Abstract
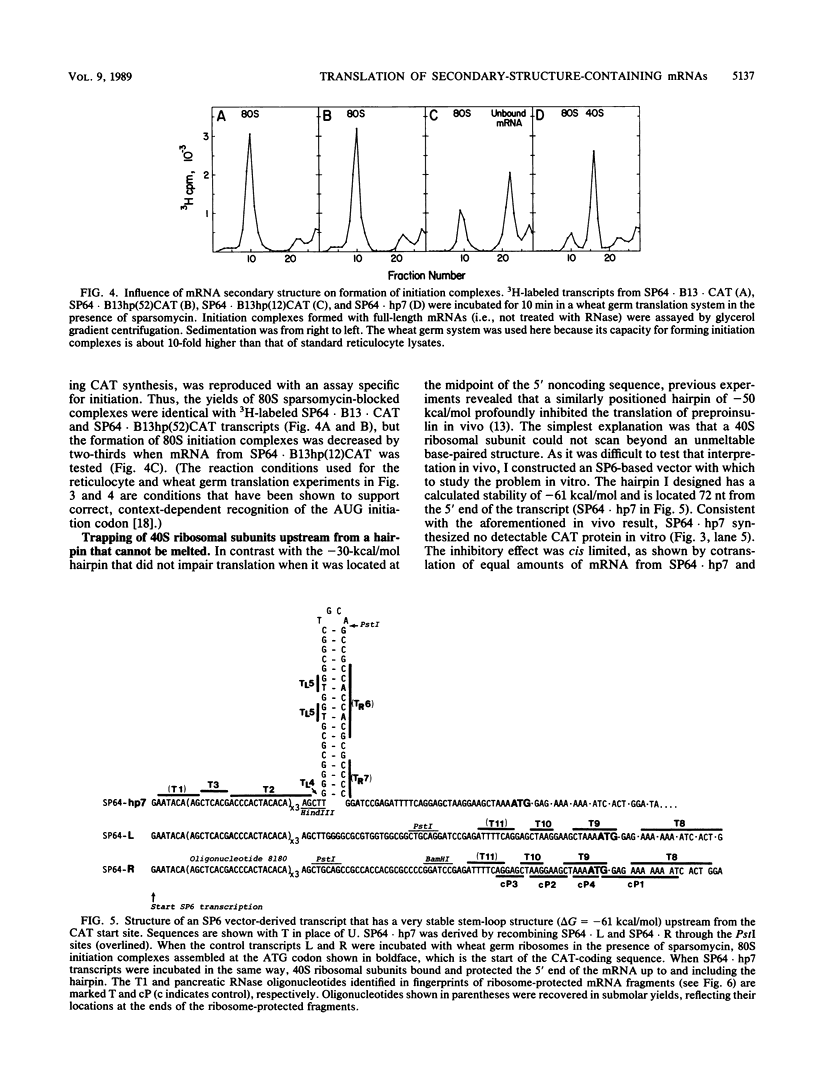
This paper describes in vitro experiments with two types of intramolecular duplex structures that inhibit translation in cis by preventing the formation of an initiation complex or by causing the complex to be abortive. One stem-loop structure (delta G = -30 kcal/mol) prevented mRNA from engaging 40S subunits when the hairpin occurred 12 nucleotides (nt) from the cap but had no deleterious effect when it was repositioned 52 nt from the cap. This result confirms prior in vivo evidence that the 40S subunit-factor complex, once bound to mRNA, has considerable ability to penetrate secondary structure. Consequently, translation is most sensitive to secondary structure at the entry site for ribosomes, i.e., the 5' end of the mRNA. The second stem-loop structure (hp7; delta G = -61 kcal/mol, located 72 nt from the cap) was too stable to be unwound by 40S ribosomes, hp7 did not prevent a 40S ribosomal subunit from binding but caused the 40S subunit to stall on the 5' side of the hairpin, exactly as the scanning model predicts. Control experiments revealed that 80S elongating ribosomes could disrupt duplex structures, such as hp7, that were too stable to be penetrated by the scanning 40S ribosome-factor complex. A third type of base-paired structure shown to inhibit translation in vivo involves a long-range interaction between the 5' and 3' noncoding sequences.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bornstein P., McKay J., Devarayalu S., Cook S. C. A highly conserved, 5' untranslated, inverted repeat sequence is ineffective in translational control of the alpha 1(I) collagen gene. Nucleic Acids Res. 1988 Oct 25;16(20):9721–9736. doi: 10.1093/nar/16.20.9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bram R. J., Young R. A., Steitz J. A. The ribonuclease III site flanking 23S sequences in the 30S ribosomal precursor RNA of E. coli. Cell. 1980 Feb;19(2):393–401. doi: 10.1016/0092-8674(80)90513-9. [DOI] [PubMed] [Google Scholar]
- Carter-Muenchau P., Wolf R. E., Jr Growth-rate-dependent regulation of 6-phosphogluconate dehydrogenase level mediated by an anti-Shine-Dalgarno sequence located within the Escherichia coli gnd structural gene. Proc Natl Acad Sci U S A. 1989 Feb;86(4):1138–1142. doi: 10.1073/pnas.86.4.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen A. K., Kahn L. E., Bourne C. M. Circular polysomes predominate on the rough endoplasmic reticulum of somatotropes and mammotropes in the rat anterior pituitary. Am J Anat. 1987 Jan;178(1):1–10. doi: 10.1002/aja.1001780102. [DOI] [PubMed] [Google Scholar]
- Cigan A. M., Feng L., Donahue T. F. tRNAi(met) functions in directing the scanning ribosome to the start site of translation. Science. 1988 Oct 7;242(4875):93–97. doi: 10.1126/science.3051379. [DOI] [PubMed] [Google Scholar]
- Donahue T. F., Cigan A. M., Pabich E. K., Valavicius B. C. Mutations at a Zn(II) finger motif in the yeast eIF-2 beta gene alter ribosomal start-site selection during the scanning process. Cell. 1988 Aug 26;54(5):621–632. doi: 10.1016/s0092-8674(88)80006-0. [DOI] [PubMed] [Google Scholar]
