Abstract
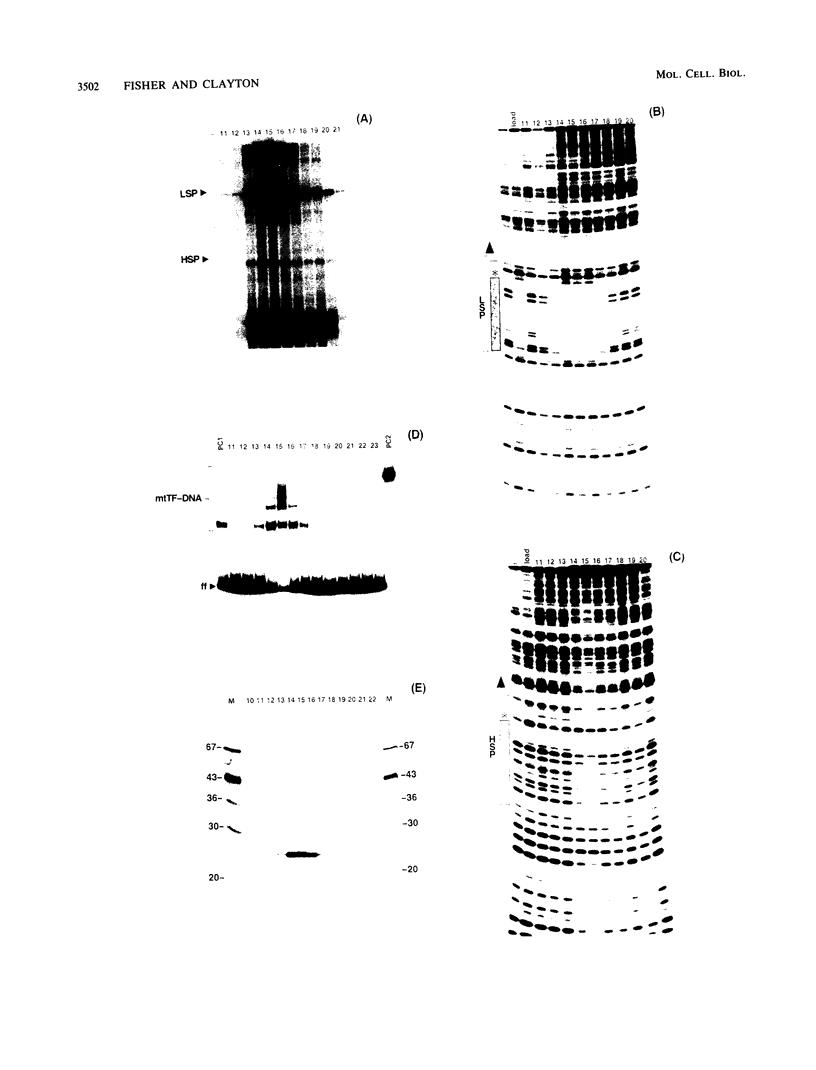
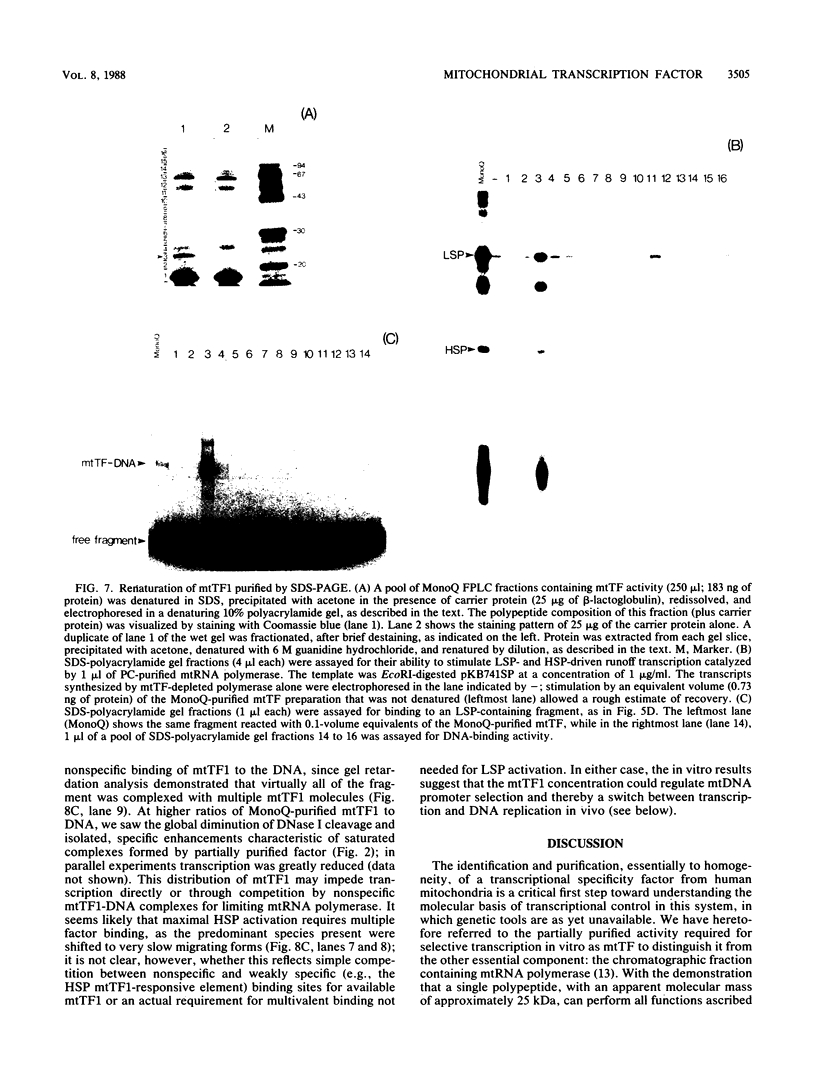
We purified to near homogeneity a transcription factor from human KB cell mitochondria. This factor, designated mitochondrial transcription factor 1 (mtTF1), is required for the in vitro recognition of both major promoters of human mitochondrial DNA by the homologous mitochondrial RNA polymerase. Furthermore, it has been shown to bind upstream regulatory elements of the two major promoters. After separation from RNA polymerase by phosphocellulose chromatography, mtTF1 was chromatographed on a MonoQ anion-exchange fast-performance liquid chromatography column. Analysis of mtTF1-containing fractions by sodium dodecyl sulfate-polyacrylamide gel electrophoresis revealed a single major polypeptide with an Mr of approximately 25,000. Centrifugation in analytical glycerol gradients indicated a sedimentation coefficient of approximately 2.5 S, consistent with a monomeric 25-kilodalton protein. Finally, when the 25-kilodalton polypeptide was excised from a stained sodium dodecyl sulfate-polyacrylamide gel and allowed to renature, it regained DNA-binding and transcriptional stimulatory activities at both promoters. Although mtTF1 is the only mitochondrial DNA-binding transcription factor to be purified and characterized, its properties, such as a high affinity for random DNA and a weak specificity for one of its target sequences, may typify this class of regulatory proteins.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981 Apr 9;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Baldacci G., Chérif-Zahar B., Bernardi G. The initiation of DNA replication in the mitochondrial genome of yeast. EMBO J. 1984 Sep;3(9):2115–2120. doi: 10.1002/j.1460-2075.1984.tb02099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen D. F., Applegate E. F., Yoza B. K. Identification of a promoter for transcription of the heavy strand of human mtDNA: in vitro transcription and deletion mutagenesis. Cell. 1984 Apr;36(4):1105–1113. doi: 10.1016/0092-8674(84)90061-8. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Yoza B. K. Accurate in vitro transcription of Xenopus laevis mitochondrial DNA from two bidirectional promoters. Mol Cell Biol. 1986 Jul;6(7):2543–2550. doi: 10.1128/mcb.6.7.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen D., Clayton D. A. The number of mitochondrial deoxyribonucleic acid genomes in mouse L and human HeLa cells. Quantitative isolation of mitochondrial deoxyribonucleic acid. J Biol Chem. 1974 Dec 25;249(24):7991–7995. [PubMed] [Google Scholar]
