In plants, AMT-type ammonium transporters are posttranslationally regulated by phosphorylation. This study provides evidence that allosteric regulation is functional in planta and that C-terminal phosphorylation mediates trans-inactivation also in heteromeric AMT complexes containing different AMT isoforms.
Abstract
Ammonium acquisition by plant roots is mediated by AMMONIUM TRANSPORTERs (AMTs), ubiquitous membrane proteins with essential roles in nitrogen nutrition in all organisms. In microbial and plant cells, ammonium transport activity is controlled by ammonium-triggered feedback inhibition to prevent cellular ammonium toxicity. Data from heterologous expression in yeast indicate that oligomerization of plant AMTs is critical for allosteric regulation of transport activity, in which the conserved cytosolic C terminus functions as a trans-activator. Employing the coexpressed transporters AMT1;1 and AMT1;3 from Arabidopsis thaliana as a model, we show here that these two isoforms form functional homo- and heterotrimers in yeast and plant roots and that AMT1;3 carrying a phosphomimic residue in its C terminus regulates both homo- and heterotrimers in a dominant-negative fashion in vivo. 15NH4+ influx studies further indicate that allosteric inhibition represses ammonium transport activity in roots of transgenic Arabidopsis expressing a phosphomimic mutant together with functional AMT1;3 or AMT1;1. Our study demonstrates in planta a regulatory role in transport activity of heterooligomerization of transporter isoforms, which may enhance their versatility for signal exchange in response to environmental triggers.
INTRODUCTION
Ammonium is the predominant N form in most soils, supporting the highest growth rates when present in a mixed supply with nitrate. Ammonium requires less energy for its assimilation than does nitrate (Bloom et al., 1992) and is preferentially taken up by Arabidopsis thaliana roots in particular when N supplies are low (Gazzarrini et al., 1999). However, supply of ammonium as the sole N form has a negative impact on root growth: The elongation of the primary and lateral roots is severely repressed, resulting in a stunted root phenotype while lateral root branching is stimulated (Lima et al., 2010; Rogato et al., 2010). In addition, excess ammonium intake unbalances cellular pH homeostasis and primary metabolism (Hachiya et al., 2012). Therefore, plant roots repress ammonium uptake at elevated ammonium supplies (Wang et al., 1993; Rawat et al., 1999; Lanquar et al., 2009), which implies the existence of ammonium sensing mechanisms and feedback regulation of ammonium uptake.
Uptake of ammonium into plant roots is mediated by AMMONIUM TRANSPORTERs (AMTs), which belong to the AMT/MEP/Rh protein superfamily that includes bacterial, fungal, and human homologs (Loqué and von Wirén, 2004). Among the five root-expressed AMT proteins in Arabidopsis, AMT1;1, AMT1;2, AMT1;3, and most likely also AMT1;5 are responsible for the high-affinity ammonium uptake capacity in roots, with 60 to 70% of it being conferred by AMT1;1 and AMT1;3 (Yuan et al., 2007a). In addition to a transport function, at least bacterial and fungal AMTs have been shown to act as transceptors with a dual function as ammonium transporters and receptors, mediating ammonium-triggered changes in morphology or the transcription of target genes (Tremblay and Hallenbeck, 2009). Plant AMTs may also be transceptors: AMT1;3, although highly similar in structure and transport properties to AMT1;1, has the ability to regulate lateral root branching in response to localized ammonium supplies (Lima et al., 2010).
At elevated external ammonium supplies, AMT proteins are posttranslationally modified by phosphorylation (Lanquar et al., 2009). While AMT1;1 transport activity relies on the interaction between its nonphosphorylated, cytosolic C terminus and the pore region of a neighboring subunit in a quaternary AMT protein complex, C-terminal phosphorylation leads to trans-inactivation of the whole complex (Loqué et al., 2007). Inactivation can also be mimicked by truncation of the C terminus or introduction of a negatively charged amino acid residue just adjacent to the last transmembrane helix (Marini et al., 2000; Ludewig et al., 2003; Loqué et al., 2007; Neuhäuser et al., 2007). This is explained by loss of contact between the cytosolic C terminus and cytosolic loops of the pore region in an adjacent neighboring subunit, which has been proposed as a prerequisite for opening of the ammonia-conducting pore (Loqué et al., 2007, 2009).
Functional studies with mutated AMTs have so far been conducted only in heterologous systems, raising the question of to what extent AMT protein phosphorylation and related interactions modulate ammonium transport activity in planta. Previous attempts to ectopically express AMT1;1 in Arabidopsis were hampered most likely by the unique sensitivity of AMT1;1 to posttranscriptional gene silencing (Yuan et al., 2007b). Therefore, AMT1;3 was used in this study to investigate whether a phosphorylation mimic in the C terminus of plant AMTs also inhibits ammonium transport activity in planta. Since AMT1;1 and AMT1;3 are coexpressed in the rhizodermis (Loqué et al., 2006), they provide the unique opportunity to study the biochemical function of their oligomerization in intact plants. Hence, we further investigated whether AMTs form heteromeric complexes and whether the allosteric regulation of AMT complexes is retained in heteromers. For this purpose, gel blot analyses of green fluorescent protein (GFP)–tagged AMT proteins were conducted and a phosphomimic variant of AMT1;3 was expressed in Arabidopsis wild-type plants or a quadruple amt mutant with reconstituted expression of AMT1;1 or AMT1;3. Influx studies using 15N-labeled ammonium showed that C-terminal phosphorylation can mediate intermonomeric trans-inhibition of ammonium transport in plant roots and that the allosteric regulation of transporter activities even extends to heteromeric AMT protein complexes.
RESULTS
Functional Interaction of AMT1;3 and AMT1;1 in Yeast
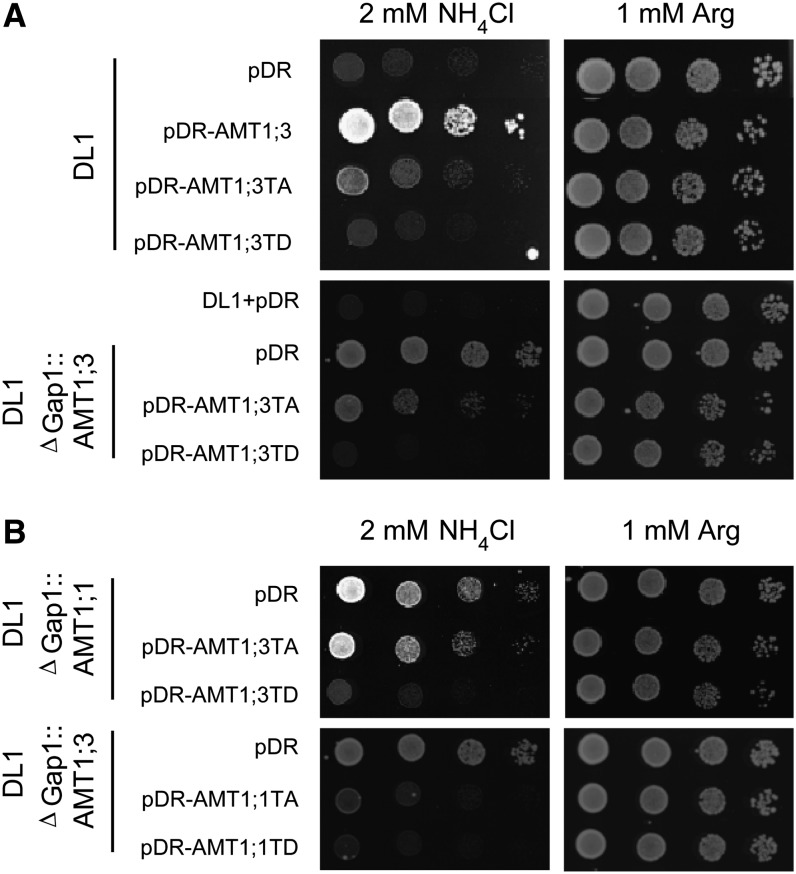
Previous studies showed that a phosphorylation mimic of a highly conserved Thr residue in the cytosolic C terminus of Arabidopsis AMT1;1 (T460D) or AMT1;2 (T472D) abolishes transport activity when heterologously expressed in yeast or oocytes (Loqué et al., 2007; Neuhäuser et al., 2007). To verify whether the corresponding position in the C terminus of AMT1;3 (see Supplemental Figure 1 online) possesses a similar function in the allosteric regulation of transport activity, we expressed two AMT1;3 variants, T464A (TA) and T464D (TD), in the yeast strain DL1 (Δmep1-3 Δgap1), which is defective in high-affinity ammonium uptake (Marini et al., 1997; Loqué et al., 2007). Wild-type AMT1;3 and AMT1;3TA were able to complement the growth of the yeast mutant on 2 mM ammonium as a sole N source (Figure 1A). By contrast, the AMT1;3TD mutant did not confer ammonium uptake in yeast, indicating that the introduction of a negatively charged amino acid residue at Thr-464 in AMT1;3 abolished transport activity, similar to a phosphorylation mimic of the corresponding Thr residues in AMT1;1 or AMT1;2 (Loqué et al., 2007; Neuhäuser et al., 2007). The stable integration of wild-type AMT1;3 into the yeast genome (Δgap1::AMT1;3) restored the growth defect of the yeast mutant DL1 to a slightly weaker extent compared with expression from the plasmid (Figure 1A), which may have been caused by different gene expression levels. However, episomal coexpression of AMT1;3TD led to inhibition of ammonium transport activity by wild-type AMT1;3, suggesting that AMT1;3TD provoked a trans-dominant negative effect on the activity of the wild-type AMT1;3 protein.
Figure 1.
Functional Interaction between AMT1;3 and AMT1;1 When Coexpressed in Yeast.
(A) The yeast strain DL1 (Δmep1-3 ΔGap1) and DL1 with stable integration of AMT1;3 (Δmep1-3 ΔGap1::AMT1;3) were transformed with the empty vector pDR, pDR-AMT1;3, pDR-AMT1;3TA(T464A), or pDR-AMT1;3TD(T464D).
(B) DL1 strains with stable integration of either AMT1;1 (Δmep1-3 ΔGap1::AMT1;1) or AMT1;3 (Δmep1-3 ΔGap1::AMT1;3) were transformed with the empty vector pDR, pDR-AMT1;3TA(T464A), pDR-AMT1;3TD(T464D), pDR-AMT1;1TA(T460A), or pDR-AMT1;1TD(T460D).
