genes in Arabidopsis (Li et al., 2005; Perruc et al., 2007), increased in control seeds during imbibition at 35°C, but initially increased and then declined in RNAi seeds.
Expression of NCED Family Members and Other Genes during Lettuce Seed Development
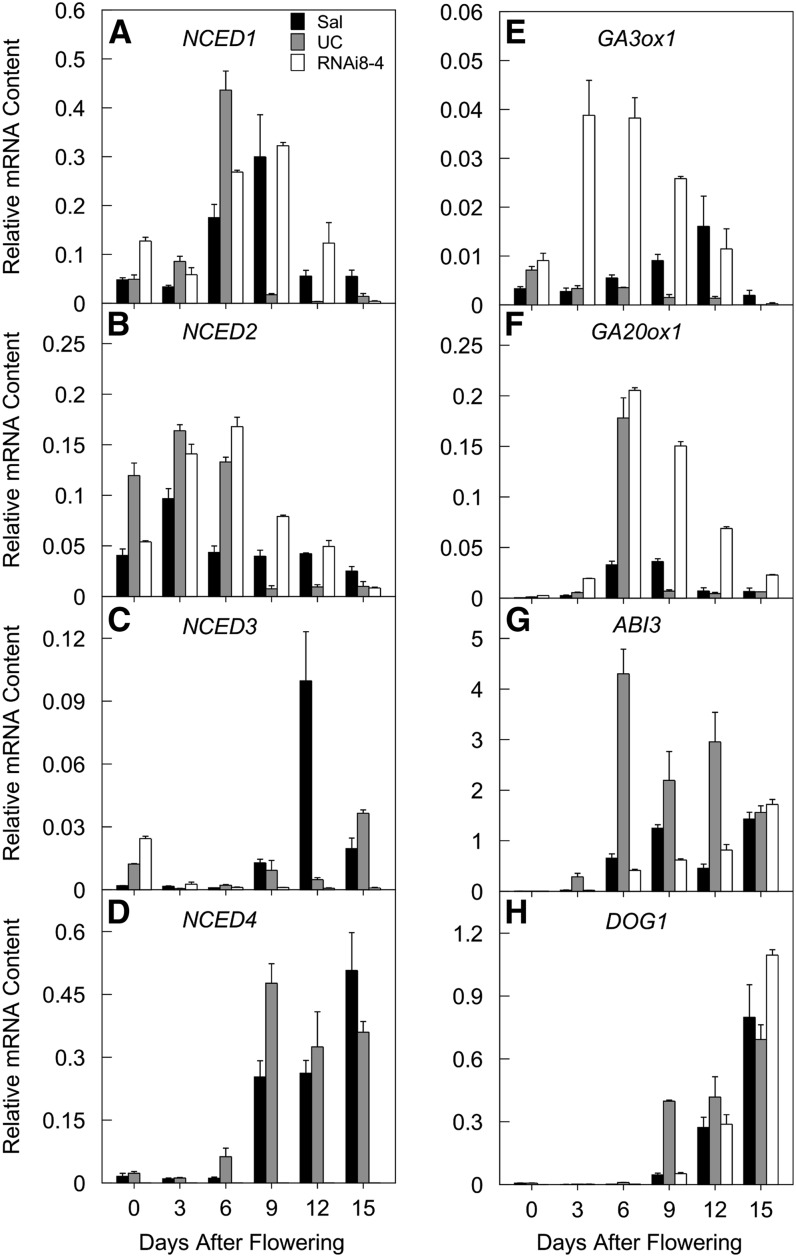
In Arabidopsis, NCED6 and NCED9 are highly expressed specifically in seeds: NCED6 is exclusively expressed in the endosperm, whereas NCED9 is expressed in both embryo and endosperm tissues (Lefebvre et al., 2006). Specific induction of NCED6 during Arabidopsis seed development is sufficient to enhance seed dormancy (Martínez-Andújar et al., 2011). In mature lettuce seeds, NCED4 mRNA is present in the embryo, but not in the endosperm (Argyris et al., 2008); information on NCED4 expression during lettuce seed development has not been reported. We therefore assayed mRNA dynamics in developing seeds of the four NCED genes identified in lettuce (NCED1-4) (Sawada et al., 2008). NCED2 expression increased first during seed development and then declined (Figure 4B), followed by a transient increase in NCED1 expression to two- to threefold higher levels (Figure 4A). NCED4 expression then increased to high levels and its mRNA remained abundant until seed maturity (Figure 4D). NCED3 mRNA was detected at much lower levels during development except for a transient increase at 12 d after pollination in Sal seeds (Figure 4C). NCED4 mRNA abundance was somewhat greater in Sal than in UC seeds at maturity, in agreement with their relative ABA contents in dry seeds (Table 1). These results suggest that NCED1 and NCED2 are responsible for ABA biosynthesis in early seed development, followed by NCED4 expression during late seed development and maturation.
Figure 4.
Transcriptional Analyses in Developing Seeds of Sal, UC, and a Sal-RNAi-NCED4 Line.
Normalized mRNA abundances of: NCED1-4 ([A] to [D]), GA3ox1 (E), GA20ox1 (F), ABI3 (G), and DOG1 (H) in developing seeds of Sal, UC, and a homozygous RNAi-NCED4 line (RNAi8-4). Error bars denote se of three biological replicates.
GAs are involved in seed development but generally decrease during maturation as dormancy is initiated (Holdsworth et al., 2008). We assayed expression of two GA biosynthetic genes, GA3ox1 and GA20ox1, during lettuce seed development (Figures 4E and 4F). Expression levels were generally quite low, with a transient rise in GA3ox1 mRNA in Sal seeds and of GA20ox1 in UC seeds during mid-development. The abundance of mRNAs of both genes was very low at maturity (15 d after flowering [DAF]).
Several regulatory genes are involved in the imposition of dormancy during seed maturation, including ABI3 and DOG1 (Bentsink et al., 2006; Holdsworth et al., 2008). The mRNA levels of ABI3 in UC seeds are higher than those in Sal seeds at all stages of development; the rise of ABI3 in UC seeds starts very early with its peak at 6 DAF, while its maximum expression in Sal developing seeds did not occur until 15 DAF (Figure 4G). DOG1 transcripts increased abruptly at 9 DAF in UC developing seeds and continued to accumulate until seed maturity (Figure 4H). In Sal, DOG1 mRNA abundance at 9 DAF was much lower than in UC seeds; however, the maximum levels at 15 DAF were similar in seeds of the two genotypes (Figure 4H).
The availability of RNAi-NCED4 Sal plants provided the opportunity to assess the specificity of gene silencing and the effect of NCED4-dependent ABA synthesis during seed maturation on the expression of other genes. The silencing of NCED4 by expressing an RNAi construct under its own promoter was extremely effective, reducing its mRNA abundance to almost undetectable levels (Figures 3A and 4D). However, this resulted in only minor effects on NCED1 and NCED2 mRNA abundances compared with Sal control seeds, except for somewhat higher amounts of NCED2 mRNA during the mid-phases of development (3 to 12 d after pollination) (Figures 4A and 4B). In addition, the rise in NCED3 expression at 9 to 12 DAF observed in Sal seeds was not evident in the RNAi seeds (Figure 4C). This is unlikely to be due to nonspecific silencing (see results for leaves below). The expression of GA3ox1 and GA20ox1 in NCED4-RNAi developing seeds was significantly altered compared with Sal seeds. GA3ox1 expression increased markedly at 3 to 9 DAF, whereas expression of GA20ox1 subsequently increased at 6 to 12 DAF (Figures 4E and 4F), indicating that GA biosynthesis might be suppressed by NCED4-dependent ABA biosynthesis. Interestingly, the elevation of expression of both of these genes occurred just prior to the normal initiation of increased NCED4 expression at 9 DAF (Figure 4D). The expression pattern of ABI3 in RNAi developing seeds is comparable to that of Sal seeds (Figure 4G). Similarly, silencing of NCED4 did not significantly affect expression of DOG1 (Figure 4H). Thus, the expression of ABI3 and DOG1 during lettuce seed development does not require NCED4-dependent ABA production.
While silencing of NCED4 had no obvious effect on seed morphology, inflorescences on RNAi plants appeared to be smaller than those on control plants (see Supplemental Figures 6A and 6B online). In fact, the average number of seeds per inflorescence was reduced by almost 50% in RNAi plants, while weight per seed was ∼20% greater (Table 2; see Supplemental Figure 6B online). The reduced seed set could result in the increased weight per seed due to less competition for resources among seeds. Undeveloped floret numbers were similar in inflorescences of RNAi and control lines (Table 2; see Supplemental Figure 6B online), indicating that the reduced seed set in RNAi lines was due to fewer florets per inflorescence, rather than to failure of pollination or abortion of seed development. This suggests that NCED4, and therefore ABA, may have a role in the differentiation of lettuce inflorescences.
