SUMMARY
Multiple eukaryotic ribosomal proteins (RP) are co-opted for extraribosomal “moonlighting” activities, but paradoxically, RPs exhibit rapid turnover when not ribosome-bound. In one illustrative case of a functional extraribosomal RP, interferon (IFN)-γ induces ribosome release of L13a and assembly into the IFN-Gamma-Activated Inhibitor of Translation (GAIT) complex for translational control of a subset of inflammation-related proteins. Here we show GAPDH functions as a chaperone, shielding newly released L13a from proteasomal degradation. However, GAPDH protective activity is lost following cell treatment with oxidatively-modified low density lipoprotein and IFN-γ. These agonists stimulate S-nitrosylation at Cys247 of GAPDH which fails to interact with L13a, causing proteasomal degradation of essentially the entire cell complement of L13a, and defective translational control. Evolution of extraribosomal RP activities might require co-evolution of protective chaperones, and pathological disruption of either protein, or their interaction, presents an alternative mechanism of diseases due to RP defects, and targets for therapeutic intervention.
INTRODUCTION
Eukaryotic cells from yeast to mammals contain precise, stoichiometric amounts of ribosomal proteins (RP) maintained by transcriptional and translational mechanisms. However, as an additional safeguard, excess unassembled RPs are degraded rapidly, with a half-time ranging from seconds to minutes (Warner, 1999). The discovery of important extraribosomal, “moonlighting” functions by multiple eukaryotic RPs (Warner and McIntosh, 2009) presents an apparent paradox, namely, how are free RPs stabilized to permit these persistent activities? As a corollary, regulation (or dysregulation) of the stabilization process can influence RP extraribosomal activity, and possibly contribute to pathology. RP L13a, a component of the large ribosomal subunit conserved in bacteria, archaea, and eukarya, exhibits an extraribosomal function as a critical component of the IFN-Gamma-Activated Inhibitor of Translation (GAIT) complex in human myeloid cells (Mazumder et al., 2003; Mukhopadhyay et al., 2008). Elucidation of physiological and pathophysiological mechanisms regulating L13a survival outside the ribosome can shed light on these fundamental questions.
The GAIT complex inhibits translation of a select group of inflammation-related transcripts in human myeloid cells (Mazumder et al., 2003; Yao et al., 2012). Translational silencing requires binding of the heterotetrameric GAIT complex to the 3′-UTR of target mRNAs including ceruloplasmin (Cp), vascular endothelial growth factor (VEGF)-A, and several chemokines and their receptors (Mukhopadhyay et al., 2009; Ray and Fox, 2007; Sampath et al., 2003; Vyas et al., 2009). Other GAIT complex constituents are glutamyl-prolyl-tRNA synthetase (EPRS), NS1-associated protein-1 (NSAP1), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (Sampath et al., 2004). Remarkably, two GAIT complex proteins are inducibly released from parent macromolecular complexes following IFN-γ-stimulated phosphorylation: EPRS from the aminoacyl-tRNA multisynthetase complex (Sampath et al., 2004) and L13a from the 60S, large ribosomal subunit (Mazumder et al., 2003). L13a release from the ribosome requires phosphor-ylation of Ser77 by zipper-interacting protein kinase (ZIPK) (Mukhopadhyay et al., 2008). GAIT complex assembly occurs in two stages. Initially, phospho-EPRS binds NSAP1 to form a non-functional, pre-GAIT complex. About 16 hr later, phospho-L13a and GAPDH join the pre-GAIT complex to form the functional GAIT complex.
Inflammation is an orchestrated response to trauma, e.g., injury or microbial invasion, characterized by induced expression and activation of signaling cascades and defensive molecules, and gradually diminished upon injury repair or elimination of the trauma stimulus (Anderson, 2010; Lindemann et al., 2005; Medzhitov and Horng, 2009; Nathan, 2002). We have proposed that delayed activation of the GAIT complex permits initial high-level expression of certain inflammation-related proteins, but subsequently silences their expression to prevent unchecked injury to host tissues (Mukhopadhyay et al., 2009). Thus, the GAIT system, via transcript-selective, delayed translational repression activity, has the potential to restrict the duration and magnitude of the inflammatory response, and contribute to the resolution of inflammation. Conversely, condition-dependent or genetic defects in the GAIT system or its upstream activation pathways, might permit persistent over-expression of injurious GAIT system-regulated products and thereby contribute to chronic inflammatory disorders. However, conditions or effectors that inactivate L13a or other GAIT complex constituents, and prevent translational silencing of GAIT system target mRNAs have not been identified.
A principal component of the inflammatory response is cellular overproduction of reactive oxygen and nitrogen species that promote pathological modification of lipids and proteins, implicated in cardiovascular and neurodegenerative disease progression (Libby et al., 2002). Oxidatively-modified LDL (LDLox) is one such factor detected in plasma, cerebrospinal fluid, and inflammatory tissues, and is associated with risk for Alzheimer’s disease (Kankaanpaa et al., 2009), end-stage renal disease (Shoji et al., 2002), and atherosclerosis (Podrez et al., 2002; Stocker and Keaney, 2004). Here we show that LDLox prevents formation of an active GAIT complex in myeloid cells thereby prolonging production of VEGF-A and Cp. Our results provide evidence for physiological regulation and pathological dysregulation of the stability of an extraribosomal RP.