Transformants were selected on YNB medium supplemented with 1 mM Arg. Five microliters of yeast cell suspensions from overnight cultures were spotted in one- to fivefold dilutions on YNB medium supplemented with 2 mM NH4Cl or 1 mM Arg at pH 5.2, and growth was recorded after 4 d at 28°C.
The integration of either wild-type AMT1;1 or wild-type AMT1;3 into the yeast genome restored the growth defect of the yeast mutant DL1 on low ammonium supply. In both cases, coexpression of the nonfunctional AMT1;3TD protein conferred loss of ammonium transport activity (Figures 1A and 1B). Moreover, expression of a nonfunctional AMT1;1TD also inhibited growth when coexpressed with the wild-type AMT1;1 (Loqué et al., 2007) or AMT1;3 protein (Figure 1B). Since functional interaction between the AMT1;1 and AMT1;3 proteins occurred in a heterologous expression system, the involvement of a third, regulatory Arabidopsis protein in this interaction was not further considered.
In an attempt to quantify the relative strength of protein interactions, we performed a mating-based split-ubiquitin assay (Lalonde et al., 2010). AMT1;1 and AMT1;3 were fused C-terminally either to Nub (N-terminal ubiquitin domain carrying a Gly mutation) or to Cub (N-terminal ubiquitin domain regulated by the Met-repressible MET25 promoter and fused to the artificial protease A-LexA-VP16 (PLV) transcription factor). Growth of yeast colonies resulted in the reconstitution of ubiquitin and release of a transcription factor complementing the His auxotrophy. Growth assays showed that AMT1;1 or AMT1;3 can interact with themselves, while reciprocal assays showed that they also interacted with each other (Figure 2). According to colony sizes, interactions with AMT1;3-Cub appeared to be stronger. However, AMT1;3-Cub also showed a higher level of autoactivation, thus leading to a higher background. Colony growth resulting from the interaction between AMT1;1-Cub and AMT1;1- or AMT1;3-fused Nub was comparable, supporting the assumption that complexes were formed with similar efficacy. Likewise, AMT1;3-mediated interaction assays did not appear to reveal a particular preference for either AMT1 isoform.
Figure 2.
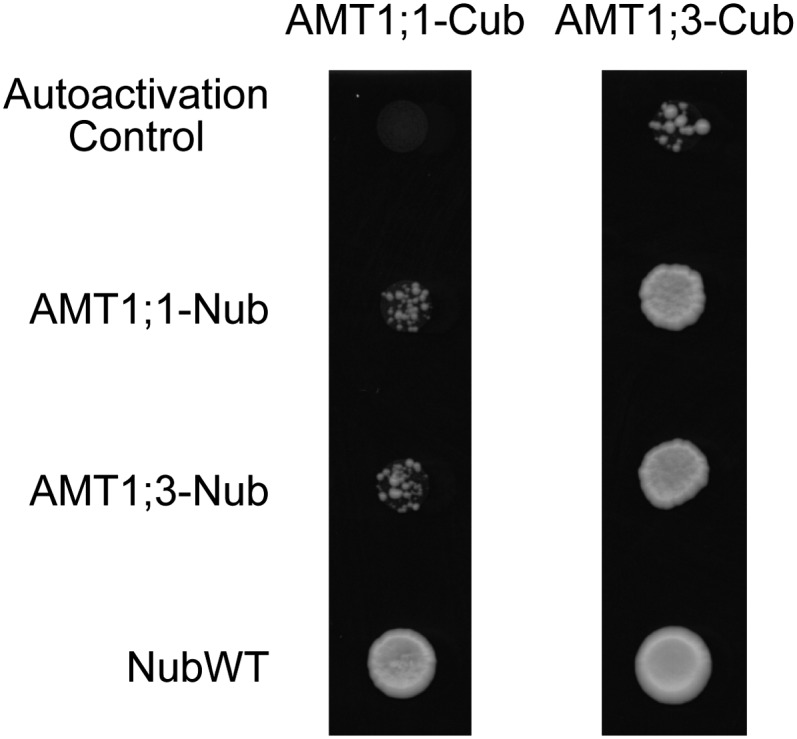
Split-Ubiquitin Assay for AMT1;1 and AMT1;3 Protein Interactions.
Split-ubiquitin binary assay of AMT1;1 and AMT1;3 on SC medium supplemented with 75 µM Met. Diploid strains containing AMT1;1- and AMT1;3-Cub fusions were tested with a AMT1;1- or AMT1;3-Nub fusion or a wild-type variant of Nub (NubWT).
Oligomerization of AMT1;3 and AMT1;1 in Root Membrane Fractions
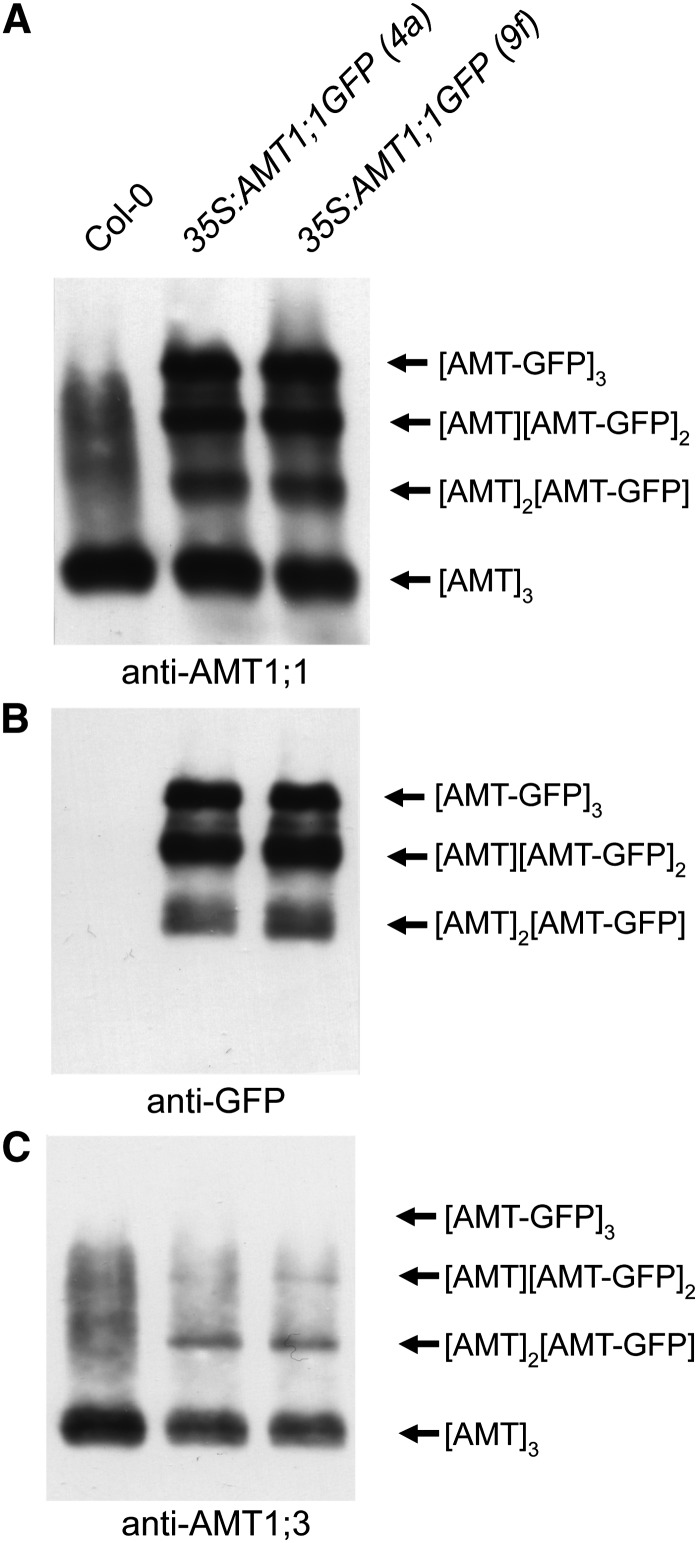
Based on the observations that endogenous AMT proteins form high molecular weight complexes (Ludewig et al., 2003; Yuan et al., 2007b; Graff et al., 2011), we anticipated that coexpression of GFP-tagged AMTs should allow determination of the oligomerization status of Arabidopsis AMTs in protein gel blot analyses. In microsomal membrane proteins from N-deficient roots separated on a 10% SDS-PAGE gel, a high molecular mass form of AMT1;3 was detected at >120 kD when protein samples were preincubated under nonreducing conditions (see Supplemental Figure 2 online). Under reducing conditions, AMT1;3 proteins were detected at a molecular mass of ∼40 kD, corresponding to the expected size of their monomers. A similar observation was previously made for AMT1;1 (Yuan et al., 2007b), suggesting the presence of a protein complex of AMT1;3 or AMT1;1 in Arabidopsis roots that is sensitive to the reduction of disulfide bonds, as recently shown for AMTs in tomato (Solanum lycopersicum; Graff et al., 2011). After preincubation under nonreducing conditions and separation in a 6% SDS-PAGE gel, the anti-AMT1;1 antibody detected the high molecular mass AMT complex as a major band at <100 kD in Columbia-0 roots due to a differential migration of the hydrophobic AMT complex and the hydrophilic marker (see Supplemental Figure 3A online). In transgenic lines expressing GFP-tagged AMT1;1, the anti-AMT1;1 antibody detected three additional bands (Figure 3A). These represented a stepwise increase in molecular weight with each further band corresponding to a difference in molecular weight of 20 to 30 kD. Using an anti-GFP antibody, these three additional bands were identified to carry a GFP tag (Figure 3B). We conclude that AMT1;1 assembled as a trimer of AMT1;1 and AMT1;1-GFP moieties, representing a population of the following protein complexes: [AMT1;1]3, [AMT1;1]2[AMT1;1-GFP], [AMT1;1][AMT1;1-GFP]2, and [AMT1;1-GFP]3.
Figure 3.
Heterotrimerization of AMT1;1 and AMT1;3 in Roots.
