Summary
Cells of the budding yeast Saccharomyces cerevisiae select a site for polarized growth in a specific pattern that depends on their cell type. Haploid a and α cells bud in the axial budding pattern, which requires assembly of a landmark that includes the Bud4 protein. To understand how an axial bud site is established, we performed a structure–function analysis of Bud4. Bud4 contains DUF1709 (domain of unknown function), which is similar to a part of the anillin-homology domain, and a putative Pleckstrin homology (PH) domain near to its C terminus. Although its localization depends on septins, a conserved family of GTP-binding proteins, Bud4 is necessary for the stable inheritance of septin rings during cell division. Although some anillins interact directly with septins, we find that neither DUF1709 nor the PH domain is necessary for targeting Bud4 to the mother-bud neck. Instead, this C-terminal region is crucial for association of Bud4 with Bud3 and other components of the axial landmark. Remarkably, septins colocalize with Bud4 mutant proteins that lack these C-terminal domains, forming an arc or a single ring instead of a double ring during and after cytokinesis. Interestingly, overexpression of Bud4 also induces formation of extra Bud4 rings and arcs that are associated with septins. Analyses of a series of bud4 truncation mutants suggest that at least two domains in the central region play a redundant role in targeting Bud4 to the mother-bud neck and are thus likely to interact with septins. Taken together, these results indicate that Bud4 functions as a platform that links septins to the axial landmark.
Key words: Anillin-related protein, Axial budding pattern, Budding yeast, Septins
Introduction
Organization and positioning of a variety of macromolecular structures at specific subcellular locations are critical for cellular processes. During vegetative growth, haploid a and α cells of the budding yeast Saccharomyces cerevisiae bud in the axial pattern, in which both mother and daughter cells select a bud site immediately adjacent to their previous division site (Chant and Herskowitz, 1991; Chant and Pringle, 1995; Freifelder, 1960; Hicks et al., 1977). The axial budding pattern is dependent on Bud3, Bud4, Axl1 and Axl2, which are thought to assemble as a spatial landmark (Chant et al., 1995; Halme et al., 1996; Lord et al., 2002; Roemer et al., 1996; Sanders and Herskowitz, 1996). This axial landmark dictates a new bud site by recruiting the Rsr1 GTPase module (Kang et al., 2001), which is comprised of Rsr1 (also known as Bud1), its GTPase activating protein (GAP) Bud2 and its GDP-GTP exchange factor (GEF) Bud5 (Bender, 1993; Bender and Pringle, 1989; Chant et al., 1991; Chant and Herskowitz, 1991; Park et al., 1993). The Rsr1 GTPase module then likely links the spatial cues to the polarity establishment machinery, including the Cdc42 GTPase (Bi and Park, 2012; Kozminski et al., 2003; Park et al., 1997).
Bud4 appears to play a pivotal role in the assembly of the axial landmark. Bud4 localizes to the mother-bud neck first by interacting with septins and then recruits other components of the axial landmark including Bud3 (Kang et al., 2012; Sanders and Herskowitz, 1996). Localization of Bud3 to the mother-bud neck is diminished greatly, although not completely, in the absence of Bud4. In contrast, Bud4 localizes to the neck normally in the absence of Bud3, but its ring structure becomes unstable during/after cytokinesis (Kang et al., 2012). Bud4 also interacts with Axl1 and Axl2 to mediate the ordered assembly of the axial landmark and its cell-type-specific interaction with Bud5 (Gao et al., 2007; Kang et al., 2012; Lord et al., 2002).
Septins are conserved GTP-binding proteins and are involved in a variety of cellular processes including cytokinesis. In budding yeast, mitotic septins (Cdc3, Cdc10, Cdc11, Cdc12 and Shs1/Sep7) form heteromeric complexes that assemble into filaments. Septins localize to the mother-bud neck, forming a tubular ‘collar’ throughout the cell cycle and a split double ring during cytokinesis that is inherited by mother and daughter cells as a single ring marking the division site (Oh and Bi, 2011; Versele and Thorner, 2005). Septins are also important for the axial budding pattern, as cdc10-10 and cdc11-6 are defective in axial budding (Chant et al., 1995; Flescher et al., 1993). Bud4 localizes poorly to the neck in septin mutants such as cdc10Δ and cdc11Δ (Frazier et al., 1998; Sanders and Herskowitz, 1996). BUD4 on a low copy plasmid partially suppresses the temperature-sensitive growth of the cdc12-6 mutant, supporting the idea that Bud4 and the septin proteins interact with each other (Sanders and Herskowitz, 1996).
How does Bud4 mediate the assembly of the axial landmark at the proper bud site? Bud4 contains DUF1709 (domain of unknown function), which is found in the anillin-related proteins. This domain has similarity to the C-terminal part of the conserved anillin-homology (AH) domain and is adjacent to the putative Pleckstrin homology (PH) domain in Bud4 as in other anillin-related proteins such as Mid1 and Mid2 in S. pombe (Berlin et al., 2003; Tasto et al., 2003). Anillins are highly conserved proteins with multiple domains that interact with cytoskeletal components and their regulators (Piekny and Maddox, 2010). The C-terminal region containing the AH and PH domains of vertebrate anillins interacts directly with septins (Kinoshita et al., 2002). Mid2 in fission yeast is involved in septin ring organization (Berlin et al., 2003; Tasto et al., 2003). Both the DUF1709 and PH domains overlap with the GTP-binding motif of Bud4. Because of this anillin-related C-terminal region, Bud4 has been considered as an anillin-related protein in budding yeast (Wloka et al., 2011). However, there was little information available about the functional significance of these domains of Bud4.
Here, we report that Bud4 is involved in linking septins to the axial landmark through its multiple domains. Bud4 is necessary for the integrity of the septin rings during cell division. To our surprise, the anillin-related C-terminal region of Bud4 is important for its association with Bud3 and other components of the axial landmark rather than with septins. A series of mutational analyses indicate that the central domains of Bud4 are involved in its localization to the mother-bud neck and are thus likely to interact with septins.
Results
Overexpression of BUD4 leads to the assembly of extra septin rings or arcs
Because Bud4 shares sequence similarity with anillin-related proteins, we wondered whether Bud4 is involved in the organization of septins. We first examined how overexpression of Bud4 affects septins by monitoring Cdc12-GFP localization. Cells overexpressing Bud4 from the inducible GAL1 promoter often exhibited more than one septin ring (Fig. 1). A new septin ring typically forms 5–8 minutes before the disappearance of the old septin ring in mother cells of rapidly growing wild-type cells, but the old ring is rarely seen in cells after the emergence of a new bud (Iwase et al., 2006; Tong et al., 2007). To distinguish whether these extra septin rings resulted from a failure in disassembly after cytokinesis or from formation of additional septin rings during one cell cycle, cells were stained with Calcofluor (to mark bud scars) before induction with galactose. While Cdc12-GFP co-localized with a single bud scar or the base of a bud in cells with a vector control (100%, n = 80) (Fig. 1Ad), cells overexpressing Bud4 showed extra septin rings in three different patterns after growing in galactose-containing media for 6 hours (n = 136) (Fig. 1A): (a) two or more Cdc12-GFP rings co-localized with bud scars (9.6%); (b) a normal Cdc12-GFP ring co-localized with a bud scar plus one or two fainter Cdc12-GFP rings that were not associated with bud scars (14%); and (c) an abnormally large and less tightly organized Cdc12-GFP ring with several ‘satellite’ rings or arcs (15%). The percentage of cells with abnormal septin structures increased as Bud4 expression was induced for longer time. Thus, extra septin rings or arcs can result from both slow disassembly of the old septin ring after division (Fig. 1Aa) and formation of extra septin rings or arcs independent of cell division (Fig. 1Ab,c).
Fig. 1.
