Abstract
A capillary zone electrophoresis (CZE) electrospray ionization (ESI) tandem mass spectrometry (MS/MS) system was integrated with an immobilized trypsin microreactor. The system was evaluated and then applied for online digestion and analysis of picogram loadings of RAW 264.7 cell lysate. Protein samples were dissolved in a buffer containing 50% (v/v) acetonitrile (ACN), and then directly loaded into the capillary for digestion, followed by CZE separation and MS/MS identification. The organic solvent (50% (v/v) ACN) assisted the immobilized trypsin digestion and simplified the protein sample preparation protocol. Neither protein reduction nor alkylation steps were employed, which minimized sample loss and contamination. The integrated CZE-ESI-MS/MS system generated confident identification of bovine serum albumin (BSA) with 19% sequence coverage and 14 peptide IDs when 20 fmole was loaded. When only 1 fmole BSA was injected, one BSA peptide was consistently detected. For the analysis of a standard protein mixture, the integrated system produced efficient protein digestion and confident identification for proteins with different molecular weights and isoelectric points when low fmole amount was loaded for each protein. We further applied the system for triplicate analysis of a RAW 264.7 cell lysate; 2 ± 1 and 7 ± 2 protein groups were confidently identified from only 300 pg and 3 ng loadings, respectively. The 300 pg sample loading corresponds to the protein content of three RAW 264.7 cells. In addition to high sensitivity analysis, the integrated CZE-ESI-MS/MS system produces good reproducibility in terms of peptide and protein IDs, peptide migration time, and peptide intensity.
Introduction
Bottom-up proteomics is routinely used for characterization of complex samples. As an extreme example of the depth of sequencing, the identification of more than 10,000 proteins from mammalian cell lysates has been reported [1]. However, most bottom-up sequencing protocols require microgram to milligram amounts of sample, which limits its applications for material-limited biological samples, such as circulating tumor cells [2], where the sample size is only a few nanograms or less. In order to improve the performance of bottom-up proteomics for analysis of mass-limited biological samples, improvements in both instrumentation and sample preparation are required for high efficient enzymatic digestion of trace amounts of proteins, high capacity peptide separation, and sensitive peptide detection.
Several groups have developed efficient enzymatic digestion of trace amounts of proteins from hundreds of cells [3–6], where the initial protein sample amounts were on the order of 100 ng. These protocols employ free trypsin to digest proteins. It is likely that the system performance is limited by the combination of low protein and trypsin concentrations. Immobilized trypsin can generate better digestion performance for proteins with very low concentration compared with free trypsin [7, 8], due to the high trypsin concentration and reduced auto-digestion of immobilized trypsin [9, 10].
We recently reported a protein sample preparation method based on methanol denaturation and immobilized trypsin digestion; we employed that system for analysis of low nanogram amounts of RAW 264.7 cell lysate [11]. In that protocol, proteins were first denatured with 50% (v/v) methanol. The denatured proteins were reduced in a buffer containing 50% (v/v) methanol and then alkylated. Finally, the treated proteins were digested using trypsin immobilized on magnetic microspheres. The whole process was performed at room temperature in 200 μL Eppendorf tubes. After ultra-performance liquid chromatography (UPLC)-electrospray ionization-tandem mass spectrometry (ESI-MS/MS) analysis, 2 ± 1 and 23 ± 2 proteins could be identified from only 6 ng and 30 ng initial cell lysate, respectively. An additional 16 proteins could be detected from the 6 ng cell lysate using an accurate mass and time tag method. The results suggest that organic solvent and immobilized trypsin based protocol may be useful for ultra-trace level protein sample preparation.
As an example of high capacity peptide separation and sensitive peptide detection, Shen et al. [12, 13] coupled high efficient (peak capacities of ~103) 15 μm i.d. packed LC capillary column to a mass spectrometer for analysis of low-nanogram amounts of proteins from complex samples. Ultrahigh sensitivity (<75 zmol for individual proteins) was obtained. Waanders et al. [14] developed a high sensitivity LC system based on splitting gradient effluents into a capture capillary and providing an inherent technical replicate for nanogram complex sample analysis with high resolution. More than 2 400 proteins could be identified from kidney glomeruli isolated by laser capture microdissection in a single analysis.
Capillary electrophoresis (CE)-ESI-MS/MS has attracted attention as an alternative tool for large-scale, high sensitivity proteomics analysis [15–22]. This group has focused on the use of an electrokinetically pumped sheath-flow electrospray interface. This system has generated low amole peptide detection limit with an LTQ-Orbitrap Velos as detector [19, 20], and yielded high zmole level peptide detection limit with triple-quadrupole mass spectrometer employing multiple reaction monitoring [21]. For analysis of low nanogram and high picogram amounts of digest, CE-MS/MS generated more peptide and protein identifications than LC-MS/MS [17, 20]. For mid-nanogram samples, CE-MS/MS and LC-MS/MS produced complementary peptide identifications [17, 22]. In addition, CE-MS also produced much higher peptide intensities than LC-MS [16].
Online integration of an immobilized enzyme microreactor with a CE-ESI-MS/MS system should be a facile method for analysis of trace-level protein samples. Several research groups have developed integrated microreactor-CE-MS systems for online protein digestion, peptide separation, and detection [23–27]. However, the sensitivity of those systems was limited by the immobilized enzyme microreactor, the CE-MS interface, and the mass spectrometer. In those reports, only one or two small standard proteins were analyzed. In addition, denaturation, reduction and alkylation steps were performed offline, followed by online protein digestion, separation and detection, which reduced the throughput and increased the potential for protein sample loss and contamination.
As we show in this paper, integration of organic solvent denaturation and immobilized trypsin digestion in a nanoliter volume microreactor further improves sample preparation efficiency. In this work, we integrate an immobilized trypsin microreactor with a CZE-ESI-MS/MS system for online digestion and analysis of picogram amounts of a RAW 264.7 cell lysate.
This system has a number of innovations.
