Abstract
The flavoprotein tryptophan 2-monooxygenase catalyzes the oxidative decarboxylation of tryptophan to yield indole-3-acetamide. This is the initial step in the biosynthesis of the plant growth hormone indole-acetic-acid by bacterial pathogens that cause crown gall and related diseases. The structure of the enzyme from Pseudomonas savastanoi has been determined by X-ray diffraction methods to a resolution of 1.95 Å. The overall structure of the protein shows that it has the same fold as the monoamine oxidase family of flavoproteins, with the greatest similarities to the L-amino acid oxidases. The location of bound indole-3-acetamide in the active site enables identification of residues responsible for substrate binding and specificity. Two residues in the enzyme are conserved in all members of the monoamine oxidase family, Lys365 and Trp466. The K365M mutation decreases the kcat and kcat/KTrp values by 60,000 and 2 million-fold, respectively. The deuterium kinetic isotope effect increases to 3.2, consistent with carbon-hydrogen bond cleavage becoming rate-limiting in the mutant enzyme. The W466F mutation decreases the kcat value less than 2-fold and the kcat/KTrp value only 5-fold, while the W466M mutation results in enzyme lacking flavin and detectable activity. This is consistent with a role for Trp466 in maintaining the structure of the flavin binding site in the more conserved FAD domain.
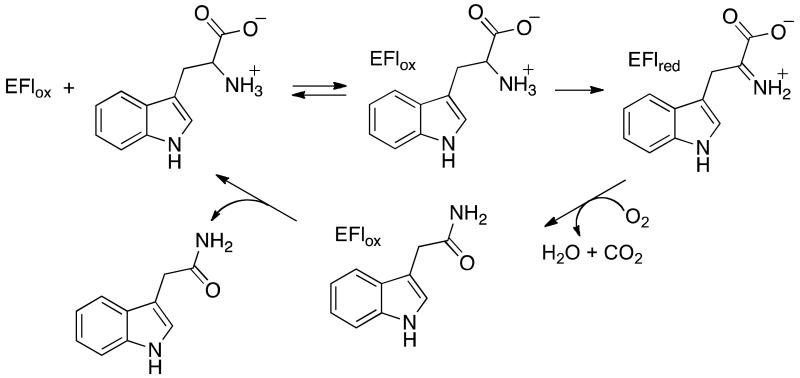
The wide-spread plant diseases crown gall and olive knot are caused by infections by Agrobacter and Pseudomonas savastanoi, respectively.1, 2 These and related3, 4 plant diseases are characterized by the formation of tumorous growths on the plants. Critical to this process is the production at the site of infection of the plant hormone indole-3-acetic acid by a two-step metabolic pathway involving bacterial proteins.5-8 In the first step, the flavoprotein tryptophan 2-monooxygenase (TMO)a catalyzes the oxidative decarboxylation of tryptophan to indole-3-acetamide (IAM) (Scheme 1).9, 10 The amide of IAM is then hydrolyzed by a protein encoded by the iah gene to yield indole-3-acetic acid.
Scheme 1.
Based on its sequence, TMO has been suggested to be a member of the larger monoamine oxidase (MAO) family of enzymes, with the greatest similarity to the L-amino acid oxidases (LAAOs).11 The MAO family is a group of FAD-containing amine oxidases from a diverse group of organisms that have a similar structural fold.12 These enzymes include MAO, which oxidizes several neurotransmitters,13 the histone lysine demethylase LSD1,14, 15 the polyamine oxidases,16 and the LAAOs,17 and catalyze the oxidation of a variety of primary, secondary and tertiary amines and amino acids. In each case a carbon-nitrogen bond is oxidized by transfer of a hydride equivalent to the flavin cofactor; the oxidized amine is typically hydrolyzed nonenzymatically to yield the respective aldehyde or ketone. In contrast, TMO catalyzes the oxidative decarboxylation of the oxidized amine to form an amide. The sequence similarities of TMO to other flavin amine oxidases is much greater in the more conserved FAD-binding domain, whereas the similarity of the substrate-binding domain to those of other flavoproteins has been too low to provide insight into the structure of TMO.
Mechanistically, TMO is the best-characterized of the flavoenzymes catalyzing the oxidation of L-amino acids. The kinetic mechanism of the enzyme has been determined using steady-state and rapid-reaction methods.18, 19 Turnover by TMO consists of two half-reactions, a reductive half-reaction in which a hydride is transferred from the substrate to the flavin cofactor and an oxidative half-reaction in which the flavin is reoxidized and the amide product is formed.18 The functions of several active-site residues in TMO have been analyzed by site-directed mutagenesis, allowing proposals to be made regarding their roles in catalysis even in the absence of a structure.11, 20, 21 Finally, TMO has served as a model for the MAO family in the application of kinetic isotope effects to establish that oxidation of the carbon-hydrogen bond in the amine substrate occurs by hydride transfer.22 Despite these extensive mechanistic studies and its biological role as a key enzyme in plant pathogens, a structure of the protein has not been described. We present here the structure of TMO from P. savastanoi.