Table 2. Effect of Silencing of NCED4 on Lettuce Seed Set and Seed Weight.
| Line | Seeds per Flower | Weight per Seed | Undeveloped Seeds per Flower |
|---|---|---|---|
| RNAi2-CTL | 10.93 ± 0.79 | 1.11 ± 0.01 | 6.12 ± 0.29 |
| RNAi8-CTL | 12.20 ± 0.79 | 1.32 ± 0.02 | 6.05 ± 0.36 |
| RNAi8-7 | 6.57 ± 0.35** | 1.55 ± 0.02** | 6.10 ± 0.28 |
| RNAi8-4 | 6.40 ± 0.38** | 1.54 ± 0.03** | 6.15 ± 0.29 |
For seed set, seeds (full and undeveloped) of 15 buds for each replicate were collected for four replicates each from two homozygous RNAi lines (RNAi8-4 and RNAi8-7) and two RNAi control lines (RNAi2-CTL and RNAi8-CTL). For seed weight, 50 mature seeds were weighed for each replicate for each line. Statistical analysis by a one-tailed Student’s t test indicated significant differences (**P < 0.01).
NCED4 Expression Is Associated with Natural Variation in Thermoinhibition of Lettuce Seeds
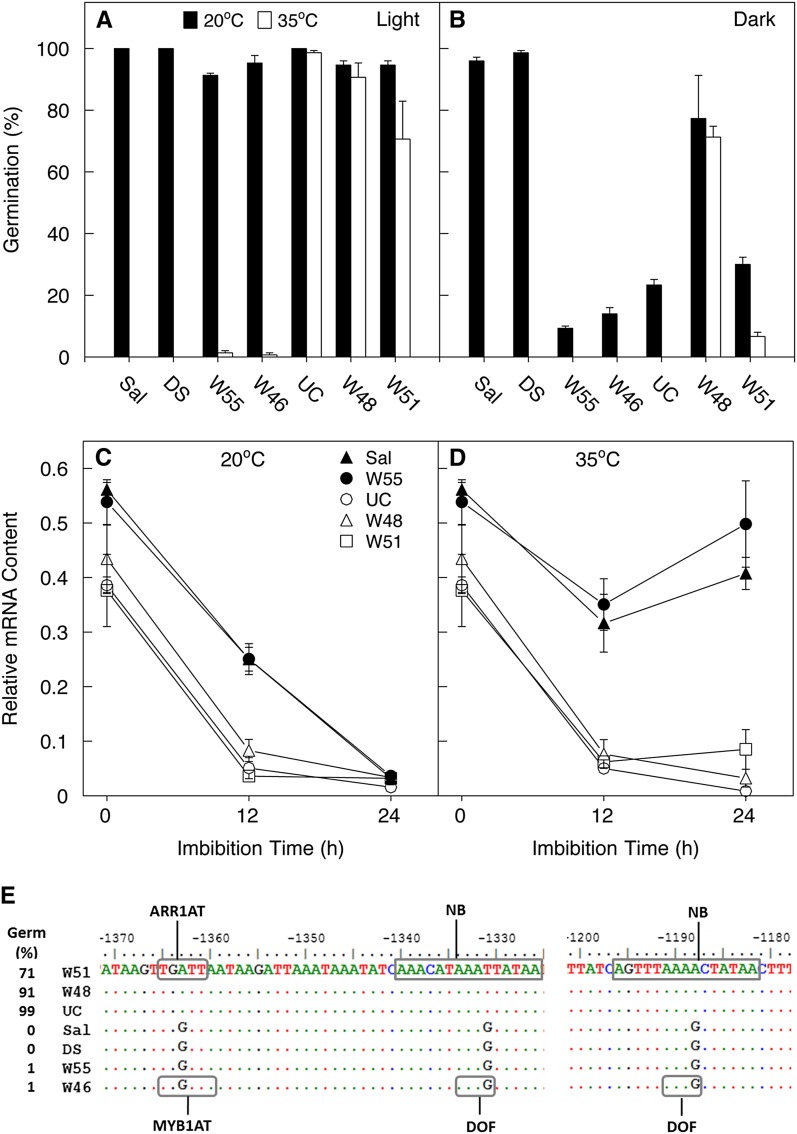
As the UC accession of L. serriola exhibits germination thermotolerance (Argyris et al., 2011), we screened 29 additional L. serriola accessions to assess their temperature sensitivity for seed germination (Argyris, 2008). Only two accessions (W48 [accession SAL-90] and W51 [accession UC99TRK18-4]) exhibited germination capacity at 35°C comparable to UC (Figure 5A), indicating that germination thermotolerance is rare even in L. serriola. Other accessions, such as W46 (SAL-28) and W55 (CGN14263), were selected to represent L. serriola accessions exhibiting thermoinhibition similar to cultivated varieties such as Sal and DS (Figure 5A). Light requirements for seed germination of these genotypes were also examined at both 20 and 35°C (Figure 5B). Seeds of both Sal and DS germinated well at 20°C even in the dark, although both are thermoinhibited in light or dark. Seeds of W46 and W55, however, required light to germinate above 15% even at 20°C. Seeds of W48, on the other hand, had germination percentages of ∼70% in the dark at both temperatures, while seeds of W51 required light to germinate well at either temperature. These results suggest that while light and temperature interact to regulate seed germination, they may act through genetically separable pathways.
Figure 5.
High NCED4 Expression Is Associated with Thermoinhibition.
(A) and (B) Seed germination of thermosensitive (Sal, DS, W46, and W55) and thermotolerant (UC, W48, and W51) varieties and accessions at 20 and 35°C in the light (A) or in the dark (B) for 3 d. Closed bars, 20°C; open bars, 35°C.
(C) and (D) Normalized NCED4 mRNA abundance in dry (0 h) and imbibed seeds at 20 or 35°C, respectively. Error bars denote se (n = 3).
(E) Alignment of NCED4 promoters from lettuce genotypes. Boxed sequences are predicted cis-elements based on the PLACE database. NB, no binding site; see the text for descriptions of MYB1AT, ARR1AT, and DOF. The seed germination percentages (Germ) of the genotypes at 35°C are shown at left; W51, W48, and UC are thermotolerant accessions.
[See online article for color version of this figure.]
We subsequently examined the NCED4 expression patterns in the two groups of genotypes (thermosensitive, Sal and W55; and thermotolerant, UC, W48, and W51) upon imbibition at 20 and 35°C in the light. At 20°C, NCED4 mRNA decreased to very low levels within 24 h in the imbibed seeds of all five genotypes (Figure 5C); at 35°C, the mRNA content also declined to low levels in thermotolerant lines, but first decreased and then became elevated in thermosensitive lines (Figure 5D). Thus, elevated expression of NCED4 in seeds imbibed at high temperature is conserved between the cultivated varieties and thermosensitive genotypes of L. serriola, while reduced expression of NCED4 at high temperature is conserved among L. serriola lines exhibiting thermotolerance.
To test whether this variation in expression patterns of NCED4 in response to high temperature could be due to polymorphisms among their genomic sequences, we isolated and sequenced the coding and upstream promoter regions of NCED4 alleles from Sal, DS, UC, W55, W51, W46, and W48 (see Supplemental Data Sets 2 and 3 online). The sequences of the promoter and coding regions of Sal and DS are identical. Similarly, W48 has identical NCED4 coding and promoter region sequences to that of UC. The alignments of all seven NCED4 protein coding sequences showed that there are only five polymorphic sites, but these sites are not conserved within the two distinct groups (i.e., among thermotolerant and thermosensitive genotypes) (see Supplemental Figure 1 online). However, 19 polymorphic sites are conserved in the promoter and 5′ untranslated regions within each group (see Supplemental Data Set 3 online). Some of these polymorphic sites are predicted to change binding sites for regulatory proteins (Figure 5E). For example, at positions −1331 and −1188, guanines in promoters of the thermosensitive lines are changed to adenine (−1188) or thymine (−1331) in thermotolerant line promoters, resulting in the predicted loss of binding sites for the DOF class of DNA binding proteins. Similarly, a polymorphism at −1363 caused the replacement of a MYB recognition site by an ARR1 binding site. Further study is required to determine whether such polymorphisms underlie the differential temperature responses of these promoters.
NCED4 Expression Is Temperature Responsive but not Drought Responsive in Leaves
As NCED4 is functionally homologous to the seed-specific NCED9 in Arabidopsis, we asked whether NCED4 expression is seed specific in lettuce. If NCED4 is expressed in other plant tissues, its mutation or silencing could have pleiotropic effects on other aspects of plant development, as noted above for floret number. In particular, as ABA is involved in plant responses to water stress, such as stomatal closure, silencing of NCED4 to improve germination could adversely affect stress tolerance. On the other hand, if NCED4 is specifically regulated by temperature, exploring its expression in other tissues could shed light on the mechanism by which temperature is sensed. We therefore assayed NCED4 mRNA levels in lettuce leaves and roots and in response to drought and high temperature stresses.
NCED4 mRNA abundance is high in roots and leaves of 5-week-old plants of both Sal and UC, indicating that NCED4 expression is not seed specific (see Supplemental Figures 7A and 7B online). The other NCED family members are also expressed in leaves and roots, but with mRNA levels much lower than that of NCED4 in both tissues (see Supplemental Figures 8A and 8B online).