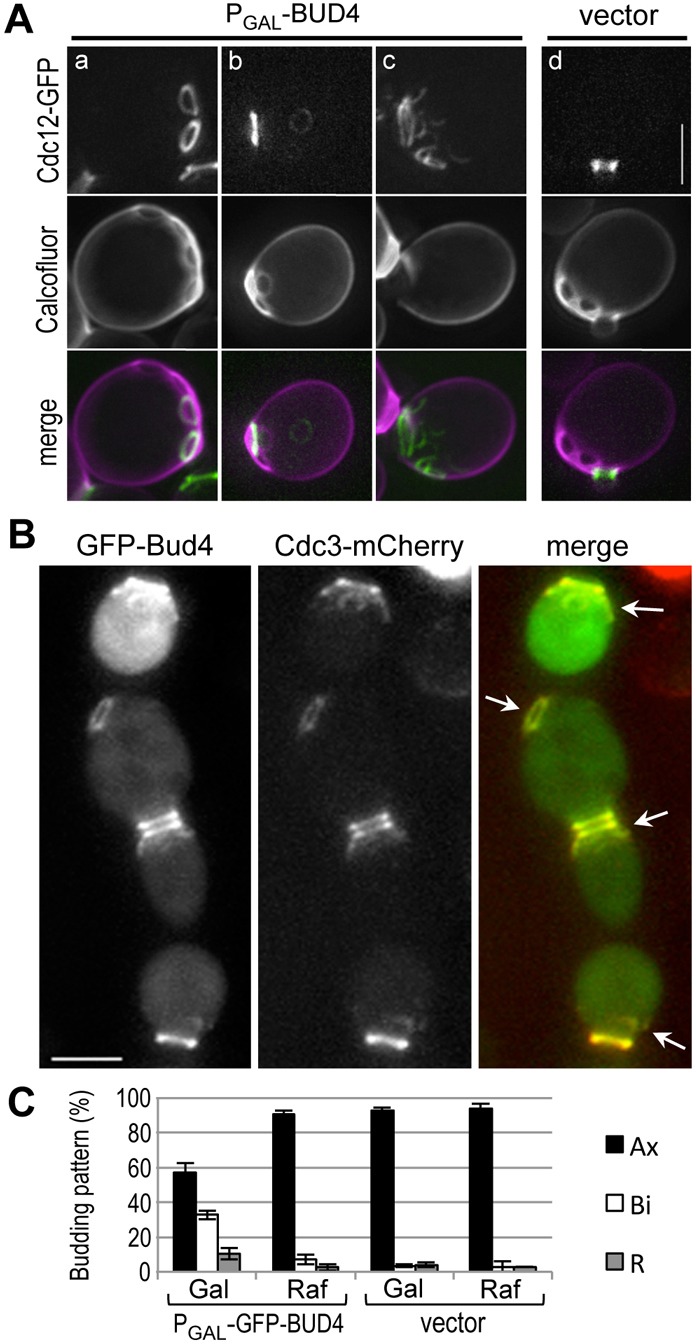
Overexpression of Bud4 from the GAL1 promoter. (A) Localization of Cdc12-GFP (pLP17) in cells (HPY210) carrying (Aa–c) the PGAL-BUD4 plasmid or (Ad) an empty vector. Cells were grown in SRaf-Ura, Leu stained with Calcofluor, and then grown in SGal-Ura, Leu for 8 hours before imaging. (B) Colocalization of Cdc3-mCherry and GFP-Bud4 in cells (HPY2008) carrying the PGAL-GFP-BUD4 plasmid. Cells were grown in SRaf-Ura and then shifted to SGal-Ura for 7 hours. Arrows indicate colocalized rings or arcs of GFP-Bud4 and Cdc3-mCherry. (C) Budding patterns of HPY2008 carrying the PGAL-GFP-BUD4 plasmid or an empty vector. Cells were either continuously grown in SRaf-Ura (Raf) or shifted to SGal-Ura for 7 hours (Gal). Cells with ≥3 bud scars were counted in three independent counts (n = 300 for PGAL-GFP-BUD4; n = 200 for the vector control). Mean (%) ±s.d. of the axial, bipolar, and random patterns are shown. Scale bars: 3 μm.
To test whether Bud4 was associated with the extra septin rings and arcs that resulted when it was overexpressed, we examined localization of Cdc3-mCherry in cells overexpressing GFP-Bud4 from the GAL1 promoter. Overexpression of GFP-Bud4 also induced formation of extra rings and fragmented Cdc3-mCherry, all of which co-localized with GFP-Bud4 (Fig. 1B). This phenotype was milder than that seen with overexpression of untagged Bud4. This difference is likely due to the lower level of GFP-Bud4 accumulation than that of untagged Bud4 upon induction with galactose (supplementary material Fig. S1A). Despite the presence of these extra rings or arcs of septins, cells overexpressing Bud4 did not exhibit any obvious cytokinesis defect or abnormal morphology. These cells did, however, exhibit a partial defect in the axial budding pattern upon overexpression of GFP-Bud4 (Fig. 1C). Taken together, these results suggest that Bud4 has a direct role in disassembly/assembly of septin rings, which are important for proper bud-site selection.
Bud4 is necessary for stable segregation of septin rings during cytokinesis
Next, we asked how septins are affected in the absence of Bud4. We determined the localization of Cdc3-mCherry in a bud4Δ mutant that also expressed GFP-Tub1 (α-tubulin) as a cell cycle marker. Unlike wild-type cells, bud4Δ cells often did not show a double ring of Cdc3-mCherry (compare a and b in Fig. 2A). After anaphase, a faint Cdc3-mCherry signal appeared only on the bud side of the mother-bud neck (87.5%; n = 9) and rapidly de-localized in bud4Δ cells (Fig. 2Ab, time 13–21), although little defect was evident until the end of mitosis. These results suggest that septins fail to maintain the intact ring structure in the absence of Bud4 during/after cytokinesis, consistent with recent reports (Eluère et al., 2012; Wloka et al., 2011).
Fig. 2.
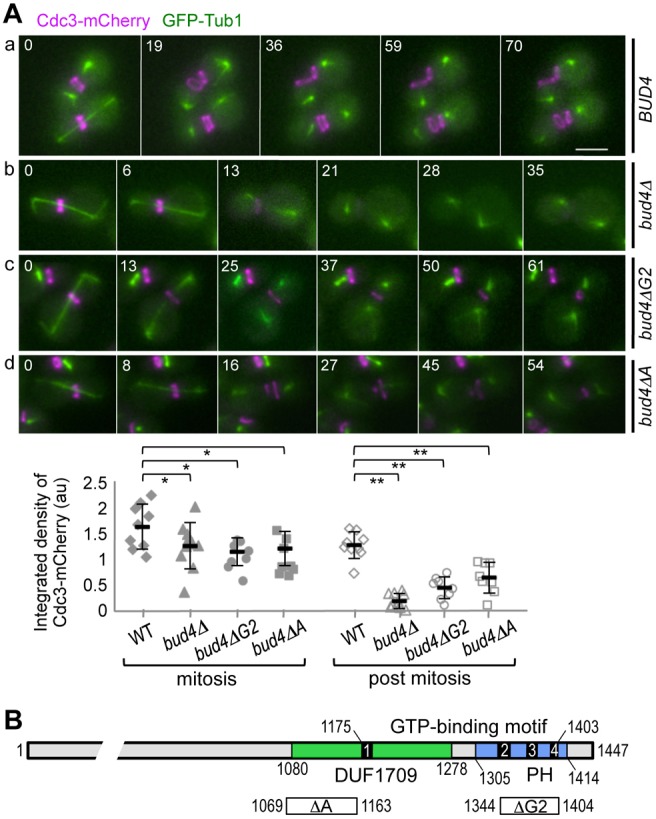
Localization of Cdc3-mCherry in bud4 mutants. (A) Time-lapse images of (Aa) wild-type (HPY2053), (Ab) bud4Δ (HPY2088), (Ac) bud4ΔG2 (HPY2099) and (Ad) bud4ΔA (HPY2139), which express Cdc3-mCherry and GFP-Tub1. Numbers indicate time (in minutes) after the first image. Below, the integrated density of Cdc3-mCherry at the mother-bud neck is quantified from large-budded cells, either with long spindles (mitosis) or with short spindles (post mitosis). Mean pixel intensity (in a.u.) ±s.d. is plotted for wild type (n = 9), bud4Δ (n = 9), bud4ΔG2 (n = 8) and bud4ΔA (n = 9) (*P>0.1, **P<0.001). Scale bar: 3 μm. (B) Diagram of Bud4 protein with the GTP-binding motif (black bars with numbers for the G1-G4 boxes), DUF1709 (green bar) and the PH domain (blue bar). Below, the regions deleted in bud4ΔA and bud4ΔG2 are depicted with white bars marked with ΔA and ΔG2, respectively.