- Carthew R. W., Chodosh L. A., Sharp P. A. An RNA polymerase II transcription factor binds to an upstream element in the adenovirus major late promoter. Cell. 1985 Dec;43(2 Pt 1):439–448. doi: 10.1016/0092-8674(85)90174-6. [DOI] [PubMed] [Google Scholar]
- Chang D. D., Clayton D. A. Precise identification of individual promoters for transcription of each strand of human mitochondrial DNA. Cell. 1984 Mar;36(3):635–643. doi: 10.1016/0092-8674(84)90343-x. [DOI] [PubMed] [Google Scholar]
- Chang D. D., Clayton D. A. Priming of human mitochondrial DNA replication occurs at the light-strand promoter. Proc Natl Acad Sci U S A. 1985 Jan;82(2):351–355. doi: 10.1073/pnas.82.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D. D., Hauswirth W. W., Clayton D. A. Replication priming and transcription initiate from precisely the same site in mouse mitochondrial DNA. EMBO J. 1985 Jun;4(6):1559–1567. doi: 10.1002/j.1460-2075.1985.tb03817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodosh L. A., Carthew R. W., Sharp P. A. A single polypeptide possesses the binding and transcription activities of the adenovirus major late transcription factor. Mol Cell Biol. 1986 Dec;6(12):4723–4733. doi: 10.1128/mcb.6.12.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton D. A. Transcription of the mammalian mitochondrial genome. Annu Rev Biochem. 1984;53:573–594. doi: 10.1146/annurev.bi.53.070184.003041. [DOI] [PubMed] [Google Scholar]
- Fisher R. P., Clayton D. A. A transcription factor required for promoter recognition by human mitochondrial RNA polymerase. Accurate initiation at the heavy- and light-strand promoters dissected and reconstituted in vitro. J Biol Chem. 1985 Sep 15;260(20):11330–11338. [PubMed] [Google Scholar]
- Fisher R. P., Topper J. N., Clayton D. A. Promoter selection in human mitochondria involves binding of a transcription factor to orientation-independent upstream regulatory elements. Cell. 1987 Jul 17;50(2):247–258. doi: 10.1016/0092-8674(87)90220-0. [DOI] [PubMed] [Google Scholar]
