Background: PfEBA-140 recognizes sialic acid on its receptor glycophorin C during erythrocyte invasion.
Results: PfEBA-140 contains two sialic acid-binding pockets distinct from other sialic acid-binding proteins and with divergent roles in receptor recognition.
Conclusion: The glycan-binding pockets define receptor recognition, specificity, and putative switching.
Significance: This study provides the first detailed molecular view of receptor recognition by a critical P. falciparum invasion ligand.
Keywords: Adhesion, Carbohydrate Glycoprotein, Erythrocyte, Infectious Diseases, Invasion, Malaria, Parasite, Receptor Structure-Function, Sialic Acid, X-ray Crystallography
Abstract
Plasmodium falciparum erythrocyte invasion is dependent on high affinity recognition of sialic acid on cell surface receptors. The erythrocyte binding-like (EBL) family of invasion ligands mediates recognition of sialic acid on erythrocyte glycoproteins. Erythrocyte-binding antigen-140 (PfEBA-140/BAEBL) is a critical EBL ligand that binds sialic acid on its receptor glycophorin C. We present here the crystal structure of the two-domain receptor-binding region of PfEBA-140 in complex with a glycan containing sialic acid. The structure identifies two glycan-binding pockets unique to PfEBA-140 and not shared by other EBL ligands. Specific molecular interactions that enable receptor engagement are identified and reveal that the glycan binding mode is distinct from that of apicomplexan and viral cell surface recognition ligands as well as host immune factors that bind sialic acid. Erythrocyte binding experiments elucidated essential glycan contact residues and identified divergent functional roles for each receptor-binding site. One of four polymorphisms proposed to affect receptor binding was localized to a glycan-binding site, providing a structural basis for altered erythrocyte engagement. The studies described here provide the first full description of sialic acid-dependent molecular interactions at the P. falciparum erythrocyte invasion interface and define a framework for development of PfEBA-140-based therapeutics, vaccines, and diagnostics assessing vaccine efficacy and natural immunity to infection.
Introduction
Plasmodium falciparum erythrocyte invasion requires targeted recognition of cell surface receptors by merozoite ligands (1, 2). High affinity binding to erythrocyte receptors is critical to the invasion process and is mediated by the erythrocyte binding-like (EBL)3 family of proteins (3). In P. falciparum, the EBL family is composed of four known functional membrane-embedded ligands that each target a specific erythrocyte receptor in a sialic acid-dependent manner (3). Receptor engagement by the merozoite allows the formation of an irreversible tight junction with the host membrane and subsequent formation of the parasitophorous vacuole (4, 5). Binding interactions mediated by the EBL family are vital to the survival of the parasite and are thus targets of therapeutics and vaccines (6, 7).
Each member of the P. falciparum EBL family contains an extracellular cysteine-rich region composed of two Duffy binding-like (DBL) domains designated F1 and F2 (8). These two domains comprise the minimal binding region of the P. falciparum EBL ligands and have been designated region II (RII). P. falciparum erythrocyte-binding antigen 140 (PfEBA-140/BAEBL) is a member of the EBL family that binds erythrocytes via its cell surface receptor glycophorin C (GPC) (9). Studies examining the erythrocyte binding capability of the individual RII PfEBA-140 DBL domains (F1 and F2) demonstrated that neither domain is sufficient to engage erythrocytes (10, 11). In addition, single amino acid mutations in either DBL domain severely disrupt erythrocyte binding, demonstrating an essential role for both DBL domains during invasion (10).
Receptor recognition by P. falciparum invasion ligands is also characterized by the sensitivity of the binding interaction to specific enzyme treatments of red blood cells (12, 13). Studies examining the binding profile of PfEBA-140 to enzyme-treated erythrocytes have provided a basic understanding of how this ligand engages its receptor. PfEBA-140 receptor binding is trypsin- and neuraminidase-sensitive, but chymotrypsin-resistant (12). In addition, soluble sialic acid is not capable of inhibiting PfEBA-140 erythrocyte binding (12). These results demonstrated that receptor glycans containing sialic acid are essential for receptor recognition and that the protein backbone of GPC also plays a role in binding (12, 14, 15). GPC possesses several putative O-linked glycans and one known N-linked glycan, all containing the sialic acid sugar moiety required for PfEBA-140 binding. The solitary N-linked glycan is essential for GPC engagement, but the role of individual O-linked glycans is not clear (16).
Numerous and widespread polymorphisms have been identified in Plasmodium invasion ligands, specifically PfEBA-175 and Plasmodium vivax Duffy-binding protein (PvDBP) (17–19). In contrast, only four polymorphic mutants have been identified in RII PfEBA-140, all of which are present in the F1 domain: I185V, N239S, K261R/K261T, and K285E (11). Polymorphic residue changes in PfEBA-140 modify the binding profile of this ligand to enzyme-treated erythrocytes. The altered binding profile suggests that PfEBA-140 is capable of interacting with other erythrocyte cell surface molecules and may mediate an invasion process independent of GPC (11, 20). In addition to altering the binding profile, it has been shown that polymorphisms reduce the affinity of PfEBA-140 for erythrocytes (21). Recent evidence suggests that binding to glycosaminoglycans (GAGs) on the erythrocyte surface promotes merozoite invasion (22). PfEBA-140 is capable of binding heparin, and it has been proposed that cell surface GAGs may function as a secondary interaction element during PfEBA-140-mediated invasion (23).