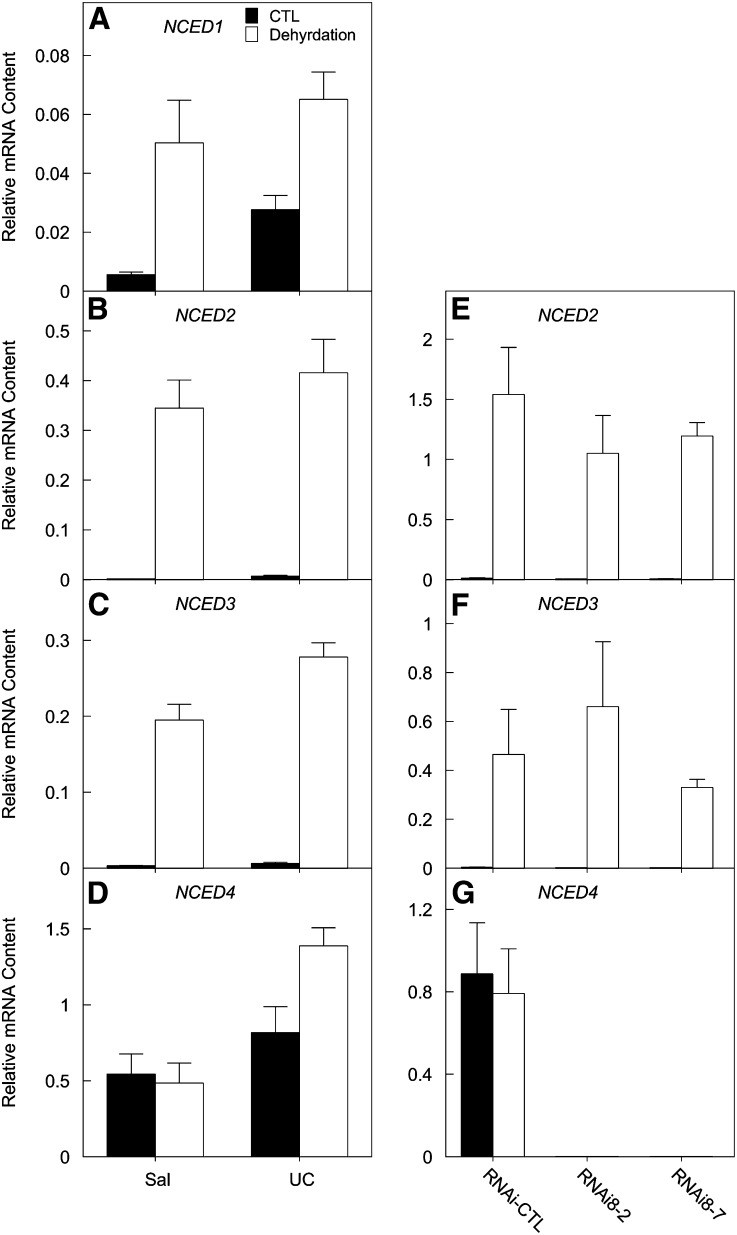
In Arabidopsis and other plants, specific NCED genes are induced by water stress (Tan et al., 2003). We therefore tested the responses to dehydration of NCED1-4 expression in lettuce leaves. Leaves from 8-week-old Sal and UC plants were collected and dehydrated at room temperature until reaching 15% water loss (∼2 h); in parallel, leaves were placed in a sealed plastic bag containing wet paper towels for the same period of time to control for excision or senescence effects. NCED2 and NCED3 expression increased markedly in response to water stress: 215- and 62-fold in Sal and 60- and 45-fold in UC, respectively (Figures 6B and 6C). NCED1 mRNA levels were very low and responded to water stress to a much smaller extent in both Sal and UC (Figure 6A). By contrast, NCED4 mRNA increased ∼50% in UC but did not change in Sal leaves in response to water stress (Figure 6D), suggesting that despite its relatively high constitutive expression in leaves, enhanced NCED4 expression is not involved in drought responses. This was confirmed in short-term drought experiments with both RNAi-NCED4 and MIS1 plants. There were no obvious differences between these NCED4-deficient lines and their respective control plants in either their exhibition of water stress symptoms during stress or their recovery following rewatering (see Supplemental Figure 8 online). NCED4 transcript was undetectable in leaves of the RNAi plants under both watered and water stress conditions, while its level in control leaves was very high and not responsive to dehydration (Figure 6G); by contrast, expression of NCED2 and NCED3 increased more than two orders of magnitude in both control and RNAi leaves due to dehydration (Figures 6E and 6F). These results indicate that NCED2 and NCED3 are specialized to respond to water stress and that despite the high level of its transcript in leaves, NCED4 expression is not essential to lettuce drought responses.
Figure 6.
NCED1-4 Responses to Dehydration Stress.
(A) to (D) NCED1-4 expression in response to dehydration. Eight-week-old leaf tissues were allowed to lose 15% of their initial fresh weight at room temperature (Dehydration). In parallel, leaf tissues were incubated in sealed plastic bags containing wet paper towels as controls (CTL). Error bars denote se (n = 3).
(E) to (G) NCED2-4 expression in leaves of RNAi-NCED4 lines (RNAi8-2 and RNAi8-7) and controls (CTL) in response to dehydration. Dehydration treatment was as described for (A) to (D) except that 6-week-old leaves were used. Error bars denote se (n = 3).
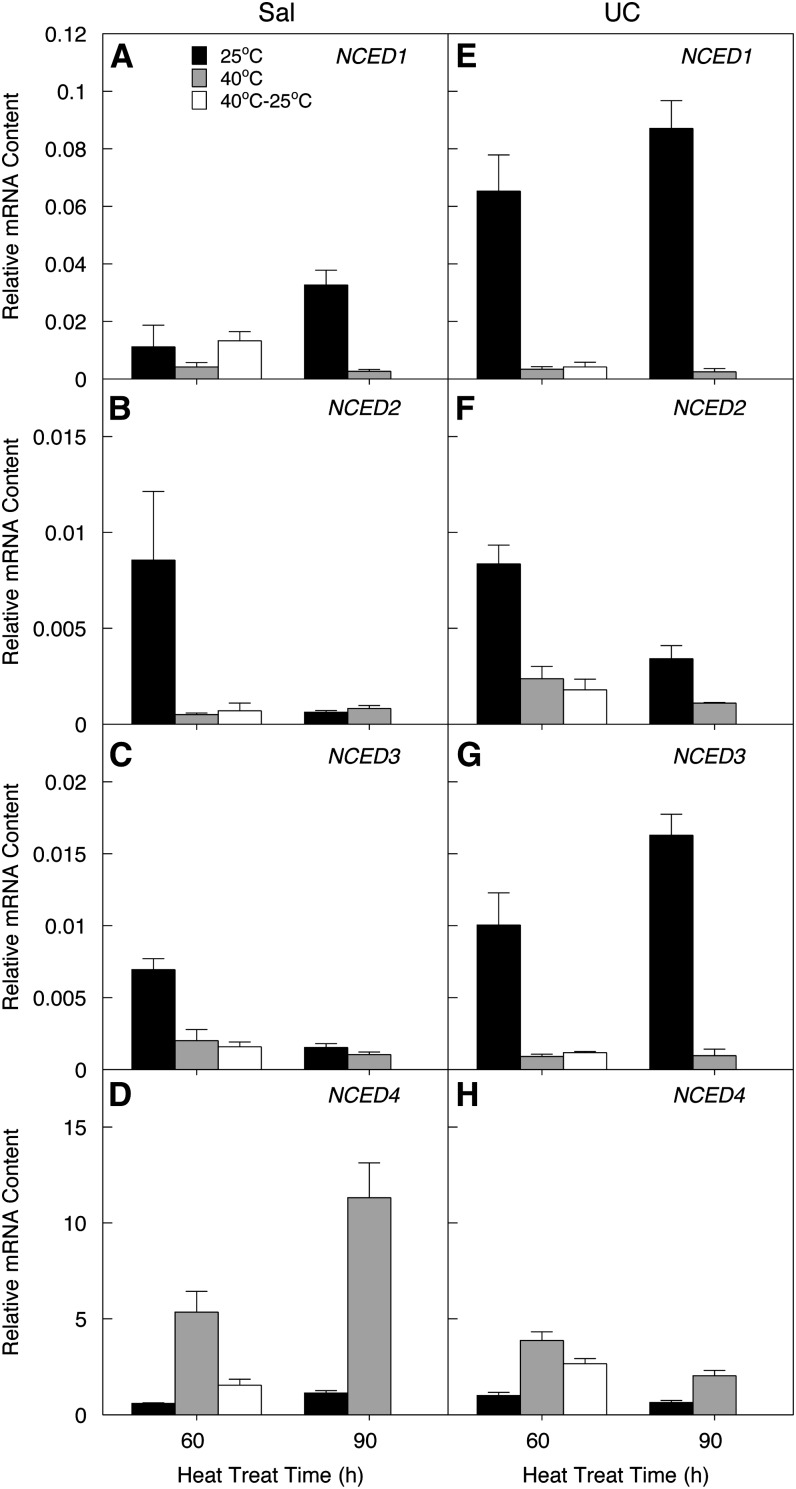
We further tested whether NCED4 expression in leaves is responsive to high temperature as it is in imbibed seeds. Leaves collected from 8-week-old Sal plants were placed into plastic bags containing wet paper towels to prevent dehydration and were exposed to a temperature of 40°C for 60 or 90 min; control leaves were treated similarly but were maintained at 25°C. In addition, following the 60-min heat stress, some leaves were returned to 25°C for 30 min to test for recovery from heat stress. NCED4 expression was upregulated ninefold and 3.8-fold compared with its expression at 25°C after 60 min at 40°C in Sal and UC, respectively, and increased further in Sal leaves after 90 min (Figure 7D). However, NCED4 mRNA in UC leaves decreased upon extension of temperature stress treatment, indicating that the Sal allele of NCED4 is more responsive to temperature than the UC allele (Figure 7H). After transfer back to 25°C, NCED4 mRNA levels decreased within 30 min (Figure 7D). In contrast with NCED4, NCED1-3 mRNA levels fell markedly in both Sal and UC leaves due to heat stress (Figures 7A to 7C and 7E to 7G), as occurs with many genes following a sudden heat shock (Busch et al., 2005). The marked contrast between the expression patterns of NCED4 and the other lettuce NCED family members in response to drought and heat suggests that the Sal-NCED4 promoter is particularly adapted to sense and respond to elevated temperatures, while NCED2 and NCED3 are specialized to respond to water stress.