RESULTS AND DISCUSSION
To explore the influence of pathological conditions on GAIT-mediated translational control we investigated the potential effect of LDLox on GAIT activity in myeloid cells. Human peripheral blood monocytes (PBM) were incubated with IFN-γ in the presence of human LDLox generated by in vitro exposure of freshly isolated LDL to the physiological, leukocyte oxidation system consisting of myeloperoxidase (MPO), NO2-, and H2O2 (Podrez et al., 2000). The effect of LDLox on endogenous expression of GAIT targets was investigated. Immunoblot analysis of cell lysates revealed induction of VEGF-A and ceruloplasmin (Cp) protein after 8 hr of IFN-γ treatment, followed by marked suppression after 24 hr (Fig. 1A, top 3 panels). Delayed suppression of both proteins is due to GAIT-mediated translational silencing (Mazumder and Fox, 1999; Ray and Fox, 2007). In contrast, treatment of cells with IFN-γ in the presence of LDLox caused high-level expression of VEGF-A and Cp even after 24 hr. The enhancement of GAIT target expression at 24 h was specific as shown by the absence of any effect on β-actin expression, or total protein synthesis as determined by [35S]Met/Cys labeling of PBM protein (Fig. 1A, bottom). Persistent high-level expression of VEGF-A and Cp in lysates from LDLox-treated cells was not due to increased mRNA, consistent with the GAIT-mediated post-transcriptional control mechanism (Fig. 1B). The effect of LDLox on transcript-selective translation was determined by in vitro translation of a GAIT element-bearing Luc reporter in rabbit reticulocyte lysate (RRL). Cytosolic lysates from U937 cells treated with IFN-γ for 24 hr inhibited Luc RNA translation, as expected (Fig. 1C). Co-treatment of cells with IFN-γ and LDLox prevented the translation inhibition completely. Translation of T7 gene 10 RNA, a control transcript that lacks the GAIT element, was unaffected indicating target specificity.
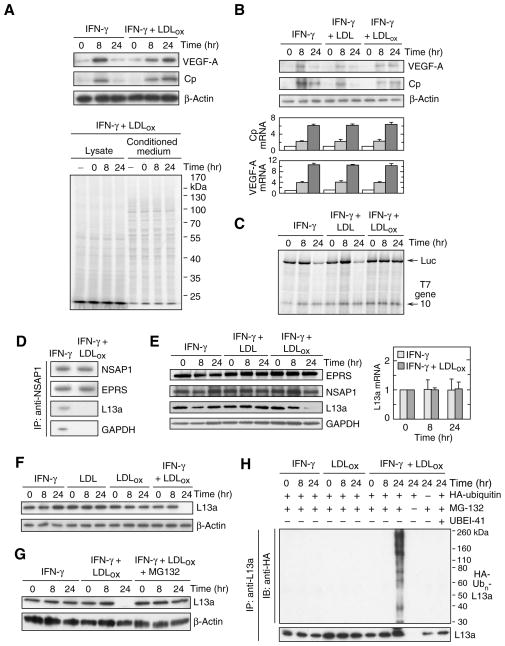
Figure 1. LDLox suppresses GAIT activity by selective degradation of ribosomal protein L13a.
(A) Effect of IFN-γ and LDLox on GAIT target gene expression. Human PBM were incubated with IFN-γ (500 unit/ml) in the presence of LDLox (50 μg/ml). Cell lysates (40 μg protein) were immunoblotted with anti-Cp, anti-VEGF-A and anti-β-actin antibodies (upper 3 panels). PBM were subjected to metabolic labeling for up to 24 h in the presence of [35S]Met/Cys and IFN-γ plus LDLox. Conditioned media and lysates were analyzed by SDS-PAGE and autoradiography (lower panel).
(B) Lack of effect of IFN-γ and LDLox on GAIT target mRNA expression. Human U937 monocytic cells were treated with IFN-γ in presence of unmodified LDL or LDLox, and lysates immunoblotted as above (top 3 panels). Cp, VEGF-A, and β-actin mRNA were determined by quantitative RT-PCR with appropriate primers; results were normalized to β-actin mRNA and expressed as mean ± SEM (n = 3 experiments).
(C) LDLox suppresses translational silencing activity of the GAIT pathway. U937 cells were incubated with IFN-γ (500 unit/ml) in the presence of LDL or LDLox (50 μg/ml) for 8 or 24 hr. Cp GAIT element-bearing Luc reporter and T7 gene 10 RNA (200 ng of each) were subjected to in vitro translation in rabbit reticulocyte lysate (RRL) in the presence of cell lysate (1 μg) and 35S-methionine.
(D) LDLox represses GAIT complex assembly. After cell treatment for 24 hr with IFN-γ or IFN-γ plus LDLox, lysates (1 mg) were immunoprecipitated with anti-NSAP1 antibody and immunoblotted with anti-NSAP1, -EPRS, -L13a and -GAPDH antibodies.
(E) IFN-γ plus LDLox represses L13a expression. U937 cells were incubated with IFN-γ and LDL or LDLox for up to 24 hr. Expression of GAIT complex proteins was determined by immunoblot analysis (left). L13a mRNA (right) was determined by quantitative RT-PCR, normalized to expression in untreated cells (mean ± SEM, n = 3 experiments).
(F) LDLox by itself does not degrade L13a. Cells were treated as in (A) and L13a and β-actin determined by immunoblot.
(G) Proteasome degradation of L13a. Cells were incubated with IFN-γ plus LDLox in presence or absence of MG132 (5 μM); MG132 was added after 4 and 12 hr for the 8- and 24-hr incubations, respectively. Cell lysates were immunoblotted with anti-L13a or anti-β-actin antibodies.
(H) Poly-ubiquitinylation of L13a. U937 cells were transfected with plasmid encoding HA-ubiquitin and then treated with IFN-γ, LDLox, MG132, and UBEI-41 (25 μM) as indicated. Lysates were subjected to immunoprecipitation with anti-L13a antibody and immunoblot analysis with anti-HA tag and -L13a antibodies.
