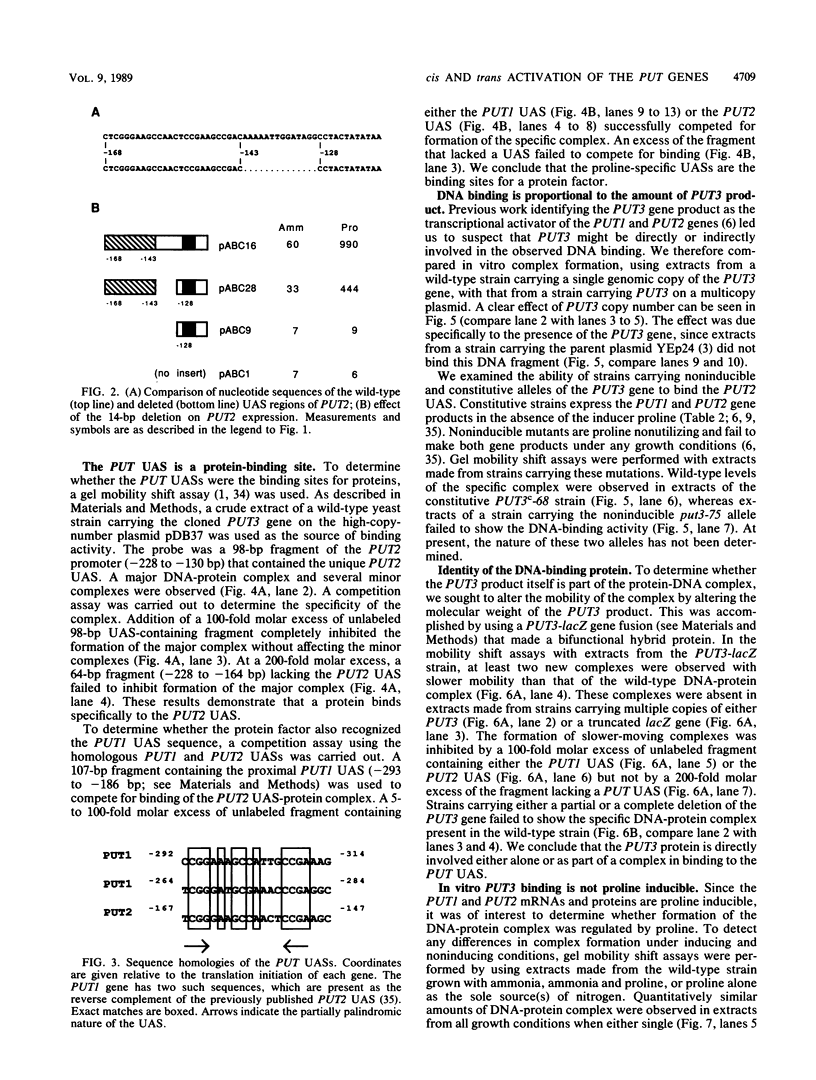
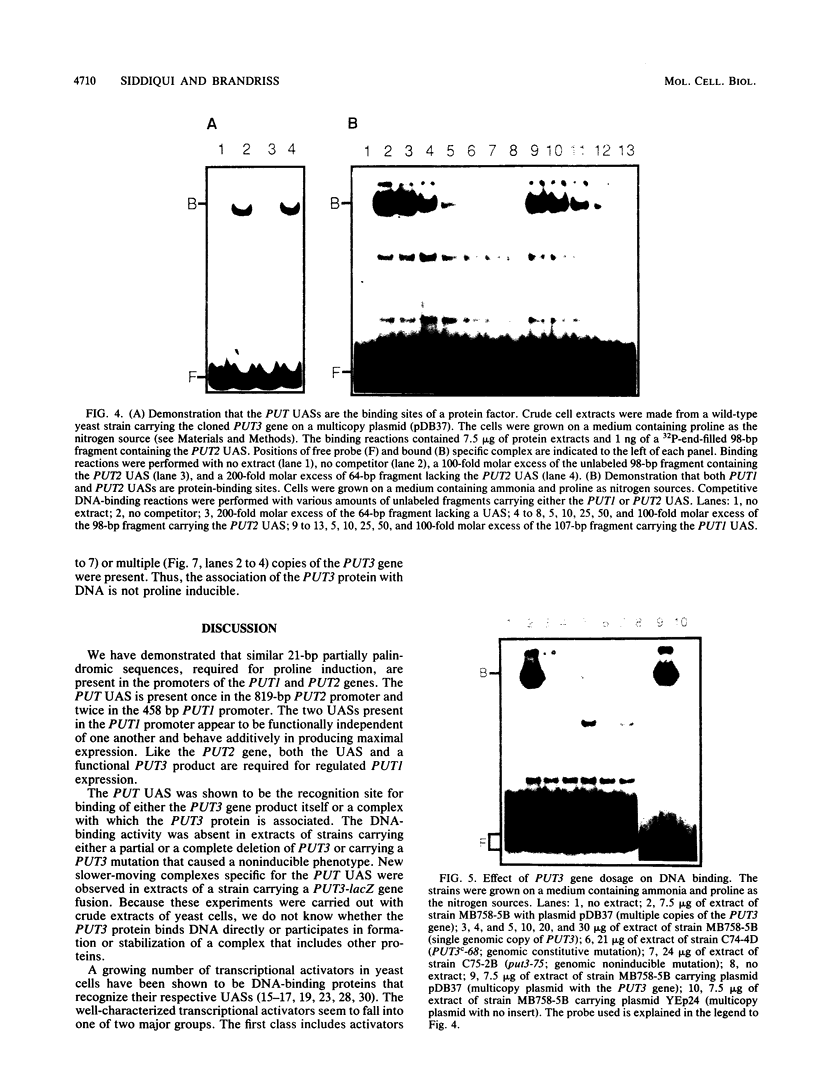
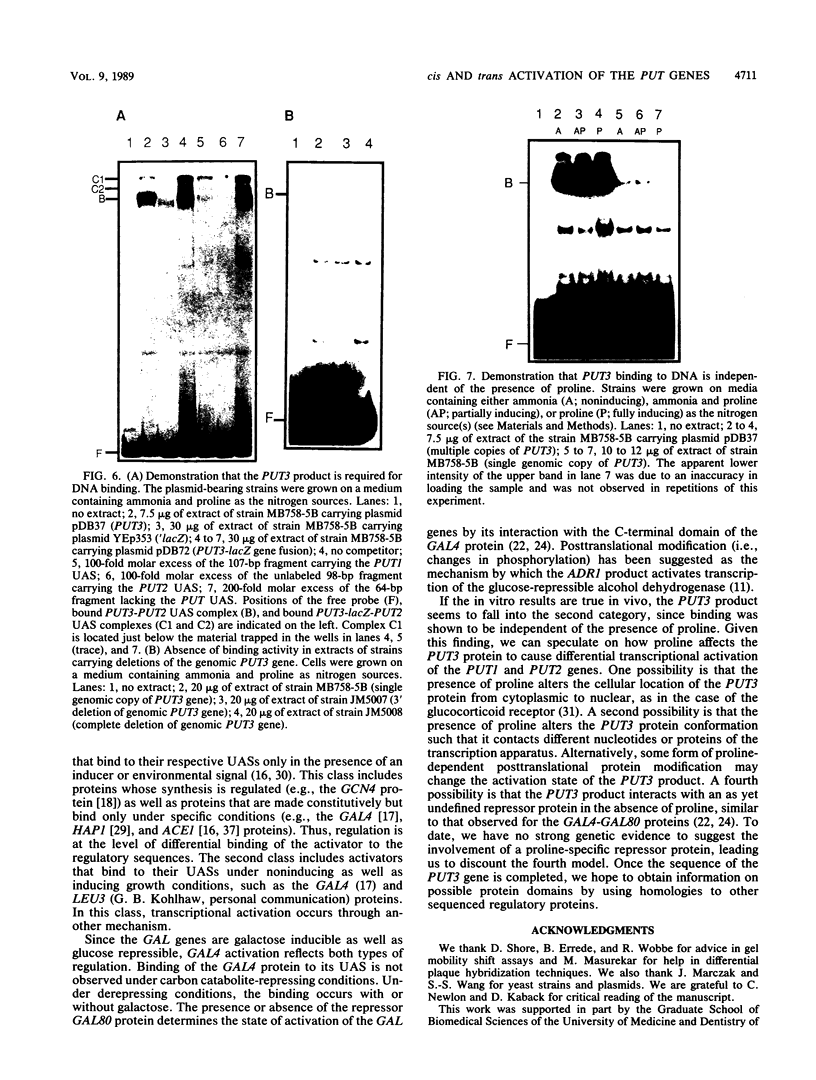
Abstract
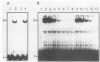
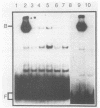
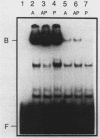
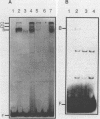
The PUT1 and PUT2 genes encoding the enzymes of the proline utilization pathway of Saccharomyces cerevisiae are induced by proline and activated by the product of the PUT3 gene. Two upstream activation sequences (UASs) in the PUT1 promoter were identified by homology to the PUT2 UAS. Deletion analysis of the two PUT1 UASs showed that they were functionally independent and additive in producing maximal levels of gene expression. The consensus PUT UAS is a 21-base-pair partially palindromic sequence required in vivo for induction of both genes. The results of a gel mobility shift assay demonstrated that the proline-specific UAS is the binding site of a protein factor. In vitro complex formation was observed in crude extracts of yeast strains carrying either a single genomic copy of the PUT3 gene or the cloned PUT3 gene on a 2 microns plasmid, and the binding was dosage dependent. DNA-binding activity was not observed in extracts of strains carrying either a put3 mutation that caused a noninducible (Put-) phenotype or a deletion of the gene. Wild-type levels of complex formation were observed in an extract of a strain carrying an allele of PUT3 that resulted in a constitutive (Put+) phenotype. Extracts from a strain carrying a PUT3-lacZ gene fusion formed two complexes of slower mobility than the wild-type complex. We conclude that the PUT3 product is either a DNA-binding protein or part of a DNA-binding complex that recognizes the UASs of both PUT1 and PUT2. Binding was observed in extracts of a strain grown in the presence or absence of proline, demonstrating the constitutive nature of the DNA-protein interaction.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arcangioli B., Lescure B. Identification of proteins involved in the regulation of yeast iso- 1-cytochrome C expression by oxygen. EMBO J. 1985 Oct;4(10):2627–2633. doi: 10.1002/j.1460-2075.1985.tb03980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D., Falco S. C., Stewart S. E., Brennan M., Scherer S., Stinchcomb D. T., Struhl K., Davis R. W. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979 Dec;8(1):17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Brandriss M. C. Evidence for positive regulation of the proline utilization pathway in Saccharomyces cerevisiae. Genetics. 1987 Nov;117(3):429–435. doi: 10.1093/genetics/117.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandriss M. C., Krzywicki K. A. Amino-terminal fragments of delta 1-pyrroline-5-carboxylate dehydrogenase direct beta-galactosidase to the mitochondrial matrix in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Oct;6(10):3502–3512. doi: 10.1128/mcb.6.10.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandriss M. C., Magasanik B. Genetics and physiology of proline utilization in Saccharomyces cerevisiae: enzyme induction by proline. J Bacteriol. 1979 Nov;140(2):498–503. doi: 10.1128/jb.140.2.498-503.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandriss M. C., Magasanik B. Genetics and physiology of proline utilization in Saccharomyces cerevisiae: mutation causing constitutive enzyme expression. J Bacteriol. 1979 Nov;140(2):504–507. doi: 10.1128/jb.140.2.504-507.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandriss M. C., Magasanik B. Subcellular compartmentation in control of converging pathways for proline and arginine metabolism in Saccharomyces cerevisiae. J Bacteriol. 1981 Mar;145(3):1359–1364. doi: 10.1128/jb.145.3.1359-1364.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandriss M. C. Proline utilization in Saccharomyces cerevisiae: analysis of the cloned PUT2 gene. Mol Cell Biol. 1983 Oct;3(10):1846–1856. doi: 10.1128/mcb.3.10.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry J. R., Johnson T. R., Dollard C., Shuster J. R., Denis C. L. Cyclic AMP-dependent protein kinase phosphorylates and inactivates the yeast transcriptional activator ADR1. Cell. 1989 Feb 10;56(3):409–419. doi: 10.1016/0092-8674(89)90244-4. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Company M., Errede B. A Ty1 cell-type-specific regulatory sequence is a recognition element for a constitutive binding factor. Mol Cell Biol. 1988 Dec;8(12):5299–5309. doi: 10.1128/mcb.8.12.5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue T. F., Daves R. S., Lucchini G., Fink G. R. A short nucleotide sequence required for regulation of HIS4 by the general control system of yeast. Cell. 1983 Jan;32(1):89–98. doi: 10.1016/0092-8674(83)90499-3. [DOI] [PubMed] [Google Scholar]