Protein gel blot analysis of microsomal membrane fractions from Arabidopsis roots of wild-type (Columbia-0 [Col-0]) or transgenic plants expressing an AMT1;1-GFP fusion construct (35S:AMT1;1GFP) using the following antibodies: anti-AMT1;1 (A), anti-GFP (B), or anti-AMT1;3 (C). According to the shift in their molecular weight, possible combinations of oligomers are indicated at the right. Protein samples were pretreated at 0°C in the absence of β-mercaptoethanol (nonreducing conditions), and a 6% SDS-PAGE gel was used to separate proteins. Protein samples were extracted from 6-week-old Arabidopsis plants that were precultured in nutrient solution containing 2 mM ammonium nitrate and harvested after a 4-d period of N deficiency.
Since AMT1;1 is coexpressed with AMT1;3 in outer root cells (Loqué et al., 2006), we used the anti-AMT1;3 antibody and assayed for coexpressed AMT1;3 moieties in the same protein complexes. The major band detected under nonreducing conditions in Columbia-0 plants represented the oligomeric AMT1;3 protein (Figure 3C; see Supplemental Figure 2 online). Above that band, there was a faint protein smear that did not allow differentiation of bands of a concrete size, perhaps representing unspecific binding of the antibody or posttranslational protein modifications. In the two protein samples of the GFP-tagged lines, two additional weaker protein bands appeared with a size matching the two lower bands of the GFP-tagged AMT1;1 trimers (Figure 3C). The lower of these two weak bands most likely represented [AMT1;3][AMT1;1/1;3][AMT1;1-GFP] and the upper [AMT1;3][AMT1;1-GFP]2. In addition, we verified heterotrimer formation in a reciprocal approach by expression of AMT1;3:GFP under control of the endogenous AMT1;3 promoter in wild-type plants (see Supplemental Figure 3B online). In these lines, two concrete additional bands appeared above the wild-type homotrimer, most likely representing the presence of one or two GFP-tagged AMT1;3 subunits in the protein complex. The [AMT1;3-GFP]3 complex remained undetected because the employed anti-AMT1;3 antibody could not bind to the AMT1;3 peptide due to a disturbance of the C-terminal epitope by the GFP fusion (see Supplemental Figure 3B online). Probing this blot with anti-AMT1;1 revealed only one additional band above the trimer, most likely representing a [AMT1;1][AMT1;3][AMT1;3-GFP] complex. In particular, in the wild-type background, the detection of these higher molecular weight complexes was accompanied by a protein smear, suggesting that further protein modifications may have taken place. Considering the fainter appearance of this smear in transgenic lines expressing AMT1-GFP fusion proteins, these modifications may have been impaired by C-terminal GFP tags. Taken together, these results indicated that both AMT1;1 and AMT1;3 assemble as homotrimers in roots and additionally form heterotrimers in a 2:1 and 1:2 ratio.
The Mechanism of Allosteric Regulation by C-Terminal Phosphorylation within Homomeric AMT Complexes Is Conserved in Planta
Supposing that ammonium transport through AMT protein complexes relies on the trans-activation of pore regions by the C terminus of neighboring subunits and phosphorylation of the C terminus inhibits this interaction (Figure 1; Loqué et al., 2007), this type of allosteric regulation should not only work in heterologous expression systems but also be retained in planta. To verify the mechanistic basis of this regulatory model, we first expressed T464A- and T464D-modified AMT1;3 variants under control of the cauliflower mosaic virus 35S (CaMV35S) promoter in the quadruple insertion line for AMTs, qko (Yuan et al., 2007a). This genetic background is characterized by a reduction in the high-affinity ammonium uptake capacity by >90% due to disruption of the AMT1;1, AMT1;2, AMT1;3, and AMT2;1 genes and allows examination of the roles of individual transporters, transporter mutants, and coexpressed functional or defective transporters in planta. Methylammonium (MeA) toxicity was used as a tool to monitor the effect of mutations on long-term ammonium uptake activity. While qko is rather insensitive to MeA (Yuan et al., 2007a), independent transformants of qko ectopically expressing wild-type AMT1;3 (qko-35S:AMT1;3) or AMT1;3TA (qko-35S:AMT1;3TA) showed severely inhibited root growth and shoot biomass production (see Supplemental Figure 4 online). Growth of qko lines expressing AMT1;3TD (qko-35S:AMT1;3TD) was unaffected by MeA, indicating that the AMT1;3TD transporter variant was nonfunctional in planta.
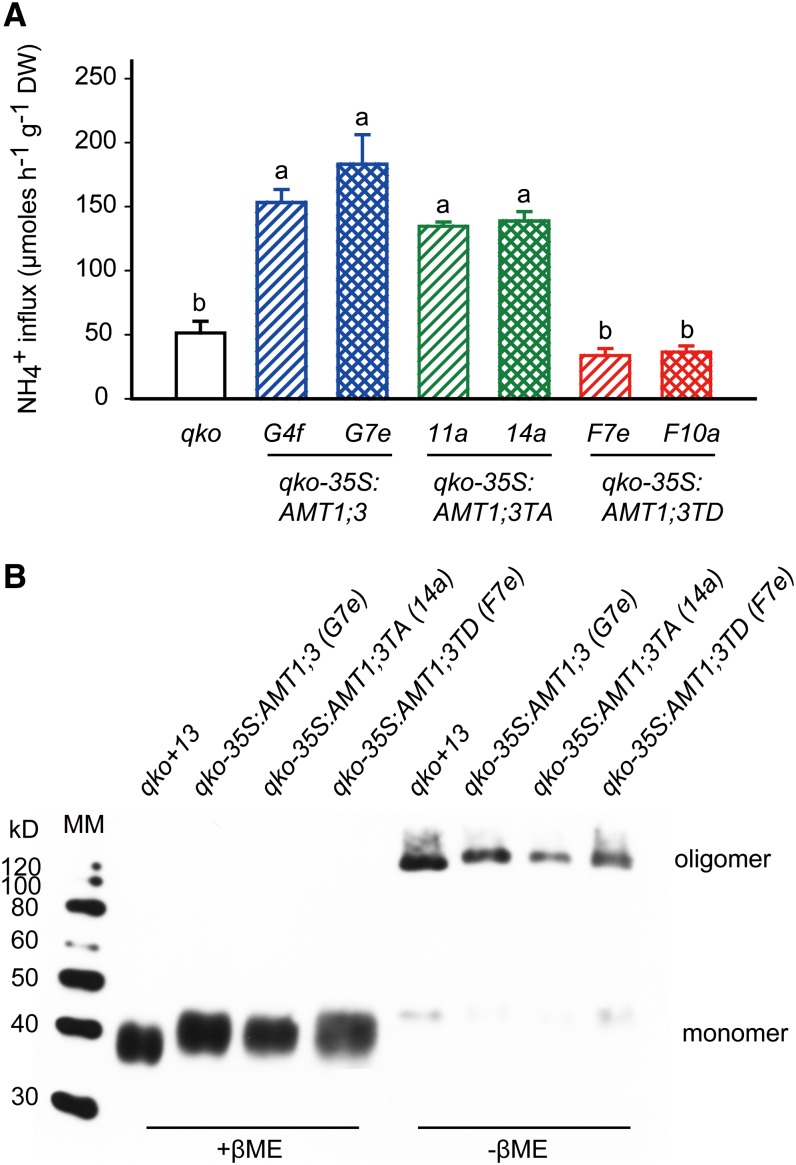
To determine the high-affinity ammonium transport capacity in roots, short-term influx studies were conducted in N-deficient plants at an external concentration of 200 µM 15N-labeled ammonium (Figure 4A). Relative to qko plants, expression of wild-type AMT1;3 conferred an ammonium uptake capacity of 100 to 120 µmol g−1 h−1, while the capacity of the TA-substituted variant was ∼90 µmol g−1 h−1. By contrast, the ammonium uptake capacity of transgenic lines expressing AMT1;3TD was not significantly different from that of qko, supporting inactivity of the AMT1;3TD protein in roots as indicated by its MeA insensitivity (see Supplemental Figure 4 online). A subsequent protein gel blot analysis using microsomal membrane fractions of the same plants showed that the TA- or TD-substituted AMT1;3 variant was expressed at a similar level as the wild type in the monomeric and oligomeric form (Figure 4B). Thus, substitution of Thr-464 by a negatively charged phosphomimicking Asp residue in the C terminus of AMT1;3 abolished transport activity without altering protein levels, protein stability, or protein oligomerization.
Figure 4.
Functional Expression of C-Terminally Modified AMT1;3 Variants in the Quadruple Insertion Line qko.
(A) Influx of 15N-labeled ammonium into roots of qko and two independent lines of qko-35S:AMT1;3, qko-35S:AMT1;3TA, or qko-35S:AMT1;3TD. Six-week-old Arabidopsis plants were precultured in nutrient solution containing 2 mM ammonium nitrate and subjected to N deficiency for a period of 4 d prior to influx analysis.15N-labeled ammonium was supplied at a concentration of 200 μM. Bars indicate means ± sd; n = 8 to 10. Significant differences at P < 0.01 according to Tukey’s test are indicated by different letters. DW, dry weight.
(B) Protein gel blot analysis of microsomal membrane fractions from roots of qko and one independent line of qko-35S:AMT1;3, qko-35S:AMT1;3TA, or qko-35S:AMT1;3TD using the anti-AMT1;3 antibody. Protein samples were pretreated either at 37°C in the presence of β-mercaptoethanol (reducing conditions; +βME) or at 0°C in the absence of β-mercaptoethanol (nonreducing conditions; −βME). MM, Magic Marker.
[See online article for color version of this figure.]
As AMT1;3 wild-type and phosphomimicking proteins functionally interacted when expressed in yeast (Figure 1), we investigated whether the C-terminal phosphomimic could trans-inhibit wild-type AMT1;3 in planta. Thus, AMT1;3TD was expressed in qko+13, in which gene expression of AMT1;3 has been reconstituted genetically in the qko background and high-affinity ammonium uptake is mediated predominantly by AMT1;3 (Yuan et al., 2007a). As expected, qko+13 was more sensitive to MeA than was qko (see Supplemental Figure 5 online), and the high-affinity uptake capacity of the native AMT1;3 protein (∼110 µmol g−1 h−1; Figure 5A) corresponded to the contribution of the ectopically expressed AMT1;3 protein in the qko background (Figure 4A). Expression of AMT1;3TD in the qko+13 background increased MeA tolerance (see Supplemental Figure 5 online) and decreased the ammonium uptake capacity by ∼90 µmol g−1 h−1 (Figure 5A). Interestingly, this decrease in ammonium uptake did not completely reach the level of qko plants but remained ∼20 µmol g−1 h−1 higher than in qko, indicating that the trans-inhibitory effect of AMT1;3TD did not fully repress the endogenous AMT1;3 protein activity. Protein gel blot analysis indicated that expression levels of the AMT1;3TD protein in both examined lines were stable and much higher than that of the endogenous AMT1;3 protein (Figure 5B). Therefore, the decrease in ammonium influx by 35S-driven expression of AMT1;3TD was not caused by cosuppression or RNA silencing of endogenous AMT1;3.