To explore the mechanism underlying GAIT inhibition by LDLox, we examined GAIT complex assembly. After stimulation with IFN-γ and LDLox for 24 hr, NSAP1 was immunoprecipitated and the interaction with the other three GAIT complex components determined by immunoblot analysis. The pre-GAIT complex, formed by interaction of EPRS with NSAP1 about 2 hr after IFN-γ treatment, assembled normally. However, L13a and GAPDH, components unique to the mature GAIT complex, did not bind NSAP1 in cells treated with IFN-γ and LDLox (Fig. 1D). Remarkably, LDLox caused a near-complete disappearance of L13a after 24 hr, but levels of other GAIT complex proteins were unaltered; native LDL did not alter expression of GAIT proteins (Fig. 1E, left). LDLox did not reduce L13a mRNA expression, indicating a post-transcriptional regulatory mechanism (Fig. 1E, right). Neither IFN-γ nor LDLox by itself reduced L13a expression, indicating an absolute requirement for both agonists (Fig. 1F). To establish degradation of L13a, IFN-γ and LDLox were added to cells in the presence of MG132, a proteasome inhibitor. MG132 prevented L13a depletion at 24 hr, substantiating proteasomal degradation (Fig. 1G). Proteins are generally targeted for proteasome degradation by polyubiquitinylation at one or more Lys residues by the coordinated action of ubiquitin activating enzyme E1, conjugating enzyme E2, and E3 ligase. Cells were transfected with cDNA encoding HA-tagged ubiquitin in the presence of MG132 to prevent proteasomal degradation of ubiquitinylated protein. Ubiquitinylation was detected with anti-HA tag antibody following immunoprecipitation with anti-L13a antibody. Cell treatment with LDLox for 24 hr induced robust polyubiquitinylation of L13a that was blocked by UBEI-41, an E1 inhibitor (Fig. 1H). Treatment of cells with LDLox or IFN-γ alone did not induce L13a ubiquitinylation confirming the requirement for both.
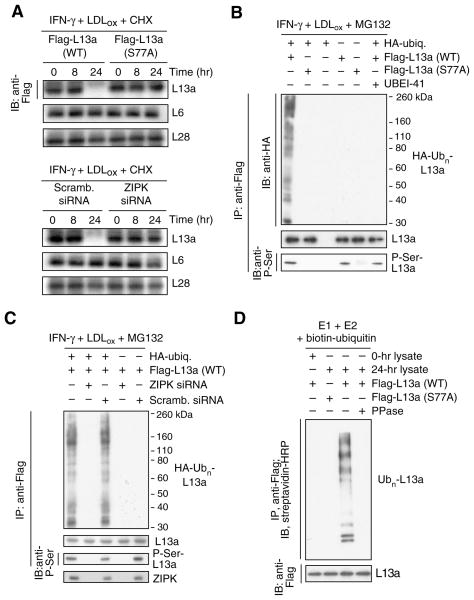
IFN-γ induces ZIPK-mediated L13a phosphorylation at Ser77, and consequent release from the large ribosome subunit (Mukhopadhyay et al., 2008). We investigated whether L13a phosphorylation is required for ubiquitinylation and degradation. Cells were transfected with plasmid encoding Flag-tagged L13a bearing a non-phosphorylatable Ser-to-Ala mutation at the Ser77 phospho-site. The wild-type transfectant was almost completely degraded whereas mutated L13a was not susceptible to LDLox-mediated degradation (Fig. 2A, top 3 panels). Endogenous RPs L6 and L28 were not degraded revealing exceptional target selectivity. Similar results were observed when ZIPK, the proximal L13a kinase, was specifically inhibited by siRNA (Fig. 2A, bottom 3 panels). To investigate the specific role of phosphorylation in L13a ubiquitinylation, U937 cells were co-transfected with HA-tagged ubiquitin and Flag-tagged wild-type or S77A mutant L13a. L13a phosphorylation was determined by immunoprecipitation with anti-Flag antibody and immunoblot analysis with anti-phosphoserine. Wild-type L13a was phosphorylated and robustly polyubiquitinylated by IFN-γ and LDLox (in the presence of MG132 to prevent degradation), but the S77A mutant was not susceptible to either covalent modification (Fig. 2B). Similarly, L13a phosphorylation and ubiquitinylation were inhibited by siRNA-mediated ZIPK suppression (Fig. 2C). Phosphorylation is required for L13a release from the ribosome, but might independently be required for L13a recognition by the E1–E2–E3 system. The latter possibility was investigated in a cell-free system. Flag-tagged L13a was incubated with biotin-ubiquitin and a reconstituted ubiquitinylation system containing recombinant E1 and E2, and cell lysate to provide E3 ligase (and L13a kinase activity in the 24-hr lysate). Wild-type L13a was robustly polyubiquitinylated by the E1–E2–E3 cascade; however, modification was prevented by substitution of the S77A mutant, by pre-treatment with alkaline phosphatase, or after addition of a 0-hr lysate lacking L13a kinase activity (Fig. 2D). Thus, L13a phosphorylation is required not only for ribosome release, but also for recognition by the ubiquitinylation system.
Figure 2. Phosphorylation-dependence of L13a ubiquitinylation and degradation.
(A) L13a degradation is phosphorylation-dependent. U937 cells were transfected with plasmids encoding Flag-tagged wild-type (WT) or phospho-null (S77A) L13a. After recovery, cells were treated with IFN-γ plus LDLox in the presence of cycloheximide (CHX) to inhibit de novo synthesis. Lysates were immunoblotted with anti-Flag, -L6, and -L28 antibodies (top 3 panels). Cells were transfected with ZIP kinase (or scrambled) siRNA and then incubated and immunoblotted as above (bottom 3 panels).
(B) L13a poly-ubiquitinylation is phosphorylation-dependent. U937 cells were co-transfected with plasmids encoding HA-ubiquitin and Flag-tagged wild-type or S77A mutant L13a. After recovery, cells were treated with IFN-γ plus LDLox and with MG132 and UBEI-41 as in Figure 1H. L13a in cell lysates was immunoprecipitated with anti-Flag antibody, and immunoblotted with anti-HA, -L13a, and -phospho-Ser antibodies.