The role of individual EBL invasion ligands during in vivo erythrocyte invasion and blood stage growth is not fully understood. However, several studies have demonstrated the importance of PfEBA-140 during natural infection as both an invasion ligand and an antigen. Individuals in malaria endemic regions mount a strong antibody response to PfEBA-140, and RII was the most immunoreactive element (24). This result provides evidence of PfEBA-140 expression and immune recognition in natural infections and supports the significance of this ligand as a member of a combinatorial vaccine. In addition to natural immunogenicity, antibodies targeting RII PfEBA-140 are capable of inhibiting invasion, which is strongly supportive of the functional role and antigenic properties of PfEBA-140 (25, 26). Furthermore, Gerbich negativity is observed at high frequencies in regions of endemic malaria in Papua New Guinea (26). This phenotype results from the loss of exon 3 within the GPC gene. The absence of this portion of GPC prevents PfEBA-140 from engaging erythrocytes and thus inhibits the invasion process. The prevalence of Gerbich negativity provides evidence that severe malaria has selected for this mutation and supports the importance of PfEBA-140 during natural infection (26).
To identify receptor-binding sites in PfEBA-140 and define molecular interactions at the merozoite-erythrocyte invasion interface, we solved the crystal structure of RII PfEBA-140 in complex with a glycan containing sialic acid, the essential sugar component of GPC that is recognized during erythrocyte engagement. Two glycan-binding pockets are identified, one in each DBL domain. Mutation of individual sialic acid contact residues disrupts erythrocyte binding by RII PfEBA-140, confirming the functional role of the sugar-binding pockets. Stark differences in the mutant erythrocyte binding phenotypes for the F1 and F2 domains suggest that each DBL domain performs a distinct function during erythrocyte engagement. One of the four polymorphic residues that affect PfEBA-140 receptor binding is found in the base of the F1 glycan-binding pocket. The localization of this residue provides insight into the structural basis of altered receptor binding observed for polymorphic variants. The structure also allowed for mapping of putative sulfate and GAG-binding sites, which may represent true interaction elements that promote red blood cell binding and invasion. This study provides the first complete molecular and structural description of sialic acid-dependent interactions critical to the formation of the tight junction at the merozoite-erythrocyte invasion interface. Our results will thus aid in the design of novel therapeutics and diagnostics targeting this essential step in the life cycle of the parasite.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
RII PfEBA-140 was expressed and purified as described previously (10). Briefly, RII PfEBA-140 was expressed in Escherichia coli and recovered from inclusion bodies using 6 m guanidinium hydrochloride. After overnight denaturing, 100 mg/liter of denatured protein was rapidly diluted in 50 mm Tris, pH 8, 10 mm EDTA, 200 mm arginine, 0.1 mm PMSF, 2 mm reduced glutathione, and 0.2 mm oxidized glutathione. The protein was refolded for 48 h at 4 °C. After refolding, RII PfEBA-140 was purified by ion-exchange and size exclusion chromatography and concentrated using Amicon centrifugal filters for crystallization.
Crystallization, Data Collection, and Structure Determination
Crystals of RII PfEBA-140 were grown using the hanging drop vapor diffusion method. The drops were produced by mixing 1 μl of protein at 7.5 mg/ml with 1 μl of reservoir containing 20% PEG 8000 and 0.1 m HEPES, pH 7.5. Crystals of RII PfEBA-140 alone were soaked with 10 mm sialyllactose for 30 min to obtain the complex. Crystals were sent for remote data collection in cryoprotectant composed of 30% glycerol, 17.5% PEG 8000, and 0.1 m HEPES, pH 7.5. The cryoprotectant was introduced gradually into the drop by pipetting, and crystals were isolated with nylon loops and stream-frozen prior to transport. Data were collected at beamline 19-ID at the Advanced Photon Source, Argonne National Laboratory, and processed with XDS (27). The structure of RII PfEBA-140 bound to sialyllactose was solved using the unbound PfEBA-140 structure Protein Data Bank (PDB) 4GF2 as a model in Phaser (10, 28). Refinement and model building were performed with PHENIX (29) and Coot (30), and refinement was completed once low R-factors and good geometry were obtained (see Table 1).
TABLE 1.
Data collection and refinement statistics for PfEBA-140 RII sialyllactose complex
Values in parentheses are for highest resolution shell.
| Data collection | |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 64.95, 76.03, 81.43 |
| α, β, γ (°) | 90.00, 96.93, 90.00 |
| Resolution (Å) | 20-3.0 (3.09-3.0) |
| Rmerge | 7.8% (65.5%) |
| I/σI | 14.06 (2.05) |
| Completeness (%) | 96.2% (97.7%) |
| Redundancy | 3.00 (3.00) |
| Refinement | |
| Resolution (Å) | 20.0-3.0 |
| No. of reflections | 15,336 |
| Rwork/Rfree (%) | 21.82/25.48 |
| No. of atoms | |
| Protein | 5266 |
| Ligand/ion | 42 |
| Water | 22 |
| B-factors | |
| Protein | 87.18 |
| Ligand/ion | 64.23 |
| Water | 42.74 |
| r.m.s.d.a | |
| Bond lengths (Å) | 0.001 |
| Bond angles (°) | 0.437 |
| Ramachandran | |
| Favored | 95.52% |
| Allowed | 4.48% |
| Disallowed | 0.00% |
a r.m.s.d., root mean square deviation.