Since the C-terminal region of Bud4 carries the DUF1709 and PH domains (Fig. 2B), we next tested whether these domains of Bud4 are necessary for the integrity of septin rings. We examined localization of Cdc3-mCherry in the bud4ΔG2 mutant, which lacks a part of the PH domain, and in the bud4ΔA mutant, which lacks a part of DUF1709 (see Fig. 2B). Despite its normal localization to the mother-bud neck prior to cytokinesis, Cdc3-mCherry failed to form an intact double ring during cytokinesis in these bud4 mutants (Fig. 2Ac,d). Instead, Cdc3-mCherry appeared as an arc or a tiny ring. This defect was evident initially at the mother side of the neck and subsequently at the bud side. The Cdc3-mCherry fluorescence in a fixed rectangle region at the mother-bud neck was reduced particularly in post-mitotic cells of bud4Δ (15%), bud4ΔG2 (36%), and bud4ΔA (50%) compared to wild type (see quantification in Fig. 2A). The steady-state levels of Cdc3-mCherry were about the same in wild type and these bud4 mutants (supplementary material Fig. S1B), suggesting that these defects in Cdc3 localization are due to delocalization rather than instability of Cdc3 in these bud4 mutants. Thus, the C-terminal region of Bud4 is necessary for the integrity of septin ring during/after cytokinesis.
The anillin-related region of Bud4 is unlikely to be involved in association with septins
GFP-Bud4 arrives initially at the mother side and then at the bud side of the septin ‘collar’ and co-localizes with the septin double ring at/after cytokinesis (supplementary material Fig. S2A). We wondered whether abnormal localization of Cdc3 in the bud4ΔA and bud4ΔG2 mutants is due to the failure of these Bud4 truncation mutants to associate with Cdc3. To test this possibility, we examined localization of Cdc3-mCherry and GFP-Bud4ΔA in the same cells by time-lapse microscopy. Remarkably, GFP-Bud4ΔA also formed an arc or a tiny ring after cytokinesis, and these structures overlapped with Cdc3-mCherry (Fig. 3A), suggesting that Bud4ΔA is unlikely to be defective in association with septins. Since the G1 box of the GTP-binding motif of Bud4 resides within DUF1709 (see Fig. 2B), we also examined localization of Cdc3-mCherry in cells expressing GFP-Bud4ΔG1, which lacks the G1 box. Localization of GFP-Bud4ΔG1 was similar to that of the wild-type GFP-Bud4, overlapping with Cdc3-mCherry particularly from the onset of cytokinesis when the septin ring splits (see cells marked with an arrow in supplementary material Fig. S2A,B).
Fig. 3.
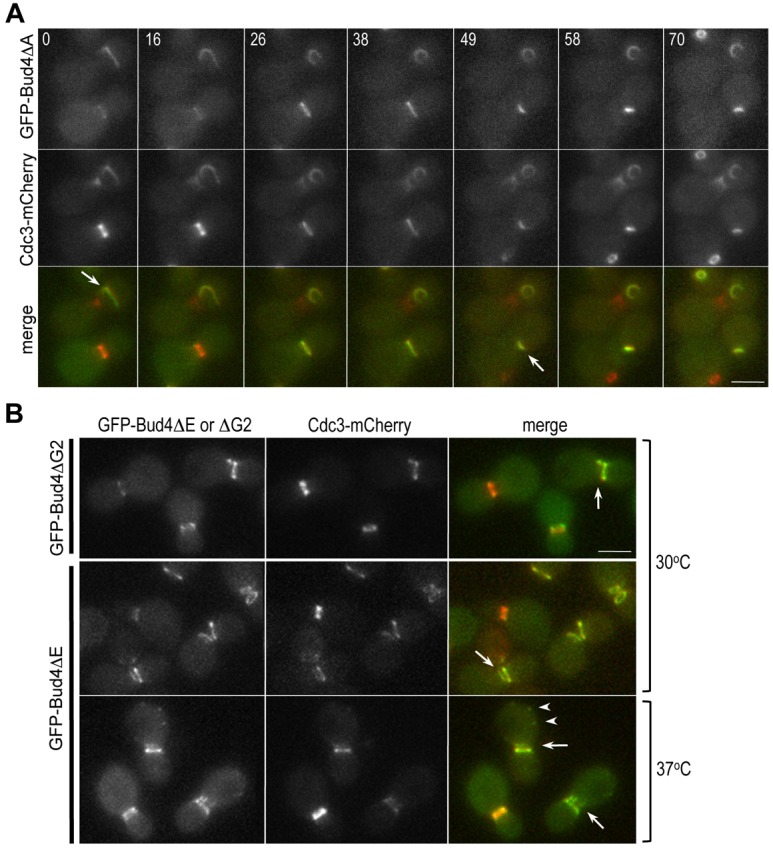
Localization of Cdc3-mCherry in cells expressing GFP fusions of the Bud4 mutant proteins lacking a part of DUF1709 and the PH domain. (A) Time-lapse images of the cells (HPY2152) expressing GFP-Bud4ΔΑ and Cdc3-mCherry. Arrows mark GFP-Bud4ΔΑ and Cdc3-mCherry starting to form abnormal shapes after cytokinesis. Numbers indicate time (in minutes) after the first image. (B) Localization of Cdc3-mCherry in cells expressing GFP-Bud4ΔG2 (HPY2265) (grown at 30°C) or GFP-Bud4ΔE (HPY2218) (grown at 30°C or 37°C). Arrows mark cells with defective double rings of Bud4 and Cdc3 in post-mitotic cells. Arrowheads indicate abnormal patches of GFP-Bud4ΔE. Scale bars: 3 μm.
We then asked whether the PH domain is involved in interaction with septins. We examined co-localization of Cdc3-mCherry and GFP-Bud4ΔG2 (see Fig. 2B) or GFP-Bud4ΔE, which lacks the predicted β-sheet structure of the PH domain. GFP-Bud4ΔG2 localized to the mother-bud neck but often failed to form a discrete double ring during/after cytokinesis, similar to GFP-Bud4ΔA. Although the defect was less severe at 30°C, GFP-Bud4ΔE also failed to form an intact double ring when cells were grown at 37°C (Fig. 3B). However, even when GFP-Bud4ΔG2 and GFP-Bud4ΔE formed the ‘broken rings’ during/after cytokinesis, these mutant proteins closely associated with Cdc3-mCherry as in the case of GFP-Bud4ΔA. Taken together, none of these Bud4 C-terminal truncation mutants is likely to be defective in interaction with septins. Thus, these data suggest that the anillin-related region of Bud4, including DUF1709 and the PH domain, is not involved in interaction with septins, contrary to our initial prediction.
DUF1709 and PH domains of Bud4 are necessary for interaction with Bud3
What, then, could be the biochemical defects of these bud4 mutants lacking the C-terminal domains? We noticed that the localization patterns of GFP-Bud4ΔA and GFP-Bud4ΔG2 were strikingly similar to that of GFP-Bud4 in bud3Δ cells (Kang et al., 2012). We thus speculated that Bud4ΔA and GFP-Bud4ΔG2 might be defective in interaction with Bud3. Consistent with this idea, we found that Bud3-mCherry co-localized poorly with GFP-Bud4ΔA and GFP-Bud4ΔG2 in most cells, with the exception of some pre-cytokinesis cells (Fig. 4Ab,c), whereas it co-localized with GFP-Bud4 at various cell cycle stages (Fig. 4Aa). Quantification of fluorescence intensity along the mother-bud axis of many large-budded cells confirmed that Bud3-mCherry co-localized poorly with GFP-Bud4ΔA and GFP-Bud4ΔG2 (see line-intensity plots in Fig. 4A, right). Since the Bud3 protein level was about the same in wild type and in these bud4 mutants (see Fig. 4B), this result suggests that Bud3 is de-localized in the bud4ΔA and bud4ΔG2 mutants.
Fig. 4.