(C) Poly-ubiquitinylation requires ZIP kinase (ZIPK). U937 cells were co-transfected with plasmids encoding HA-ubiquitin and Flag-L13a, and with ZIPK (or scrambled) siRNA. Cells were treated as above, immunoprecipitated with anti-Flag antibody, and immunoblotted with anti-HA (top), anti-L13a (second panel), or anti-phospho-Ser (third panel) antibodies. ZIPK was detected in 16 hr lysates by immunoblot with anti-ZIPK antibody (bottom).
(D) L13a phosphorylation is required for E3 ligase recognition. Flag-tagged L13a was incubated with biotin-ubiquitin and a reconstituted ubiquitinylation system containing recombinant E1 and E2 and lysate from cells treated with IFN-γ for 24 hr (or 0 hr as negative control) to provide E3 ligase and L13a kinase activity.
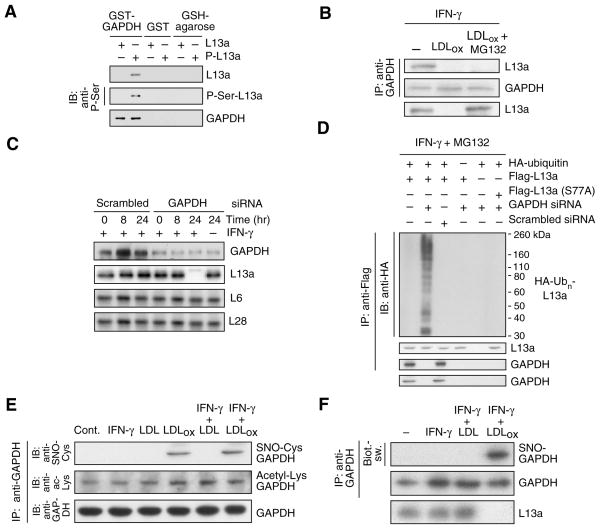
The second phase of GAIT complex assembly requires both GAPDH and L13a. We considered the possible role of GAPDH in determining L13a stability. To investigate their interaction in the absence of other GAIT components, L13a and phospho-L13a were incubated with immobilized glutathione-S-transferase (GST)–GAPDH fusion protein, and the interaction detected by immunoblot. GAPDH bound phospho-L13a, but not unmodified L13a (Fig. 3A). Importantly, treatment of cells with LDLox for 24 hr (in the presence of MG132 to prevent L13a degradation) completely blocked the IFN-γ-driven interaction of GAPDH and L13a, suggesting that the interaction might be critical for maintenance of free cellular L13a (Fig. 3B). We considered the possibility that GAPDH functions as a shield that binds phospho-L13a and prevents its degradation. GAPDH-deficient cells were generated using siRNA targeted against GAPDH in the presence of pyruvate, the 3-carbon end-product of glycolysis, to minimize bioenergetic imbalance (Hara et al., 2005). GAPDH depletion, even in the absence of LDLox, caused specific and near-complete degradation of L13a after 24-h treatment with IFN-γ (Fig. 3C). Similarly, depletion of GAPDH induced phosphorylation-dependent polyubiquinylation of L13a in the absence of LDLox (Fig. 3D).
Figure 3. GAPDH binds phospho-L13a and protects it from degradation.
(A) GAPDH binds phospho-L13a in absence of other GAIT components. Unmodified and phosphorylated L13a (P-L13a, prepared by treatment with ZIPK) were incubated with GST-GAPDH or GST immobilized to glutathione (GSH)-agarose beads. After washing, binding was detected by immunoblot analysis with anti-L13a, -phospho-Ser and -GAPDH antibodies.
(B) LDLox blocks GAPDH binding to phospho-L13a. Cells were treated with IFN-γ, LDLox, and MG132 as shown. Cell lysates were immunoprecipitated with anti-GAPDH antibody, and immunoblotted with anti-L13a and -GAPDH antibodies. Total L13a was detected by immunoblot with anti-L13a antibody.
(C) GAPDH protects L13a from degradation. U937 cells were transfected with GAPDH (or scrambled) siRNA. After recovery, cells were treated with IFN-γ, and lysates immunoblotted with anti-GAPDH, -L13a, -L6, and -L28 antibodies.
(D) GAPDH prevents ubiquitinylation of phospho-L13a in cells. U937 cells were co-transfected with GAPDH (or scrambled) siRNA and with plasmids encoding HA-ubiquitin and Flag-tagged L13a (wild-type or S77A mutant). After recovery, cells were incubated with IFN-γ and MG132 for 24 hr. Lysates were immunoprecipitated with anti-Flag antibody, and immunoblotted with anti-HA, -L13a, and -GAPDH antibodies. Total GAPDH was determined by immunoblot with anti-GAPDH antibody.
(E) LDLox induces S-nitrosylation of GAPDH. Cells were treated with IFN-γ, LDL, and LDLox. Lysates were immunoprecipitated with anti-GAPDH antibody and immunoblotted with anti-GAPDH, -S-nitrosocysteine (SNO-Cys), and -acetyl-Lys antibodies.
(F) Determination of GAPDH S-nitrosylation by biotin-switch assay. S-nitrosylation of endogenous GAPDH and L13a degradation were determined in human PBM treated with IFN-γ in presence of LDL or LDLox for 24 hr. GAPDH was immunoprecipitated and S-nitrosylation detected with avidin-HRP in the biotin-switch assay. Efficiency of immunoprecipitation was shown by immunoblot analysis with anti-GAPDH antibody.