- Fried M. G., Crothers D. M. Kinetics and mechanism in the reaction of gene regulatory proteins with DNA. J Mol Biol. 1984 Jan 25;172(3):263–282. doi: 10.1016/s0022-2836(84)80026-1. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand R., Attardi G. Synthesis and turnover of mitochondrial ribonucleic acid in HeLa cells: the mature ribosomal and messenger ribonucleic acid species are metabolically unstable. Mol Cell Biol. 1981 Jun;1(6):497–511. doi: 10.1128/mcb.1.6.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman M. Z., Wilson R. N., Weinberg R. A. Multiple protein-binding sites in the 5'-flanking region regulate c-fos expression. Mol Cell Biol. 1986 Dec;6(12):4305–4316. doi: 10.1128/mcb.6.12.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager D. A., Burgess R. R. Elution of proteins from sodium dodecyl sulfate-polyacrylamide gels, removal of sodium dodecyl sulfate, and renaturation of enzymatic activity: results with sigma subunit of Escherichia coli RNA polymerase, wheat germ DNA topoisomerase, and other enzymes. Anal Biochem. 1980 Nov 15;109(1):76–86. doi: 10.1016/0003-2697(80)90013-5. [DOI] [PubMed] [Google Scholar]
- Hixson J. E., Clayton D. A. Initiation of transcription from each of the two human mitochondrial promoters requires unique nucleotides at the transcriptional start sites. Proc Natl Acad Sci U S A. 1985 May;82(9):2660–2664. doi: 10.1073/pnas.82.9.2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack W. E., Terry B. J., Modrich P. Involvement of outside DNA sequences in the major kinetic path by which EcoRI endonuclease locates and leaves its recognition sequence. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4010–4014. doi: 10.1073/pnas.79.13.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Levens D., Howley P. M. Novel method for identifying sequence-specific DNA-binding proteins. Mol Cell Biol. 1985 Sep;5(9):2307–2315. doi: 10.1128/mcb.5.9.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Prywes R., Roeder R. G. Inducible binding of a factor to the c-fos enhancer. Cell. 1986 Dec 5;47(5):777–784. doi: 10.1016/0092-8674(86)90520-9. [DOI] [PubMed] [Google Scholar]
- Sawadogo M., Roeder R. G. Interaction of a gene-specific transcription factor with the adenovirus major late promoter upstream of the TATA box region. Cell. 1985 Nov;43(1):165–175. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- Schaffner W., Weissmann C. A rapid, sensitive, and specific method for the determination of protein in dilute solution. Anal Biochem. 1973 Dec;56(2):502–514. doi: 10.1016/0003-2697(73)90217-0. [DOI] [PubMed] [Google Scholar]