Figure 5.
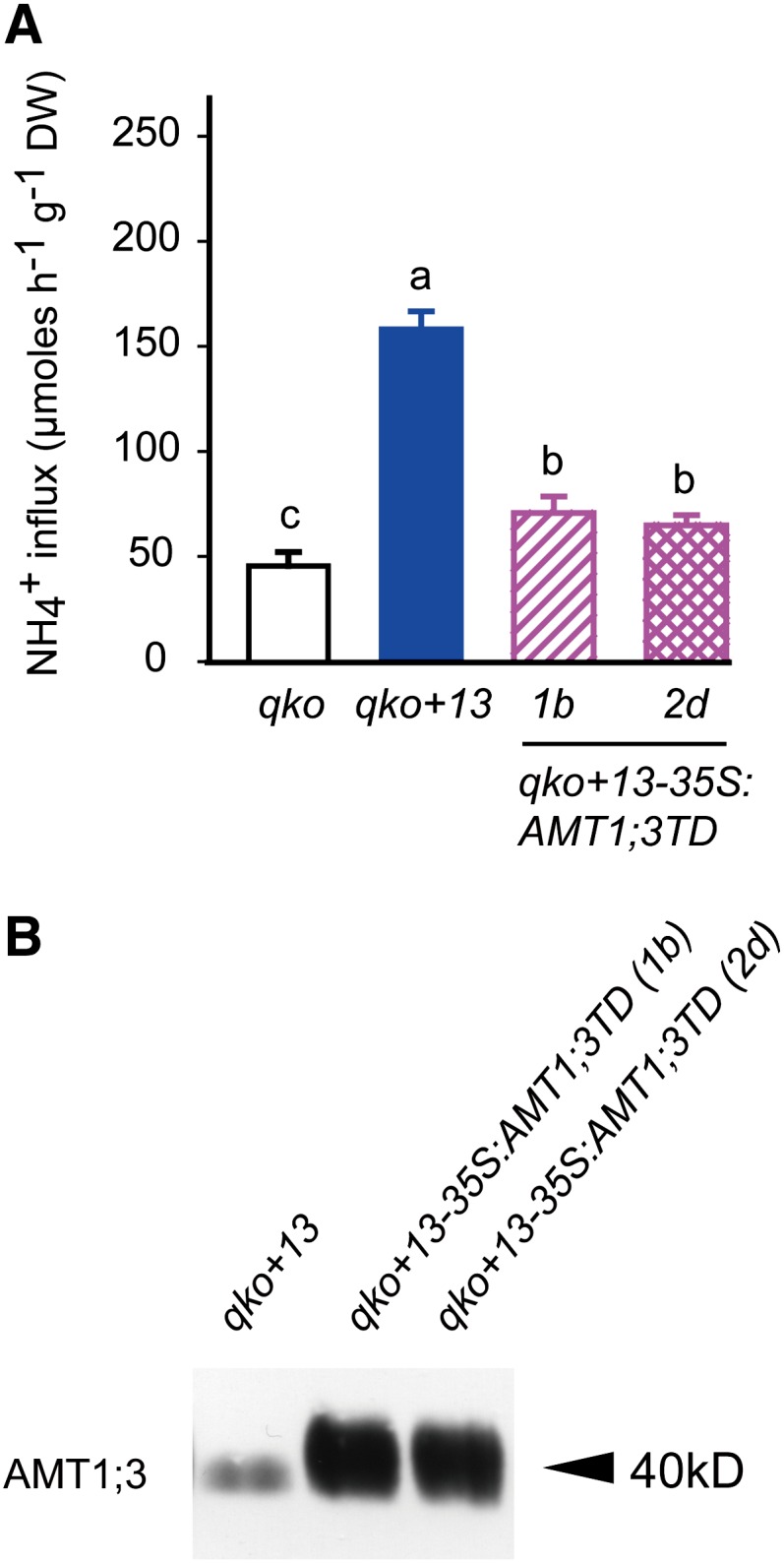
Inhibition of Ammonium Transport Capacities in Roots of the Triple Insertion Line qko+AMT1;3 (qko+13) by Ectopic Expression of AMT1;3TD.
(A) Influx of 15N-labeled ammonium into roots of the qko, qko+13, and two independent lines of qko+13-35S:AMT1;3TD. Six-week-old Arabidopsis plants were precultured in nutrient solution containing 2 mM ammonium nitrate and subjected to N deficiency for a period of 4 d prior to influx analysis. 15N-labeled ammonium was supplied at a concentration of 200 μM. Bars indicate means ± sd; n = 8 to 10. Significant differences at P < 0.01 according to Tukey’s test are indicated by different letters. DW, dry weight.
(B) Protein gel blot analysis of microsomal membrane fractions from roots of the same lines as in (A) using the anti-AMT1;3 antibody.
[See online article for color version of this figure.]
Dominant-Negative Regulation of Ammonium Transport Activity in Heteromeric AMT Complexes
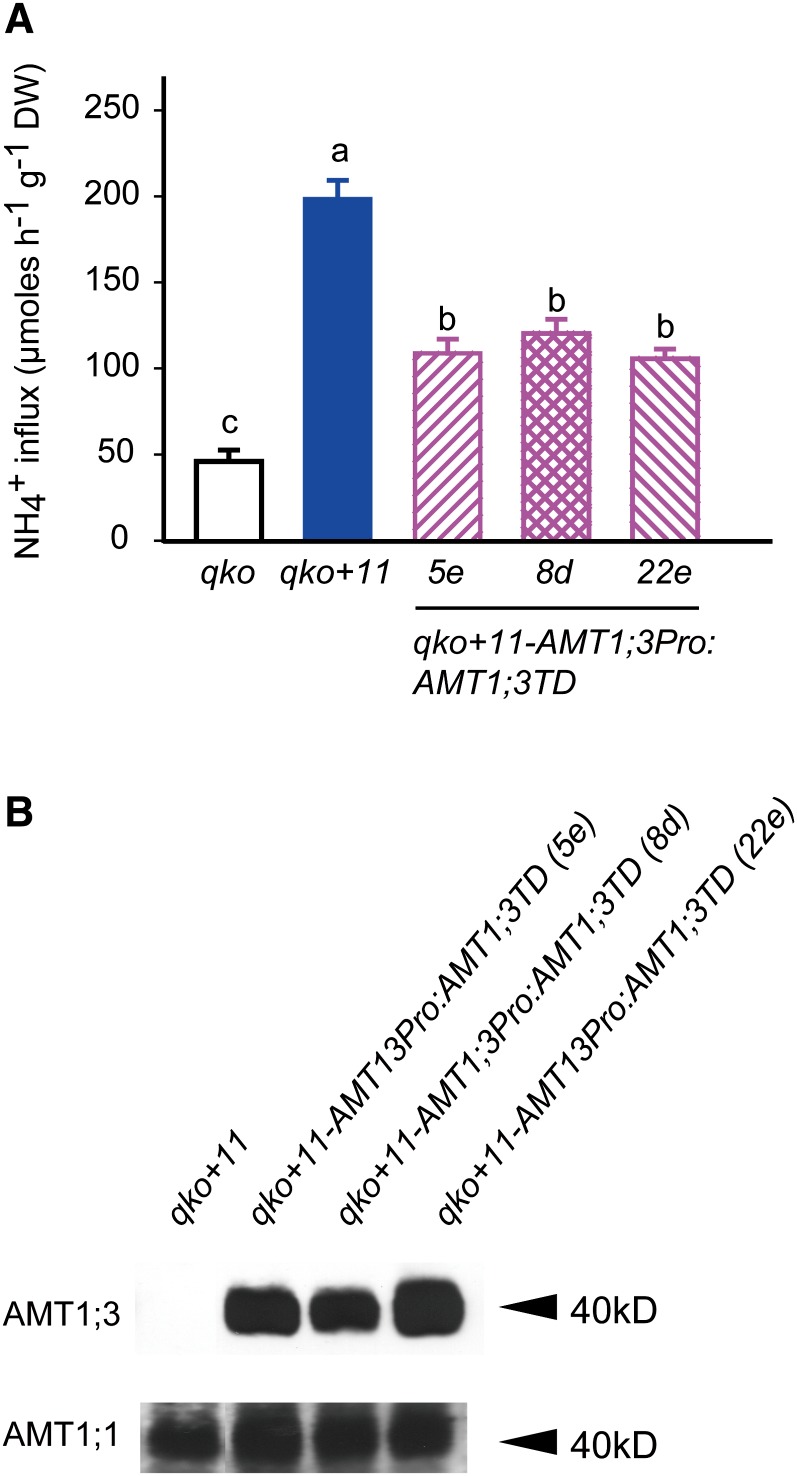
Based on the observations that AMT1;1 and AMT1;3 form heteromeric complexes in roots (Figure 3) and functionally interact in yeast cells (Figures 1 and 2), we hypothesized that a phosphomimic version of AMT1;3 should repress AMT1;1-dependent ammonium transport in roots. We therefore generated transgenic lines expressing AMT1;3TD in the qko background with reconstituted expression of AMT1;1 (qko+11) (Yuan et al., 2007a). Relative to qko, reconstituted expression of AMT1;1 conferred an ammonium transport capacity of ∼150 µmol g−1 h−1 (Figure 6A), which is 40 µmol g−1 h−1 larger than that of qko+13 (Figure 5A). To test for any artifacts possibly arising from ectopic expression from the constitutive CaMV35S promoter, the native promoter of AMT1;3 also was used to drive expression of AMT1;3TD, but ultimately both constructs yielded similar results (see Supplemental Figure 6 online). Compared with qko+11, the resulting three independent transformants showed a lower MeA sensitivity (see Supplemental Figure 6 online) and a consistent decrease in ammonium influx by ∼85 µmol g−1 h−1 (Figure 6A). Again, none of the transgenic lines showed signs of altered stability or synthesis of endogenous AMT1;1 protein (Figure 6B). These observations suggest that there is inhibition of AMT1;1-dependent ammonium transport activity resulting from protein–protein interactions between AMT1;3TD and the endogenous AMT1;1 protein.