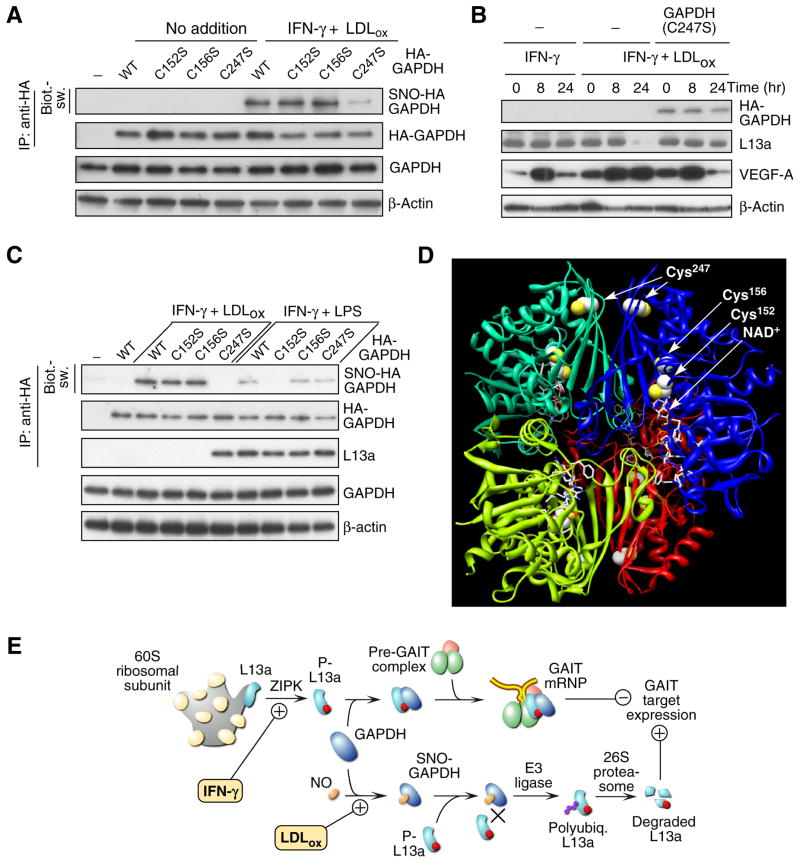
We examined the possibility that LDLox induced a post-translational modification of GAPDH that blocked its ability to shield L13a. Cell treatment with LDLox induced formation of S-nitrosylated GAPDH (SNO-GAPDH) as determined by immunoblot analysis with anti-SNO-Cys antibody (Fig. 3E). Modification did not require IFN-γ. Lys-acetylation was also detected, but this modification was not stimulus-dependent. Lys- and Arg-methylation, and Ser-, Tyr-, and Thr-phosphorylation were not detected (not shown). S-nitrosylation was confirmed by the “biotin-switch” procedure that replaces Cys nitrosyl groups with biotin for subsequent detection by avidin (Jaffrey et al., 2001). IFN-γ plus LDLox induced robust S-nitrosylation of GAPDH in human PBM (Fig. 3F). To determine the Cys residue S-nitrosylated by LDLox, U937 cells were transfected with human HA-GAPDH cDNAs containing Cys-to-Ser mutations at each of the three positions 152, 156, and 247. LDLox plus IFN-γ induced S-nitrosylation of GAPDH bearing mutations at Cys152 and Cys156 but not at Cys247, indicating Cys247 is the primary LDLox-inducible target (Fig. 4A, Supplemental Fig. S1A). S-nitrosylation of Cys247 was confirmed by mass spectrometric analysis of GAPDH isolated from cells treated with LDLox plus IFN-γ (Supplemental Fig. S1B). Transfection of the C247S GAPDH mutant protected L13a from degradation and restored IFN-γ-mediated translational silencing of VEGF-A in the presence of LDLox; this result also shows that other constituents of the GAIT complex are not impaired (Fig. 4B). Importantly, LDLox S-nitrosylates an amino acid distinct from the Cys150 residue (the rat equivalent of human Cys152) S-nitrosylated by lipopolysaccharide (LPS) and other apoptotic stimuli (Hara et al., 2005). Indeed, treatment of human PBM with IFN-γ plus LPS induced specific S-nitrosylation of Cys152 and failed to induce L13a degradation (Fig. 4C). Together, these results indicate that GAPDH protects L13a from degradation, and that LDLox-induced S-nitrosylation of GAPDH inactivates the protective function.
Figure 4. Effect of LDLox on GAIT target gene expression and schematic.
(A) LDLox induces S-nitrosylation of GAPDH at Cys247. Mouse BMDM were transfected with plasmids encoding wild-type HA-GAPDH, or C152S, C156S, or C247S mutants. Cells were treated with IFN-γ and LDLox for 24 hr. S-nitrosylation of transfected GAPDH was detected in lysates by immunoprecipitation with anti-HA tag antibody followed by biotin-switch analysis. Total transfected HA-GAPDH was determined by immunoprecipitation with anti-HA tag antibody followed by immunoblot analysis with anti-GAPDH antibody. Total GAPDH and β-actin were determined by immunoblot analysis.
(B) Expression of GAPDH C247S mutant restores translational silencing of VEGF-A. Cells were transfected with plasmid encoding HA-tagged C247S mutant GADPH and treated with IFN-γ or IFN-γ plus LDLox for up to 24 hr. Expression of HA-GAPDH, L13a, VEGF-A, and β-actin was detected by immunoblot analysis.
(C) LDLox and LPS Induce GAPDH S-nitrosylation at different sites. Human PBM were transfected with plasmids encoding wild-type HA-GAPDH, or C152S, C156S, or C247S mutants. S-nitrosylation of transfected GAPDH in presence of IFN-γ plus LDLox or IFN-γ plus LPS (10 μg/ml) was detected by biotin-switch. Transfected HA-GAPDH and GAPDH-bound L13a were determined by immunoprecipitation with anti-HA tag antibody followed by immunoblot analysis with anti-GAPDH and anti-L13a antibodies. Total GAPDH and β-actin were determined by immunoblot analysis.