First, the sample was prepared in a 50% ACN buffer. Organic solvents are used to unfold the protein structure [28, 29] and assist in tryptic protein digestion [30–32].
Second, no protein reduction and alkylation steps were performed. Eliminating these steps reduced potential sample loss and contamination. Eliminating the steps also improved the throughput.
Third, the immobilized microreactor was based on an acrylamide-based monolithic. This material is hydrophilic and tends to reduce non-specific adsorption of peptides/proteins.
Fourth, the total volume of the prepared trypsin microreactor was reduced to less than 40 nL. This small volume aids in handling small volume protein samples.
Fifth, the 50% (v/v) ACN sample preparation buffer has lower conductivity than the separation buffer of CZE. This low-conductivity buffer results in sample stacking at the beginning of CZE separation, which improved the CZE separation and MS detection.
Finally, an electrokinetically pumped sheath-flow electrospray interface [18] was used to couple the CZE to a LTQ-Orbitrap Velos mass spectrometer. The combination results in high sensitive peptide detection.
This integrated CZE-ESI-MS/MS system confidently identified 2 ± 1 protein groups from 300 pg of RAW 264.7 cell lysate. 7 ± 2 protein groups were confidently identified from 3 ng of the lysate.
Experimental section
Materials and Chemicals
All chemicals were from Sigma–Aldrich (St. Louis, MO, USA), unless specified. Poly (ethylene glycol) (PEG, MW 10, 000) was ordered from Polysciences, Inc. (Warrington, PA, USA). Dimethyl sulfoxide (DMSO) was ordered from Electron Microscopy Sciences (Hatfield, PA, USA). Acetonitrile (ACN) was purchased from Fisher Scientific (Pittsburgh, PA, USA). Methanol was purchased from Honeywell Burdick & Jackson (Wicklow, IE, USA). Water was deionized by a Nano Pure system from Thermo scientific (Marietta, OH, USA). Fused capillaries (50 μm i.d. ×150 μm o.d.) were purchased from Polymicro Technologies (Phoenix, AZ, USA).
Dulbecco’s Modified Eagle’s Medium (DMEM) with L-glutamine and fetal bovine serum (FBS) were purchased from ATCC (Manassas, VA, USA). Mammalian Cell-PE LB™ Buffer for cell lysis was purchased from G-Biosciences (St. Louis, MO, USA). Complete, mini protease inhibitor cocktail (provided in EASYpacks) was purchased from Roche (Indianapolis, IN, USA).
Preparation of immobilized trypsin microreactor
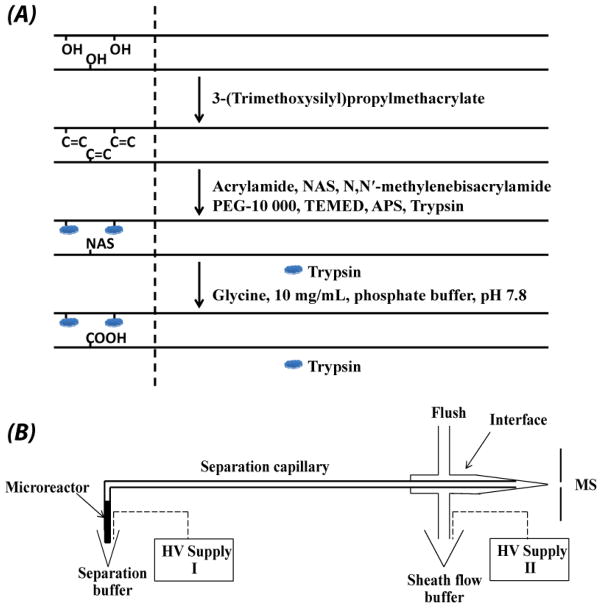
A diagram outlining the preparation of the immobilized trypsin microreactor is shown in Fig. 1A. A fused capillary (50 μm i.d. ×150 μm o.d., 50 cm) was successively washed with methanol, sodium hydroxide, water, hydrochloric acid, water, and methanol. The capillary was then dried with nitrogen at room temperature. Subsequently, a ~10 cm portion of the capillary was filled with 50% (v/v) 3-(trimethoxysilyl)propylmethacrylate in methanol, and reacted at room temperature for 24 h. After that, the capillary was washed with methanol to remove the unreacted components, and dried again with nitrogen. The treated capillary was stored at room temperature for use.
Figure 1.
Procedure for preparation of the trypsin immobilized microreactor (A) and schematic diagram of the integrated CZE-ESI-MS/MS system (B).
The details of the polymerization mixture and process were similar to reference [33] with some modifications. A mixture of 20 mg of acrylamide, 30 mg of N, N′-methylenebisacrylamide, and 30 mg of PEG was dissolved in 1 mL of 0.2 M sodium bicarbonate/0.5 M sodium chloride (pH ~8.0) buffer. The mixture was vortexed for 30 s and then heated at ~50 °C for 15 min to completely dissolve the monomers. Then, 2 μL of 20% (v/v) TEMED (N, N, N′, N′-tetramethylethylene diamine) was added into 0.5 mL of the prepared mixture, and the mixture was degassed for 15 min using nitrogen. Subsequently, 7 μL of N-acryloxysuccinimide (NAS) [140 mg/mL, dissolved in DMSO] was added on top of the solution, and degassed for 1 min. After that, 2 μL of 20% (w/v) ammonium persulfate (APS) was added into the middle part of the solution to initiate polymerization. After vortexing for several seconds, 190 μL of the solution was mixed quickly with 10 μL freshly prepared trypsin (20 mg/mL in a buffer containing 0.5 M benzamidine, pH 8.0). Finally, the vinylized part of the treated capillary (~10 cm) was filled with the mixture through capillary action, and reacted at room temperature for 30 min.
After the reaction, ~7 cm microreactor was discarded. The remaining portion of the capillary including a few centimeters of the microreactor was successively washed with deionized water, glycine (10 mg/mL in phosphate buffer, pH 8.0), and 5 mM NH4HCO3 (pH ~8.0) for 30 min. The capillary was stored at 4 °C before use.