Figure 7.
NCED1-4 Responses to Heat Stress.
Relative mRNA abundances of NCED1-4 in leaves of Sal ([A] to [D]) and UC ([E] to [H]) under heat stress. Leaves were sealed in plastic bags containing wet paper towels and were held at 25 or 40°C for 60 (black bars) or 90 (gray bars) min or were held at 40°C for 60 min followed by 25°C for 30 min (white bars). Results are representative of three independent experiments. Error bars denote se (n = 3) of biological replicates within an experiment.
DISCUSSION
Thermoinhibition of Germination by Temperature Regulation of NCED4 Expression in Lettuce Seeds
We previously identified one accession (UC) of the progenitor species (L. serriola) of cultivated lettuce (L. sativa) that exhibits thermotolerance during seed germination (Argyris et al., 2005). A major QTL for the high temperature germination trait (Htg6.1) collocated with NCED4, and fine mapping along with physiological and gene expression data supported the identification of NCED4 as a candidate gene for this QTL (Argyris et al., 2008, 2011). There are six single-nucleotide polymorphisms (SNPs) in the protein coding regions between Sal and UC alleles of NCED4, resulting in four amino acid differences (see Supplemental Figure 1 and Supplemental Data Set 2 online). However, mutant complementation studies in Arabidopsis showed that expression of NCED4 coding sequences from both Sal and UC could equally rescue nced6-1 nced9-1 double mutant seeds (Figure 1), indicating that both NCED4 alleles encode functional NCED proteins and that the thermotolerance of UC seeds is not due to its dysfunction but more likely to its reduced expression in response to high temperature. That NCED4 is required for thermoinhibition was demonstrated by silencing NCED4 in Sal seeds (Figure 2) and by mutations in NCED4 near the active site of the encoded enzyme (Figure 3), both of which resulted in increased germination at high temperatures. In addition, transformation of UC plants with Sal-NCED4 driven by the Sal promoter resulted in seeds that exhibited thermoinhibition (Figure 2). This result further indicates that the temperature-sensing mechanisms and trans-elements for regulating NCED4 expression are present and responsive to high temperature in UC seeds. Together, our results demonstrate that the elevated transcription of NCED4 in Sal seeds, but not in UC seeds, in response to imbibition at high temperature is responsible for their difference in germination thermosensitivity and that the differential regulation of NCED4 alleles might be derived from interactions between their promoters and trans-regulatory elements.
Our analysis of the promoter sequences of NCED4 alleles among both wild and cultivated genotypes revealed that 19 SNPs are conserved within thermotolerant or thermosensitive groups, some of which would be predicted to result in changes in binding specificities for trans-acting factors (Figure 5E; see Supplemental Data Set 3 online). A recent study showed that the wheat homolog of MOTHER OF FT (MFT) regulates seed germination in response to temperature and that the high MFT expression in the dormant wheat line was due to a single SNP in the promoter and a polymorphic single-sequence repeat in its third intron (Nakamura et al., 2011). The binding site for a basic domain/Leu zipper transcription factor in the less dormant line might be lost in the dormant line due to the SNP, resulting in the differential expression of MFT in the two lines. Similarly, genes encoding α-amylase enzymes in cereal grains are GA inducible and ABA repressible through binding of different combinatorial transcription factors to the same cis-regulatory elements in response to different environmental/ hormonal signals (Rushton et al., 2010). This mechanism may also apply to the regulation of NCED4 by temperature. At low temperature, one set of transcription factors may bind cis-elements containing no SNPs between Sal and UC or can tolerate the SNPs that are present to bind both cis-elements and downregulate NCED4 transcription. At high temperature, the polymorphic cis-elements could be bound by a different set of transcription factors, or the binding capacity may be lost in some variants, resulting in differential expression of NCED4 in Sal and UC seeds. For example, DOF binding sites at the positions of −1331 and −1188 of NCED4 promoters were conserved in the thermosensitive lines but were lost in the thermotolerant group (Figure 5E). Two DOF proteins, DOF AFFECTING GERMINATION1 (DAG1) and DAG2, have been reported to regulate seed germination in opposite ways: DAG1 is a negative regulator of Arabidopsis seed germination, whereas DAG2 positively regulates germination (Gualberti et al., 2002; Papi et al., 2002). Mutation of DAG1 also improved seed germination at high temperature, particularly under salinity stress (Rizza et al., 2011). Loss of thermoinhibition in dag1 mutants of Arabidopsis could be due to the coincident downregulation of several ABA biosynthetic genes, including ABA1, NCED6, and NCED9 (Gabriele et al., 2010). Thus, it is possible that modification of DOF protein binding could prevent the upregulation of NCED4 in thermotolerant lettuce lines.
FUSCA3 (FUS3), a B3-domain transcription factor and regulator of seed maturation, was recently reported to be responsive to high temperature during seed germination (Chiu et al., 2012). Although no evidence was provided for direct regulation of At-NCED9 by FUS3 in the seeds imbibed at high temperature, the promoter of At-NCED9 contains RY elements (CATGCATG) enabling DNA binding by FUS3 (Reidt et al., 2000; Mönke et al., 2004; Toh et al., 2008; Chiu et al., 2012). Our microarray data (unpublished) showed that FUS3 mRNA decreased in UC and Sal seeds imbibed at 20°C but remained at a high level when seeds were imbibed at 35°C for 24 h. RY elements were present in the promoter regions of all lettuce genotypes sequenced (e.g., position −390; see Supplemental Data Set 3 online), and a conserved SNP occurred just upstream of one RY site (position −398). Whether this SNP could affect the binding of B3-domain transcription factors and result in differential regulation of NCED4 remains to be investigated. The differences in temperature responses between the promoters of the Sal and UC alleles of NCED4 could be used to further explore temperature-sensitive signaling pathways and trans-acting factors.
NCED4 Shows Distinct Expression from Other NCED Genes in Lettuce
Different NCED gene family members from the same species differ with respect to their localization of expression and response to environmental signals (Tan et al., 2003; Lefebvre et al., 2006). In Arabidopsis, five NCED genes (NCED2, NCED3, NCED5, NCED6, and NCED9) have been proposed to be involved in ABA biosynthesis (Schwartz et al., 2003). NCED3 together with NCED5 are important regulators of ABA levels during water stress in Arabidopsis, and their transcripts are induced by drought (Tan et al., 2003; Ruggiero et al., 2004; Frey et al., 2012). Arabidopsis NCED5, NCED6, and NCED9 play important roles in ABA synthesis in the embryo and endosperm during dormancy induction (Lefebvre et al., 2006; Seo et al., 2006; Martínez-Andújar et al., 2011; Frey et al., 2012). NCED6 is regulated by light and its mutation promoted seed germination in the dark, whereas NCED9 and NCED5 are induced by high temperature in the imbibed seeds and their mutation resulted in loss of thermoinhibition (Seo et al., 2006; Toh et al., 2008). These results suggest that although NCED family members share high sequence similarity, they have distinct functions in responding to different environmental conditions.
In lettuce, four different NCED genes were identified (Sawada et al., 2008), but their specific expression patterns have not been reported. During seed development, NCED1 was expressed first, followed by NCED2 and then NCED4, while expression of NCED3 was much lower throughout (Figure 4). Expression of both NCED2 and NCED3 was strongly upregulated in leaves by water stress and reduced by heat, while NCED4 exhibited the opposite response (Figures 6 and 7). Clearly, multiple members of a related gene family provide considerable developmental and environmental flexibility in regulating ABA biosynthesis. Unlike At-NCED6 and At-NCED9 that are mainly expressed in reproductive organs, the embryo and endosperm in particular (Lefebvre et al., 2006), Ls-NCED4 mRNA is highly abundant constitutively in vegetative tissues, including roots and leaves (see Supplemental Figure 7 online), although translation and enzyme activity have not been assessed. Gene regulation can differ between seeds and vegetative tissues, as when temperature-responsive transcripts of Arabidopsis seeds and of seedlings grown at two temperatures were compared, there was very little overlap between the two developmental stages (Kendall et al., 2011). For example, HSP70 expression is strongly regulated in the seedling in response to temperature change, but HSP70 is not temperature regulated in seeds (Kumar and Wigge, 2010; Kendall et al., 2011). Upregulation of Ls-NCED4 by high temperature (40°C) in leaves was rapidly reversed by a short incubation at low temperature (25°C) (Figure 7D), indicating that its mRNA abundance in both leaves and seeds is modulated by temperature. Ls-NCED4 may play roles in heat stress in vegetative tissues as well as in seed thermoinhibition, as does At-NCED5 in both drought tolerance in vegetative tissues and seed dormancy induction in Arabidopsis (Frey et al., 2012).