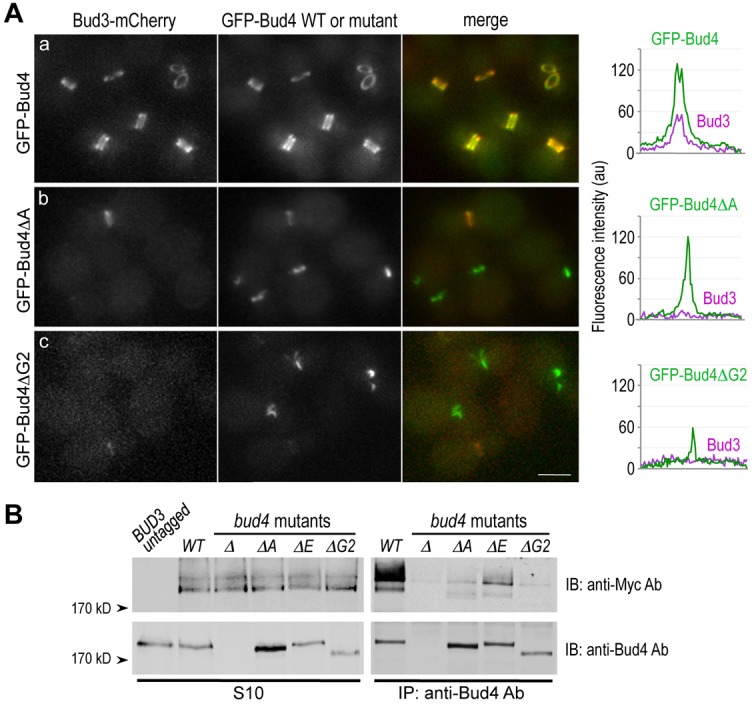
DUF1709 and the PH domain of Bud4 are necessary for interaction with Bud3. (A) Localization of Bud3-mCherry in cells expressing (Aa) GFP-Bud4 (HPY2217), (Ab) GFP-Bud4ΔA (HPY2202) and (Ac) GFP-Bud4ΔG2 (HPY2249). A representative line scan of a cell with a large bud is shown along the mother-bud axis for each strain on the right. (B) Co-immunoprecipitation of Bud3-Myc with Bud4. Extracts of the BUD3-Myc strains [wild-type (HPY1511), bud4Δ (HPY1514), bud4ΔA (HPY2254), bud4ΔE (HPY2255) and bud4ΔG2 (HPY1991)] were subjected to immunoprecipitation using polyclonal anti-Bud4 antibodies. Bud4 and Bud3-Myc recovered by immunoprecipitation (right panels) and from the S10 fractions (left panels) were analyzed by immunoblotting with anti-Bud4 antibodies (lower two panels) and anti-Myc antibody (top two panels). The first lane shows the S10 fraction from the wild-type strain (HPY16) expressing untagged Bud3.
Next, we assessed the physical association of Bud3 and these Bud4 C-terminal mutant proteins by immunoprecipitation assays using anti-Bud4 antibodies (which cross-react with wild type and the C-terminal truncations of Bud4 equally well). Unlike wild type, Bud4ΔA indeed associated poorly with Bud3-Myc (Fig. 4B). Similarly, we tested the two PH domain truncation mutants of Bud4, and found that Bud3-Myc associated poorly with Bud4ΔE and even less with Bud4ΔG2 (Fig. 4B). The level of Bud4ΔG2 was about 70% of the wild-type Bud4 (Kang et al., 2012), but even when normalized to the wild-type Bud4 level, association of Bud3 with Bud4ΔG2 was negligible. These results thus indicate that both DUF1709 and the PH domain of Bud4 are necessary for the association of Bud4 with Bud3. Bud3-Myc appeared as a broad band on SDS-PAGE gels and, intriguingly, the slower-mobility form of Bud3-Myc was enriched after immunoprecipitation with anti-Bud4 antibodies (Fig. 4B). These observations suggest a possibility that Bud3 undergoes post-translational modification and that the modified Bud3 preferentially associates with Bud4 (see Discussion).
Central region of Bud4 is necessary for its localization to the mother-bud neck
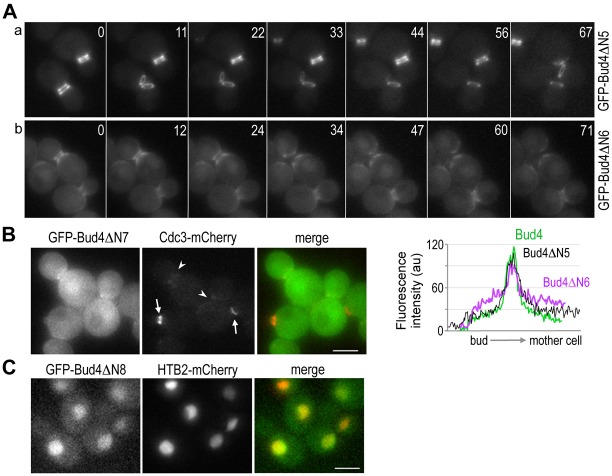
Since localization of Bud4 to the mother-bud neck depends on septins (Frazier et al., 1998; Sanders and Herskowitz, 1996) but not any other components of the axial landmark (Kang et al., 2012), a Bud4 region that is necessary for its localization to the neck is likely to be involved in interaction with septins. Which region of Bud4 is then necessary for its localization to the neck? To address this question, we first generated a series of N-terminal deletion mutants of BUD4 (see later), and determined the localization of each Bud4 mutant protein fused to GFP. Several GFP-Bud4 fusions with N-terminal deletions upto the residue 510 of Bud4 (ΔN1–ΔN5) did not exhibit any obvious defect in localization (Fig. 5Aa; data not shown), suggesting that most of the N-terminal region of Bud4 is dispensable for its localization. While GFP-Bud4ΔΝ6 was enriched at the mother-bud neck, it formed a less tightly organized ring structure and also existed diffusely in the cytoplasm (Fig. 5Ab). Quantification of the GFP fusions of the wild-type and mutant Bud4 proteins in many cells confirmed this conclusion (n>20 for each strain; see line-intensity plots of representative cells in Fig. 5). In contrast, GFP-Bud4ΔN7 appeared diffuse in the cytoplasm with slight enrichment at the neck (Fig. 5B), indicating that the region containing residues 884–1000 is necessary for targeting Bud4 to the mother-bud neck. In the GFP-Bud4ΔN7 strain, Cdc3-mCherry localized properly in cells undergoing bud emergence or with a small bud but poorly in cells with large buds (see cells marked with arrows and arrowheads, respectively, in Fig. 5B), as in bud4Δ cells. Surprisingly, GFP-Bud4ΔN8, which carries the DUF1709 and PH domains, localized to the nucleus, as judged by colocalization with Htb2 (histone H2B)-mCherry (Fig. 5C) (see Discussion). Taken together, analyses of these N-terminal deletion mutants indicate that the mid-region of Bud4 is involved in its localization to the mother-bud neck. Consistent with this conclusion, GFP-Bud4ΔC exhibited little defect in its initial localization to the neck, but failed to form a double ring at the neck and a single ring at the division site (Fig. 6B).
Fig. 5.
Localization of GFP fusions of Bud4 N-terminal truncation mutants. (A) Time-lapse images of cells expressing (Aa) GFP-Bud4ΔN5 (HPY2159) and (Ab) GFP-Bud4ΔN6 (HPY2158) from the BUD4 locus. Numbers indicate relative time (in minutes) after the first image. Below, a representative line scan of a cell with a large bud is shown along the mother-bud axis: GFP-Bud4 (green line), GFP-Bud4ΔN5 (black line), and GFP-Bud4ΔN6 (purple line). (B) Localization of GFP-Bud4ΔN7 and Cdc3-mCherry (HPY2226). Arrows mark the Cdc3-mCherry rings in cells with a tiny bud and an emerging bud; arrowheads mark the mother-bud neck of cells with large buds that failed to maintain the intact Cdc3-mCherry ring. (C) Localization of GFP-Bud4ΔN8 and Htb2-mCherry expressed from the BUD4 locus (HPY2132) and from a CEN plasmid (pFJ115), respectively. Scale bars: 3 μm.
Fig. 6.