Sample preparation
Insulin chain b oxidized was dissolved in 5 mM NH4HCO3 for analysis with the integrated CZE-ESI-MS/MS system. Several standard protein standards were dissolved in 50% (v/v) ACN and 5 mM NH4HCO3 for analysis with the integrated CZE-ESI-MS/MS system. The first sample was bovine serum albumin (BSA). The second was a three protein mixture of BSA, myoglobin (myo), and cytochrome c (cyto.c). The third was a seven protein mixture (BSA, myo, cyto.c, α-casein, β-casein, β-lactoglobulin, insulin chain b oxidized).
RAW 264.7 cells were cultured in a T25 flask at 37 °C and 5% CO2 in DMEM with L-glutamine and 10% FBS. The cells were lysed with 1 mL mammalian cell-PE LB™ buffer (pH 7.5) supplemented with complete protease inhibitor for 30 min on ice. The cell lysate was centrifuged at 18,000 g for 15 min, and the supernatant was collected for measurement of protein concentration with the BCA method. After that, a 300 μL aliquot of the cell lysate was precipitated with 1.2 mL cold acetone at −20 °C for 24 h, followed by centrifugation at 18,000 g for 15 min. The protein pellet was washed with cold acetone again and dried at room temperature. A sixty micrograms aliquot of the proteins was dissolved in 30 μL 0.5% (w/v) sodium deoxycholate (SDC) and 10 mM NH4HCO3 buffer (pH 8.0). After centrifugation, a 15 μL aliquot of the protein solution was mixed with 15 μL ACN, which resulted in a ~1 mg/mL protein solution dissolved in 50% ACN, 0.25% SDC and 5 mM NH4HCO3 buffer (pH 8.0). In addition, 5 μL of the 1 mg/mL protein solution was further diluted to 0.1 mg/mL with 50% ACN and 5 mM NH4HCO3 buffer (pH 8.0). The 1 mg/mL and 0.1 mg/mL protein samples were analyzed by the integrated CZE-ESI-MS/MS system in triplicate.
On-line protein digestion, peptide separation, and identification with the integrated CZE-ESI-MS/MS system
The integrated CZE-ESI-MS/MS system included a separation capillary (50 μm i.d. ×150 μm o.d.) with an immobilized trypsin microreactor at the injection end, an electrokinetically driven sheath-flow electrospray interface [18], two Spellman CZE 1000R power supplies, and a LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific), Fig. 1B.
The protocol had three steps. Protein samples were injected into the microreactor end of the separation capillary by air pressure. Proteins were then digested by the microreactor at room temperature. Finally, the digests were separated with CZE and analyzed by the LTQ-Orbitrap Velos.
For oxidized insulin chain b analysis, a 34 cm long separation capillary with a 1.5 cm long microreactor was used, and the digestion time was 1 min. For analysis of BSA and the standard protein mixtures, a 34 cm long separation capillary with a 2 cm microreactor was used, and the digestion time was 2 min. For analysis of the RAW 264.7 cell lysate, a 34 cm separation capillary with 2 cm microreactor and 3 min digestion time was used. In all cases, the separation buffer was 5 mM NH4HCO3 (pH 8.0), and the sheath flow buffer was 50% (v/v) methanol and 0.05% (v/v) FA. For standard proteins analysis, 19.5 kV was applied at the injection end of the capillary for separation and 1.5 kV was applied for electrospray. For RAW 264.7 cell lysate analysis, 15 kV was applied at the injection end of the capillary for separation, and 1 kV was applied for electrospray. The size of the tip for electrospray was ~8 μm.
For LTQ-Orbitrap Velos analysis, the ion transfer tube temperature was held at 300 °C. The mass spectrometer was programmed in data dependent mode. Full MS scans were acquired in the Orbitrap mass analyzer over m/z 395–1900 range with resolution 60,000 (m/z 400). Ten most intense peaks (for standard protein analysis) and 20 most intense peaks (for cell lysate analysis) with charge state ≥ 2 were selected for sequencing and fragmented in the ion trap with a normalized collision energy of 35%, activation q = 0.25, activation time of 20 ms, and one microscan. For standard protein analysis, peaks selected for fragmentation more than twice within 60 s were excluded from selection for 60 s. For the standard protein mixtures and cell lysate analysis, peaks selected for fragmentation more than once within 30 s were excluded from selection for 30 s.
Data analysis
Database searching of raw files was performed in Proteome Discoverer 1.3 with MASCOT 2.2.4. For standard proteins from bovine, the insulin chain b sequence was added into ipi.bovin.v3.68.fasta to form the database. For myoglobin, equine.fasta was used for database searching. For RAW 264.7 cell lysate, Swiss-Prot database with taxonomy as Mus. (16 252 sequences) was used. The database searching parameters included up to four missed cleavages allowed for full tryptic digestion, precursor mass tolerance 20 ppm, fragment mass tolerance 1.0 Da. For analysis of standard proteins from bovine, oxidation of cysteine (+47.9847 Da) was set as dynamic modification. Database searching against the corresponding decoy database was also performed to evaluate the false discovery rate (FDR) of peptide identification.
For standard protein analysis, MASCOT significance threshold 0.05 (95% confidence) was used to filter the peptide identification. For RAW 264.7 cell lysate data, MASCOT significance threshold of 0.01 (99% confidence) was applied to filter the peptide identification. In addition, manual evaluation was also used for the cell lysate peptide identifications. At the protein level, protein grouping was enabled, and each protein group had at least one unique peptide. Leucine and isoleucine were considered as equal, and the strict maximum parsimony principle was applied.
Results and discussions
Integrated CZE-ESI-MS/MS system
The system was composed of a separation capillary with an immobilized trypsin microreactor, an electrokinetically driven sheath-flow CE-MS interface [18], two high voltage (HV) supplies, and an LTQ-Orbitrap Velos mass spectrometer, Fig. 1B.