(D) Three-dimensional structure of GAPDH. Ribbon diagram of tetrameric GAPDH structure with individual subunits shown in different colors. Cys residues are shown in spacefill with sulfur atoms highlighted (yellow). NAD+ in active site is shown (stick model).
(E) Schematic of GAIT pathway activation and its disruption by LDLox. IFN-γ induces GAIT complex assembly that causes translational repression of inflammation-related, GAIT target genes (upper pathway). LDLox induces GAPDH S-nitrosylation and consequent L13a degradation, causing GAIT pathway dysregulation and prolonging GAIT target gene expression (lower pathway).
See also Figure S1.
Protein S-nitrosylation is a ubiquitous, post-translational effector mechanism. Dysregu-lation of protein S-nitrosylation is associated with diverse pathological conditions (Foster et al., 2009). A concept is emerging in which altered modification of a specific disease-related protein, rather than a global nitrosative effect, contributes to disease phenotype. In a well-studied example, apoptotic stimulation of murine macrophages with lipopoly-saccharide (LPS) triggers S-nitrosylation of GAPDH to facilitate binding and nuclear translocation of Siah1, contributing to cytotoxicity. (Hara et al., 2005). In our experiments LDLox induces S-nitrosylation at human GAPDH Cys247 (equivalent to rat Cys245), a residue distant from the active-site Cys152 (equivalent to rat Cys150) residue modified following apoptotic stimulation by LPS (Hara et al., 2005). Differential cysteine S-nitrosylation suggests that LDLox and LPS, in presence of IFN-γ, trigger distinct proinflammatory signaling pathways. Sequence motifs associated with S-nitrosylation are not well-defined; however, a modified acid-base motif has been proposed in which charged residues are concentrated at the solvent-exposed surface of the protein near the NO-Cys site, which may not be surface-exposed (Marino and Gladyshev, 2010). The X-ray crystal structure of human GAPDH indicates the Cys247 site is not exposed to the surface, but charged residues nearby are exposed (Fig. 4D). Moreover, because mutation of human Cys247, in contrast to Cys152, is far from the active-site and does not alter GAPDH enzymatic activity (Nakajima et al., 2009), even extensive modification is unlikely to negatively influence cell energy balance.
GAPDH is an archetype multi-functional protein; in addition to its principal role in glycolysis, GAPDH acts as reversible metabolic sensor that responds to oxidative stress (Zheng et al., 2003). Our new results suggest an unexpected protective function of GAPDH. In its native form, GAPDH binds phosphorylated L13a upon release from the 60S ribosomal subunit, and protects it from degradation by the ubiquitin-proteasome system (Fig. 4E). The stoichiometry of ribosomal proteins is generally enforced by rapid degradation of newly synthesized ribosomal proteins not bound to ribosome sub-complexes during assembly (Nomura, 1999). Several ribosomal proteins are protected and controlled by heat shock proteins during ribosome biogenesis (Kim et al., 2006). However, one can envisage that a specialized mechanism is required for protection of a free ribosomal protein released from a mature ribosome as in the case of L13a. Possibly, the mechanism is generalizable and GAPDH binds and protects other ribosomal proteins with extra-ribosomal functions (Warner and McIntosh, 2009). S-nitrosylation of GAPDH results in complete loss of the protective function, and consequent ubiquitinylation and degradation of phospho-L13a and loss of GAIT complex activity.
Ribosomal proteins are implicated in multiple pathologies including developmental impairment (Warner and McIntosh, 2009), malignant peripheral nerve tumors (Amsterdam et al., 2004), glucose tolerance and diabetes (Ruvinsky et al., 2005), Diamond-Blackfan anemia (Flygare and Karlsson, 2007; Warner and McIntosh, 2009), and cleft palate and abnormal thumbs (Draptchinskaia et al., 1999; Gazda et al., 2008). In all of these cases, the pathology can be traced to genetic modification of a ribosomal protein. In contrast, selective, stimulus-mediated degradation of L13a is remarkable in that the phenotype is not due to genetic alteration, but rather is a consequence of environment-dependent modification. Moreover, the pathophysiological significance of L13a is unique in that it is the extraribosomal function that is subject to dysregulation. The possible role of extraribosomal functions of other RPs in pathogenesis, for example in Diamond-Blackfan anemia, has been considered, but remains uncertain (Flygare and Karlsson, 2007; Warner and McIntosh, 2009). Our findings suggest that in seeking the etiological underpinnings of RP-related disease, attention should be given not just to the ribosome and their associated RPs, but also to extraribosomal RPs and their binding partners.
EXPERIMENTAL PROCEDURES
Reagents, Plasmids, Site-directed Mutagenesis, and Recombinant Proteins
See Extended Experimental Procedures.
Cell Culture and Transfection
Human U937 monocytic cells (ATCC, Rockville, MD) were cultured in RPMI 1640 medium containing 10% fetal bovine serum (FBS). BMDM were harvested and cultured from femurs of mice in C57BL/6J background under a protocol approved by the Cleveland Clinic Institutional Animal Care and Use Committee. PBM from healthy clinical donors were isolated by leukapheresis and countercurrent centrifugal elutriation under a Cleveland Clinic Institutional Review Board-approved protocol that adhered to American Association of Blood Bank guidelines. Cells were pre-incubated for 1 hr in medium containing 1% FBS before treatment with IFN-γ (500 units/ml), lipoproteins (50 μg/ml), and other reagents for up to 24 hr. In some experiments, U937 cells were transfected with plasmids (5 μg DNA) expressing HA-ubiquitin, Flag-L13a (wild-type or S77A), or HA-GAPDH (wild-type or Cys mutants) using Human Monocyte Nucleofector Kit (Lonza, Walkersville, MD). Cell transfection with siRNA against ZIPK or GAPDH was done according to manufacturer’s protocol and allowed to recover 24 hr before use.