Figure 6.
Inhibition of Ammonium Transport Capacities in Roots of the Triple Insertion Line qko+AMT1;1 (qko+11) by Ectopic Expression of AMT1;3TD.
(A) Influx of 15N-labeled ammonium into roots of qko, qko+11, and three independent lines of qko+11-AMT1;3Pro:AMT1;3TD. Six-week-old Arabidopsis plants were precultured in nutrient solution containing 2 mM ammonium nitrate and subjected to N deficiency for a period of 4 d prior to influx analysis. 15N-labeled ammonium was supplied at a concentration of 200 μM. Bars indicate means ± sd; n = 8 to 10. Significant differences at P < 0.01 according to Tukey’s test are indicated by different letters. DW, dry weight.
(B) Protein gel blot analysis of microsomal membrane fractions from roots of the same lines as in (A) using an anti-AMT1;3 or anti-AMT1;1 antibody.
[See online article for color version of this figure.]
To verify the quantitative extent of the trans-dominant inhibition of AMT1;1-mediated ammonium transport by AMT1;3TD, we expressed AMT1;3TD in the amt1;3-1 single insertion line (amt1;3-1-35S:AMT1;3TD), in which the endogenous AMT1;3 protein is absent (Loqué et al., 2006). Relative to the reference line amt1;3-1, ectopic expression of AMT1;3TD conferred increased growth on MeA (see Supplemental Figure 7 online) and reduced the ammonium transport capacity also by ∼85 µmol g−1 h−1 (Figure 7A). Thus, AMT1;3TD-mediated inhibition of AMT1;1 was independent of the genetic background and of the presence of other AMTs. Despite an almost identical ammonium uptake capacity in the two lines D2c and D9c (Figure 7A), protein gel blots revealed a much higher AMT1;3TD protein expression level in D9c (Figure 7B), suggesting that the lower amount of AMT1;3TD protein in D2c was sufficient to confer a maximum repression of ammonium influx in this background. In both lines, protein levels of AMT1;1 were stable and not affected by the presence of AMT1;3TD, indicating that AMT1;3TD did not cause posttranscriptional silencing or a loss of protein stability of endogenous AMT1;1.
Figure 7.
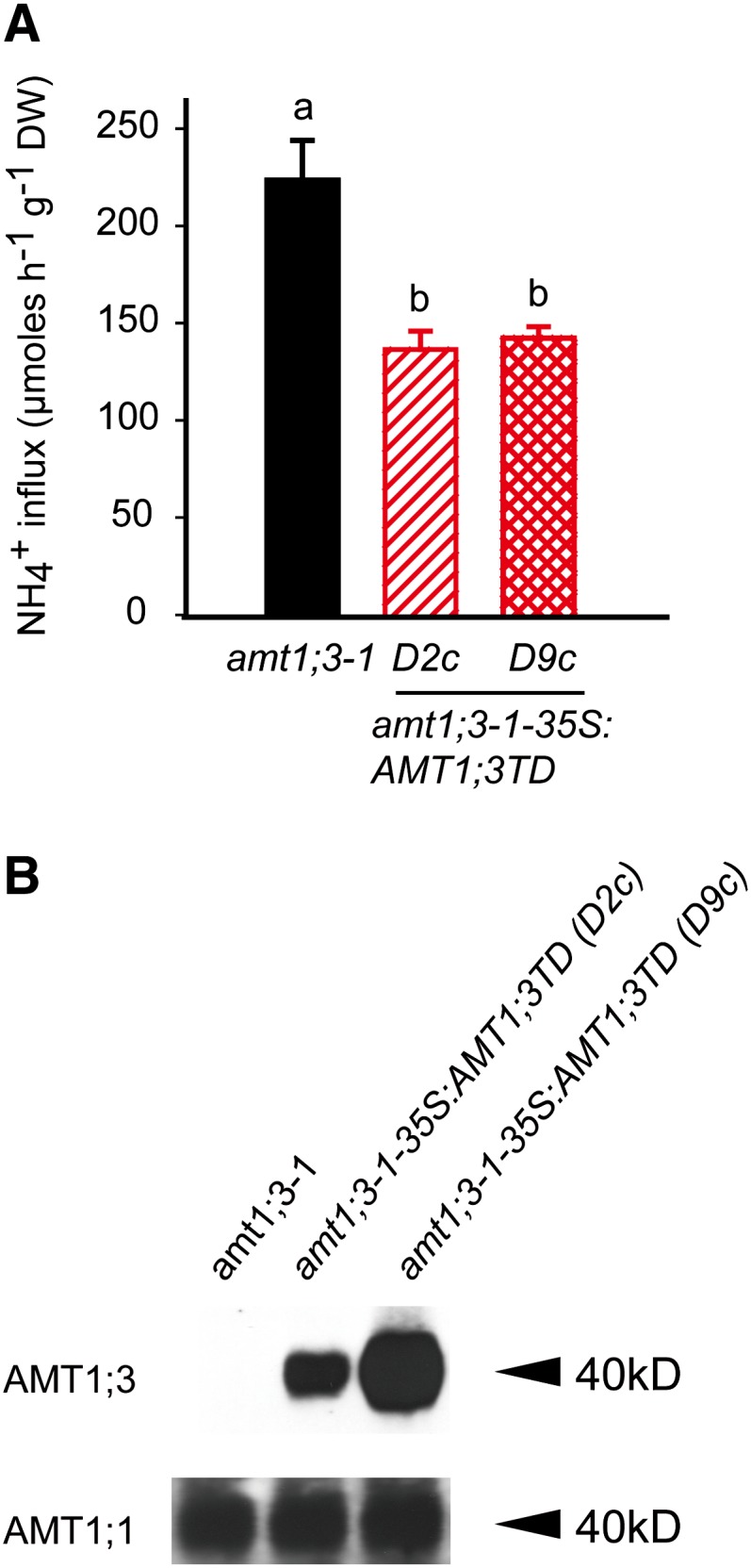
Inhibition of Ammonium Transport Capacities in Roots of the Single Insertion Line amt1;3-1 by Ectopic Expression of AMT1;3TD.
(A) Influx of 15N-labeled ammonium into roots of amt1;3-1 and two independent lines of amt1;3-1-35S:AMT1;3TD. Six-week-old Arabidopsis plants were precultured in nutrient solution containing 2 mM ammonium nitrate and subjected to N deficiency for a period of 4 d prior to influx analysis. 15N-labeled ammonium was supplied at a concentration of 200 μM. Bars indicate means ± sd; n = 8 to 10. Significant differences at P < 0.01 according to Tukey’s test are indicated by different letters. DW, dry weight.
(B) Protein gel blot analysis of microsomal membrane fractions from roots of the same lines as in (A) using the anti-AMT1;3 and anti-AMT1;1 antibody.
[See online article for color version of this figure.]
Since AMT1;3TD-mediated trans-inhibition appeared specific for AMT1;1 and AMT1;3, we anticipated both transport capacities to be repressed when expressing CaMV35S-AMT1;3TD in Wassilewskija (WS-35S:AMT1;3TD), which is the wild-type accession to amt1;3-1. In both transgenic lines, total protein levels of AMT1;3 were severalfold higher than in the wild type, confirming stability of the ectopically expressed AMT1;3TD variant (Figure 8B). Upon expression of AMT1;3TD, MeA sensitivity considerably decreased (see Supplemental Figure 8 online) and high-affinity ammonium influx was ∼120 µmol g−1 h−1 lower (Figure 8A). This loss of transport capacity was clearly higher than that conferred by AMT1;1 or AMT1;3 alone in qko, even though it remained below the sum of both transport capacities (175 µmol g−1 h−1). This was most likely due to the larger amount of AMT target proteins interacting with AMT1;3TD in Wassilewskija. Irrespective of this dose-dependent effect, this experiment showed that AMT1;3TD was able to trans-inactivate both AMT1;1 or AMT1;3 at the same time.
Figure 8.
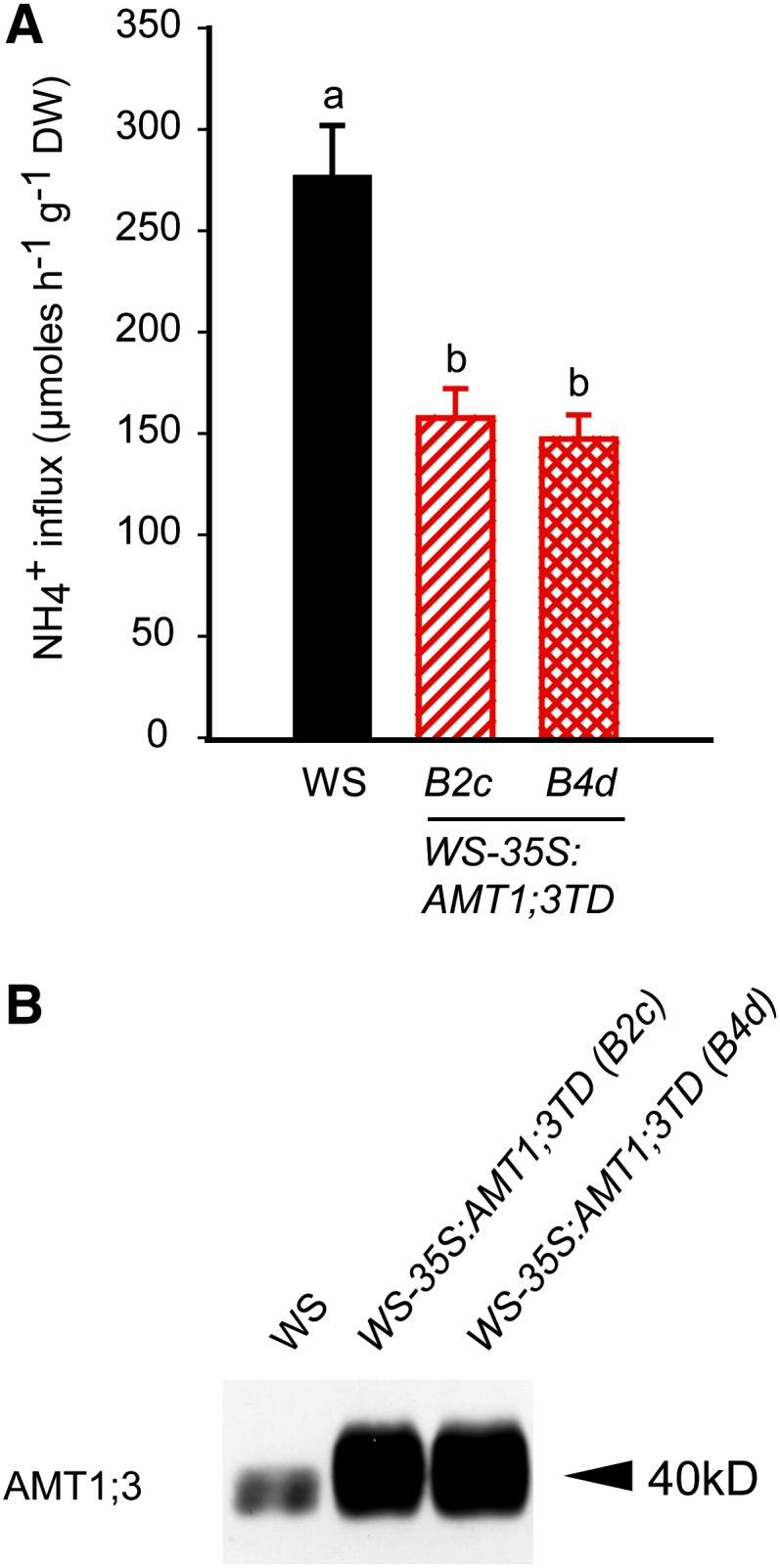
Inhibition of Ammonium Transport Capacities in the Roots of Arabidopsis Wild Type by Ectopic Expression of AMT1;3TD.