The microreactor was synthesized in-situ at the injection end of the separation capillary. The integrated system does not require an additional interface to connect the microreactor and separation capillary. The trypsin microreactor was prepared from an acrylamide monolithic due to its good permeability, fast mass transfer, high stability, and easy modification [34]. The hydrophilic nature of the acrylamide microreactor reduced non-specific adsorption of proteins/peptides. The total volume of the trypsin microreactor used in this work was less than 40 nL, which was efficient for trace protein sample digestion. In order to keep high electroosmotic flow (EOF) in the separation capillary, only a short portion of the separation capillary was vinylized for preparation of the microreactor, Fig. 1A. Excess succinimide groups on the monolithic material were blocked with glycine. Glycine’s amine group reacted with the succinimide groups, leaving the carboxyl groups of glycine exposed to the separation buffer, which was useful for improving the EOF in the separation capillary.
An electrokinetically driven sheath-flow interface, which was developed by our group [18], was used due to its very high sensitivity for peptide detection [19, 20]. Two power supplies were used in the experiment. HV supply I was applied at the injection end of the capillary. HV supply II was applied to the interface to produce electrospray. The voltage difference between the power supplies drove the capillary electrophoretic separation, while the potential provided by the second power supply drove the electrospray interface. An LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) was used for peptide identification.
In conventional studies of microgram or larger sample amounts, proteins are denatured, reduced and alkylated, followed by trypsin digestion. However, these sample pre-treatment steps can lead to sample losses that are significant when dealing with low nanogram or picogram protein homogenates. We applied a simplified sample preparation protocol when analyzing samples smaller than a few nanograms. We used a buffer containing 50% (v/v) ACN and 5 mM NH4HCO3 to dissolve the proteins, which was then directly loaded into the integrated CZE-ESI-MS/MS system for protein digestion in the trypsin microreactor, peptide separation in the capillary, and identification by tandem mass spectrometry.
This protocol has at least two advantages. First, the one-step sample preparation reduces the sample loss and also avoids the contamination from the reducing and alkylating reagents. Second, the sample buffer (50% (v/v) ACN and 5 mM NH4HCO3) has lower conductivity than the separation buffer (5 mM NH4HCO3). This difference in buffer conductivity results in sample stacking at the beginning of the separation [35, 36], which improves separation efficiency. The resulting peaks are sharper and of higher amplitude, which assists detection. This protocol also has potential limitations for analysis of highly structured proteins with many disulfide bonds due to the elimination of reduction and alkylation steps. The 50% (v/v) ACN helps to denature non-crosslinked proteins, but the denaturation performance will be limited for highly structured proteins with many disulfide bonds.
Evaluation of the integrated CZE-ESI-MS/MS system
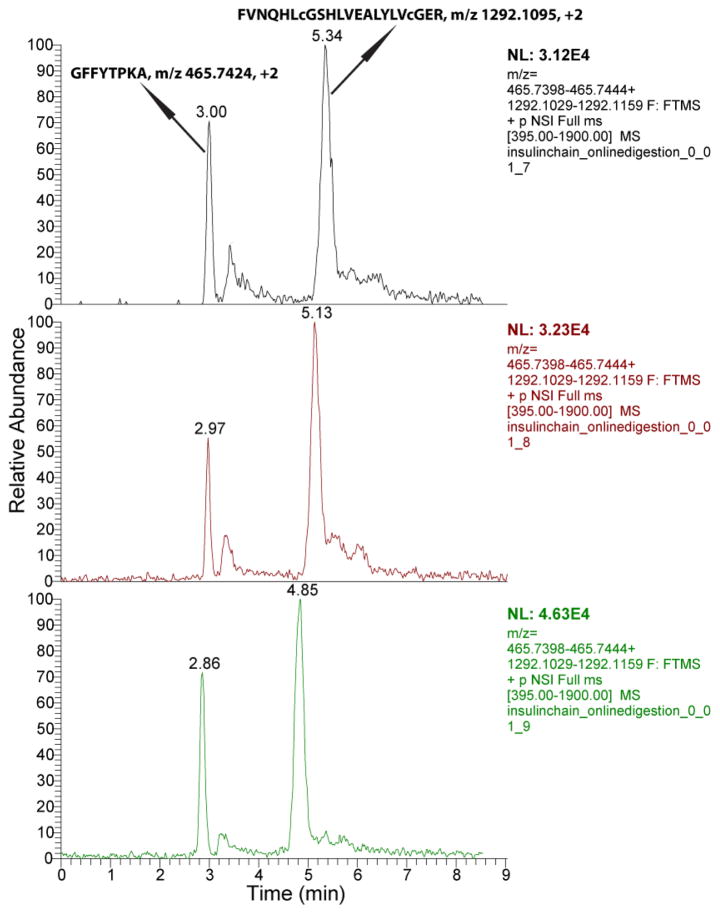
To evaluate the system for protein analysis, we used two model proteins, insulin chain b oxidized and BSA. Insulin chain b oxidized (30 amine acids with sequence FVNQHLCGSHLVEALYLVCGERGFFYTPKA and 3 494 Da molecular weight) was first used to evaluate the system. Because it is a small protein, the protein was directly dissolved in the separation buffer (5 mM NH4HCO3) for analysis. After on-line digestion, separation, and identification with the integrated CZE-ESI-MS/MS system, 100% sequence coverage was obtained with a 7 fmole sample. Two identified peptides, GFFYTPKA and FVNQHLcGSHLVEALYLVcGER, were extracted from the raw files of triplicate runs with mass tolerance as 5 ppm, Fig. 2. The two peptides were well separated and their migration time was reasonably reproducible (relative standard derivation (RSD) less than 5%) in triplicate runs. In addition, RSD of the peptide intensity in triplicate runs was less than 30% for 7 fmole loading amount, and was about 20% for 70 fmole loading amount. These results indicate that the integrated CZE-ESI-MS/MS system is efficient and reproducible for analysis of low fmole amounts of proteins.
Figure 2.