Experimental procedures
Materials
l-Tryptophan, IAM, indole-pyruvate, and glucose oxidase were purchased from Sigma (St. Louis, MO). Escherichia coli strain M15 (pREP4) was from Qiagen (Valencia, CA). [α-2H]-l-Tryptophan was synthesized via the method of Kiick and Phillips.23
Cloning
Mutations were introduced using the standard QuikChange Kit (Agilent Technologies, Santa Clara, CA) with the following forward primers and the complementary reverse primers: K365M TMO: CTGACAGGTTCATCGATGCTTTTCATTCTCAC, W466F TMO: GTATTGCACCATGACTTCCTCACCGATCCCCATTC, and W466M TMO: GTATTGCACCATGACATGCTCACCGATCCCC.
Protein expression and purification
Wild-type and mutant forms of TMO were expressed in E. coli and purified as reported previously.20 A HiPrep DEAE FF 16/10 column (GE Healthcare Life Sciences, Piscataway, NJ) was used instead of DEAE-Sephacel for anion exchange chromatography.
Enzyme assays
Assays were performed at 25 °C in 50 mM Tris buffer, pH 8.3, containing 1.0 mM EDTA and 0.5 mM dithiothreitol and were initiated by the addition of enzyme. The activities of the wild-type and Trp466 mutant enzymes were determined by measuring oxygen consumption during the reaction using an oxygen electrode (Yellow Springs model 5300, Yellow Springs, OH).24 Assays contained 25-1000 μM tryptophan and ~0.125 μM TMO. The effect of the concentration of oxygen on the initial rate was determined by varying the oxygen concentration from 25-600 μM at saturating concentrations of tryptophan (3.0 mM), as previously described.18 The steady-state activity of the TMO K365M enzyme was measured using an endpoint HPLC-based assay. l-Tryptophan concentrations were varied from 0.125 to 6 mM; reactions were initiated by the addition of enzyme (~1.5 μM) and quenched with an equal volume of 1 M HCl at varying time points over 20 minutes. The substrate tryptophan and the products IAM and indole-pyruvate were separated on a Phenomenex (Torrance, CA) C18 column (250 mm × 4.6 mm) using an isocratic elution of 25% acetonitrile containing 0.1% TFA. Detection was by fluorescence, with excitation at 278 nm and emission at 350 nm; the limit of detection was 50 nM for IAM. The values of the steady state kinetic parameters kcat, KM, and kcat/KM were determined by fitting kinetic data to the Michaelis-Menten equation using the program KaleidaGraph (Synergy Software.) Substrate deuterium kinetic isotope effects on kcat were calculated by directly comparing kcat values obtained with protiated and deuterated substrate at a concentration of 5 mM.
Protein identification by mass spectrometry
Confirmation of the identity of the purified W466M TMO was done by mass spectrometric analysis of tryptic peptides of the protein in the UTHSCSA Institutional Mass Spectrometry Laboratory. W466M TMO was purified by the protocol for the wild-type enzyme, yielding a colorless protein with the appropriate molecular weight that was 90-95 % pure by SDS-polyacrylamide gel electrophoresis. The prominent band was excised from the gel using a razor blade and digested in situ with trypsin according to standard protocols.25 The resulting peptides were resuspended in 0.5% TFA and analyzed by capillary-HPLC-electrospray tandem mass spectrometry on a Thermo Fisher LTQ ion trap mass spectrometer coupled to an Eksigent NanoLC micro HPLC by means of a PicoView (New Objective) nanospray interface. Capillary on-line HPLC separation of tryptic peptides was conducted using the following conditions: column, New Objective PicoFrit, 75 μm id, packed to 11 cm with C18 adsorbent (Vydac 218MSB5); mobile phase A, 0.5% acetic acid/0.005% trifluoroacetic acid in water; mobile phase B, 90% acetonitrile/0.5% acetic acid/0.005% trifluoroacetic acid in water; gradient, 2% B to 42% B in 30 min; flow rate, 0.4 μl/min. The data-dependent acquisition consisted of one survey scan followed by 7 collision-induced dissociation spectra. The resulting CID spectra were searched against the NCBInr or SwissProt database using Mascot (Matrix Science). A 95% confidence level threshold was used for Mascot peptide scores. Scaffold software version 3.6.5 was used to validate MS/MS based peptide and protein identifications. Protein probabilities were assigned by the Protein Prophet algorithm Nesvizhskii.26
Crystallization, structure determination and refinement
Preliminary crystals of TMO were grown in the UTHSCSA X-ray Crystallography Core Laboratory from commercial crystallization screen kits (Qiagen Inc., Valencia, CA) using a Phoenix crystallization robot (Art Robbins Instruments, Sunnyvale, CA). Optimized crystals were grown within 1 week by the sitting drop vapor diffusion method by mixing the protein solution in 50 mM Tris-HCl, pH 8.3, 1 mM EDTA, 3 mM tris(2-carboxyethyl)phosphine (TCEP), and 100 μM IAM in a 1:1 ratio with buffer containing 1.6 M sodium/potassium phosphate, pH 7.5, and 12% ethylene glycol. The crystals were flash-cooled using liquid nitrogen prior to data collection. Data were collected in the UTHSCSA X-ray Crystallography Core Laboratory with a Rigaku MicroMax-007 HF X-ray generator and R-AXIS HTC imaging plates. The data were processed using HKL-2000. 27 Phases were obtained by the molecular replacement method implemented in Phaser28 using the l-phenylalanine oxidase coordinates in Protein Data Bank (PDB) entry 2YR5 as the search model, which shares 25% sequence identity with TMO. TMO coordinates were refined against the data using PHENIX,29 including simulated annealing, alternating with manual rebuilding using COOT.30 Data collection and refinement statistics are listed in Table 1. The refined coordinates have been deposited in the PDB with accession code 4IV9.