Quantitative RT-PCR, in Vitro Transcription, and in Vitro Translation
Total RNA (10 ng) from U937 cells was reverse-transcribed, and analyzed by quantitative RT-PCR using primers specific for VEGF-A, Cp, β-actin, or L13a. Capped, poly(A)-tailed Luc reporter and T7 gene 10 RNAs were prepared using Message Machine SP6 and T7 Kit (Ambion). DNA template preparation and mRNA purification were as described (Sampath et al., 2004). For determination of GAIT activity, cell lysates (1 μg) were pre-incubated with reporter and control mRNAs, and in vitro translation determined by incorporation of [35S]methionine in RRL, followed by autoradiography (Jia et al., 2008).
Determination of L13a Ubiquitinylation
To determine L13a polyubiquitinylation, U937 cells (1 × 107 cells) were transfected with plasmid encoding HA-ubiquitin (5 μg, or 2.5 μg when co-transfected with 2.5 μg Flag-L13a), and then incubated with IFN-γ and LDL for up to 24 hr in the presence of MG132 (5 μM) to prevent proteasomal degradation. Cell lysates (1 mg) were immunoprecipi-tated with anti-L13a or -Flag antibody, and immunoblotted with antibodies against HA-tag, L13a, and P-Ser. For in vitro reconstitution of L13a ubiquitinylation, purified Flag-tagged L13a (1 μg of wild-type or S77A mutant) was pre-incubated with U937 cell lys-ate, and then incubated with a mixture E1 and E2 enzymes, biotin-ubiquitin, and cell lysate as a source of L13a kinase and E3 ligase. Recombinant L13a was immunopre-cipitated with anti-Flag tag antibody, and biotin-ubiquitin detected with streptavidin-HRP.
Determination of GAPDH-L13a Interaction
Purified GST-GAPDH was immobilized on glutathione-agarose beads, and incubated with L13a (unmodified or phosphorylated) expressed and phosphorylated as described (Mukhopadhyay et al., 2008). After washing, bound proteins were heat-denatured in SDS gel-loading solution and analyzed by immunoblotting.
Biotin-Switch Determination of S-Nitrosylation of GAPDH
U937 cell (1 × 107 cells) lysates were labeled by the biotin-switch method (Jaffrey et al., 2001) with the S-Nitrosylation Protein Detection Assay Kit, immunoprecipitated with anti-GAPDH antibody, and detected with avidin-HRP.
Supplementary Material
HIGHLIGHTS.
GAPDH acts as a shield, protecting free ribosomal protein L13a from degradation
Condition-dependent degradation of an extraribosomal ribosomal protein
S-Nitrosylation of GAPDH at Cys247 suppresses its protective activity
Degradation of unprotected L13a inactivates GAIT system and induces VEGF-A expression
Acknowledgments
This work was supported by NIH grants P01 HL029582, P01 HL076491, and R01 GM086430, (to P.L.F.). A.A. was supported by National Center Scientist Development Grant 10SDG3930003 from the American Heart Association. We are grateful to Dr. Ephraim Sehayek for helpful discussions.
Footnotes
None of the authors have any financial conflict of interest with the information in this manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Jie Jia, Email: jiaj@ccf.org.
Abul Arif, Email: arifaa@ccf.org.
Dennis J. Stuehr, Email: stuehrd@ccf.org.
Stanley L. Hazen, Email: hazens@ccf.org.
References
- Amsterdam A, Sadler KC, Lai K, Farrington S, Bronson RT, Lees JA, Hopkins N. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol. 2004;2:E139. doi: 10.1371/journal.pbio.0020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P. Post-transcriptional regulons coordinate the initiation and resolution of inflammation. Nat Rev Immunol. 2010;10:24–35. doi: 10.1038/nri2685. [DOI] [PubMed] [Google Scholar]
- Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Willig TN, Dianzani I, Ball S, Tchernia G, Klar J, Matsson H, et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet. 1999;21:169–175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- Flygare J, Karlsson S. Diamond-Blackfan anemia: erythropoiesis lost in translation. Blood. 2007;109:3152–3154. doi: 10.1182/blood-2006-09-001222. [DOI] [PubMed] [Google Scholar]
- Foster MW, Hess DT, Stamler JS. Protein S-nitrosylation in health and disease: a current perspective. Trends Mol Med. 2009;15:391–404. doi: 10.1016/j.molmed.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazda HT, Sheen MR, Vlachos A, Choesmel V, O’Donohue MF, Schneider H, Darras N, Hasman C, Sieff CA, Newburger PE, et al. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am J Hum Genet. 2008;83:769–780. doi: 10.1016/j.ajhg.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, Takahashi M, Cheah JH, Tankou SK, Hester LD, et al. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat Cell Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- Jia J, Arif A, Ray PS, Fox PL. WHEP domains direct noncanonical function of glutamyl-prolyl tRNA synthetase in translational control of gene expression. Mol Cell. 2008;29:679–690. doi: 10.1016/j.molcel.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankaanpaa J, Turunen SP, Moilanen V, Horkko S, Remes AM. Cerebrospinal fluid antibodies to oxidized LDL are increased in Alzheimer’s disease. Neurobiol Dis. 2009;33:467–472. doi: 10.1016/j.nbd.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Kim TS, Jang CY, Kim HD, Lee JY, Ahn BY, Kim J. Interaction of Hsp90 with ribosomal proteins protects from ubiquitination and proteasome-dependent degradation. Mol Biol Cell. 2006;17:824–833. doi: 10.1091/mbc.E05-08-0713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby P, Ridker PM, Maseri A. Inflammation and atherosclerosis. Circulation. 2002;105:1135–1143. doi: 10.1161/hc0902.104353. [DOI] [PubMed] [Google Scholar]