Extracted ion electropherogram of two peptides of insulin chain b oxidized after analysis by the integrated CZE-ESI-MS/MS system in triplicate with 7 fmole injection amount per run. Five-point Gaussian smoothing was applied to the spectra, and the peptides were extracted with mass tolerance of 5 ppm.
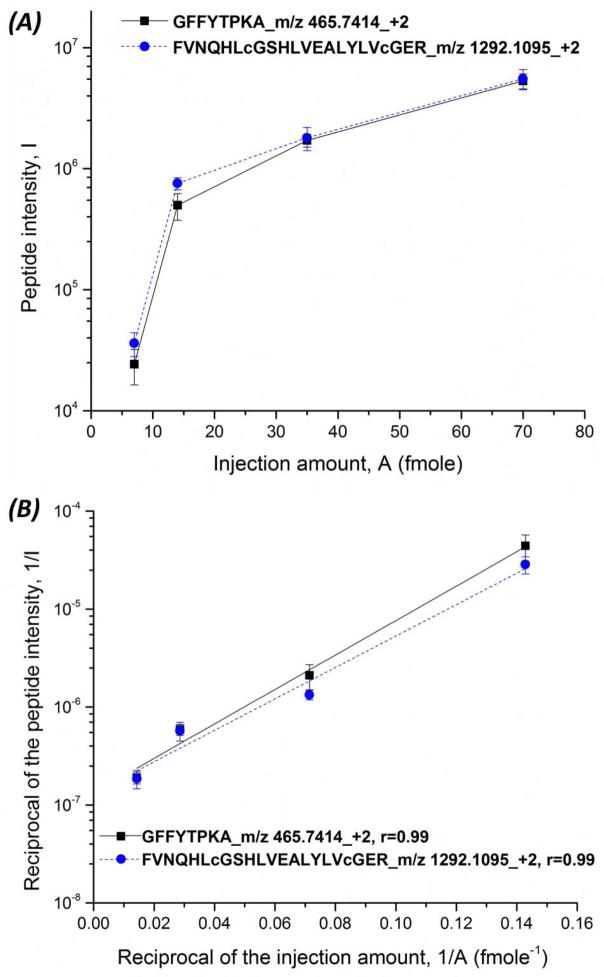
We further determined the relationship between the injection amount (A) of insulin chain b oxidized and the peptide intensity (I), Fig. 3A. When the sample injection amount increased from 7 fmole to 70 fmole, the peptide intensity also increased, but the relationship is not linear. In our experiment, the injection volume was fixed, and the concentration of insulin chain b oxidized ranged from 0.01 to 0.1 mg/mL, resulting in 7 to 70 fmole injection amounts. The digestion time was fixed, so the generated peptide amount directly related to the trypsin digestion rate. The peptide intensity from the mass spectrometer is a good parameter to estimate the peptide amount. Protease digestion can be described by the Michaelis-Menten kinetics. We linearlized the classical Michaelis-Menton equation, which was expressed in terms of peptide intensity (I), the Michaelis constant (Km), and loaded amount (A) (Eq. 1),
Figure 3.
Relationship of the injection amount of insulin chain b oxidized and the peptide intensity (GFFYTPKA and FVNQHLcGSHLVEALYLVcGER) (A) and the relationship between the reciprocal of the injection amount and the reciprocal of the peptide intensity, Equation 1 (B) produced by the integrated CZE-ESI-MS/MS system in triplicate. The peptide intensity was obtained by peak extraction with mass tolerance of 5 ppm. Gaussian smoothing with 5 points was applied.
| (Eq. 1) |
A plot of 1/I vs. 1/A followed this linearlized equation (R = 0.99), Fig. 3B.
To check the system for the analysis of larger and more complex proteins, BSA with 66 kDa molecular weight and 17 disulfide bonds was also analyzed by the system. BSA was dissolved in 50% (v/v) ACN and 5 mM NH4HCO3 (pH 8.0), and directly injected into the microreactor end of the separation capillary and digestion at room temperature for 2 min without reduction and alkylation, followed by CZE-ESI-MS/MS analysis in duplicate. When about 4 fmole BSA was injected and analyzed by the system, two peptides and 4.6% sequence coverage were consistently produced, which suggest that the integrated system is also efficient for the analysis of proteins with large molecular weight and many disulfide bonds without reduction and alkylation. When the injection amount was increased to 20 fmole, the peptide number and sequence coverage dramatically increased to an average of 14 and 19%, respectively.
No peptides were detected from the tandem spectra when 1 fmol of BSA was injected. However, we generated an extracted ion electropherogram corresponding to a BSA peptide (KVPQVSTPTLVEVSR, m/z 820.4744, +2) with mass tolerance of 2 ppm. This extracted ion electropherogram had an average intensity of 1.6E+04, which is consistent with the average intensity produced by 4 fmol sample (7.55 E+04). In addition, the charge state of the peptide from 1 fmol was the same as that from 4 fmol, and the migration time of the peptide between the 1 fmol and 4 fmol results were reasonably reproducible. These results indicate that BSA could be detected from injection of only 1 fmol.
Application for analysis of standard protein mixtures
A three-protein mixture (BSA - 0.12 mg/mL, cytochrome c - 0.01 mg/mL, and myoglobin - 0.01 mg/mL) dissolved in 50% (v/v) ACN and 5 mM NH4HCO3 (pH 8.0) was analyzed by the integrated CZE-ESI-MS/MS system in triplicate. Injection amounts were 7 fmole (BSA), 3 fmole (cytochrome c) and 2 fmole (myoglobin) for each run. The three proteins could be confidently and reproducibly identified by the system with the number of peptide IDs as 10 ± 2 (Cytochrome c), 11 ± 1 (myoglobin), and 1 ±1 (BSA), Table 1. Besides the three proteins, transthyretin was also identified with the number of peptide IDs as 1 ±1, Table 1. Transthyretin might be an impurity from BSA, because transthyretin was also identified from the 20 fmole BSA data with 6 peptides. Interestingly, we also identified transthyretin as an impurity in BSA using top-down analysis of intact proteins (data not shown).
Table 1.