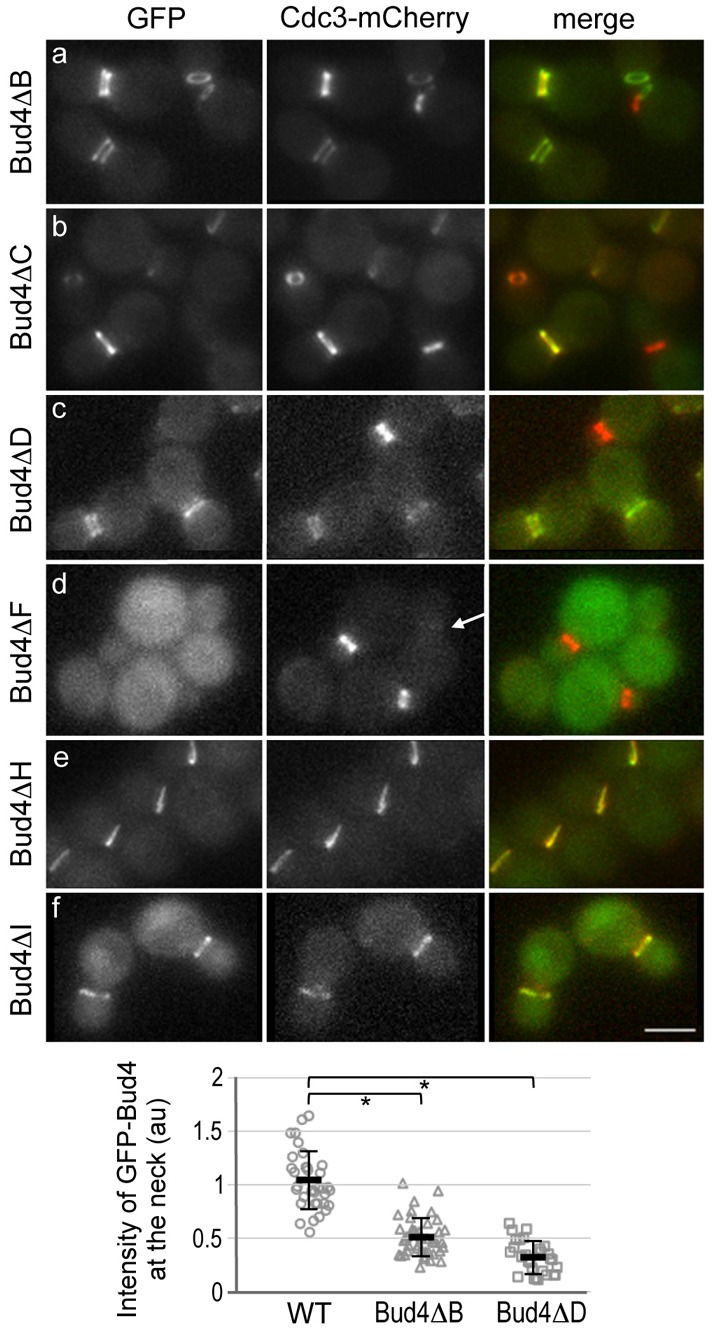
Localization of GFP fusions of Bud4 truncation mutants and Cdc3-mCherry expressed from the chromosomal loci. Note that although there was little difference in budding pattern between the BUD4 and GFP-BUD4 strains, the GFP-bud4ΔF mutant exhibited slightly less axial pattern (10%) than bud4ΔF (23%). Thus, the localization defect of GFP-Bud4ΔF might be slightly more pronounced than that of Bud4ΔF. Below, the integrated density of GFP-Bud4, GFP-Bud4ΔB and GFP-Bud4ΔD at the mother-bud neck is quantified from static images of large-budded cells. Mean pixel intensity (in a.u.) ±s.d. is plotted for wild type (n = 35), Bud4ΔΒ (n = 43) and Bud4ΔD (n = 28) (*P<1×10−5). Scale bar: 3 μm.
To define more precisely the central region of Bud4 that is involved in targeting of Bud4 to the mother-bud neck, we then generated several internal deletions (see Fig. 7A). To our surprise, GFP-Bud4ΔB and GFP-Bud4ΔD were able to localize to the neck (Fig. 6A,C). However, the double ring signal was weaker in these cells, and the cytoplasmic diffuse signal was particularly evident in cells expressing GFP-Bud4ΔD. When many static images of large budded cells were analyzed, the integrated density of GFP-Bud4ΔB and GFP-Bud4ΔD in a fixed rectangle region of the neck was significantly lower than that of the wild-type GFP-Bud4 (see graph in Fig. 6). In contrast, GFP-Bud4ΔF, which lacks both regions (residues 530–997) deleted in Bud4ΔB and Bud4ΔD, appeared completely diffuse in the cytoplasm, and Cdc3-mCherry also localized poorly to the neck in large-budded cells in this strain (see a cell marked with an arrow, Fig. 6D), as in bud4Δ cells. The steady-state levels of these Bud4 truncation mutant proteins (with the exception of Bud4ΔC and Bud4ΔG2; see below) were about equal to or higher than the wild-type Bud4 (Fig. 7B) (Kang et al., 2012), suggesting that their localization defects are not due to lower expression or instability of the mutant proteins.
Fig. 7.
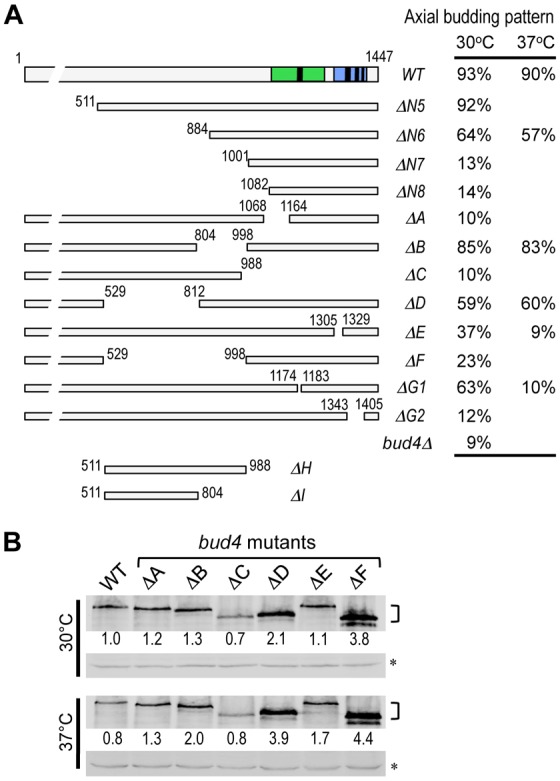
Budding patterns and the protein levels of bud4 mutants. (A) Budding patterns of the wild type and bud4 mutants. The average percentage of axially budding cells is shown after growing at the indicated temperatures. Cells with ≥3 bud scars were counted in 3 or 5 independent counts (n = 300, s.d.<3% for cells grown at 30°C; n = 500, s.d. = 3–5% for cells grown at 37°C). (B) Steady-state level of the wild-type and mutant Bud4 proteins. Extracts were prepared from cells grown in YPD at 30°C or 37°C, and then analyzed by immunoblotting using polyclonal anti-Bud4 antibodies. A non-specific cross-reacting band (marked with an asterisk) was used as a loading control; numbers represent the relative protein levels normalized against the wild-type Bud4 at 30°C.
Finally, we tested whether the central region (lacking in GFP-Bud4ΔF) is sufficient for its targeting to the mother-bud neck. Consistent with the deletion analyses described above, GFP-Bud4ΔH (carrying amino acids 511–988) and GFP-Bud4ΔI (carrying amino acids 511–804) were able to localize to the neck, although some cytoplasmic and nuclear fluorescence was also observed in cells expressing GFP-Bud4ΔI (Fig. 6E,F). Taken together, these results suggest that the central region (residues 530–988) of Bud4 contains at least two domains that have a partially redundant role in targeting Bud4 to the mother-bud neck.
Both the C-terminal and central regions of Bud4 are necessary for proper bud-site selection
We next determined the budding pattern of the bud4 mutants by introducing each bud4 allele into a haploid bud4Δ strain. As expected from the localization patterns described above, the N-terminal deletions, bud4ΔN1–bud4ΔN5, had little effect on the budding pattern, whereas bud4N6 resulted in a partial defect in the axial budding pattern. In contrast, bud4ΔN7 and bud4ΔN8 resulted in almost complete defects in the axial budding pattern (Fig. 7A; data not shown). Deletions of a part of the central region that is involved in targeting Bud4 to the mother-bud neck (bud4ΔB and bud4ΔD) resulted in partial axial budding defects, while a deletion of the whole central region (bud4ΔF) resulted in a more severe defect in the axial budding pattern (Fig. 7A).
The C-terminal truncation mutants were also defective in axial budding: The budding patterns of bud4ΔA and bud4ΔG2 were similar to that of a bud4 null mutant (Fig. 7A). Although the bud4ΔG1 and bud4ΔE mutants were partially defective at 30°C, these mutants were almost completely defective in the axial budding pattern at 37°C. Because these bud4 mutations did not affect overall protein stability at 30 and 37°C (with an exception of bud4ΔC and bud4ΔG2, which are reduced to 70% of wild type) (Fig. 7B) (Kang et al., 2012), their phenotypes are unlikely due to any global structural change or instability of the proteins (see Discussion). Taken together, the phenotypes of these bud4 mutants thus indicate that both the C-terminal and central regions of Bud4 are essential for the axial budding pattern.
Discussion
DUF1709 and PH domains of Bud4 are necessary for the assembly of the axial landmark
A number of pieces of data presented in this study suggest that the anillin-related C-terminal region of Bud4 is likely to be involved in the assembly of the axial landmark rather than the interaction with septins. While localization of Bud4 to the mother-bud neck depends on septins (Frazier et al., 1998; Sanders and Herskowitz, 1996), partial deletion of DUF1709 (bud4ΔA) or the overlapping G1 box (bud4ΔG1) did not disrupt targeting of Bud4 to the bud neck and its association with Cdc3. Instead, bud4ΔA and bud4ΔG1 disrupted the interaction of Bud4 with Bud3 (this study) and Axl1 and Axl2 (Kang et al., 2012), respectively.