Hormonal Crosstalk in Lettuce Seed Thermoinhibition
During seed germination, ABA is known to antagonize the promoting effect of GA, and ABA and GA mutually affect the metabolism of the other hormone (Bewley et al., 2013). The GA biosynthetic gene GA3ox1 was upregulated in lettuce seeds imbibed at 35°C in which NCED4 was silenced (Figure 3G). By contrast, a gene encoding a GA degrading enzyme, GA2ox1, remained highly expressed at high temperature but was reduced in RNAi-NCED4 seeds (Figure 3H), in a pattern similar to that for ABI5, ABI3, and SNF4. The upregulation of GA3ox1 and downregulation of GA2ox1 were likely a result of lower ABA content (Table 1), as GA3ox1 and GA20ox1 (also a GA biosynthesis gene) were also upregulated in RNAi-NCED4 developing seeds (Figures 4E and 4F). Similar results were observed in the Arabidopsis ABA biosynthetic mutants aba2-1 and aba2-2 (Seo et al., 2006; Toh et al., 2008). Thus, NCED4-dependent ABA biosynthesis appears to be involved in regulating GA biosynthesis and metabolism genes during both seed development and following imbibition of lettuce seeds.
Ethylene can promote seed germination, possibly through enhancing endosperm cap weakening (Kepczynski and Kepczynska, 1997; Linkies et al., 2009). Exogenous application of ethylene or its precursor ACC can alleviate thermoinhibition in lettuce seeds, whereas ethylene biosynthesis or action inhibitors can enhance thermoinhibition (Saini et al., 1986, 1989; Nascimento et al., 2000, 2004; Kozarewa et al., 2006). Silencing of NCED4 in Sal seeds increased transcript levels of two ethylene biosynthetic genes, ACS1 and ACO2 (Figures 3J and 3K), both of which also have high transcript levels in UC seeds imbibed at high temperature (Argyris et al., 2008). Similarly, At-ACO1 was also upregulated in Arabidopsis mutants that are deficient in ABA biosynthesis and signal transduction (Nakabayashi et al., 2005; Penfield et al., 2006; Carrera et al., 2008; Cheng et al., 2009). In Lepidium sativum, ABA inhibited the increase in transcript levels of ACO-encoding genes upon germination (Linkies et al., 2009). GA can promote ACO gene expression in Arabidopsis seeds (Ogawa et al., 2003), and GA and ethylene biosynthetic genes exhibited parallel expression patterns in lettuce seeds (Argyris et al., 2008). However, PAC did not suppress the germination of RNAi-NCED4 seeds (see Supplemental Figure 5C online), suggesting that ethylene might promote germination even in the absence of GA. Thus, ethylene may act antagonistically to ABA, and perhaps independently of GA, to regulate seed thermoinhibition in lettuce.
In general, the effects of silencing NCED4 in Sal seeds on the expression of other hormonal and dormancy-related genes were similar to those observed in UC seeds imbibed at high temperature (Argyris et al., 2008), further supporting the identification of the Htg6.1 QTL with reduced expression of the UC allele of NCED4 in response to imbibition at high temperature. Whether these expression differences are due directly to a reduction of ABA biosynthesis and content or are associated indirectly with the derepression of germination requires further investigation.
METHODS
Plant Materials
Lettuce (Lactuca sativa) cultivars Salinas and Desert Storm and Lactuca serriola accessions UC96US23, SAL-28 (W46), SAL-90 (W48), UC99TRK18-4 (W51), and CGN14263 (W55) were used in this study. Three TILLING mutants (STOP1, MIS1, and MIS2) of Desert Storm were identified and recovered for this study. Arabidopsis thaliana Columbia and its single mutants nced6-1 and nced9-1 were gifts from Eiji Nambara, while the double mutant nced6-1 nced9-1 was kindly provided by Annie Marion-Poll. Lettuce plants were grown in a greenhouse at 25°C under long days (16 h light/8 h dark). Arabidopsis plants were grown in a growth chamber at 22°C under long days (16 h light/8 h dark).
Seed Germination Assays
Lettuce seed germination was conducted in 4.7-cm Petri dishes using the methods described previously (Argyris et al., 2008). Thermoinhibition tests were performed in the light or dark at 35°C unless stated. For germination sensitivity to PAC, lettuce seeds were imbibed in 200 μM PAC (Sigma-Aldrich) for 3 d at 4°C and then transferred to 31°C in the light.
Arabidopsis germination tests used the method described by Toh et al. (2008). For germination sensitivity to PAC, Arabidopsis seeds were imbibed in 30 μM PAC for 3 d at 4°C before transfer to a germination chamber at 22°C.
Isolation of Ls-NCED4 and Its Promoters
End-to-end PCR primers were designed from the NCED4 sequence of Grand Rapids (AB120110) and used for isolation of NCED4 from either genomic DNA or cDNA. For isolation of the NCED4 promoter, PCR was used to screen a L. sativa cv Diana BAC library and identify a BAC containing NCED4 (KC676791). All primers for isolating NCED4 and its promoters are listed in Supplemental Table 1 online.
TILLING
Mutations in NCED4 were identified by TILLING using a population of ethyl methanesulfonate–mutagenized L. sativa cv Desert Storm (D. Facciotti, C. McGuire, and C.M. McCallum, unpublished data). Gene-specific TILLING primers were designed based on the NCED4 sequence of cv Grand Rapids (AB120110). Two PCR fragments covering most of the open reading frame were screened using the primer pairs 5′-CCGCCTAAGATAAAACCGACATCCAAATC-3′ with 5′-CCTCACGTGACTTCTTACCGTTTTTCTCG-3′ and 5′-CCAACAATGATCCATGACTTCGCTATCAC-3′ with 5′-ACCATGAAAACCATACGGCACTCTACCAG-3′. Mutations were identified as previously described (Colbert et al., 2001). The exact nature of each mutation was determined by sequencing and the severity of the change was predicted using the bioinformatic programs PARSESNP and SIFT (Ng and Henikoff, 2003; Taylor and Greene, 2003). Plants containing alleles predicted to severely affect gene function were recovered for analysis.
Gene Expression Assays
Total RNA was extracted from leaves, roots, and developing and imbibed seeds and analyzed by a real-time PCR system (StepOne; Applied Biosystems) as previously described (Argyris et al., 2008), except that a 15-μL reaction with 200 nM primer each was used for PCR detection. Abundance of mRNA for all genes was normalized against three constitutively expressed genes (18S1, UBQ, and PP2A2) as previously described (Vandesompele et al., 2002; Argyris et al., 2008). Genes and primers used for quantitative RT-PCR analyses can be found in Supplemental Table 2 online.
ABA Extraction and Measurement
ABA extraction from mature seeds was performed as described by Contreras et al. (2009) with slight modifications. Seeds (65 mg) were frozen in liquid nitrogen, ground into fine powder, and then lyophilized for 24 h using a SpeedVac at 4°C (Heto Vacuum Centrifuge; Thermo Scientific). Methanol containing 500 mg L−1 citric acid monohydrate and 100 mg L−1 butylated hydroxytoluene was added to samples at a ratio of 1.0 mL for each 10 mg of dry tissue. Samples were incubated with shaking (100 rpm) at 4°C for 20 h and centrifuged at 1500g for 10 min to collect the supernatant. The extracts were assayed using the Phytodetek ABA test kit (Agdia).
Plasmid Construction
For molecular complementation of Arabidopsis mutants, full-length Ls-NCED4 was amplified from genomic DNA as it contains no intron. Amplification was performed with the following primers: (NcoI) 5′-CCATGGACCACCATTAAAGCTATCAC-3′ and (AfeI) 5′-AGCGCTTAATTAGCCAAGTCATGGG-3′. The amplified fragments were subcloned into PCR 2.1-TOPO vectors for sequence confirmation. The amplified fragment was inserted into the binary vector pCAMBIA1301 to replace the β-glucuronidase (GUS) cassette that is driven by the CaMV35S promoter. The original plasmid pCAMBIA1301 was used as a control vector in the plant transformation.
For overexpression of Sal-NCED4 in UC plants, full-length NCED4 and its promoter was amplified with the primers (SalI) 5′-GTCGACATGGACAC CTCTGTGACTCTCAC-3′ and 5′-ATTAGCCAAGTCATGGGTACTAAC-3′. The Sal-NCED4 promoter was amplified with the following primers: 5′-TAACCGATCTAGTAACATAGATGACACCG-3′ and (SalI) 5′-GTCGACTGGAGGCGGTGGTAGTGATG-3′. Two amplified fragments were separately subcloned into PCR 2.1-TOPO vectors for sequence confirmation. The cassette, ProSal:Sal-NCED4, was obtained from the ligation of the Sal promoter and Sal-NCED4 through SalI digestion. The binary expression vector (pPIPRA558) was modified from a binary vector (pPIPRA539) and a shuttle vector (pPIPRA477), both of which were kindly provided by the Public Intellectual Property Resource for Agriculture. A cassette containing a constitutive FMV34S (34S RNA promoter from figwort mosaic virus) and nptII gene from pPIPRA477 was assembled into pPIPRA539 as a plant selective marker. This modified expression vector (pPIPRA558) containing a uidA (GUS) gene driven by a FMV34S promoter was used as a control vector, whereas Sal-NCED4 expression construct was derived from the replacement of FMV34S:GUS with the ProSal:Sal-NCED4 cassette.