Results of the triplicate analysis of the three-protein mixture.
| MW (kDa) | pI | Injection amount (pg) | Injection amount (fmole) | Peptide IDs | Sequence coverage (%) | |
|---|---|---|---|---|---|---|
| Cytochrome c | 11.7 | 9.5 | 40 | 3 | 10 ± 2 | 69 ± 2 |
| Myoglobin | 16.9 | 7.8 | 40 | 2 | 11 ± 1 | 74 ± 17 |
| BSA | 69.5 | 6.2 | 480 | 7 | 1 ± 1 | 2 ± 2 |
| Transthyretin | 15.7 | 6.3 | Un# | Un | 1 ± 1 | 6±5 |
The injection amount of transthyretin was unknown (Un).
We further analyzed a seven-protein mixture (BSA - 0.11 mg/mL, cytochrome c - 0.009 mg/mL, myoglobin - 0.009 mg/mL, α-casein - 0.015 mg/mL, β-casein - 0.015 mg/mL, β-lactoglobulin - 0.015 mg/mL, and insulin chain b oxidized - 0.01 mg/mL) dissolved in 50% (v/v) ACN and 5 mM NH4HCO3 (pH 8.0) in triplicate. The injection amount was 6 fmole (BSA), 3 fmole (cytochrome c), 2 fmole (myoglobin), 2 fmole (α-casein), 2 fmole (β-casein), 3 fmole (β-lactoglobulin), and 12 fmole (insulin chain b oxidized) for each run. Six proteins (all but β-lactoglobulin) were identified from triplicate runs, and their sequence coverage ranged from 6% to 100% and peptide IDs ranged from 2 to 16, Table 2. Besides the identification of the six proteins, three additional proteins (transthyretin, uncharacterized protein, and BARX2 protein) were also identified with the number of peptide IDs as 1, 4 and 1, respectively, Table 2. These proteins might be impurities present in the seven standard proteins.
Table 2.
Results of the triplicate analysis of the seven-protein mixture.
| MW (kDa) | Injection amount (pg) | Injection amount (fmole) | Peptide IDs# | Sequence coverage (%)# | |
|---|---|---|---|---|---|
| BSA | 69.5 | 440 | 6 | 3 | 6 |
| Cytochrome c | 11.7 | 36 | 3 | 16 | 73 |
| Myoglobin | 16.9 | 36 | 2 | 13 | 96 |
| α-casein | 24.5 | 60 | 2 | 2 | 20 |
| β-casein | 25.1 | 60 | 2 | 4 | 19 |
| Insulin chain b oxidized | 3.4 | 40 | 12 | 4 | 100 |
| β-lactoglobulin | 18.4 | 60 | 3 | 0 | 0 |
| Transthyretin | 15.7 | Un* | Un | 1 | 7 |
| Uncharacterized protein | 18.3 | Un | Un | 4 | 27 |
| BARX2 protein | 31.0 | Un | Un | 1 | 2 |
Peptide IDs and sequence coverage were from the combined triplicate runs.
The injection amounts of these proteins were unknown (Un).
The protein mixtures analyzed in the experiments contained basic and acidic proteins, large and small proteins, and phosphorylated and unphosphorylated proteins, which suggest that the integrated CZE-ESI-MS/MS system is of high sensitivity and efficiency for analysis of complex mixtures of proteins.
Application for analysis of picogram range of RAW 264.7 cell lysate
The integrated system was used to analyze RAW 264.7 cell lysate in triplicate. Two buffers were used to prepare the protein samples. In one case, a sample containing 0.1 mg/mL proteins was dissolved in 50% (v/v) ACN, 0.025% (w/v) SDC, and 5 mM NH4HCO3. In a second case, a 1 mg/mL protein sample was dissolved in 50% (v/v) ACN, 0.25% (w/v) SDC, and 5 mM NH4HCO3. The injection amounts per run for the 0.1 mg/mL and 1 mg/mL samples were ~300 pg and ~3 ng, respectively.
After triplicate analysis, 2 ± 1 and 7 ± 2 protein groups were identified from the 300 pg and 3 ng RAW 264.7 cell lysates, respectively. Information of the identified protein groups is listed in supporting material I. The protein groups (except one protein group, Glial fibrillary acidic protein) obtained from the 300 pg sample were also identified from the 3 ng sample. For glial fibrillary acidic protein, one unique peptide was identified, and its Exp value and mass error were 1.4E-06 and 1 ppm, respectively, which confirmed the protein group identification. Most of the proteins identified from the 300 pg and 3 ng samples were relatively high abundant in RAW 264.7 cell lysate according to the protein spectral count information from the large-scale proteome data generated previously in our group. In addition, the peptide identifications from the 300 pg and 3 ng samples were also manually evaluated, and the annotated tandem spectra of those peptides are listed in supporting material II. We also analyzed the molecular weight and pI of proteins generated by triplicate analysis of the 3 ng RAW 264.7 cell lysate. The molecular weight ranged from 5.7 kDa to 58.8 kDa, and pI ranged from 4.5 to 11.0, which suggest that the integrated CZE-ESI-MS/MS system is efficient for small and large proteins, and also acidic and basic proteins. We further checked the disulfide bonds of the identified proteins from 3 ng RAW 264.7 cell lysate, and the identified proteins tend to have few or no disulfide bonds, which is expected due to the much easier denaturation of these proteins with 50% (v/v) ACN. The result indicates that the integrated CZE-ESI-MS/MS system might have potential limitations for identification of highly structured proteins with many disulfide bonds. On the other hand, the system might be useful for selective identification of the proteins with few disulfide bonds in complex samples.