- Friden P., Schimmel P. LEU3 of Saccharomyces cerevisiae encodes a factor for control of RNA levels of a group of leucine-specific genes. Mol Cell Biol. 1987 Aug;7(8):2708–2717. doi: 10.1128/mcb.7.8.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürst P., Hu S., Hackett R., Hamer D. Copper activates metallothionein gene transcription by altering the conformation of a specific DNA binding protein. Cell. 1988 Nov 18;55(4):705–717. doi: 10.1016/0092-8674(88)90229-2. [DOI] [PubMed] [Google Scholar]
- Giniger E., Varnum S. M., Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985 Apr;40(4):767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G. Evidence for translational regulation of the activator of general amino acid control in yeast. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6442–6446. doi: 10.1073/pnas.81.20.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. GCN4 protein, synthesized in vitro, binds HIS3 regulatory sequences: implications for general control of amino acid biosynthetic genes in yeast. Cell. 1985 Nov;43(1):177–188. doi: 10.1016/0092-8674(85)90022-4. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis E. E., Hagen D. C., Sprague G. F., Jr Identification of a DNA segment that is necessary and sufficient for alpha-specific gene control in Saccharomyces cerevisiae: implications for regulation of alpha-specific and a-specific genes. Mol Cell Biol. 1988 Jan;8(1):309–320. doi: 10.1128/mcb.8.1.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston S. A., Salmeron J. M., Jr, Dincher S. S. Interaction of positive and negative regulatory proteins in the galactose regulon of yeast. Cell. 1987 Jul 3;50(1):143–146. doi: 10.1016/0092-8674(87)90671-4. [DOI] [PubMed] [Google Scholar]
- Keegan L., Gill G., Ptashne M. Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein. Science. 1986 Feb 14;231(4739):699–704. doi: 10.1126/science.3080805. [DOI] [PubMed] [Google Scholar]
- Ma J., Ptashne M. The carboxy-terminal 30 amino acids of GAL4 are recognized by GAL80. Cell. 1987 Jul 3;50(1):137–142. doi: 10.1016/0092-8674(87)90670-2. [DOI] [PubMed] [Google Scholar]
- Myers A. M., Tzagoloff A., Kinney D. M., Lusty C. J. Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene. 1986;45(3):299–310. doi: 10.1016/0378-1119(86)90028-4. [DOI] [PubMed] [Google Scholar]
- Olesen J., Hahn S., Guarente L. Yeast HAP2 and HAP3 activators both bind to the CYC1 upstream activation site, UAS2, in an interdependent manner. Cell. 1987 Dec 24;51(6):953–961. doi: 10.1016/0092-8674(87)90582-4. [DOI] [PubMed] [Google Scholar]