Table 1. Data collection and refinement statistics.
| Data Collection | |
|---|---|
| space group | P21212 |
| cell dimensions | |
| a, b, c (Å) | 111.1, 122.2, 76.6 |
| α, β, γ(°) | 90, 90, 90 |
| wavelength (Å) | 1.5418 |
| resolution (Å) | 30.0 - 1.95 (2.02-1.95)a |
| Rsym* | 0.142 (0.577) |
| σ | 11.0 (2.1) |
| completeness (%) | 99.5 (96.5) |
| redundancy | 5.4 (4.1) |
| Refinement | |
|---|---|
| resolution (Å) | 28.0 - 1.95 |
| no. of reflections | 76,161 |
| Rwork / Rfree | 0.187/0.245 |
| no. of dimers per asymmetric unit | 1 |
| no. of atoms | |
| protein | 8644 |
| ligands | 146 |
| solvent | 747 |
| B-factors (Å2) | |
| protein | 14.6 |
| ligands | 12.8 |
| solvent | 18.6 |
| r.m.s.d | |
| bond lengths (Å) | 0.008 |
| bond angles (°) | 1.096 |
Values in parentheses are for the highest-resolution shell.
Structure analysis
Protein structures were visualized with Chimera31 and Pymol (Schrödinger, Inc.). The locations of internal cavities in the structure were determined using CASTp.32
Results and Discussion
Overall structure
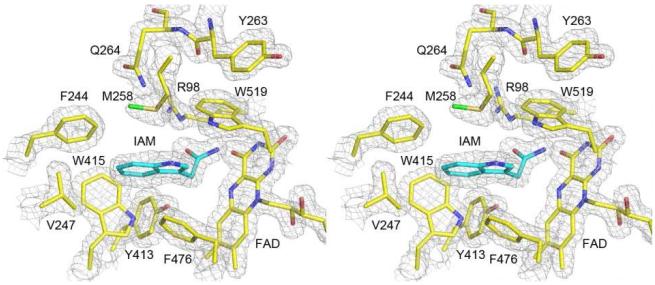
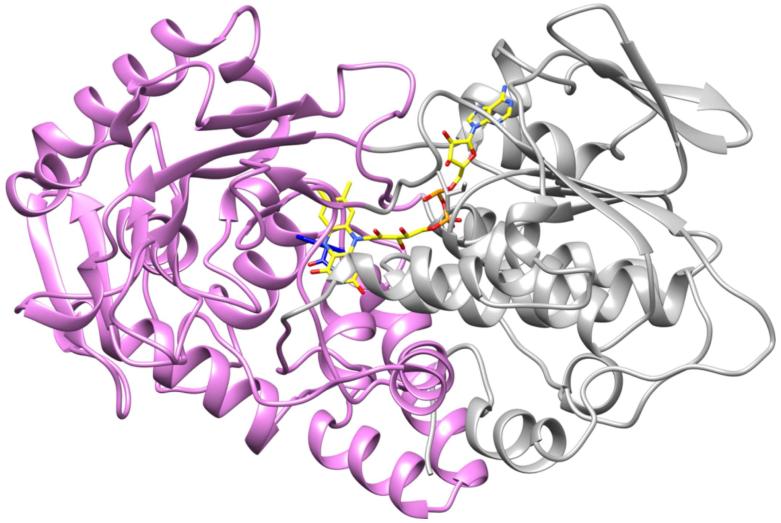
TMO is purified with the product IAM bound (Figure 1) as a result of its being present during the purification to prevent partial loss of the FAD.24 The IAM-containing enzyme crystallizes in space group P21212, and a 99.5% complete diffraction data set was taken at a resolution of 1.95 Å. The asymmetric unit contains 1 TMO dimer of identical ~60 kDa subunits (Table 1, Figure S1). Each monomer contains 17 α-helices (6 residues or longer) and 16 β-strands (3 residues or longer) that form distinct FAD-binding (residues 6-78, 271-360, 483-554) and substrate-binding domains (Figure 2). In addition to the molecule of IAM in the active site, there are two phosphate molecules and an ethylene glycol molecule on the side of each monomer opposite the dimer interface. The overall structure of the TMO monomer shares significant similarity with other members of the MAO family, with the greatest similarity to a number of L-amino acid oxidases and L-phenylalanine oxidase from Pseudomonas sp. P-501,33,34 despite sequence identities of less than 20%. Alignment of the structures of TMO and LAAO from the pit viper C. rhodostoma (PDB file 1F8S17) yields an overall r.m.s.d. of 1.6 Å for 1577 atoms (Figure S1), while alignment of TMO with L-phenylalanine oxidase (PDB file 2YR435) yields an overall r.m.s.d. of 2.0 Å for 1761 atoms.
Figure 1.
Refined TMO structure superimposed onto a σA weighted 2mFo-DFc electron density map contoured at 1.5 σ.
Figure 2.
Overall ribbon structure of the TMO monomer with the FAD-binding domain colored gray and the substrate-binding domain colored magenta.
A PSI-BLAST search of the available bacterial genomic sequences identifies a large number of related proteins that are annotated as tryptophan monooxygenase, monoamine oxidase, or amine oxidase. These include the tryptophan monooxygenase from the Ti plasmid of Agrobacter tumafaciens, the causative agent in plant crown gall disease,5 which is 69% identical to P. savastanoi TMO. The closest paralog to TMO is lysine monooxygenase, which catalyzes a similar oxidative decarboxylation of lysine,36 and is 50% identical.