PH domains are found in a variety of proteins and are involved in interaction with phosphoinositides in some cases, allowing membrane recruitment of proteins, or in protein-protein interactions in other cases (Lemmon, 2004). Although the isolated Bud4 fragment carrying the PH domain can bind to PtdIns(3)P, it appears to do so with very low affinity, and its GFP fusion localizes to the cytoplasm (Yu et al., 2004). Consistent with this previous finding, we found that the Bud4 C-terminal fragments (GFP-Bud4ΔN7 and GFP-Bud4ΔN8) fail to localize to the plasma membrane; instead, these proteins localize mostly to the cytoplasm and the nucleus, respectively. The C-terminal region of Bud4 contains four putative nuclear localization sequences (NLS) residing within Bud4ΔN8 and a putative nuclear export sequence (NES) located at residues 1075–1085. Disruption of this NES may account for the nuclear localization of GFP-Bud4ΔN8, but further mutational studies are necessary to test this idea and to address whether there is any functional significance of these putative NLS and NES motifs in Bud4.
Despite some potential caveats of our mutational study (such as possible global structural changes), our data indicate that the PH domain of Bud4 is involved in the assembly of the axial landmark. Deletions of a part of the PH domain of Bud4, bud4ΔE and bud4ΔG2, result in defects in association with Bud3, which accounts for their axial bud-site-selection defect. The region deleted in Bud4ΔG2 overlaps the G2-G4 boxes of the GTP-binding motif. However, its failure to associate with Bud3 is unlikely due to the lack of GTP binding because Bud4ΔG1, which is more severely defective in GTP binding than Bud4ΔG2, is not defective in association with Bud3 and septins (Kang et al., 2012; this study). The PH domain of Bud4 is thus involved in association with Bud3 (this study), which is prerequisite for the assembly of Axl1 and Axl2 (Kang et al., 2012). Interestingly, Bud4 appears to associate preferentially with the Bud3 species that run more slowly on SDS-PAGE gels. Since several high-throughput studies have indicated that Bud3 is phosphorylated (Archambault et al., 2004; Bloom et al., 2011; Chi et al., 2007; Holt et al., 2009; Smolka et al., 2007) and ubiquitinated (Peng et al., 2003; Seyfried et al., 2008; Starita et al., 2012; Ziv et al., 2011), these modifications of Bud3 might be necessary for its interaction with Bud4. However, we cannot rule out the possibility that the faster-migrating Bud3 band represents cleaved Bud3 fragment(s). Further investigation is necessary to determine the functional significance of these potential Bud3 modifications.
Interestingly, budding patterns of bud4ΔG1 and bud4ΔE are partially defective at 30°C but more severely so at 37°C. The temperature-sensitive defect in the axial budding pattern appears to be specific to bud4ΔG1 and bud4ΔE, as other bud4 mutants did not exhibit such a temperature-sensitive phenotype (see Fig. 7A). This phenotype of bud4ΔG1 correlates with the temperature-dependent defect of Axl1 localization in this mutant despite the normal localization pattern of GFP-Bud4ΔG1 (Kang et al., 2012). Localization of Bud4ΔE is more severely defective at a higher temperature (see Fig. 3), likely affecting the assembly of the axial landmark.
Although Bud4 is necessary for the integrity of the septin ring particularly during/after cytokinesis (this study; Eluère et al., 2012; Wloka et al., 2011), similar to Mid2 in S. pombe (Berlin et al., 2003; Tasto et al., 2003), our findings indicate that the anillin-related region of Bud4 is important for the assembly of the axial landmark. Bud4 might be a specialized anillin-related protein that coordinates cytokinesis and selection of a growth site, as these two processes are closely linked in haploid budding yeast (see below). The AH domains are also involved in interaction with proteins other than septins, including the actin cytoskeleton and RhoA (Piekny and Maddox, 2010), although vertebrate anillins interact directly with septins (Kinoshita et al., 2002).
Central domains of Bud4 are necessary and sufficient for targeting Bud4 to the mother-bud neck
A series of mutational analyses uncovered a central region that is necessary and sufficient for targeting Bud4 to the mother-bud neck. Interestingly, a smaller fragment of Bud4 (amino acids 511–804) can also localize to the neck (see GFP-Bud4ΔI, Fig. 6), whereas deletion of this region (bud4ΔD) only partially affects localization and the axial budding pattern. Similarly, despite the drastic difference between the localization patterns of Bud4ΔN6 and Bud5ΔN7, Bud5ΔB has only minor defects in localization and budding pattern. Thus, the central region (amino acids 511–988) appears to have two domains that have a partially redundant role in targeting Bud4 to the mother-bud neck. Since localization of Bud4 to the neck depends on septins but not any other components of the axial landmark (Frazier et al., 1998; Sanders and Herskowitz, 1996), this central region of Bud4 is likely to be involved in interaction with septins. A close examination of this central region does not reveal any obvious common motif between these two domains. Several questions remain: What are the exact boundaries of each domain or the residues involved in targeting Bud4 to the mother-bud neck? Do both or any of these central domains of Bud4 interact directly with septins? Which components of septins interact with Bud4? Further investigation will be required to answer these questions.
Role of Bud4 in linking septins to the axial landmark
A ‘cytokinesis tag’ model has been previously proposed to explain the axial budding pattern in haploid cell types (Chant and Herskowitz, 1991; Snyder et al., 1991). In this model, a tag remaining from cytokinesis is thought to direct assembly of components for bud formation at the cortex in the subsequent cell cycle. This model has been supported by a number of observations including localization patterns of the axial-budding-specific proteins to the mother-bud neck (and the subsequent division site) and their genetic interactions with septins (Bi and Park, 2012). Our findings in this study further support the cytokinesis tag model and identify Bud4 as a key player for the faithful inheritance of the axial spatial cue from the cytokinesis tag septins. Bud4 may function as a platform to link septins to the axial landmark through its multiple domains, as modeled in Fig. 8. Septins recruit Bud4 to the mother-bud neck (Frazier et al., 1998; Sanders and Herskowitz, 1996) by interacting with the central domains of Bud4 (this study). Bud4 then mediates the assembly of the axial landmark via DUF1709 and the PH domain (Kang et al., 2012; this study). Bud4 is also necessary for the integrity of the septin ring during and after cytokinesis (this study; Eluère et al., 2012; Wloka et al., 2011). The stable anchoring of both the septin and Bud4 rings at the division site depends on the interaction between Bud3 and the C-terminal domains of Bud4 and is likely to contribute to the fidelity of the axial budding pattern (this study). The exact residues or domains of Bud4 that are involved in interaction with these proteins remain unknown. Further analyses are necessary to fully understand the spatiotemporal regulation of the axial landmark and the role of septins in axial budding.
Fig. 8.
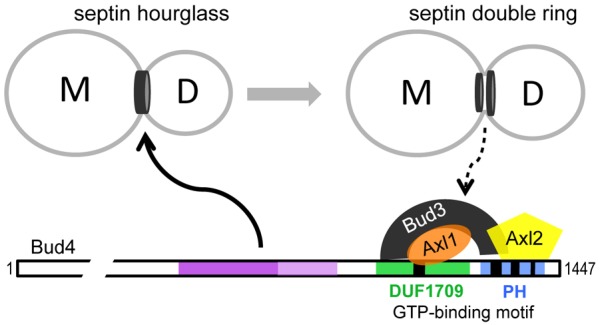
Model for the role of Bud4 in linking the cytokinesis tag septins to the axial landmark. Bud4 is likely to interact directly with septin hourglass via its central region in the G2 phase and mediate the assembly of the axial landmark via its C-terminal domains. The split septin rings then interact with the axial landmark either directly or indirectly. The exact domains or residues of Bud4 involved in interaction with septins, Bud3, Axl1 or Axl2 are not known (see text for details).
Materials and Methods
Strains, plasmids, genetic methods and growth conditions
Standard methods of yeast genetics, DNA manipulation, and growth conditions were used (Ausubel et al., 1999; Guthrie and Fink, 1991), unless indicated otherwise. The strains and plasmids used in this study are listed in supplementary material Tables S1, S2. Details of plasmid construction are provided below supplementary material Table S2.