- Schinkel A. H., Groot Koerkamp M. J., Van der Horst G. T., Touw E. P., Osinga K. A., Van der Bliek A. M., Veeneman G. H., Van Boom J. H., Tabak H. F. Characterization of the promoter of the large ribosomal RNA gene in yeast mitochondria and separation of mitochondrial RNA polymerase into two different functional components. EMBO J. 1986 May;5(5):1041–1047. doi: 10.1002/j.1460-2075.1986.tb04320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinkel A. H., Koerkamp M. J., Touw E. P., Tabak H. F. Specificity factor of yeast mitochondrial RNA polymerase. Purification and interaction with core RNA polymerase. J Biol Chem. 1987 Sep 15;262(26):12785–12791. [PubMed] [Google Scholar]
- Treisman R. Identification of a protein-binding site that mediates transcriptional response of the c-fos gene to serum factors. Cell. 1986 Aug 15;46(4):567–574. doi: 10.1016/0092-8674(86)90882-2. [DOI] [PubMed] [Google Scholar]
- Walberg M. W., Clayton D. A. In vitro transcription of human mitochondrial DNA. Identification of specific light strand transcripts from the displacement loop region. J Biol Chem. 1983 Jan 25;258(2):1268–1275. [PubMed] [Google Scholar]
- Walberg M. W., Clayton D. A. Sequence and properties of the human KB cell and mouse L cell D-loop regions of mitochondrial DNA. Nucleic Acids Res. 1981 Oct 24;9(20):5411–5421. doi: 10.1093/nar/9.20.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. C., Revzin A., Butler A. P., von Hippel P. H. Binding of E.coli lac repressor to non-operator DNA. Nucleic Acids Res. 1977;4(5):1579–1593. doi: 10.1093/nar/4.5.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkley C. S., Keller M. J., Jaehning J. A. A multicomponent mitochondrial RNA polymerase from Saccharomyces cerevisiae. J Biol Chem. 1985 Nov 15;260(26):14214–14223. [PubMed] [Google Scholar]
- Winter R. B., Berg O. G., von Hippel P. H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 3. The Escherichia coli lac repressor--operator interaction: kinetic measurements and conclusions. Biochemistry. 1981 Nov 24;20(24):6961–6977. doi: 10.1021/bi00527a030. [DOI] [PubMed] [Google Scholar]
- Wray W., Boulikas T., Wray V. P., Hancock R. Silver staining of proteins in polyacrylamide gels. Anal Biochem. 1981 Nov 15;118(1):197–203. doi: 10.1016/0003-2697(81)90179-2. [DOI] [PubMed] [Google Scholar]
- deHaseth P. L., Lohman T. M., Record M. T., Jr Nonspecific interaction of lac repressor with DNA: an association reaction driven by counterion release. Biochemistry. 1977 Nov 1;16(22):4783–4790. doi: 10.1021/bi00641a004. [DOI] [PubMed] [Google Scholar]