TMO dimerizes with the monomers in two-fold symmetry with the FAD cofactors oriented in opposite directions (Figure S2). Several interactions occur between the two TMO monomers, including salt bridges between Arg134 and Asp350, Arg227 and Asp203, Arg391 and Glu357, and His359 and Asp417. Additional hydrogen bonds between Tyr207 and Asp203 and Arg227 and Tyr557 are also present. Of these interactions, only that between His359 and Asp417 is conserved in phenylalanine oxidase and C. rhodostoma LAAO. While PISA analysis37 supports a dimeric structure with 4385 Å2 of buried surface area per monomer, gel-filtration analyses of enzyme at a lower concentration (~50 μM) than found in crystals show only a monomer, so it is likely that the active form of TMO is a monomer rather than a dimer.
The FAD-binding domain is composed of a series of β-sheets and alpha helices that form the nucleotide-binding Rossmann-like fold that is typically found in FAD-containing enzymes;38 discontinuous regions of the peptide chain form a four-stranded parallel β-sheet sandwiched between an additional three-stranded β-sheet and a series of α-helices (Figure 2). There is an extensive series of interactions between the protein and the cofactor. All of the interactions are conserved in C. rhodostoma LAAO (PDB file 2IID) and phenylalanine oxidase (PDB file 2YR6) with the exception of that between Cys511 and the C2′ hydroxyl oxygen of the FAD ribityl. This residue is replaced by an aspartate in phenylalanine oxidase and a glutamate in all LAAOs. With this one exception the same protein-cofactor interactions are also conserved in human MAO A and B, demonstrating the conservation of the FAD-binding domain in this family of flavoprotein amine oxidases. The lack of conservation of Cys511 is consistent with the small effects of the TMO C511S mutation, for which the largest effect is an ~8-fold decrease in the second-order rate constant for binding of the substrate tryptophan to the enzyme.11
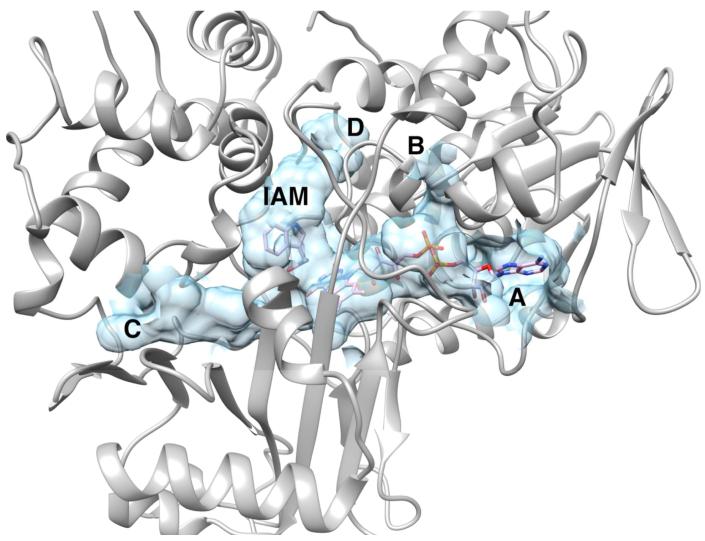
The FAD cofactor and the bound indole-3-acetamide occupy a large internal cavity with a volume of ~2500 Å3 (Figure 3). This cavity has three openings to the surface. Two provide access to the cofactor, one (A) to the adenosine and the second (B) to the pyrophosphate. This second opening provides a rationale for the sensitivity of the enzyme to thiol reagents24 and 2-oxo-3-pentynoate,39 since the side chain of Cys511, a target of the latter, forms one side of this cavity. The third opening (C) is appropriately placed to allow substrates access to the FAD N5, suggesting that this is the access tunnel for the substrate. The large cavity also extends from the active site toward the opposite side of the protein (D). Access to the surface from that side is blocked by the side chains of Tyr263 and Gln261. The access to the active site via opening C does not appear to be conserved in closely related members of the MAO structural family. The access in C. rhodostoma LAAO is well-defined by three aminobenzoate molecules,40 and does not match any of the cavities in TMO. In the structures of phenylalanine oxidase with bound ligands, there is no direct access from the active site to the surface, but one is seen in the pro-enzyme; this cavity overlays that in LAAO rather than in TMO.35
Figure 3.
The internal cavity in TMO containing the FAD and indole-3-acetamide (IAM).
Active site
The substrate-binding domain of TMO consists of residues 79-270 and 361-482. The IAM is bound on the re face of the isoalloxazine ring, as is commonly found with members of the MAO structural family (Figures 1 and S3). The amide carbon of IAM is 3.9 Å from the flavin N5; a similar arrangement for the amino acid substrate would be appropriate for hydride transfer from that carbon to the FAD. The carbonyl oxygen of the bound product forms a hydrogen bond (3.1 Å) with NH1 of Arg98. Based on site-directed mutagenesis, Arg98 and Tyr413 were previously proposed to interact with the carboxylate of the amino acid substrate in the active site of TMO, since mutation of either residue in TMO dramatically reduces activity.20,21 The interaction with Arg98 seen here is consistent with that conclusion. In addition, both Arg98 and Tyr413 occupy similar locations in the active site of TMO to the arginine and tyrosine in the LAAOs that bind the substrate carboxylate in those enzymes (Figure S4).