Immunoblotting and immunoprecipitation
Yeast cells were grown to mid-log phase (OD600 ∼1.0) in YPD at 30°C, and extracts were prepared as previously described (Kang et al., 2004a). Immunoprecipitation and immunoblotting were carried as previously described (Kang et al., 2010) using anti-Bud4 antibodies, which were raised against the N-terminal fragment of Bud4 (a.a. 1–398) (Kang et al., 2012), anti-Myc antibody 9E10 (kindly provided by M. Bishop, University of California-San Francisco), and anti-DsRED antibodies (Clontech Laboratories, Mountain View, CA). Protein bands were then detected with Alexa Fluor 680 goat anti-rabbit IgG (Molecular Probes, Eugene, OR) or IRDye 800CW conjugated goat anti-mouse IgG (LI-COR Biosciences, Lincoln, NE) secondary antibodies using the LI-COR Odyssey system (LI-COR Biosciences, Lincoln, NE). The amount of protein was quantified using the software of the LI-COR Odyssey system, and the relative amount of proteins was estimated based on an internal control on immunoblots.
Fluorescence microscopy and image processing
To determine budding patterns, cells were grown in YPD at 30°C or 37°C (where indicated) overnight, and bud scars were visualized by staining cells with Calcofluor, as previously described (Pringle, 1991). For live-cell imaging, cells were grown at 30°C in synthetic complete (SC) or SD-His (to select for the HTB2-mCherry plasmid). For imaging of cells overexpressing Bud4, cells were grown overnight in raffinose (Raf)-based synthetic media, either SRaf-Ura (Fig. 1B) or SRaf-Ura, Leu (Fig. 1A), and then induced by adding galactose (2% final) for 6–9 hours. Cells were then concentrated by centrifugation and spotted on a slab of the same media containing 2% agarose for imaging. GFP- and mCherry-fusion proteins were visualized in exponentially growing cells, essentially as previously described (Kang et al., 2012; Kang et al., 2004b), using a Nikon E800 microscope (Nikon, Tokyo, Japan) fitted with a 100× oil-immersion objective (NA 1.30) and FITC/GFP and mCherry/TexasRed filters from Chroma (Brattleboro, VT). Optical Z-sections (0.3 μm intervals) were collected using Slidebook software (Intelligent Imaging Innovations, Denver, CO) with a Hamamatsu ORCA-2 CCD camera (Hamamatsu Photonics, Bridgewater, NJ) at room temperature (22–24°C) unless indicated otherwise. A maximum projection was created for 2D images with Slidebook software for all figures. A single optical section was used for quantification of fluorescence intensities either along the line crossing cells or in a defined area using ImageJ software (NIH). For line scan analyses, relative intensity of fluorescence was analyzed as described in figure legends and the representative cells are shown. To compare fluorescence intensity at the mother-bud neck region, the integrated density was calculated by subtracting the fluorescence intensity in the cytosol from the total intensity in an ImageJ-drawn polygon covering the neck (Fig. 2A; Fig. 6). Mean±s.d. was calculated and statistical significance was determined using Student's t-test.
Supplementary Material
Acknowledgments
We thank E. Bi, A. Straight, M. Longtine, T. Davis, R. Deshaies, R. Li, O. Egriboz, J. Hopper and M. Bishop for providing plasmids and antiserum.
Footnotes
Author contributions
P.J.K. and H.-O.P. conceived and designed the experiments; P.J.K., J.K.H. and H.-O.P. performed the experiments; P.J.K. and H.-O.P. analyzed the data; P.J.K. and J.K.H. contributed reagents and materials; P.J.K. and H.-O.P. wrote the paper.
Funding
This work was supported National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) [grant number R01-GM76375 to H.-O.P.]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.118521/-/DC1
References
- Archambault V., Chang E. J., Drapkin B. J., Cross F. R., Chait B. T., Rout M. P. (2004). Targeted proteomic study of the cyclin-Cdk module. Mol. Cell 14, 699–711 10.1016/j.molcel.2004.05.025 [DOI] [PubMed] [Google Scholar]
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Struhl K. (1999). Current Protocols in Molecular Biology. New York, NY: John Wiley & Sons [Google Scholar]
- Bender A. (1993). Genetic evidence for the roles of the bud-site-selection genes BUD5 and BUD2 in control of the Rsr1p (Bud1p) GTPase in yeast. Proc. Natl. Acad. Sci. USA 90, 9926–9929 10.1073/pnas.90.21.9926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A., Pringle J. R. (1989). Multicopy suppression of the cdc24 budding defect in yeast by CDC42 and three newly identified genes including the ras-related gene RSR1. Proc. Natl. Acad. Sci. USA 86, 9976–9980 10.1073/pnas.86.24.9976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin A., Paoletti A., Chang F. (2003). Mid2p stabilizes septin rings during cytokinesis in fission yeast. J. Cell Biol. 160, 1083–1092 10.1083/jcb.200212016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E., Pringle J. R. (1996). ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 5264–5275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E., Park H-O. (2012). Cell polarization and cytokinesis in budding yeast. Genetics 191, 347–387 10.1534/genetics.111.132886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom J., Cristea I. M., Procko A. L., Lubkov V., Chait B. T., Snyder M., Cross F. R. (2011). Global analysis of Cdc14 phosphatase reveals diverse roles in mitotic processes. J. Biol. Chem. 286, 5434–5445 10.1074/jbc.M110.205054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chant J., Herskowitz I. (1991). Genetic control of bud site selection in yeast by a set of gene products that constitute a morphogenetic pathway. Cell 65, 1203–1212 10.1016/0092-8674(91)90015-Q [DOI] [PubMed] [Google Scholar]
- Chant J., Pringle J. R. (1995). Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. J. Cell Biol. 129, 751–765 10.1083/jcb.129.3.751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chant J., Corrado K., Pringle J. R., Herskowitz I. (1991). Yeast BUD5, encoding a putative GDP-GTP exchange factor, is necessary for bud site selection and interacts with bud formation gene BEM1. Cell 65, 1213–1224 10.1016/0092-8674(91)90016-R [DOI] [PubMed] [Google Scholar]
- Chant J., Mischke M., Mitchell E., Herskowitz I., Pringle J. R. (1995). Role of Bud3p in producing the axial budding pattern of yeast. J. Cell Biol. 129, 767–778 10.1083/jcb.129.3.767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi A., Huttenhower C., Geer L. Y., Coon J. J., Syka J. E. P., Bai D. L., Shabanowitz J., Burke D. J., Troyanskaya O. G., Hunt D. F. (2007). Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc. Natl. Acad. Sci. USA 104, 2193–2198 10.1073/pnas.0607084104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egriboz O., Jiang F., Hopper J. E. (2011). Rapid GAL gene switch of Saccharomyces cerevisiae depends on nuclear Gal3, not nucleocytoplasmic trafficking of Gal3 and Gal80. Genetics 189, 825–836 10.1534/genetics.111.131839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eluère R., Varlet I., Bernadac A., Simon M. N. (2012). Cdk and the anillin homolog Bud4 define a new pathway regulating septin organization in yeast. Cell Cycle 11, 151–158 10.4161/cc.11.1.18542 [DOI] [PubMed] [Google Scholar]
- Flescher E. G., Madden K., Snyder M. (1993). Components required for cytokinesis are important for bud site selection in yeast. J. Cell Biol. 122, 373–386 10.1083/jcb.122.2.373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier J. A., Wong M. L., Longtine M. S., Pringle J. R., Mann M., Mitchison T. J., Field C. (1998). Polymerization of purified yeast septins: evidence that organized filament arrays may not be required for septin function. J. Cell Biol. 143, 737–749 10.1083/jcb.143.3.737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freifelder D. (1960). Bud position in Saccharomyces cerevisiae. J. Bacteriol. 80, 567–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X. D., Sperber L. M., Kane S. A., Tong Z., Tong A. H., Boone C., Bi E. (2007). Sequential and distinct roles of the cadherin domain-containing protein Axl2p in cell polarization in yeast cell cycle. Mol. Biol. Cell 18, 2542–2560 10.1091/mbc.E06-09-0822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C., Fink G. R. (1991). Guide to Yeast Genetics and Molecular Biology. San Diego, CA: Academic Press [Google Scholar]