For RNAi, a 350-bp fragment was amplified from Sal genomic DNA with the primers (SfiI) 5′-ATGGCCAGAGAGGCCATCTCCCTTGCCCCCTCGTA-3′ and (SfiI) 5′-ATGGCCATGTAGGCCTGAACCGCTGGATCAACCTG-3′. The fragments were digested with SfiI and assembled into a binary RNAi vector modified from the vector pGSA1165 (Wroblewski et al., 2007). The RNAi construct that contained the CaMV35S promoter driving the inverted NCED4 fragment repeats was digested with EcoRI and XhoI, and both larger (∼9.8 kb, containing inverted NCED4 repeats) and smaller (∼1.6 kb, containing MAS 2' promoter-nptII-MAS 3′ terminator-CaMV35S promoter) fragments were gel purified. The smaller fragment was subsequently ligated to EcoRI-XhoI–digested Zero Blunt TOPO PCR cloning vector (Invitrogen) to form a shuttle cloning vector and then digested with BglII and XhoI to remove the CaMV35S promoter. The BglII-XhoI–digested shuttle cloning vector (containing the MAS 2' promoter-nptII-MAS 3′ terminator cassette) was used for ligation with a BglII-XhoI–digested Sal-NCED4 promoter fragment that was amplified from genomic DNA with primers (BglII) 5′-TTAAAGATCTAACAGACAAAAGTCAACGGAGTTAG-3′ and (XhoI) 5′-TTCTCGAGTGGAGGCGGTGGTAGTGATG-3′. The shuttle cloning vector containing Sal-NCED4 promoter was cut by EcoRI and XhoI to release a fragment containing MAS 2' promoter-nptII-MAS 3′ terminator-Sal-NCED4 promoter, and the fragment was gel purified and ligated with the previously purified 9.8-kb fragment (containing inverted NCED4 repeats) to form the final RNAi construct. The RNAi vector that has no Sal-NCED4 promoter and inverted NCED4 repeats was used as a control in the lettuce transformation.
Generation of Transgenic Plants
For molecular complementation of Arabidopsis, the plasmid construct was transferred into Agrobacterium tumefaciens strain GV3101 using the heat shock method. Stable transgenic Arabidopsis plants were generated using the flower dipping method (Clough and Bent, 1998). Transgenic plants were selected on 0.5× Murashige and Skoog plates with 25 mg L−1 hygromycin B and further screened by PCR using hygromycin-specific primers (see Supplemental Table 1 online). The F4 generations of transgenic Arabidopsis seeds were used for experiments unless otherwise noted.
For lettuce transformation, the expression construct and RNAi construct were electroporated into Agrobacterium strain LBA4404. Transgenic lettuce plants were generated in the Ralph M. Parsons Foundation Plant Transformation Facility at UC Davis. Plants expressing ProSal:Sal-NCED4 were genotyped using TaqMan allelic discrimination (Applied Biosystems), while PCR screening for RNAi plants was performed using nptII gene-specific primers (see Supplemental Table 1 online).
Heat Stress Assay
Three 8-week-old plants were chosen for leaf sampling. Three leaves of similar age were collected from each plant for different treatments. Each leaf was cut into six pieces; three pieces were randomly picked from the three different leaves and placed in a plastic bag containing wet paper towel. Five bags of leaf samples were collected from each plant. In general, three bags of leaves from three different plants were used for each treatment. Heat stress treatment was performed at 40°C for 60 or 90 min or 40°C for 60 min and then 25°C for 30 min; controls were incubated at 25°C for 60 and 90 min. After treatment, leaves were immediately immersed in liquid nitrogen and stored at −80°C.
Dehydration Assay and Drought Tolerance Test
Three 8-week-old wild-type plants or three 6-week-old RNAi-NCED4 and control (RNAi-CTL) plants were chosen for leaf sampling for dehydration treatments. Leaf sampling was as described for heat stress except that each leaf was cut in half. Halves of three leaves from each plant were dehydrated until 15% loss of fresh weight and then pooled together as a biological replicate. For controls, the other halves of the leaves were placed into bags containing wet paper towels to avoid water loss. When the stressed leaf halves had dehydrated to the desired level, both stressed and control leaves were collected as described for the heat stress treatment.
For the dehydration tolerance tests, RNAi-CTL, RNAi-2, RNAi-8, MIS-1, and DS plants were grown in 60-cell (6 × 10) trays in potting soil. In each tray, there were two rows of six seedlings from each line, with three trays for each treatment. Water was withheld from 5-week-old seedlings for 7 and 10 d followed by watering for 10 d.
Seed Size and Number
Seeds from 15 buds were collected from each line as one biological replicate. Four replicates were collected from two RNAi controls (RNAi2-CT1 and RNAi8-CT4) and RNAi lines (RNAi8-4 and RNAi8-7). Average numbers of filled and unfilled seeds per bud and average seed weights of filled seeds were determined.
Sequence and Phylogenetic Analyses
Lettuce homologs were obtained by BLASTX search of Arabidopsis genes whose accession numbers are listed in Supplemental Table 2 online against the Compositae Genome Project Database (http://cgpdb.ucdavis.edu/cgpdb2/blast_search/).
Sequence alignment was conducted with ClustalW using the Bioedit V 7.09 (Hall, 1999). Primers were designed using Primer 3.0 (http://frodo.wi.mit.edu/primer3/) unless stated. Prediction of cis-elements was performed using the PLACE program (Higo et al., 1999).
For phylogenetic analysis, amino acid sequences (see Supplemental Data Set 1 online) were aligned and the phylogenetic tree was constructed using the phylogenetic tree function of MEGA (version 5.05; www.megasoftware.net) with the neighbor-joining method and default parameters.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under accession numbers listed in Supplemental Tables 2 and 3 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of NCED4 Amino Acid Sequences from Different Lettuce Varieties.
Supplemental Figure 2. Phylogenetic Analysis of NCED Proteins from Arabidopsis and Lettuce.
Supplemental Figure 3. Germination at High Temperature of Seeds of UC Lines Transformed with ProSal:Sal-NCED4.
Supplemental Figure 4. Comparison of NCED4 Amino Acid Sequences and Structure to VP14 from Maize.
Supplemental Figure 5. Germination of Heterozygous and Homozygous RNAi-NCED4 Lines.
Supplemental Figure 6. Effect of NCED4 Silencing on Inflorescence and Seed Development.
Supplemental Figure 7. Transcriptional Analysis of NCED1-4 in Roots and Leaves of Sal and UC Plants.
Supplemental Figure 8. Drought Tolerance Assay.
Supplemental Table 1. Primer Combinations Used for Isolation of Ls-NCED4 and Its Promoters.
Supplemental Table 2. Lettuce Genes and Primer Combinations Used for qRT-PCR.
Supplemental Table 3. Accession Numbers for Sequences Used in This Study.
Supplemental Data Set 1. Alignments Used to Generate the Phylogeny in Supplemental Figure 2.
Supplemental Data Set 2. Alignment of DNA Sequences of NCED4 Coding Regions from Thermotolerant (UC, W48, and W51) and Thermosensitive (Sal, Ds, W55, and W46) Lactuca Genotypes.
Supplemental Data Set 3. Alignment of DNA Sequences of NCED4 Promoter Regions from Thermotolerant (UC, W48, and W51) and Thermosensitive (Sal, W46, and W55) Lactuca Genotypes.
Supplementary Material
Acknowledgments
We thank interns and visiting students working in our lab, including Jacson Zuchi, Luís Eduardo Panozzo, and Camilla Sediyama. We thank Pedro Bello and Fei-Yian Yoong for suggestions and kind help. Richard Michelmore provided BAC clones and RNAi vectors. We also thank Daniel Facciotti, Cate McGuire, and Aaron Holm for contributing TILLING library construction and screening. This study was supported by USDA-National Institute of Food and Agriculture Award 2008-35304-0472 and National Science Foundation Award 0820451.
AUTHOR CONTRIBUTIONS
H.H. and K.J.B. designed experiments. H.H. performed most of the experiments. P.D. performed BAC clone screening and characterization of TILLING mutants. K.K. contributed to mutant characterization. C.M.M. identified and provided the TILLING mutants used for analysis. H.H. and K.J.B. wrote the article with input from other authors.