MAO A and B contain a pair of tyrosine residues located on opposite sides of the substrate amine that have been proposed to play critical roles in orienting the substrate for oxidation.41 Residues Trp519 and Phe 476 occupy similar positions in TMO (Figure 1). The amide nitrogen of IAM is 3.1 Å from the benzene ring of Trp519, consistent with a similar interaction of this residue with the neutral amino group of the substrate during catalysis. This tryptophan residue is conserved in the available LAAO structures and in phenylalanine oxidase. Phe476 corresponds to the other tyrosine residue in MAO; the aromatic ring is stacked on the indole ring of IAM and too far from its amide nitrogen for a significant interaction. The corresponding residue in phenylalanine oxidase is also a phenylalanine, but it is replaced with isoleucine in the LAAOs (Figure S4). Mutagenesis to phenylalanine of the corresponding tyrosine in MAO B has a much smaller effect on activity than does the same mutation of the tyrosine in that enzyme corresponding to Trp519 in TMO.42 Taken together, these results suggest that the interaction with Trp519 is more important than that with Phe476 in TMO and possibly in all members of the MAO family.
The indole ring of the IAM is in a hydrophobic pocket made up of Phe244, Val 247, Met258, Trp415, Phe476, Leu478 (Figures S3 and S5). These residues are not conserved among TMO, the LAAOs, and phenylalanine oxidase, although all of these enzymes prefer aromatic amino acids as substrates. The identity of the residue that corresponds to Val247 at the bottom of the substrate binding pocket in TMO does correlate with the substrate specificity of these enzymes. TMO prefers tryptophan as a substrate over phenylalanine by two orders of magnitude,24 while phenylalanine oxidase prefers phenylalanine over tryptophan by at least an order of magnitude.43 In the latter enzyme, Val247 is replaced by Leu319. Comparison of the active sites of the two enzymes shows that this replacement results in a smaller active site in which Leu319 would have to move to accommodate the larger indole ring of tryptophan. Lysine monooxygenase carries out a similar reaction to TMO, but prefers lysine and arginine as substrates; in that enzyme Val247 is replaced by Asp238.
Site-directed mutagenesis
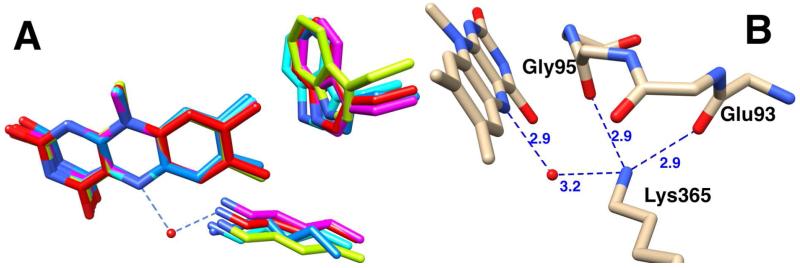
Despite the similar folds and catalytic reactions of members of the MAO structural family, there is low sequence identity across the family. The majority of the identical residues are in the N-terminal 77 residues of the highly conserved FAD-binding domain of TMO. Outside of this region, TMO contains two other residues conserved in the MAO family, Lys365 and Trp466, both of which are within 5 Å of the isoalloxazine ring (Figure 4A). The conserved lysine in the MAO family forms a water-mediated hydrogen bond with the N5 atom of the flavin cofactor.44, 45 In TMO the amino moiety of Lys365 also forms hydrogen bonds (2.9 Å) with the carbonyl oxygens of Glu93 and Gly95 (Figure 4B). Both residues are in a loop that packs against the si side of the flavin isoalloxazine ring; Gly95 is conserved in the MAO family and Glu93 shows only conservative replacements.
Figure 4.
A: Overlay of the conserved active site Lys and Trp residues in TMO (red carbons and water), Pseudomonas phenylalanine oxidase (magenta carbons, PDB file 2YR6), C. rhodostoma LAAO (light blue carbons, PDB file 2IID), human MAO B (blue carbons, PDB file 1GOS), and human LSD1 (light green carbons, PDB file 2H94). B: Lys365 interactions in TMO.
The effect of mutating the conserved lysine depends upon the enzyme. The K300M mutation in maize polyamine oxidase decreases the rate constant for flavin reduction by three orders of magnitude46 and that for flavin oxidation by 30-fold.47 Based on the structure of the mutant protein, the effect on flavin reduction was attributed to unproductive binding of the substrate spermine to the mutant enzyme.47 In contrast, the K315M mutation in mouse polyamine oxidase does not affect the rate constant for reduction of the flavin by spermine,48 but does decrease the rate constant for flavin oxidation by 25-fold.49 pH and solvent isotope effects on the mutant polyamine oxidase support a role for this residue in assisting in proton transfer during flavin oxidation. In LSD1 the K661A mutation decreases activity 100-fold or more, although the specific step(s) affected was not identified.50
To gain insight into the role of Lys 365 in the reaction catalyzed by TMO, the K365M enzyme was characterized. The mutation resulted in a 60,000-fold decrease in the kcat value and a 2 million-fold decrease in the kcat/KTrp value compared to the wild-type enzyme (Table 2). The large decrease in the kcat /KTrp value establishes that the residue plays a critical role in the reductive half reaction. The deuterium kinetic isotope effect on the kcat value was determined to gain further insight into the effects of the mutation, yielding a Dkcat value of 3.2 ± 0.3. The Dkcat value for the wild-type enzyme is only 1.1, due to rate-limiting product release.19 The substantial change in the Dkcat value establishes that the low level of activity detected with the mutant protein is not due to contamination by wild-type enzyme. The Dkcat value for the mutant enzyme is comparable to the isotope effect on the rate constant for flavin reduction of 2.4 for the wild-type enzyme,18 indicating that C-H bond cleavage has become rate-limiting for turnover by the mutant enzyme, and providing additional evidence that this residue plays a role in the reductive half-reaction of TMO. If the kcat value for the mutant enzyme is equal to the rate constant for flavin reduction as the isotope effect suggests, the mutation results in a decrease of 600,000-fold from the wild-type value of 140 s−1.18 Given the location of Lys365 in the active site, the most likely role of this residue is to maintain the protein structure around the FAD cofactor and properly position it relative to the amino acid substrate rather than to play a catalytic role in the reaction.