- Halme A., Michelitch M., Mitchell E. L., Chant J. (1996). Bud10p directs axial cell polarization in budding yeast and resembles a transmembrane receptor. Curr. Biol. 6, 570–579 10.1016/S0960-9822(02)00543-2 [DOI] [PubMed] [Google Scholar]
- Hicks J. B., Strathern J. N., Herskowitz I. (1977). Interconversion of yeast mating types III. Action of the homothallism (HO) gene in cells homozygous for the mating type locus. Genetics 85, 395–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt L. J., Tuch B. B., Villén J., Johnson A. D., Gygi S. P., Morgan D. O. (2009). Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325, 1682–1686 10.1126/science.1172867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase M., Luo J., Nagaraj S., Longtine M., Kim H. B., Haarer B. K., Caruso C., Tong Z., Pringle J. R., Bi E. (2006). Role of a Cdc42p effector pathway in recruitment of the yeast septins to the presumptive bud site. Mol. Biol. Cell 17, 1110–1125 10.1091/mbc.E05-08-0793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang P. J., Sanson A., Lee B., Park H-O. (2001). A GDP/GTP exchange factor involved in linking a spatial landmark to cell polarity. Science 292, 1376–1378 10.1126/science.1060360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang P. J., Angerman E., Nakashima K., Pringle J. R., Park H-O. (2004a). Interactions among Rax1p, Rax2p, Bud8p, and Bud9p in marking cortical sites for bipolar bud-site selection in yeast. Mol. Biol. Cell 15, 5145–5157 10.1091/mbc.E04-07-0600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang P. J., Lee B., Park H-O. (2004b). Specific residues of the GDP/GTP exchange factor Bud5p are involved in establishment of the cell type-specific budding pattern in yeast. J. Biol. Chem. 279, 27980–27985 10.1074/jbc.C400175200 [DOI] [PubMed] [Google Scholar]
- Kang P. J., Béven L., Hariharan S., Park H-O. (2010). The Rsr1/Bud1 GTPase interacts with itself and the Cdc42 GTPase during bud-site selection and polarity establishment in budding yeast. Mol. Biol. Cell 21, 3007–3016 10.1091/mbc.E10-03-0232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang P. J., Angerman E., Jung C. H., Park H. O. (2012). Bud4 mediates the cell-type-specific assembly of the axial landmark in budding yeast. J. Cell Sci. 125, 3840–3849 10.1242/jcs.103697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita M., Field C. M., Coughlin M. L., Straight A. F., Mitchison T. J. (2002). Self- and actin-templated assembly of Mammalian septins. Dev. Cell 3, 791–802 10.1016/S1534-5807(02)00366-0 [DOI] [PubMed] [Google Scholar]
- Kozminski K. G., Beven L., Angerman E., Tong A. H. Y., Boone C., Park H. O.(2003). Interaction between a Ras and a Rho GTPase couples selection of a growth site to the development of cell polarity in yeast. Mol. Biol. Cell 14, 4958–4970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon M. A. (2004). Pleckstrin homology domains: not just for phosphoinositides. Biochem. Soc. Trans. 32, 707–711 10.1042/BST0320707 [DOI] [PubMed] [Google Scholar]
- Lippincott J., Li R. (1998). Dual function of Cyk2, a cdc15/PSTPIP family protein, in regulating actomyosin ring dynamics and septin distribution. J. Cell Biol. 143, 1947–1960 10.1083/jcb.143.7.1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord M., Inose F., Hiroko T., Hata T., Fujita A., Chant J. (2002). Subcellular localization of Axl1, the cell type-specific regulator of polarity. Curr. Biol. 12, 1347–1352 10.1016/S0960-9822(02)01042-4 [DOI] [PubMed] [Google Scholar]
- Oh Y., Bi E. (2011). Septin structure and function in yeast and beyond. Trends Cell Biol. 21, 141–148 10.1016/j.tcb.2010.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H-O., Chant J., Herskowitz I. (1993). BUD2 encodes a GTPase-activating protein for Bud1/Rsr1 necessary for proper bud-site selection in yeast. Nature 365, 269–274 10.1038/365269a0 [DOI] [PubMed] [Google Scholar]
- Park H-O., Bi E., Pringle J. R., Herskowitz I.(1997). Two active states of the Ras-related Bud1/Rsr1 protein bind to different effectors to determine yeast cell polarity. Proc. Natl. Acad. Sci. USA 94, 4463–4468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H-O., Kang P. J., Rachfal A. W. (2002). Localization of the Rsr1/Bud1 GTPase involved in selection of a proper growth site in yeast. J. Biol. Chem. 277, 26721–26724 10.1074/jbc.C200245200 [DOI] [PubMed] [Google Scholar]
- Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S. P. (2003). A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21, 921–926 10.1038/nbt849 [DOI] [PubMed] [Google Scholar]
- Piekny A. J., Maddox A. S. (2010). The myriad roles of Anillin during cytokinesis. Semin. Cell Dev. Biol. 21, 881–891 10.1016/j.semcdb.2010.08.002 [DOI] [PubMed] [Google Scholar]
- Pringle J. R. (1991). Staining of bud scars and other cell wall chitin with Calcofluor. Methods Enzymol, vol. 194 (ed. Guthrie C, Fink G R.), pp. 732–735 San Diego, CA: Academic Press; [DOI] [PubMed] [Google Scholar]
- Roemer T., Madden K., Chang J., Snyder M. (1996). Selection of axial growth sites in yeast requires Axl2p, a novel plasma membrane glycoprotein. Genes Dev. 10, 777–793 10.1101/gad.10.7.777 [DOI] [PubMed] [Google Scholar]
- Sanders S. L., Herskowitz I. (1996). The BUD4 protein of yeast, required for axial budding, is localized to the mother/BUD neck in a cell cycle-dependent manner. J. Cell Biol. 134, 413–427 10.1083/jcb.134.2.413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyfried N. T., Xu P., Duong D. M., Cheng D., Hanfelt J., Peng J. (2008). Systematic approach for validating the ubiquitinated proteome. Anal. Chem. 80, 4161–4169 10.1021/ac702516a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K., Kang P. J., Park H-O. (2008). The Rho5 GTPase is necessary for oxidant-induced cell death in budding yeast. Proc. Natl. Acad. Sci. USA 105, 1522–1527 10.1073/pnas.0707359105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolka M. B., Albuquerque C. P., Chen S. H., Zhou H. (2007). Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc. Natl. Acad. Sci. USA 104, 10364–10369 10.1073/pnas.0701622104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M., Gehrung S., Page B. D. (1991). Studies concerning the temporal and genetic control of cell polarity in Saccharomyces cerevisiae. J. Cell Biol. 114, 515–532 10.1083/jcb.114.3.515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starita L. M., Lo R. S., Eng J. K., von Haller P. D., Fields S. (2012). Sites of ubiquitin attachment in Saccharomyces cerevisiae. Proteomics 12, 236–240 10.1002/pmic.201100166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight A. F., Marshall W. F., Sedat J. W., Murray A. W. (1997). Mitosis in living budding yeast: anaphase A but no metaphase plate. Science 277, 574–578 10.1126/science.277.5325.574 [DOI] [PubMed] [Google Scholar]
- Tasto J. J., Morrell J. L., Gould K. L. (2003). An anillin homologue, Mid2p, acts during fission yeast cytokinesis to organize the septin ring and promote cell separation. J. Cell Biol. 160, 1093–1103 10.1083/jcb.200211126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Z., Gao X-D., Howell A. S., Bose I., Lew D. J., Bi E. (2007). Adjacent positioning of cellular structures enabled by a Cdc42 GTPase-activating protein-mediated zone of inhibition. J. Cell Biol. 179, 1375–1384 10.1083/jcb.200705160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versele M., Thorner J. (2005). Some assembly required: yeast septins provide the instruction manual. Trends Cell Biol. 15, 414–424 10.1016/j.tcb.2005.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloka C., Nishihama R., Onishi M., Oh Y., Hanna J., Pringle J. R., Krauss M., Bi E. (2011). Evidence that a septin diffusion barrier is dispensable for cytokinesis in budding yeast. Biol. Chem. 392, 813–829 10.1515/BC.2011.083 [DOI] [PubMed] [Google Scholar]
- Yu J. W., Mendrola J. M., Audhya A., Singh S., Keleti D., DeWald D. B., Murray D., Emr S. D., Lemmon M. A. (2004). Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol. Cell 13, 677–688 10.1016/S1097-2765(04)00083-8 [DOI] [PubMed] [Google Scholar]
- Ziv I., Matiuhin Y., Kirkpatrick D. S., Erpapazoglou Z., Leon S., Pantazopoulou M., Kim W., Gygi S. P., Haguenauer–Tsapis R., Reis N.et al. (2011). A perturbed ubiquitin landscape distinguishes between ubiquitin in trafficking and in proteolysis. Mol. Cell Proteomics 10, 009753 10.1074/mcp.M111.009753 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.