(A) Influx of 15N-labeled ammonium into roots of the wild type (Wassilewskija [WS]) and two independent lines of WS-35S:AMT1;3TD. Six-week-old Arabidopsis plants were precultured in nutrient solution containing 2 mM ammonium nitrate and subjected to N deficiency for a period of 4 d prior to influx analysis. 15N-labeled ammonium was supplied at a concentration of 200 μM. Bars indicate means ± sd; n = 8 to 10. Significant differences at P < 0.01 according to Tukey’s test are indicated by different letters. DW, dry weight.
(B) Protein gel blot analysis of microsomal membrane fractions from roots of the same lines as in (A) using an anti-AMT1;3 antibody.
[See online article for color version of this figure.]
To analyze the ability of AMTs to form homo- and heterooligomers in planta, N-terminal or C-terminal halves of the yellow fluorescent protein (YFP) were fused to the C terminus of AMT1;1 and AMT1;3. Protein interactions were investigated by transient coexpression of the two fusions in Nicotiana benthamiana and by observing YFP fluorescence using confocal microscopy. Fluorescence was observed for coexpression of N-terminal halves of AMT1;1 or AMT1;3 with C-terminal halves linked to the same protein as well as for coexpression of AMT1;1 with AMT1;3 (see Supplemental Figure 9 online). No fluorescence was observed when AMT1;1 or AMT1;3 was coexpressed with a complementary free half of YFP. In all cases, reconstitution of fluorescence appeared confined to the plasma membrane. These data indicate that AMT1;1 and AMT1;3 form homo- and hetero-oligomers at the plasma membrane in planta.
DISCUSSION
Enhanced ammonium uptake capacities in crop roots could potentially improve N fertilizer use efficiency and yield formation. However, previous attempts to enhance ammonium uptake capacities by ectopic expression of AMTs largely failed because their transport capacities are tightly regulated by ammonium (Yuan et al., 2007b). To prevent cells from cytosolic ammonium overload and membrane depolarization, AMT-type ammonium transporters are posttranslationally inactivated by C-terminal phosphorylation, which represents a rapid shut-off mechanism (Lanquar et al., 2009). We show here that this type of allosteric regulation efficiently represses ammonium transport capacities of homomeric as well as heteromeric protein complexes in plant roots, thereby allowing mutual regulation and crosstalk within protein complexes consisting of coassembled AMT isoforms.
Heterotrimerization of AMT1;1 and AMT1;3
Several lines of evidence indicated that AMT1;3 assembles in homotrimers as well as in heterotrimers with AMT1;1. First, in microsomal membrane fractions, AMT1;3 appeared as a high molecular weight form of >120 kD, which could be converted to a 40-kD form after denaturation, reflecting the expected size of the AMT1;3 monomer (see Supplemental Figure 2 online). This is in full agreement with the migration of the tomato AMT1 protein in SDS-PAGE as long as the N terminus required for trimer stability remains intact (Graff et al., 2011). Second, the high molecular weight forms of both AMT1;1 and AMT1;3 generated two to three additional bands when expressed with a GFP tag, which is consistent with the formation of homotrimers (Figure 3; see Supplemental Figure 3 online). This experimental evidence for the physical assembly of AMT1 proteins in trimers fully supports predictions on AMT1 trimerization derived from homology modeling based on the structure of crystallized AmtB and Amt-1 proteins (Khademi et al., 2004; Zheng et al., 2004; Andrade et al., 2005; Loqué et al., 2007) or from the occurrence of natural tomato AMT1 homologs that differ in protein complex stabilities and were detected in putative mono-, di-, and trimeric forms in protein gel blot analyses (Graff et al., 2011). Third, as the anti-AMT1;3 antibody also detected GFP-tagged AMT1;1 moieties and vice versa (Figure 3C; see Supplemental Figure 3 online), 1:2 and 2:1 complexes of AMT1;3 with AMT1;1 must have been generated besides the two homotrimeric forms. Based on their functional expression in yeast, the physical interactions between AMT1;1 and AMT1;3 subunits appeared to be independent of the participation of a third Arabidopsis protein because trans-inhibition conferred by AMT1;3TD or AMT1;1TD remained conserved (Figure 1; Loqué et al., 2007). The physical interaction between AMT1 moieties was further corroborated by split-ubiquitin analyses in yeast (Figure 2). Taking colony size as a proxy for the interaction strength did not appear to provide evidence for a particular preference for the formation of homo- versus heterooligomers. Employing a split-YFP assay further demonstrated that the formation of the homo- and heteromers of AMT1;1 and AMT1;3 occurred at the plasma membrane (see Supplemental Figure 9 online). Based on the coexpression of AMT1;1 and AMT1;3 in rhizodermal and cortical cells (Loqué et al., 2006) four classes of trimers are expected in cells that coexpress two isoforms: [AMT1;1]3, [AMT1;1]2[AMT1;3]1, [AMT1;1]1[AMT1;3]2, and [AMT1;1]3. Provided the two proteins are equally abundant and have similar affinities to form homo- and heteromers, one would expect 25% for each homomer and 25% for each heterotrimer (see Supplemental Figure 10 online). However, neither assay allowed a quantitative estimation of relative interaction affinities between the AMT1 subunits in a homo- or heterooligomeric complex. Alternative technologies like fluorescence resonance energy transfer (FRET) assays in stably transformed plants may provide a means to evaluate the relative affinities for oligomer formation among AMT isoforms.
Quantitative Dimension of the Allosteric Regulation in AMT1 Heteromers
To date, functional expression studies on heteromerizing membrane transporters have been mostly confined to heterologous expression systems. In the case of maize (Zea mays) plasma membrane intrinsic proteins (PIPs), PIP2;5-dependent water conductivity in oocytes increased with the amount of coexpressed but nonfunctional PIP1;2, probably mediated by formation of heterotetramers (Fetter et al., 2004), as the PIP1–PIP2 interaction was required for PIP1 trafficking to the plasma membrane to modulate substrate permeability (Zelazny et al., 2007). Coexpression of Arabidopsis AKT2 and KAT2 in oocytes revealed a diversity of different types of K+ currents, which most likely reflected the activities of homo- and heterotetrameric channel populations (Xicluna et al., 2007). Another K+ inward rectifier, AKT1, coassembled with nonfunctional Shaker-like KC1 subunits in heterotetrameric channels for the fine-tuning of electrophysiological properties depending on external K supplies (Geiger et al., 2009). These examples point to protein trafficking and modulation of biochemical transport activities as mechanisms, by which heteromerization can confer posttranslational regulation of oligomeric protein complexes. This study went beyond this by addressing the functional implications of AMT1 subunit heteromerization from a quantitative perspective in planta.
The wild-type AMT1;3 protein conferred an uptake capacity of ∼110 µmol ammonium g−1 h−1 irrespective of whether expression was driven from the 35S promoter or from the native promoter as in qko+13 (Figures 4A and 5A). Despite a large excess of the introduced AMT1;3TD protein in the qko+13 background, trans-inhibition of the wild-type AMT1;3 accounted just for 90 µmol ammonium g−1 h−1 (Figure 5A), leaving another 20 µmol ammonium g−1 h−1 that most likely reflected the activity of remaining pure wild-type AMT1;3 complexes. This remaining activity corresponded to ∼15% of the overall transport capacity, suggesting that approximately one-eighth of the AMT1;3 wild-type protein still assembled in a homotrimeric complex. When tomato wild-type AMT1;1 and the inactive G458D variant or Arabidopsis wild-type AMT1;2 and the inactive T472D variant were coexpressed at an equimolar complementary RNA ratio in oocyte cells, a residual ammonium conductivity of ∼12 to 15% remained, which was attributed to the activity of pure wild-type protein complexes (Ludewig et al., 2003; Neuhäuser et al., 2007). Cosuppression effects, decreased protein stabilities, or disordered targeting of these AMT1 variants are unlikely to explain our results given the large amounts of ectopically expressed protein detected in roots (Figures 4 to 8) and the observation that the substitutions and C-terminally truncated variants of Arabidopsis or tomato AMT1;1 were still targeted to the plasma membrane when expressed in yeast or plant protoplasts, respectively (Loqué et al., 2007; Neuhäuser et al., 2007). Moreover, in the absence of MeA, biomass production of wild-type and qko plants ectopically expressing the AMT1 variants was similar (see Supplemental Figures 4 to 8 online), indicating unaffected growth in the long run and thus further supporting a specific regulatory interaction between AMT1 proteins in planta.