Table 2. Kinetic parameters for wild-type and mutant TMOa.
| Wild-typeb | K365M | W466F | |
|---|---|---|---|
| kcat (s−1) | 13.2 ± 0.7 | 0.00022 ± 0.00002 | 9.2 ± 0.4 |
| kcat/KTrp (mM−1 s−1) | 360 ± 37 | 0.00019 ± 0.00003 | 72 ± 7 |
| KTrp (mM) | 0.04 ± 0.005 | 1.2 ± 0.3 | 0.13 ± 0.02 |
| kcat/KO2 (mM−1 s−1) | 140 ± 20 | NDc | 70 ± 10 |
| KO2 (mM) | 90 ± 10 | ND | 140 ± 30 |
Kinetic parameters were measured at 25°C in 50 mM Tris buffer, pH 8.3, containing 1.0 mM EDTA and 0.5 mM dithiothreitol.
Wild-type values are from reference 18.
Not determined due to the low activity of the protein.
MAO family members also contain a conserved tryptophan residue near the methyl groups on the isoalloxazine ring. In TMO, the pyrrole ring of the side chain of Trp466 approaches within 3.5 Å of the 7-methyl substituent of the FAD (Figure 4A). The indole ring is packed in a hydrophobic environment, within van der Waals distance (3.4-3.8 Å) of backbone or side chain atoms of Arg71, Arg77, and His464; only Arg77 is conserved in the MAO family. Trp466 was mutated to phenylalanine and methionine. The W466F mutation in TMO does not have a significant effect on the kcat value, while the kcat /KO2 value decreases only two-fold, and the kcat /KTrp value only 5-fold (Table 2). Expression and purification of W466M TMO using the protocol for the wild-type enzyme yielded a colorless enzyme of the correct molecular weight but lacking FAD or activity. The identity of the purified protein as W466M TMO was established using MS/MS of peptides resulting from trypsin digestion of the protein; the peptide analysis yielded 91% sequence coverage of TMO W466M. These results suggest that the large and rigid tryptophan residue at this position is important for binding of FAD by maintaining the shape of the isoalloxazine-binding pocket.
Conclusions
The three-dimensional structure of TMO, a key enzyme in plant gall formation, establishes the enzyme as a member of the MAO structural family with a similar structure to LAAOs despite the low sequence identities among these proteins. These results justify the use of this enzyme as a model for studies of the chemical mechanisms of this large family of flavoprotein amine oxidases. The large effect of the K365M mutation is consistent with this residue playing a critical role in the reductive half-reaction of the enzyme, while the effects of mutating Trp466 suggest that this residue is important for binding the cofactor.
Supplementary Material
Acknowledgements
Support for the X-ray Crystallography Core Laboratory and the Institutional Mass Spectrometry Laboratory by the UTHSCSA Executive Research Committee and the Cancer Therapy Research Center is gratefully acknowledged.
Footnotes
This work was supported in part by NIH grants R01 GM058698 to PFF and F32 GM097762 to HMG, and by The Welch Foundation grant AQ-1399 to PJH.