- Edery I., Petryshyn R., Sonenberg N. Activation of double-stranded RNA-dependent kinase (dsl) by the TAR region of HIV-1 mRNA: a novel translational control mechanism. Cell. 1989 Jan 27;56(2):303–312. doi: 10.1016/0092-8674(89)90904-5. [DOI] [PubMed] [Google Scholar]
- Hsu M. T., Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979 Jul 26;280(5720):339–340. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- Hsu M. T., Parvin J. D., Gupta S., Krystal M., Palese P. Genomic RNAs of influenza viruses are held in a circular conformation in virions and in infected cells by a terminal panhandle. Proc Natl Acad Sci U S A. 1987 Nov;84(22):8140–8144. doi: 10.1073/pnas.84.22.8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987 Oct 26;15(20):8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J Mol Biol. 1987 Aug 20;196(4):947–950. doi: 10.1016/0022-2836(87)90418-9. [DOI] [PubMed] [Google Scholar]
- Kozak M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc Natl Acad Sci U S A. 1986 May;83(9):2850–2854. doi: 10.1073/pnas.83.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Leader length and secondary structure modulate mRNA function under conditions of stress. Mol Cell Biol. 1988 Jul;8(7):2737–2744. doi: 10.1128/mcb.8.7.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Point mutations close to the AUG initiator codon affect the efficiency of translation of rat preproinsulin in vivo. Nature. 1984 Mar 15;308(5956):241–246. doi: 10.1038/308241a0. [DOI] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986 Jan 31;44(2):283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Kozak M., Shatkin A. J. Characterization of ribosome-protected fragments from reovirus messenger RNA. J Biol Chem. 1976 Jul 25;251(14):4259–4266. [PubMed] [Google Scholar]
- Kozak M., Shatkin A. J. Migration of 40 S ribosomal subunits on messenger RNA in the presence of edeine. J Biol Chem. 1978 Sep 25;253(18):6568–6577. [PubMed] [Google Scholar]
- Kozak M., Shatkin A. J. Sequences and properties of two ribosome binding sites from the small size class of reovirus messenger RNA. J Biol Chem. 1977 Oct 10;252(19):6895–6908. [PubMed] [Google Scholar]
- Kozak M., Shatkin A. J. Sequences of two 5'-terminal ribosome-protected fragments from reovirus messenger RNAs. J Mol Biol. 1977 May 5;112(1):75–96. doi: 10.1016/s0022-2836(77)80157-5. [DOI] [PubMed] [Google Scholar]
- Kozak M. The scanning model for translation: an update. J Cell Biol. 1989 Feb;108(2):229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Translation of insulin-related polypeptides from messenger RNAs with tandemly reiterated copies of the ribosome binding site. Cell. 1983 Oct;34(3):971–978. doi: 10.1016/0092-8674(83)90554-8. [DOI] [PubMed] [Google Scholar]