A comparison of ammonium influx in the amt1;3 single mutant and in qko+11 clearly indicated that interactions of AMT1;3 with other AMT proteins in Arabidopsis roots are primarily confined to AMT1;1. Ectopic expression of AMT1;3TD in qko+11 resulted in a loss of ammonium influx by ∼85 µmol g−1 h−1 (Figure 6A). This value matched closely that achieved by expression of the same construct in the amt1;3 background (Figure 7A), suggesting that the transport capacities of AMT1;2 and AMT1;5 in amt1;3 were not significantly affected by trans-inactivation of AMT1;3TD. AMT1;3TD conferred (with 85 and 90 µmol g−1 h−1 activity, respectively) highly similar repression of transport capacity in qko+13 (Figure 5A) and in qko+11 (Figure 6A). Assuming that the higher transport capacity of AMT1;1 relative to AMT1;3 (Yuan et al., 2007a) was conferred by a higher transport rate of a single protein, this comparison may indicate a similar preference of AMT1;3TD to assemble with AMT1;1 or AMT1;3 moieties. In the case of a larger number of AMT1;1 transport proteins, AMT1;3TD would even have shown a larger preference to AMT1;1 over AMT1;3. Taken together, this quantitative approach reveals a specific interaction of AMT1;3TD with wild-type AMT1;1 and AMT1;3. Otherwise, 35S-driven overexpression of AMT1;3TD should also have affected other root-expressed AMT isoforms.
Physiological Implications of AMT Transporter Oligomerization
From a physiological perspective, an intermonomeric regulation among AMT1;1 and AMT1;3 heterotrimers enhances the versatility of AMT1 complexes to respond to external triggers. As shown for AMT1;1, extracellular ammonium triggers C-terminal phosphorylation at Thr-460 coinciding with a decrease in ammonium transport activity (Lanquar et al., 2009; see Supplemental Figure 10 online). Although the antibody recognizing the phosphorylated Thr-460 residue in AMT1;1 should in principle also recognize the phosphorylated Thr-464 residue in AMT1;3, it failed to prove phosphorylation of AMT1;3 upon ammonium resupply under our conditions. Phosphoproteomics approaches also did not detect AMT1;3T464 phosphorylation after ammonium resupply (Engelsberger and Schulze, 2012), which may indicate either that this residue is not phosphorylated in vivo or that the conditions that trigger its phosphorylation have not yet been identified. Thus, the ammonium stimulus triggering phosphorylation of Thr-460 in AMT1;1 may not be perceived by AMT1;3. By contrast, AMT1;3 triggers lateral root initiation in response to localized ammonium supplies, indicating an involvement in ammonium signaling, whereas AMT1;1 is incapable to do so (Lima et al., 2010). Thus, these two transporters apparently act in different signaling pathways, so that their colocalization and functional interaction by heteromerization in rhizodermal and cortical cells may provide a cross-point for signal exchange and regulatory fine-tuning of ammonium transport activities (see Supplemental Figure 10 online).
METHODS
Cloning of AMT1;3 Mutants and Expression in Yeast
The open reading frame of AMT1;3 was cloned into pDRf1-GW, which is a yeast shuttle vector carrying a Gateway cloning cassette (Loqué et al., 2007), resulting in the plasmid pDR-AMT1;3. The mutation was generated by site-directed mutagenesis of wild-type AMT1;3, yielding the plasmid pDR-AMT1;3TA using the primers AMT13TAF (GGGATGGATATGGCCCGTCACGGTGGC) and AMT1;3TAR (GCCACCGTGACGGGCCATATCCATCCC) or pDR-AMT1;3TD using the primers AMT13TDF (GGGATGGATATGGATCGTCACGGTGGC) and AMT1;3TDR (GCCACCGTGACGATCCATATCCATCCC). The yeast strain 31019b (Δmep1-3; Marini et al., 1997) was used to generate the strain DL1 by deletion of Gap1 or by integrating AMTs into the Gap1 gene to generate ΔGap1::AMT1;3 or ΔGap1::AMT1;1, similar as described by Loqué et al. (2007). The resulting yeast strains were transformed with different plasmids, and growth complementation was assayed on solid yeast nitrogen based (YNB) medium supplemented with 3% Glc, 2 mM ammonium chloride, or 1 mM Arg as sole N source and buffered at pH 5.2 by 50 mM MES-Tris.
Mating-Based Split-Ubiquitin System and Split-YFP Assay
For mating-based split-ubiquitin assays, AMT1;1 and AMT1;3 cDNAs were cloned into the mating-based split-ubiquitin Nub vectors pXN22_GW and pXN25_GW and Cub vector pMETYC_GW (Graff et al., 2011). Assays were performed as described by Lalonde et al. (2010). For the split-YFP assay, AMT1;1 and AMT1;3 cDNAs were cloned into the binary vectors pXNGW(-nYFP) and pXCGW (-cCFP). Assays were performed as described by Kim et al. (2009). The indicated combinations of constructs were introduced into Agrobacterium tumefaciens strains GV3101 or C58C1 and mixed with the strain harboring the tomato bushy stunt virus P19 as silencing suppressor for the transient expression in leaf cells of Nicotiana benthamiana. Plants were precultured in a growth chamber for 36 to 48 h under the continuous light, and the lower surface of leaves was infiltrated with the mixture of Agrobacterium cells using a syringe. YFP-derived fluorescence was analyzed by confocal microscopy.
Generation of Transgenic Arabidopsis Plants Expressing AMT1;3 Mutant Proteins
The DNA fragment of AMT1;3, AMT1;3TA, or AMT1;3TD was released from the corresponding constructs and cloned into the pCR-BluntII-TOPO vector (Invitrogen). Then, a XbaI-SpeI fragment containing each AMT1;3 was subcloned between the CaMV35S promoter and rbcs terminator sequence into the plant transformation vector pPT-Hyg using the restriction site XbaI. The AMT1;3 promoter fragment was released from the AMT1;3 promoter:ORF-GFP fusion construct (Loqué et al., 2006) and inserted into pTOPO-AMT1;3TD, resulting in a pTOPO-AMT1;3Pro:AMT1;3TD plasmid. A XbaI-SpeI fragment containing AMT1;3Pro:AMT1;3TD was subcloned into the plant transformation vector pT-Hyg (modified pPT-Hyg by removing the CaMV35S promoter) using the restriction site XbaI. The DNA fragment of AMT1;1ORF-GFP was released from the AMT1;1 promoter:ORF-GFP fusion construct (Loqué et al., 2006) and cloned into pTOPO, resulting in the plasmid pTOPO-AMT1;1GFP. Then, an ApaI-EcoRI fragment containing AMT1;1ORF-GFP was subcloned between the CaMV35S promoter and terminator sequence into the plant transformation vector pGreen-Hyg. These constructs were used to transform Arabidopsis thaliana lines with defective AMT gene expression (Loqué et al., 2006; Yuan et al., 2007a) by Agrobacterium-mediated transformation. The transformants were selected by hygromycin resistance, and homozygous T3 lines were further selected by segregation analysis.
Plant Culture
Arabidopsis plants were grown hydroponically for 6 weeks with full nutrient supply under a 22°C/18°C and 10-h/14-h light/dark regime and a light intensity of 280 µmol m−2 s−1 as described by Yuan et al. (2007a). To induce N deficiency, plants were maintained in N-free nutrient solution for a period of 4 d. In plate growth experiments, Arabidopsis seeds were surface sterilized and plated onto half-strength Murashige and Skoog medium (containing 5 mM nitrate as sole N source) solidified with Difco agar. The plants were precultured for 7 d and transferred to vertical plates containing half-strength Murashige and Skoog medium supplemented with MeA at different concentrations. Plants were grown under axenic conditions in a growth chamber under the above-mentioned conditions except that the light intensity was 120 µmol m−2 s−1.
15N-Ammonium Influx Analysis
Ammonium influx measurements in Arabidopsis roots were conducted after rinsing the roots of hydroponically grown plants in 1 mM CaSO4 solution for 1 min, followed by an incubation of 6 min in nutrient solution containing different concentrations of 15N-labeled NH4+ (95 atom% 15N) as the sole N source, before a final wash in 1 mM CaSO4 solution for 1 min. Roots were harvested and stored at −70°C before freeze-drying. Each sample was ground, and ∼1.6 mg powder was used for 15N determination by isotope ratio mass spectrometry (Finnigan).
Extraction of Membrane Fractions and Protein Gel Blot Analysis
Total microsomal membrane fractions were extracted from Arabidopsis roots as described by Yuan et al. (2007a). Protein concentrations were determined using the Bradford Protein Assay (Bio-Rad), and BSA served as a standard. Protein samples (5 to 10 µg per lane) were denatured in loading buffer (62.5 mM Tris-HCl, pH 6.8, 10% [v/v] glycerol, 2% [w/v] SDS, 0.01% [w/v] bromophenol blue, and 1% PMSF), incubated at 37°C for 30 min with 2.5% [v/v] β-mercaptoethanol or without β-mercaptoethanol at 0°C, and then electrophoresed in 10 or 6% SDS polyacrylamide gels and transferred to polyvinylidene fluoride membranes. Blots were developed using an ECL Advance Western Blotting Detection Kit (Amersham). Polyclonal antibodies were raised against the C terminus of AMT1;3, the loop between TM2 and TM3 of AMT1;1, and the C terminus of AMT1;1 (Loqué et al., 2006, 2007). Protein gel blots were assayed with anti-GFP (Invitrogen) and peroxidase-conjugated anti-rabbit IgG as a secondary antibody (Amersham). Magic Marker (Invitrogen) was used as molecular weight marker.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AMT1;1 (At4g13510), AMT1;2 (At1g64780), AMT1;3 (At3g24300), AMT1;4 (At4g28700), AMT1;5 (At3g24290), and AMT2;1 (At2g38290).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Amino Acid Sequence Alignments of Arabidopsis AMT1;3 and AMT1;1.
Supplemental Figure 2. Oligomerization of AMT1;3 in Roots.
Supplemental Figure 3. Heterotrimerization of AMT1;3 and AMT1;1.
Supplemental Figure 4. Functional Expression of C-Terminally Modified AMT1;3 Variants in the Quadruple Insertion Line qko as Revealed by Methylammonium Sensitivity.