Glossary
- ABA
abscisic acid
- GA
gibberellin
- QTL
quantitative trait locus
- CaMV35S
cauliflower mosaic virus 35S promoter
- PAC
paclobutrazol
- RNAi
RNA interference
- DAF
days after flowering
- SNP
single-nucleotide polymorphism
- ACC
1-aminocyclopropane-1-carboxylic acid
References
- Allen, P.S., Benech-Arnold, R.L., Batlla, D., and Bradford, K.J. (2007). Modeling of seed dormancy. In Seed Development, Dormancy and Germination, K.J. Bradford and H. Nonogaki, eds (Oxford, UK: Blackwell Publishing), pp. 72–112. [Google Scholar]
- Argyris J., Dahal P., Hayashi E., Still D.W., Bradford K.J. (2008). Genetic variation for lettuce seed thermoinhibition is associated with temperature-sensitive expression of abscisic Acid, gibberellin, and ethylene biosynthesis, metabolism, and response genes. Plant Physiol. 148: 926–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argyris J., Truco M.J., Ochoa O., Knapp S.J., Still D.W., Lenssen G.M., Schut J.W., Michelmore R.W., Bradford K.J. (2005). Quantitative trait loci associated with seed and seedling traits in Lactuca. Theor. Appl. Genet. 111: 1365–1376 [DOI] [PubMed] [Google Scholar]
- Argyris J., Truco M.J., Ochoa O., McHale L., Dahal P., Van Deynze A., Michelmore R.W., Bradford K.J. (2011). A gene encoding an abscisic acid biosynthetic enzyme (LsNCED4) collocates with the high temperature germination locus Htg6.1 in lettuce (Lactuca sp.). Theor. Appl. Genet. 122: 95–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argyris, J.M. (2008). Natural Variation and Genetic Analyses of Seed and Seedling Traits in Lettuce: Discovery, Confirmation and Proposed Role for a Quantitative Trait Locus in Regulating Seed Dormancy at High Temperature. PhD dissertation (Davis, CA: University of California). [Google Scholar]
- Barnabás B., Jäger K., Fehér A. (2008). The effect of drought and heat stress on reproductive processes in cereals. Plant Cell Environ. 31: 11–38 [DOI] [PubMed] [Google Scholar]
- Baskin, C.C., and Baskin, J.M. (1998). Seeds: Ecology, Biogeography, and Evolution of Dormancy and Germination. (New York: Academic Press). [Google Scholar]
- Bentsink L., Jowett J., Hanhart C.J., Koornneef M. (2006). Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 17042–17047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley, J.D., Bradford, K.J., Hilhorst, H.W.M., and Nonogaki, H. (2013). Seeds: Physiology of Development, Germination and Dormancy, 3rd ed. (New York: Springer). [Google Scholar]
- Bradford K.J., Downie A.B., Gee O.H., Alvarado V., Yang H., Dahal P. (2003). Abscisic acid and gibberellin differentially regulate expression of genes of the SNF1-related kinase complex in tomato seeds. Plant Physiol. 132: 1560–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch W., Wunderlich M., Schöffl F. (2005). Identification of novel heat shock factor-dependent genes and biochemical pathways in Arabidopsis thaliana. Plant J. 41: 1–14 [DOI] [PubMed] [Google Scholar]
- Carrera E., Holman T., Medhurst A., Dietrich D., Footitt S., Theodoulou F.L., Holdsworth M.J. (2008). Seed after-ripening is a discrete developmental pathway associated with specific gene networks in Arabidopsis. Plant J. 53: 214–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W.H., Chiang M.H., Hwang S.G., Lin P.C. (2009). Antagonism between abscisic acid and ethylene in Arabidopsis acts in parallel with the reciprocal regulation of their metabolism and signaling pathways. Plant Mol. Biol. 71: 61–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang G.C.K., Bartsch M., Barua D., Nakabayashi K., Debieu M., Kronholm I., Koornneef M., Soppe W.J., Donohue K., De Meaux J. (2011). DOG1 expression is predicted by the seed-maturation environment and contributes to geographical variation in germination in Arabidopsis thaliana. Mol. Ecol. 20: 3336–3349 [DOI] [PubMed] [Google Scholar]
- Chiang G.C.K., Barua D., Kramer E.M., Amasino R.M., Donohue K. (2009). Major flowering time gene, flowering locus C, regulates seed germination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 106: 11661–11666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu R.S., Nahal H., Provart N.J., Gazzarrini S. (2012). The role of the Arabidopsis FUSCA3 transcription factor during inhibition of seed germination at high temperature. BMC Plant Biol. 12: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colbert T., Till B.J., Tompa R., Reynolds S., Steine M.N., Yeung A.T., McCallum C.M., Comai L., Henikoff S. (2001). High-throughput screening for induced point mutations. Plant Physiol. 126: 480–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L., Henikoff S. (2006). TILLING: Practical single-nucleotide mutation discovery. Plant J. 45: 684–694 [DOI] [PubMed] [Google Scholar]
- Contreras S., Bennett M.A., Metzger J.D., Tay D., Nerson H. (2009). Red to far-red ratio during seed development affects lettuce seed germinability and longevity. HortScience 44: 130–134 [Google Scholar]
- Delseny M., Bies-Etheve N., Carles C., Hull G., Vicient C., Raynal M., Grellet F., Aspart L. (2001). Late Embryogenesis Abundant (LEA) protein gene regulation during Arabidopsis seed maturation. J. Plant Physiol. 158: 419–427 [Google Scholar]
- Donohue K., Dorn L., Griffith C., Kim E., Aguilera A., Polisetty C.R., Schmitt J. (2005). Environmental and genetic influences on the germination of Arabidopsis thaliana in the field. Evolution 59: 740–757 [PubMed] [Google Scholar]
- Donohue K., Rubio de Casas R., Burghardt L., Kovach K., Willis C.G. (2010). Germination, postgermination adaptation, and species ecological ranges. Annu. Rev. Ecol. Evol. Syst. 41: 293–319 [Google Scholar]
- Dutta S., Bradford K.J. (1994). Water relations of lettuce seed thermoinhibition. II. Ethylene and endosperm effects on base water potential. Seed Sci. Res. 4: 11–18 [Google Scholar]
- Finkelstein R., Reeves W., Ariizumi T., Steber C. (2008). Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 59: 387–415 [DOI] [PubMed] [Google Scholar]
- Footitt S., Douterelo-Soler I., Clay H., Finch-Savage W.E. (2011). Dormancy cycling in Arabidopsis seeds is controlled by seasonally distinct hormone-signaling pathways. Proc. Natl. Acad. Sci. USA 108: 20236–20241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey A., Effroy D., Lefebvre V., Seo M., Perreau F., Berger A., Sechet J., To A., North H.M., Marion-Poll A. (2012). Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. Plant J. 70: 501–512 [DOI] [PubMed] [Google Scholar]
- Gabriele S., Rizza A., Martone J., Circelli P., Costantino P., Vittorioso P. (2010). The Dof protein DAG1 mediates PIL5 activity on seed germination by negatively regulating GA biosynthetic gene AtGA3ox1. Plant J. 61: 312–323 [DOI] [PubMed] [Google Scholar]
- Gonai T., Kawahara S., Tougou M., Satoh S., Hashiba T., Hirai N., Kawaide H., Kamiya Y., Yoshioka T. (2004). Abscisic acid in the thermoinhibition of lettuce seed germination and enhancement of its catabolism by gibberellin. J. Exp. Bot. 55: 111–118 [DOI] [PubMed] [Google Scholar]
- Gray D., Wurr D.C.E., Ward J.A., Fellows J.R. (1988). Influence of post-flowering temperature on seed development, and subsequent performance of crisp lettuce. Ann. Appl. Biol. 113: 391–402 [Google Scholar]
- Gualberti G., Papi M., Bellucci L., Ricci I., Bouchez D., Camilleri C., Costantino P., Vittorioso P. (2002). Mutations in the Dof zinc finger genes DAG2 and DAG1 influence with opposite effects the germination of Arabidopsis seeds. Plant Cell 14: 1253–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T.A. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Series 41: 95–98 [Google Scholar]
- Higo K., Ugawa Y., Iwamoto M., Korenaga T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdsworth M.J., Bentsink L., Soppe W.J.J. (2008). Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 179: 33–54 [DOI] [PubMed] [Google Scholar]
- Kendall S.L., Hellwege A., Marriot P., Whalley C., Graham I.A., Penfield S. (2011). Induction of dormancy in Arabidopsis summer annuals requires parallel regulation of DOG1 and hormone metabolism by low temperature and CBF transcription factors. Plant Cell 23: 2568–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepczynski J., Kepczynska E. (1997). Ethylene in seed dormancy and germination. Physiol. Plant. 101: 720–726 [Google Scholar]
- Kozarewa I., Cantliffe D.J., Nagata R.T., Stoffella P.J. (2006). High maturation temperature of lettuce seeds during development increased ethylene production and germination at elevated temperatures. J. Am. Soc. Hortic. Sci. 131: 564–570 [Google Scholar]
- Kumar S.V., Wigge P.A. (2010). H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140: 136–147 [DOI] [PubMed] [Google Scholar]
- Lefebvre V., North H., Frey A., Sotta B., Seo M., Okamoto M., Nambara E., Marion-Poll A. (2006). Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 45: 309–319 [DOI] [PubMed] [Google Scholar]