Supporting Information Available
Figures S1-S4 are provided in Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
ABBREVIATIONS: TMO, tryptophan-2-monooxygenase; IAM, indole-3-acetamide; MAO, monoamine oxidase; LAAO, L-amino acid oxidase; PDB, Protein Data Bank
References
- 1.Zhu J, Oger PM, Schrammeijer B, Hooykaas PJJ, Farrand SK, Winans SC. The bases of crown gall tumorigenesis. J. Bacteriol. 2000;182:3885–3895. doi: 10.1128/jb.182.14.3885-3895.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ramos C, Matas IM, Bardaji L, Aragón IM, Murillo J. Pseudomonas savastanoi pv. savastanoi: some like it knot. Mol. Plant Pathol. 2012;13:998–1009. doi: 10.1111/j.1364-3703.2012.00816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burr TJ, Otten L. Crown gall of grape: Biology and disease management. Annu. Rev. Phytopathol. 1999;37:53. doi: 10.1146/annurev.phyto.37.1.53. [DOI] [PubMed] [Google Scholar]
- 4.Best VM, Vasanthakumar A, McManus PS. Anatomy of cranberry stem gall and localization of bacteria in galls. Phytopathology. 2004;94:1172, 1177. doi: 10.1094/PHYTO.2004.94.11.1172. [DOI] [PubMed] [Google Scholar]
- 5.Thomashow MF, Hugly S, Buchholz WG, Thomashow LS. Molecular basis for the auxin-independent phenotype of crown gall tumor tissues. Science. 1986;231:616–618. doi: 10.1126/science.3511528. [DOI] [PubMed] [Google Scholar]
- 6.Smidt M, Kosuge T. The role of indole-3-acetic acid accumulation by alpha methyl tryptophan-resistant mutants of Pseudomonas savastanoi in gall formation on oleanders. Physiol. Plant Path. 1978;13:203–214. [Google Scholar]
- 7.Comai L, Kosuge T. Involvement of plasmid deoxyribonucleic acid in indoleacetic acid synthesis in Pseudomonas savastanoi. J. Bacteriology. 1980;143:950–957. doi: 10.1128/jb.143.2.950-957.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Escobar MA, Civerolo EL, Summerfelt KR, Dandekar AM. RNAi-mediated oncogene silencing confers resistance to crown gall tumorigenesis. Proceedings of the National Academy of Sciences. 2001;98:13437–13442. doi: 10.1073/pnas.241276898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Magie AR, Wilson EE, Kosuge T. Indoleacetamide as an intermediate in the synthesis of indoleacetic acid in Pseudomonas savastanoi. Science. 1963;141:1281–1282. doi: 10.1126/science.141.3587.1281. [DOI] [PubMed] [Google Scholar]
- 10.Kosuge T, Heskett MG, Wilson EE. Microbial synthesis and degradation of indole-3-acetic acid I. The conversion of L-tryptophan to indole-3-acetamide by an enzyme system from Pseudomonas savastanoi. J. Biol. Chem. 1966;241:3738–3744. [PubMed] [Google Scholar]
- 11.Sobrado P, Fitzpatrick PF. Analysis of the roles of amino acid residues in the flavoprotein tryptophan 2-monooxygenase modified by 2-oxo-3-pentynoate: characterization of His338, Cys339, and Cys511 mutant enzymes. Arch. Biochem. Biophys. 2002;402:24–30. doi: 10.1016/S0003-9861(02)00063-2. [DOI] [PubMed] [Google Scholar]
- 12.Fitzpatrick PF. Oxidation of amines by flavoproteins. Arch. Biochem. Biophys. 2010;493:13–25. doi: 10.1016/j.abb.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edmondson DE, Binda C, Mattevi A. Structural insights into the mechanism of amine oxidation by monoamine oxidases A and B. Arch. Biochem. Biophys. 2007;464:269–276. doi: 10.1016/j.abb.2007.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forneris F, Battaglioli E, Mattevi A, Binda C. New roles of flavoproteins in molecular cell biology: Histone demethylase LSD1 and chromatin. FEBS Journal. 2009;276:4304–4312. doi: 10.1111/j.1742-4658.2009.07142.x. [DOI] [PubMed] [Google Scholar]
- 15.Gaweska H, Henderson Pozzi M, Schmidt DMZ, McCafferty DG, Fitzpatrick PF. Use of pH and kinetic isotope effects to establish chemistry as rate-limiting in oxidation of a peptide substrate by LSD1. Biochemistry. 2009;48:5440–5445. doi: 10.1021/bi900499w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Binda C, Mattevi A, Edmondson DE. Structure-function relationships in flavoenzyme dependent amine oxidations. A comparison of polyamine oxidase and monoamine oxidase. J. Biol. Chem. 2002;277:23973–23976. doi: 10.1074/jbc.R200005200. [DOI] [PubMed] [Google Scholar]
- 17.Moustafa IM, Foster S, Lyubimov AY, Vrielink A. Crystal structure of LAAO from Calloselasma rhodostoma with an L-phenylalanine substrate: insights into structure and mechanism. J. Mol. Biol. 2006;364:991–1002. doi: 10.1016/j.jmb.2006.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emanuele JJ, Jr., Fitzpatrick PF. Mechanistic studies of the flavoprotein tryptophan 2-monooxygenase. 1. Kinetic mechanism. Biochemistry. 1995;34:3710–3715. doi: 10.1021/bi00011a028. [DOI] [PubMed] [Google Scholar]
- 19.Emanuele JJ, Jr., Fitzpatrick PF. Mechanistic studies of the flavoprotein tryptophan 2-monooxygenase. 2. pH and kinetic isotope effects. Biochemistry. 1995;34:3716–3723. doi: 10.1021/bi00011a029. [DOI] [PubMed] [Google Scholar]
- 20.Sobrado P, Fitzpatrick P. Analysis of the role of the active site residue Arg98 in the flavoprotein tryptophan 2-monooxygenase, a member of the L-amino oxidase family. Biochemistry. 2003;42:13826–13832. doi: 10.1021/bi035299n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sobrado P, Fitzpatrick PF. Identification of Tyr413 as an active site residue in the flavoprotein tryptophan 2-monooxygenase and analysis of its contribution to catalysis. Biochemistry. 2003;42:13833–13838. doi: 10.1021/bi035300i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ralph EC, Anderson MA, Cleland WW, Fitzpatrick PF. Mechanistic studies of the flavoenzyme tryptophan 2-monooxygenase: Deuterium and 15N kinetic isotope effects on alanine oxidation by an L-amino acid oxidase. Biochemistry. 2006;45:15844–15852. doi: 10.1021/bi061894o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiick DM, Phillips RS. Mechanistic deductions from multiple kinetic and solvent deuterium isotope effects and pH studies of pyridoxal phosphate dependent carbon-carbon lyases: Escherichia coli tryptophan indole-lyase. Biochemistry. 1988;27:7339–7344. doi: 10.1021/bi00419a024. [DOI] [PubMed] [Google Scholar]