- Kruppa J., Clemens M. J. Differential kinetics of changes in the state of phosphorylation of ribosomal protein S6 and in the rate of protein synthesis in MPC 11 cells during tonicity shifts. EMBO J. 1984 Jan;3(1):95–100. doi: 10.1002/j.1460-2075.1984.tb01767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A. Injected anti-sense RNAs specifically block messenger RNA translation in vivo. Proc Natl Acad Sci U S A. 1985 Jan;82(1):144–148. doi: 10.1073/pnas.82.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen C. Long-range translational coupling in the rplJL-rpoBC operon of Escherichia coli. J Mol Biol. 1989 Mar 20;206(2):323–332. doi: 10.1016/0022-2836(89)90482-8. [DOI] [PubMed] [Google Scholar]
- Rao C. D., Pech M., Robbins K. C., Aaronson S. A. The 5' untranslated sequence of the c-sis/platelet-derived growth factor 2 transcript is a potent translational inhibitor. Mol Cell Biol. 1988 Jan;8(1):284–292. doi: 10.1128/mcb.8.1.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault T. A., Hentze M. W., Caughman S. W., Harford J. B., Klausner R. D. Binding of a cytosolic protein to the iron-responsive element of human ferritin messenger RNA. Science. 1988 Sep 2;241(4870):1207–1210. doi: 10.1126/science.3413484. [DOI] [PubMed] [Google Scholar]
- Saborio J. L., Pong S. S., Koch G. Selective and reversible inhibition of initiation of protein synthesis in mammalian cells. J Mol Biol. 1974 May 15;85(2):195–211. doi: 10.1016/0022-2836(74)90360-x. [DOI] [PubMed] [Google Scholar]
- Shatkin A. J. Capping of eucaryotic mRNAs. Cell. 1976 Dec;9(4 Pt 2):645–653. doi: 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- Shatkin A. J. mRNA cap binding proteins: essential factors for initiating translation. Cell. 1985 Feb;40(2):223–224. doi: 10.1016/0092-8674(85)90132-1. [DOI] [PubMed] [Google Scholar]
- Soares M. B., Schon E., Henderson A., Karathanasis S. K., Cate R., Zeitlin S., Chirgwin J., Efstratiadis A. RNA-mediated gene duplication: the rat preproinsulin I gene is a functional retroposon. Mol Cell Biol. 1985 Aug;5(8):2090–2103. doi: 10.1128/mcb.5.8.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soe L. H., Shieh C. K., Baker S. C., Chang M. F., Lai M. M. Sequence and translation of the murine coronavirus 5'-end genomic RNA reveals the N-terminal structure of the putative RNA polymerase. J Virol. 1987 Dec;61(12):3968–3976. doi: 10.1128/jvi.61.12.3968-3976.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spena A., Krause E., Dobberstein B. Translation efficiency of zein mRNA is reduced by hybrid formation between the 5'- and 3'-untranslated region. EMBO J. 1985 Sep;4(9):2153–2158. doi: 10.1002/j.1460-2075.1985.tb03909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Walder R. Y., Walder J. A. Role of RNase H in hybrid-arrested translation by antisense oligonucleotides. Proc Natl Acad Sci U S A. 1988 Jul;85(14):5011–5015. doi: 10.1073/pnas.85.14.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yager D. R., Coen D. M. Analysis of the transcript of the herpes simplex virus DNA polymerase gene provides evidence that polymerase expression is inefficient at the level of translation. J Virol. 1988 Jun;62(6):2007–2015. doi: 10.1128/jvi.62.6.2007-2015.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]