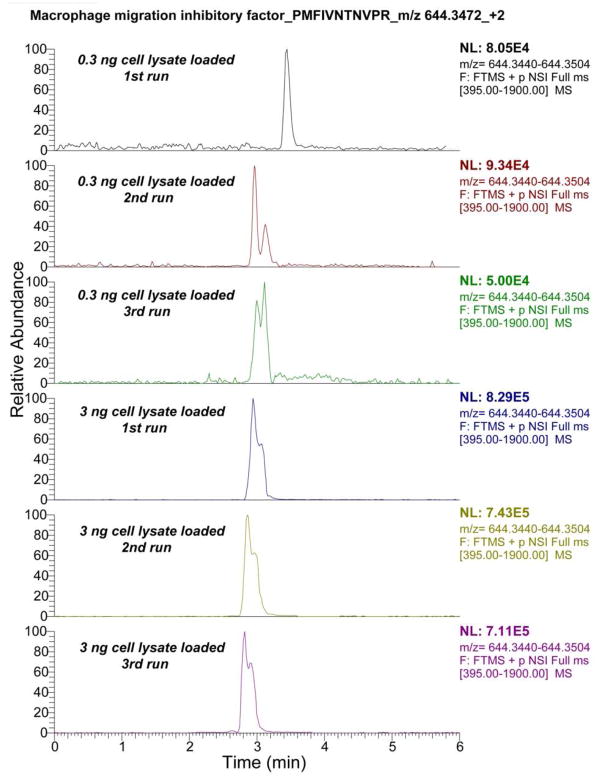
We further extracted the electropherograms from a peptide from macrophage migration inhibitory factor, which was identified from both 300 pg and 3 ng samples, Fig. 4. The integrated CZE-ESI-MS/MS system yielded reasonably reproducible migration time for the peptide for analysis of both 300 pg and 3 ng samples. In addition, the peptide intensity from triplicate analysis of the 3 ng cell lysate was quite consistent (RSD = 8%), and the peptide intensity from the 300 pg cell lysate was reasonably reproducible (RSD = 30%). In addition, when the injection amount was increased from 300 pg to 3 ng, the average peptide intensity generated by the system increased from 7.5E+04 to 7.6E+05. The reproducible peptide migration time and peptide intensity indicate that the integrated CZE-ESI-MS/MS system is reproducible for trace complex cell lysate analysis. The peptide intensities generated for 300 pg and 3 ng cell lysates reasonably reflects the difference in sample amounts, which suggests that the system might be useful for further quantitative protein analysis.
Figure 4.
Triplicate extracted ion electropherograms of one peptide (PMFIVNTNVPR) from the macrophage migration inhibitory factor that was identified from both 300 pg and 3 ng RAW 264.7 cell lysate. The electropherogram was extracted with a mass tolerance of 5 ppm, and Gaussian smoothing with 5 points was applied before plotting the results.
We also extracted two peptides of 10 kDa heat shock protein. One peptide (VLQATVVAVGSGGK) was only identified from 0.3 ng cell lysate, S-Fig. 1 in supporting material II, and another peptide (VLQATVVAVGSGGKGK) was only identified from 3 ng cell lysates, S-Fig. 2 in supporting material II. These two peptides, extracted with mass tolerance as 2 ppm, generated reasonable signal intensities from all the 300 pg and 3 ng cell lysate data. The migration time and peptide intensity of these two peptides from 300 pg and 3 ng cell lysates were also reproducible, which further confirmed the reproducibility of the integrated CZE-ESI-MS/MS system. The intensity of these two peptides also increased with loading amounts from 300 pg to 3 ng.
One obvious limitation of our analysis is that the protein homogenate was prepared from a large volume and only a small aliquot was used for analysis. However, we note that the protein content in a single RAW 264.7 cell is about 0.1 ng [37], and the protein amounts analyzed in this work correspond to the protein content in three cells (0.3 ng) and 30 cells (3 ng). The simplified sample preparation protocol used in this analysis, when coupled with the high-sensitivity electrokinetically-pumped sheath flow cuvette nanospray interface, suggests that the system could be coupled to a single-cell injection and lysis system for single-cell analysis [38, 39]. However, there are still challenges for analysis of small number of cells or of single cell with the system, including the cell injection, efficient cell lysis, removal of interfering compounds in the cell lysate, efficient and accurate control of the transfer of cell lysate to the trypsin microreactor, and the higher capacity peptide separation and deeper protein profiling. We also note that this experiment employed an uncoated capillary for separation. We have recently described the use of a coated capillary for analysis of the E. coli proteome [40]. That approach produced over 1 250 peptide IDs in a single run. Use of a coated capillary may also produce enhanced peptide detection in the analysis of minute amounts of the products generated by the on-column digestion system.
Supplementary Material
Acknowledgments
We thank Dr. William Boggess in the Notre Dame Mass Spectrometry and Proteomics Facility for his help with this project. This work was funded by the National Institutes of Health (R01GM096767).