- 24.Emanuele JJ, Jr., Heasley CJ, Fitzpatrick PF. Purification and characterization of the flavoprotein tryptophan 2-monooxygenase expressed at high levels in. E. coli, Arch. Biochem. Biophys. 1995;316:241–248. doi: 10.1006/abbi.1995.1034. [DOI] [PubMed] [Google Scholar]
- 25.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 26.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical nodel for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 27.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 28.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. 2010;D66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 32.Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34:W116–W118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer ELL, Eddy SR, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ida K, Kurabayashi M, Suguro M, Hiruma Y, Hikima T, Yamomoto M, Suzuki H. Structural basis of proteolytic activation of L-phenylalanine oxidase from Pseudomonas sp. P-501. J. Biol. Chem. 2008;283:16584–16590. doi: 10.1074/jbc.M800366200. [DOI] [PubMed] [Google Scholar]
- 36.Flashner MIS, Massey V. Purification and properties of L-lysine monooxygenase from Pseudomonas fluorescens. J. Biol. Chem. 1974;249:2579–2586. [PubMed] [Google Scholar]
- 37.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 38.Wierenga RK, Terpstra P, Hol WG. Prediction of the occurrence of the ADP-binding ßαß-fold in proteins, using an amino acid sequence fingerprint. J. Mol. Biol. 1986;187:101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]
- 39.Gadda G, Dangott LJ, Johnson WH, Jr., Whitman CP, Fitzpatrick PF. Characterization of 2-Oxo-3-pentynoate as an active-site-directed inactivator of flavoprotein oxidases: Identification of active-site peptides in tryptophan 2-monooxygenase. Biochemistry. 1999;38:5822–5828. doi: 10.1021/bi982777z. [DOI] [PubMed] [Google Scholar]
- 40.Pawelek PD, Cheah J, Coulombe R, Macheroux P, Ghisla S, Vrielink A. The structure of L-amino acid oxidase reveals the substrate trajectory into an enantiomerically conserved active site. EMBO J. 2000;19:4204–4215. doi: 10.1093/emboj/19.16.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li M, Binda C, Mattevi A, Edmondson DE. Functional role of the “aromatic cage” in human monoamine oxidase B: structures and catalytic properties of Tyr435 mutant proteins. Biochemistry. 2006;45:4775–4784. doi: 10.1021/bi051847g. [DOI] [PubMed] [Google Scholar]
- 42.Geha RM, Chen K, Wouters J, Ooms F, Shih JC. Analysis of conserved active site residues in monoamine oxidase A and B and their three-dimensional molecular modeling. J. Biol. Chem. 2002;277:17209–17216. doi: 10.1074/jbc.M110920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koyama H. Purification and characterization of a novel L-phenylalanine oxidase (deaminating and decarboxylating) from Pseudomonas sp. P-501. J. Biochem. 1982;92:1235–1240. doi: 10.1093/oxfordjournals.jbchem.a134041. [DOI] [PubMed] [Google Scholar]
- 44.Binda C, Coda A, Angelini R, Federico R, Ascenzi P, Mattevi A. A 30 Å long U-shaped catalytic tunnel in the crystal structure of polyamine oxidase. Structure. 1999;7:265–276. doi: 10.1016/s0969-2126(99)80037-9. [DOI] [PubMed] [Google Scholar]
- 45.Binda C, Li M, Hubalek F, Restelli N, Edmondson D, Mattevi A. Insights into the mode of inhibition of human mitochondrial monoamine oxidase B from high-resolution crystal structures. Proc. Natl. Acad. Sci. USA. 2003;100:9750–9755. doi: 10.1073/pnas.1633804100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Polticelli F, Basran J, Faso C, Cona A, Minervini G, Angelini R, Federico R, Scrutton NS, Tavladoraki P. Lys300 plays a major role in the catalytic mechanism of maize polyamine oxidase. Biochemistry. 2005;44:16108–16120. doi: 10.1021/bi050983i. [DOI] [PubMed] [Google Scholar]
- 47.Fiorillo A, Federico R, Polticelli F, Boffi A, Mazzei F, Di Fusco M, Ilari A, Tavladoraki P. The structure of maize polyamine oxidase K300M mutant in complex with the natural substrates provides a snapshot of the catalytic mechanism of polyamine oxidation. FEBS Journal. 2011;278:809–821. doi: 10.1111/j.1742-4658.2010.08000.x. [DOI] [PubMed] [Google Scholar]
- 48.Henderson Pozzi M, Gawandi V, Fitzpatrick PF. pH Dependence of a mammalian polyamine oxidase: Insights into substrate specificity and the role of lysine 315. Biochemistry. 2009;48:1508–1516. doi: 10.1021/bi802227m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Henderson Pozzi M, Fitzpatrick PF. A lysine conserved in the monoamine oxidase family is involved in oxidation of the reduced flavin in mouse polyamine oxidase. Arch. Biochem. Biophys. 2010;498:83–88. doi: 10.1016/j.abb.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stavropoulos P, Blobel G, Hoelz A. Crystal structure and mechanism of human lysine-specific demethylase-1. Nat. Struct. Mol. Biol. 2006;13:626–632. doi: 10.1038/nsmb1113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.