Supplemental Figure 5. Inhibition of the Ammonium Transport Capacity in Roots of the Triple Insertion Line qko+AMT1;3 (qko+13) by Ectopic Expression of AMT1;3TD as Revealed by Methylammonium Sensitivity.
Supplemental Figure 6. Inhibition of the Ammonium Transport Capacity in Roots of the Triple Insertion Line qko+AMT1;1 (qko+11) by Ectopic Expression of AMT1;3TD as Revealed by Methylammonium Sensitivity.
Supplemental Figure 7. Inhibition of the Ammonium Transport Capacity in Roots of the Single Insertion Line amt1;3-1 by Ectopic Expression of AMT1;3TD as Revealed by Methylammonium Sensitivity.
Supplemental Figure 8. Inhibition of the Ammonium Transport Capacity in Roots of Arabidopsis Wild-Type Plants by Ectopic Expression of AMT1;3TD as Revealed by Methylammonium Sensitivity.
Supplemental Figure 9. Split-YFP–Based Interaction Assay for AMT1;1 and AMT1;3.
Supplemental Figure 10. Model for the trans-Inactivation of a Trimeric AMT Complex by Phosphorylated AMT1;1 or AMT1;3.
Supplementary Material
Acknowledgments
We thank Roberto de Michele (Carnegie Institution for Science, Stanford, CA) and Ben Gruber (Leibniz Institute of Plant Genetics and Crop Plant Research [IPK] Gatersleben) for critically reading the article. We acknowledge funding for L.Y. from the National Natural Science Foundation of China (30870189 and 31121062), for N.v.W. from the Deutsche Forschungsgemeinschaft, Bonn (WI1728/13), and for the W.B.F. lab through a grant from National Science Foundation (NSF 2010: 1021677).
AUTHOR CONTRIBUTIONS
L.Y. and N.v.W. designed the research. L.Y., R.G., Y.X., E.S.-V., and D.L. performed research. L.Y., W.B.F., and N.v.W. analyzed data. L.Y., W.B.F., and N.v.W. wrote the article.
Glossary
- GFP
green fluorescent protein
- CaMV35S
cauliflower mosaic virus 35S
- MeA
methylammonium
- YFP
yellow fluorescent protein
- YNB
yeast nitrogen based
References
- Andrade S.L., Dickmanns A., Ficner R., Einsle O. (2005). Crystal structure of the archaeal ammonium transporter Amt-1 from Archaeoglobus fulgidus. Proc. Natl. Acad. Sci. USA 102: 14994–14999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom A.J., Sukrapanna S.S., Warner R.L. (1992). Root respiration associated with ammonium and nitrate absorption and assimilation by barley. Plant Physiol. 99: 1294–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelsberger W.R., Schulze W.X. (2012). Nitrate and ammonium lead to distinct global dynamic phosphorylation patterns when resupplied to nitrogen-starved Arabidopsis seedlings. Plant J. 69: 978–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetter K., Van Wilder V., Moshelion M., Chaumont F. (2004). Interactions between plasma membrane aquaporins modulate their water channel activity. Plant Cell 16: 215–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzarrini S., Lejay L., Gojon A., Ninnemann O., Frommer W.B., von Wirén N. (1999). Three functional transporters for constitutive, diurnally regulated, and starvation-induced uptake of ammonium into Arabidopsis roots. Plant Cell 11: 937–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D., Becker D., Vosloh D., Gambale F., Palme K., Rehers M., Anschuetz U., Dreyer I., Kudla J., Hedrich R. (2009). Heteromeric AtKC1⋅AKT1 channels in Arabidopsis roots facilitate growth under K+-limiting conditions. J. Biol. Chem. 284: 21288–21295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff L., Obrdlik P., Yuan L., Loqué D., Frommer W.B., von Wirén N. (2011). N-terminal cysteines affect oligomer stability of the allosterically regulated ammonium transporter LeAMT1;1. J. Exp. Bot. 62: 1361–1373 [DOI] [PubMed] [Google Scholar]
- Hachiya T., Watanabe C.K., Fujimoto M., Ishikawa T., Takahara K., Kawai-Yamada M., Uchimiya H., Uesono Y., Terashima I., Noguchi K. (2012). Nitrate addition alleviates ammonium toxicity without lessening ammonium accumulation, organic acid depletion and inorganic cation depletion in Arabidopsis thaliana shoots. Plant Cell Physiol. 53: 577–591 [DOI] [PubMed] [Google Scholar]
- Kim J.G., et al. (2009). Xanthomonas T3S effector XopN suppresses PAMP-triggered immunity and interacts with a tomato atypical receptor-like kinase and TFT1. Plant Cell 21: 1305–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khademi S., O’Connell J., III, Remis J., Robles-Colmenares Y., Miercke L.J., Stroud R.M. (2004). Mechanism of ammonia transport by Amt/MEP/Rh: Structure of AmtB at 1.35 A. Science 305: 1587–1594 [DOI] [PubMed] [Google Scholar]
- Lalonde S., et al. (2010). A membrane protein/signaling protein interaction network for Arabidopsis version AMPv2. Front. Physiol. 1: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanquar V., Loqué D., Hörmann F., Yuan L., Bohner A., Engelsberger W.R., Lalonde S., Schulze W.X., von Wirén N., Frommer W.B. (2009). Feedback inhibition of ammonium uptake by a phospho-dependent allosteric mechanism in Arabidopsis. Plant Cell 21: 3610–3622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima J.E., Kojima S., Takahashi H., von Wirén N. (2010). Ammonium triggers lateral root branching in Arabidopsis in an AMMONIUM TRANSPORTER1;3-dependent manner. Plant Cell 22: 3621–3633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loqué D., Lalonde S., Looger L.L., von Wirén N., Frommer W.B. (2007). A cytosolic trans-activation domain essential for ammonium uptake. Nature 446: 195–198 [DOI] [PubMed] [Google Scholar]
- Loqué D., Mora S.I., Andrade S.L.A., Pantoja O., Frommer W.B. (2009). Pore mutations in ammonium transporter AMT1 with increased electrogenic ammonium transport activity. J. Biol. Chem. 284: 24988–24995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loqué D., von Wirén N. (2004). Regulatory levels for the transport of ammonium in plant roots. J. Exp. Bot. 55: 1293–1305 [DOI] [PubMed] [Google Scholar]
- Loqué D., Yuan L., Kojima S., Gojon A., Wirth J., Gazzarrini S., Ishiyama K., Takahashi H., von Wirén N. (2006). Additive contribution of AMT1;1 and AMT1;3 to high-affinity ammonium uptake across the plasma membrane of nitrogen-deficient Arabidopsis roots. Plant J. 48: 522–534 [DOI] [PubMed] [Google Scholar]
- Ludewig U., Wilken S., Wu B., Jost W., Obrdlik P., El Bakkoury M., Marini A.-M., André B., Hamacher T., Boles E., von Wirén N., Frommer W.B. (2003). Homo- and hetero-oligomerization of ammonium transporter-1 NH4 uniporters. J. Biol. Chem. 278: 45603–45610 [DOI] [PubMed] [Google Scholar]
- Marini A.M., Soussi-Boudekou S., Vissers S., Andre B. (1997). A family of ammonium transporters in Saccharomyces cerevisiae. Mol. Cell. Biol. 17: 4282–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini A.M., Springael J.Y., Frommer W.B., André B. (2000). Cross-talk between ammonium transporters in yeast and interference by the soybean SAT1 protein. Mol. Microbiol. 35: 378–385 [DOI] [PubMed] [Google Scholar]
- Neuhäuser B., Dynowski M., Mayer M., Ludewig U. (2007). Regulation of NH4+ transport by essential cross talk between AMT monomers through the carboxyl tails. Plant Physiol. 143: 1651–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat S.R., Silim S.N., Kronzucker H.J., Siddiqi M.Y., Glass A.D. (1999). AtAMT1 gene expression and NH4+ uptake in roots of Arabidopsis thaliana: Evidence for regulation by root glutamine levels. Plant J. 19: 143–152 [DOI] [PubMed] [Google Scholar]
- Rogato A., D’Apuzzo E., Barbulova A., Omrane S., Parlati A., Carfagna S., Costa A., Lo Schiavo F., Esposito S., Chiurazzi M. (2010). Characterization of a developmental root response caused by external ammonium supply in Lotus japonicus. Plant Physiol. 154: 784–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay P.L., Hallenbeck P.C. (2009). Of blood, brains and bacteria, the Amt/Rh transporter family: Emerging role of Amt as a unique microbial sensor. Mol. Microbiol. 71: 12–22 [DOI] [PubMed] [Google Scholar]
- Wang M.Y., Siddiqi M.Y., Ruth T.J., Glass A.D.M. (1993). Ammonium uptake by rice roots. II. Kinetics of 13NH4+ influx across the plasmalemma. Plant Physiol. 103: 1259–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xicluna J., Lacombe B., Dreyer I., Alcon C., Jeanguenin L., Sentenac H., Thibaud J.B., Chérel I. (2007). Increased functional diversity of plant K+ channels by preferential heteromerization of the shaker-like subunits AKT2 and KAT2. J. Biol. Chem. 282: 486–494 [DOI] [PubMed] [Google Scholar]
- Yuan L., Loqué D., Kojima S., Rauch S., Ishiyama K., Inoue E., Takahashi H., von Wirén N. (2007a). The organization of high-affinity ammonium uptake in Arabidopsis roots depends on the spatial arrangement and biochemical properties of AMT1-type transporters. Plant Cell 19: 2636–2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L., Loqué D., Ye F., Frommer W.B., von Wirén N. (2007b). Nitrogen-dependent posttranscriptional regulation of the ammonium transporter AtAMT1;1. Plant Physiol. 143: 732–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelazny E., Borst J.W., Muylaert M., Batoko H., Hemminga M.A., Chaumont F. (2007). FRET imaging in living maize cells reveals that plasma membrane aquaporins interact to regulate their subcellular localization. Proc. Natl. Acad. Sci. USA 104: 12359–12364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Kostrewa D., Bernèche S., Winkler F.K., Li X.-D. (2004). The mechanism of ammonia transport based on the crystal structure of AmtB of Escherichia coli. Proc. Natl. Acad. Sci. USA 101: 17090–17095 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.