References
- 1.Geiger T, Wehner A, Schaab C, Cox J, Mann M. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp. M111.014050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang N, Xu M, Wang P, Li L. Anal Chem. 2010;82:2262–2271. doi: 10.1021/ac9023022. [DOI] [PubMed] [Google Scholar]
- 4.Tian R, Wang S, Elisma F, Li L, Zhou H, Wang L, Figeys D. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp. M110.000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 6.Wiśniewski JR, Ostasiewicz P, Mann M. J Proteome Res. 2011;10:3040–3049. doi: 10.1021/pr200019m. [DOI] [PubMed] [Google Scholar]
- 7.Ma J, Liang Z, Qiao X, Deng Q, Tao D, Zhang L, Zhang Y. Anal Chem. 2008;80:2949–2956. doi: 10.1021/ac702343a. [DOI] [PubMed] [Google Scholar]
- 8.Sun L, Li Y, Yang P, Zhu G, Dovichi NJ. J Chromatogr A. 2012;1220:68–74. doi: 10.1016/j.chroma.2011.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma J, Zhang L, Liang Z, Shan Y, Zhang Y. Trends Anal Chem. 2011;30:691–702. [Google Scholar]
- 10.Duan J, Sun L, Liang Z, Zhang J, Wang H, Zhang L, Zhang W, Zhang Y. J Chromatogr A. 2006;1106:165–174. doi: 10.1016/j.chroma.2005.11.102. [DOI] [PubMed] [Google Scholar]
- 11.Sun L, Zhu G, Li Y, Yang P, Dovichi NJ. Anal Chem. 2012;84:8715–8721. doi: 10.1021/ac3019608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen Y, Tolić N, Masselon C, Paša-Tolić L, Camp DG, II, Hixson KK, Zhao R, Anderson GA, Smith RD. Anal Chem. 2004;76:144–154. doi: 10.1021/ac030096q. [DOI] [PubMed] [Google Scholar]
- 13.Shen Y, Tolić N, Masselon C, Paša-Tolić L, Camp DG, II, Lipton MS, Anderson GA, Smith RD. Anal Bioanal Chem. 2004;378:1037–1045. doi: 10.1007/s00216-003-2329-8. [DOI] [PubMed] [Google Scholar]
- 14.Waanders LF, Chwalek K, Monetti M, Kumar C, Lammert E, Mann M. Proc Natl Acad Sci USA. 2009;106:18902–18907. doi: 10.1073/pnas.0908351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moini M. Anal Chem. 2007;79:4241–4246. doi: 10.1021/ac0704560. [DOI] [PubMed] [Google Scholar]
- 16.Faserl K, Sarg B, Kremser L, Lindner H. Anal Chem. 2011;83:7297–7305. doi: 10.1021/ac2010372. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Fonslow BR, Wong CCL, Nakorchevsky A, Yates JR., 3rd Anal Chem. 2012;84:8505–8513. doi: 10.1021/ac301091m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wojcik R, Dada OO, Sadilek M, Dovichi NJ. Rapid Commun Mass Spectrom. 2010;24:2554–2560. doi: 10.1002/rcm.4672. [DOI] [PubMed] [Google Scholar]
- 19.Wojcik R, Li Y, MacCoss MJ, Dovichi NJ. Talanta. 2012;88:324–329. doi: 10.1016/j.talanta.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun L, Zhu G, Li Y, Wojcik R, Yang P, Dovichi NJ. Proteomics. 2012;12:3013–3019. doi: 10.1002/pmic.201200100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Wojcik R, Dovichi NJ, Champion MM. Anal Chem. 2012;84:6116–6121. doi: 10.1021/ac300926h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, Champion MM, Sun L, Champion PAD, Wojcik R, Dovichi NJ. Anal Chem. 2012;84:1617–1622. doi: 10.1021/ac202899p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Licklider L, Kuhr WG, Lacoy MP, Keough T, Purdon MP, Takigiku R. Anal Chem. 1995;67:4170–4177. [Google Scholar]
- 24.Gao J, Xu J, Locascio LE, Lee CS. Anal Chem. 2001;73:2648–2655. doi: 10.1021/ac001126h. [DOI] [PubMed] [Google Scholar]
- 25.Kato M, Sakai-Kato K, Jin H, Kubota K, Miyano H, Toyo3oka T, Dulay MT, Zare RN. Anal Chem. 2004;76:1896–1902. doi: 10.1021/ac035107u. [DOI] [PubMed] [Google Scholar]
- 26.Schoenherr RM, Ye M, Vannatta M, Dovichi NJ. Anal Chem. 2007;79:2230–2238. doi: 10.1021/ac061638h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Wojcik R, Dovichi NJ. J Chromatogr A. 2011;1218:2007–2011. doi: 10.1016/j.chroma.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alonso DOV, Daggett V. J Mol Biol. 1995;247:501–520. doi: 10.1006/jmbi.1994.0156. [DOI] [PubMed] [Google Scholar]
- 29.Griebenow K, Klibanov AM. J Am Chem Soc. 1996;118:11695–11700. [Google Scholar]
- 30.Russell WK, Park ZY, Russell DH. Anal Chem. 2001;73:2682–2685. doi: 10.1021/ac001332p. [DOI] [PubMed] [Google Scholar]
- 31.Slysz GW, Schriemer DC. Rapid Commun Mass Spectrom. 2003;17:1044–1050. doi: 10.1002/rcm.1022. [DOI] [PubMed] [Google Scholar]
- 32.Strader MB, Tabb DL, Hervey WJ, Pan C, Hurst GB. Anal Chem. 2006;78:125–134. doi: 10.1021/ac051348l. [DOI] [PubMed] [Google Scholar]
- 33.Palm AK, Novotny MV. Rapid Commun Mass Spectrom. 2004;18:1374–1382. doi: 10.1002/rcm.1500. [DOI] [PubMed] [Google Scholar]
- 34.Zhu G, Zhang L, Yuan H, Liang Z, Zhang W, Zhang Y. J Sep Sci. 2007;30:792–803. doi: 10.1002/jssc.200600496. [DOI] [PubMed] [Google Scholar]
- 35.Shihabi ZK. J Chromatogr A. 2000;902:107–117. doi: 10.1016/s0021-9673(00)00743-3. [DOI] [PubMed] [Google Scholar]
- 36.Yang Y, Boysen RI, Hearn MTW. Anal Chem. 2006;78:4752–4758. doi: 10.1021/ac051735v. [DOI] [PubMed] [Google Scholar]
- 37.Cohen D, Dickerson JA, Whitmore CD, Turner EH, Palcic MM, Hindsgaul O, Dovichi NJ. Annu Rev Anal Chem. 2008;1:165–190. doi: 10.1146/annurev.anchem.1.031207.113104. [DOI] [PubMed] [Google Scholar]
- 38.Hu S, Le Z, Krylov S, Dovichi NJ. Anal Chem. 2003;75:3495–3501. doi: 10.1021/ac034153r. [DOI] [PubMed] [Google Scholar]
- 39.Krylov SN, Zhang Z, Chan NW, Arriaga E, Palcic MM, Dovichi NJ. Cytometry. 1999;37:14–20. doi: 10.1002/(sici)1097-0320(19990901)37:1<14::aid-cyto2>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 40.Zhu G, Sun L, Yan X, Dovichi NJ. Anal Chem. 2013:85,32569–2573. doi: 10.1021/ac303750g. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.