- Lindemann SW, Weyrich AS, Zimmerman GA. Signaling to translational control pathways: diversity in gene regulation in inflammatory and vascular cells. Trends Cardiovasc Med. 2005;15:9–17. doi: 10.1016/j.tcm.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Marino SM, Gladyshev VN. Structural analysis of cysteine S-nitrosylation: a modified acid-based motif and the emerging role of trans-nitrosylation. J Mol Biol. 2010;395:844–859. doi: 10.1016/j.jmb.2009.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder B, Fox PL. Delayed translational silencing of ceruloplasmin transcript in gamma interferon-activated U937 monocytic cells: Role of the 3′ untranslated region. Mol Cell Biol. 1999;19:6898–6905. doi: 10.1128/mcb.19.10.6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder B, Sampath P, Seshadri V, Maitra RK, DiCorleto P, Fox PL. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell. 2003;115:187–198. doi: 10.1016/s0092-8674(03)00773-6. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat Rev Immunol. 2009;9:692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay R, Jia J, Arif A, Ray PS, Fox PL. The GAIT system: a gatekeeper of inflammatory gene expression. Trends Biochem Sci. 2009;34:324–331. doi: 10.1016/j.tibs.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R, Ray PS, Arif A, Brady AK, Kinter M, Fox PL. DAPK-ZIPK-L13a axis constitutes a negative-feedback module regulating inflammatory gene expression. Mol Cell. 2008;32:371–382. doi: 10.1016/j.molcel.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima H, Amano W, Kubo T, Fukuhara A, Ihara H, Azuma YT, Tajima H, Inui T, Sawa A, Takeuchi T. Glyceraldehyde-3-phosphate dehydrogenase aggregate formation participates in oxidative stress-induced cell death. J Biol Chem. 2009;284:34331–34341. doi: 10.1074/jbc.M109.027698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan C. Points of control in inflammation. Nature. 2002;420:846–852. doi: 10.1038/nature01320. [DOI] [PubMed] [Google Scholar]
- Nomura M. Regulation of ribosome biosynthesis in Escherichia coli and Saccharomyces cerevisiae: Diversity and common principles. J Bacteriol. 1999;181:6857–6864. doi: 10.1128/jb.181.22.6857-6864.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podrez EA, Batyreva E, Shen Z, Zhang R, Deng Y, Sun M, Finton PJ, Shan L, Febbraio M, Hajjar DP, et al. A novel family of atherogenic oxidized phospholipids promotes macrophage foam cell formation via the scavenger receptor CD36 and is enriched in atherosclerotic lesions. J Biol Chem. 2002;277:38517–38523. doi: 10.1074/jbc.M205924200. [DOI] [PubMed] [Google Scholar]
- Podrez EA, Febbraio M, Sheibani N, Schmitt D, Silverstein RL, Hajjar DP, Cohen PA, Frazier WA, Hoff HF, Hazen SL. Macrophage scavenger receptor CD36 is the major receptor for LDL modified by monocyte-generated reactive nitrogen species. J Clin Invest. 2000;105:1095–1108. doi: 10.1172/JCI8574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray PS, Fox PL. A post-transcriptional pathway represses monocyte VEGF-A expression and angiogenic activity. EMBO J. 2007;26:3360–3372. doi: 10.1038/sj.emboj.7601774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvinsky I, Sharon N, Lerer T, Cohen H, Stolovich-Rain M, Nir T, Dor Y, Zisman P, Meyuhas O. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev. 2005;19:2199–2211. doi: 10.1101/gad.351605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath P, Mazumder B, Seshadri V, Fox PL. Transcript-selective translational silencing by gamma interferon is directed by a novel structural element in the ceruloplasmin mRNA 3′ untranslated region. Mol Cell Biol. 2003;23:1509–1519. doi: 10.1128/MCB.23.5.1509-1519.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath P, Mazumder B, Seshadri V, Gerber CA, Chavatte L, Kinter M, Ting SM, Dignam JD, Kim S, Driscoll DM, Fox PL. Noncanonical function of glutamyl-prolyl-tRNA synthetase: gene-specific silencing of translation. Cell. 2004;119:195–208. doi: 10.1016/j.cell.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Shoji T, Fukumoto M, Kimoto E, Shinohara K, Emoto M, Tahara H, Koyama H, Ishimura E, Nakatani T, Miki T, et al. Antibody to oxidized low-density lipoprotein and cardiovascular mortality in end-stage renal disease. Kidney Int. 2002;62:2230–2237. doi: 10.1046/j.1523-1755.2002.00692.x. [DOI] [PubMed] [Google Scholar]
- Stocker R, Keaney JF., Jr Role of oxidative modifications in atherosclerosis. Physiol Rev. 2004;84:1381–1478. doi: 10.1152/physrev.00047.2003. [DOI] [PubMed] [Google Scholar]
- Vyas K, Chaudhuri S, Leaman DW, Komar AA, Musiyenko A, Barik S, Mazumder B. Genome-wide polysome profiling reveals an inflammation-responsive post-transcriptional operon in IFN-gamma-activated monocytes. Mol Cell Biol. 2009;29:458–470. doi: 10.1128/MCB.00824-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol Cell. 2009;34:3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao P, Potdar AA, Arif A, Ray PS, Mukhopadhyay R, Willard B, Xu Y, Yan J, Saidel GM, Fox PL. Coding region polyadenylation generates a truncated tRNA synthetase that counters translation repression. Cell. 2012;149:88–100. doi: 10.1016/j.cell.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Roeder RG, Luo Y. S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell. 2003;114:255–266. doi: 10.1016/s0092-8674(03)00552-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.