- Li H.C., Chuang K., Henderson J.T., Rider S.D., Jr, Bai Y., Zhang H., Fountain M., Gerber J., Ogas J. (2005). PICKLE acts during germination to repress expression of embryonic traits. Plant J. 44: 1010–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkies A., Müller K., Morris K., Turecková V., Wenk M., Cadman C.S.C., Corbineau F., Strnad M., Lynn J.R., Finch-Savage W.E., Leubner-Metzger G. (2009). Ethylene interacts with abscisic acid to regulate endosperm rupture during germination: A comparative approach using Lepidium sativum and Arabidopsis thaliana. Plant Cell 21: 3803–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Andújar C., Ordiz M.I., Huang Z., Nonogaki M., Beachy R.N., Nonogaki H. (2011). Induction of 9-cis-epoxycarotenoid dioxygenase in Arabidopsis thaliana seeds enhances seed dormancy. Proc. Natl. Acad. Sci. USA 108: 17225–17229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing S.A.J., Gabelli S.B., Echeverria I., Vogel J.T., Guan J.C., Tan B.C., Klee H.J., McCarty D.R., Amzel L.M. (2010). Structural insights into maize viviparous14, a key enzyme in the biosynthesis of the phytohormone abscisic acid. Plant Cell 22: 2970–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mönke G., Altschmied L., Tewes A., Reidt W., Mock H.P., Bäumlein H., Conrad U. (2004). Seed-specific transcription factors ABI3 and FUS3: Molecular interaction with DNA. Planta 219: 158–166 [DOI] [PubMed] [Google Scholar]
- Nakabayashi K., Okamoto M., Koshiba T., Kamiya Y., Nambara E. (2005). Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: Epigenetic and genetic regulation of transcription in seed. Plant J. 41: 697–709 [DOI] [PubMed] [Google Scholar]
- Nakamura S., et al. (2011). A wheat homolog of MOTHER OF FT AND TFL1 acts in the regulation of germination. Plant Cell 23: 3215–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E., Marion-Poll A. (2005). Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56: 165–185 [DOI] [PubMed] [Google Scholar]
- Nascimento W.M., Cantliffe D.J., Huber D.J. (2000). Thermotolerance in lettuce seeds: Association with ethylene and endo-beta-mannanase. J. Am. Soc. Hortic. Sci. 125: 518–524 [Google Scholar]
- Nascimento W.M., Cantliffe D.J., Huber D.J. (2004). Ethylene evolution and endo-beta-mannanase activity during lettuce seed germination at high temperature. Sci. Agric. 61: 156–163 [Google Scholar]
- Ng P.C., Henikoff S. (2003). SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 31: 3812–3814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M., Hanada A., Yamauchi Y., Kuwahara A., Kamiya Y., Yamaguchi S. (2003). Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15: 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M., Kuwahara A., Seo M., Kushiro T., Asami T., Hirai N., Kamiya Y., Koshiba T., Nambara E. (2006). CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol. 141: 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papi M., Sabatini S., Altamura M.M., Hennig L., Schäfer E., Costantino P., Vittorioso P. (2002). Inactivation of the phloem-specific Dof zinc finger gene DAG1 affects response to light and integrity of the testa of Arabidopsis seeds. Plant Physiol. 128: 411–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S. (2008). Temperature perception and signal transduction in plants. New Phytol. 179: 615–628 [DOI] [PubMed] [Google Scholar]
- Penfield S., Hall A. (2009). A role for multiple circadian clock genes in the response to signals that break seed dormancy in Arabidopsis. Plant Cell 21: 1722–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S., Josse E.M., Kannangara R., Gilday A.D., Halliday K.J., Graham I.A. (2005). Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr. Biol. 15: 1998–2006 [DOI] [PubMed] [Google Scholar]
- Penfield S., Li Y., Gilday A.D., Graham S., Graham I.A. (2006). Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell 18: 1887–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perruc E., Kinoshita N., Lopez-Molina L. (2007). The role of chromatin-remodeling factor PKL in balancing osmotic stress responses during Arabidopsis seed germination. Plant J. 52: 927–936 [DOI] [PubMed] [Google Scholar]
- Piskurewicz U., Jikumaru Y., Kinoshita N., Nambara E., Kamiya Y., Lopez-Molina L. (2008). The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 20: 2729–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radchuk R., Radchuk V., Weschke W., Borisjuk L., Weber H. (2006). Repressing the expression of the SUCROSE NONFERMENTING-1-RELATED PROTEIN KINASE gene in pea embryo causes pleiotropic defects of maturation similar to an abscisic acid-insensitive phenotype. Plant Physiol. 140: 263–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidt W., Wohlfarth T., Ellerström M., Czihal A., Tewes A., Ezcurra I., Rask L., Bäumlein H. (2000). Gene regulation during late embryogenesis: The RY motif of maturation-specific gene promoters is a direct target of the FUS3 gene product. Plant J. 21: 401–408 [DOI] [PubMed] [Google Scholar]
- Rieu I., Eriksson S., Powers S.J., Gong F., Griffiths J., Woolley L., Benlloch R., Nilsson O., Thomas S.G., Hedden P., Phillips A.L. (2008). Genetic analysis reveals that C19-GA 2-oxidation is a major gibberellin inactivation pathway in Arabidopsis. Plant Cell 20: 2420–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizza A., Boccaccini A., Lopez-Vidriero I., Costantino P., Vittorioso P. (2011). Inactivation of the ELIP1 and ELIP2 genes affects Arabidopsis seed germination. New Phytol. 190: 896–905 [DOI] [PubMed] [Google Scholar]
- Rosnoblet C., Aubry C., Leprince O., Vu B.L., Rogniaux H., Buitink J. (2007). The regulatory gamma subunit SNF4b of the sucrose non-fermenting-related kinase complex is involved in longevity and stachyose accumulation during maturation of Medicago truncatula seeds. Plant J. 51: 47–59 [DOI] [PubMed] [Google Scholar]
- Ruggiero B., Koiwa H., Manabe Y., Quist T.M., Inan G., Saccardo F., Joly R.J., Hasegawa P.M., Bressan R.A., Maggio A. (2004). Uncoupling the effects of abscisic acid on plant growth and water relations. Analysis of sto1/nced3, an abscisic acid-deficient but salt stress-tolerant mutant in Arabidopsis. Plant Physiol. 136: 3134–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton P.J., Somssich I.E., Ringler P., Shen Q.J. (2010). WRKY transcription factors. Trends Plant Sci. 15: 247–258 [DOI] [PubMed] [Google Scholar]
- Saini H.S., Consolacion E.D., Bassi P.K., Spencer M.S. (1986). Requirement for ethylene synthesis and action during relief of thermoinhibition of lettuce seed germination by combinations of gibberellic acid, kinetin, and carbon dioxide. Plant Physiol. 81: 950–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini H.S., Consolacion E.D., Bassi P.K., Spencer M.S. (1989). Control processes in the induction and relief of thermoinhibition of lettuce seed germination: Actions of phytochrome and endogenous ethylene. Plant Physiol. 90: 311–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada Y., Aoki M., Nakaminami K., Mitsuhashi W., Tatematsu K., Kushiro T., Koshiba T., Kamiya Y., Inoue Y., Nambara E., Toyomasu T. (2008). Phytochrome- and gibberellin-mediated regulation of abscisic acid metabolism during germination of photoblastic lettuce seeds. Plant Physiol. 146: 1386–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S.H., Qin X., Zeevaart J.A. (2003). Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol. 131: 1591–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M., et al. (2006). Regulation of hormone metabolism in Arabidopsis seeds: Phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J. 48: 354–366 [DOI] [PubMed] [Google Scholar]
- Seo M., Koshiba T. (2002). Complex regulation of ABA biosynthesis in plants. Trends Plant Sci. 7: 41–48 [DOI] [PubMed] [Google Scholar]
- Tamura N., Yoshida T., Tanaka A., Sasaki R., Bando A., Toh S., Lepiniec L., Kawakami N. (2006). Isolation and characterization of high temperature-resistant germination mutants of Arabidopsis thaliana. Plant Cell Physiol. 47: 1081–1094 [DOI] [PubMed] [Google Scholar]
- Tan B.C., Joseph L.M., Deng W.T., Liu L.J., Li Q.B., Cline K., McCarty D.R. (2003). Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 35: 44–56 [DOI] [PubMed] [Google Scholar]
- Taylor N.E., Greene E.A. (2003). PARSESNP: A tool for the analysis of nucleotide polymorphisms. Nucleic Acids Res. 31: 3808–3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh S., et al. (2008). High temperature-induced abscisic acid biosynthesis and its role in the inhibition of gibberellin action in Arabidopsis seeds. Plant Physiol. 146: 1368–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdes V.M., Bradford K.J., Mayberry K.S. (1985). Alleviation of thermodormancy in coated lettuce seeds by seed priming. HortScience 20: 1112–1114 [Google Scholar]
- Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., and Speleman, F. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3: RESEARCH0034. [DOI] [PMC free article] [PubMed]
- Varshney R.K., Bansal K.C., Aggarwal P.K., Datta S.K., Craufurd P.Q. (2011). Agricultural biotechnology for crop improvement in a variable climate: hope or hype? Trends Plant Sci. 16: 363–371 [DOI] [PubMed] [Google Scholar]
- Wroblewski T., Piskurewicz U., Tomczak A., Ochoa O., Michelmore R.W. (2007). Silencing of the major family of NBS-LRR-encoding genes in lettuce results in the loss of multiple resistance specificities. Plant J. 51: 803–818 [DOI] [PubMed] [Google Scholar]
- Yamauchi Y., Ogawa M., Kuwahara A., Hanada A., Kamiya Y., Yamaguchi S. (2004). Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell 16: 367–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka T., Endo T., Satoh S. (1998). Restoration of seed germination at supraoptimal temperatures by fluridone, an inhibitor of abscisic acid biosynthesis. Plant Cell Physiol. 39: 307–312 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.