- Pfeifer K., Arcangioli B., Guarente L. Yeast HAP1 activator competes with the factor RC2 for binding to the upstream activation site UAS1 of the CYC1 gene. Cell. 1987 Apr 10;49(1):9–18. doi: 10.1016/0092-8674(87)90750-1. [DOI] [PubMed] [Google Scholar]
- Pfeifer K., Kim K. S., Kogan S., Guarente L. Functional dissection and sequence of yeast HAP1 activator. Cell. 1989 Jan 27;56(2):291–301. doi: 10.1016/0092-8674(89)90903-3. [DOI] [PubMed] [Google Scholar]
- Picard D., Yamamoto K. R. Two signals mediate hormone-dependent nuclear localization of the glucocorticoid receptor. EMBO J. 1987 Nov;6(11):3333–3340. doi: 10.1002/j.1460-2075.1987.tb02654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph H., Hinnen A. The yeast PHO5 promoter: phosphate-control elements and sequences mediating mRNA start-site selection. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1340–1344. doi: 10.1073/pnas.84.5.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore D., Stillman D. J., Brand A. H., Nasmyth K. A. Identification of silencer binding proteins from yeast: possible roles in SIR control and DNA replication. EMBO J. 1987 Feb;6(2):461–467. doi: 10.1002/j.1460-2075.1987.tb04776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui A. H., Brandriss M. C. A regulatory region responsible for proline-specific induction of the yeast PUT2 gene is adjacent to its TATA box. Mol Cell Biol. 1988 Nov;8(11):4634–4641. doi: 10.1128/mcb.8.11.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczypka M. S., Thiele D. J. A cysteine-rich nuclear protein activates yeast metallothionein gene transcription. Mol Cell Biol. 1989 Feb;9(2):421–429. doi: 10.1128/mcb.9.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. W., Ott J., Eckstein F. The rapid generation of oligonucleotide-directed mutations at high frequency using phosphorothioate-modified DNA. Nucleic Acids Res. 1985 Dec 20;13(24):8765–8785. doi: 10.1093/nar/13.24.8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. S., Brandriss M. C. Proline utilization in Saccharomyces cerevisiae: analysis of the cloned PUT1 gene. Mol Cell Biol. 1986 Jul;6(7):2638–2645. doi: 10.1128/mcb.6.7.2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. S., Brandriss M. C. Proline utilization in Saccharomyces cerevisiae: sequence, regulation, and mitochondrial localization of the PUT1 gene product. Mol Cell Biol. 1987 Dec;7(12):4431–4440. doi: 10.1128/mcb.7.12.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]