Functional Studies
RII PfEBA-140 was fused to a C-terminal GFP in plasmid pRE4 for surface expression on HEK-293T cells. Individual alanine point mutants were produced using the QuikChange method and verified by plasmid sequencing. Monolayers of HEK-293T cells grown in 3.5-cm wells were transfected with 2.7 μg of plasmid DNA in polyethyleneimine. Erythrocyte binding of individual constructs was assayed 20 h after transfection.
To assess the binding phenotypes of RII PfEBA-140 mutant constructs, transfected HEK-293 cells were incubated with normal human erythrocytes. For enzyme treatment experiments, normal erythrocytes at 50% hematocrit were treated with 0.1 mg/ml trypsin, 0.1 mg/ml chymotrypsin, or 5 milliunits of Vibrio cholerae neuraminidase at 37 °C for 2 h and washed prior to use. Erythrocytes at 2% hematocrit were then overlaid onto the transfected HEK-293 cells for 2 h. Following incubation with untreated or enzyme-treated erythrocytes, the transfected HEK-293 cells were washed three times with phosphate-buffered saline (PBS) to remove unbound erythrocytes. For the mutation studies, three individual wells of HEK-293 cells were transfected with each mutant construct. The binding percentage was calculated as the number of rosette-positive cells over the number of GFP-positive cells and was normalized to wild type binding. Binding phenotypes were quantified over 10 fields of view from each of the three wells of transfected cells (a total of 30 images for each construct). Images were obtained using a Zeiss LSM 510 META laser scanning microscope with an LD Achroplan 20× Korr differential interference contrast objective. Images were randomized prior to counting with ImageJ.
RESULTS
Identification of Two Glycan-binding Sites in RII PfEBA-140
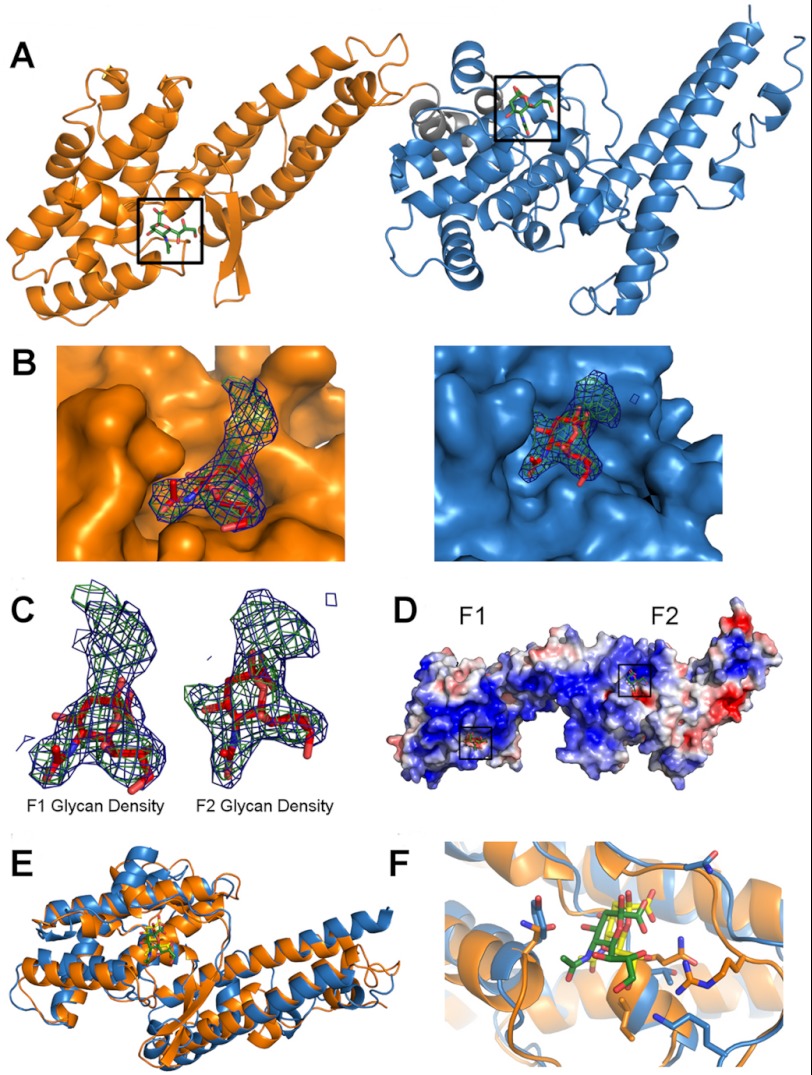
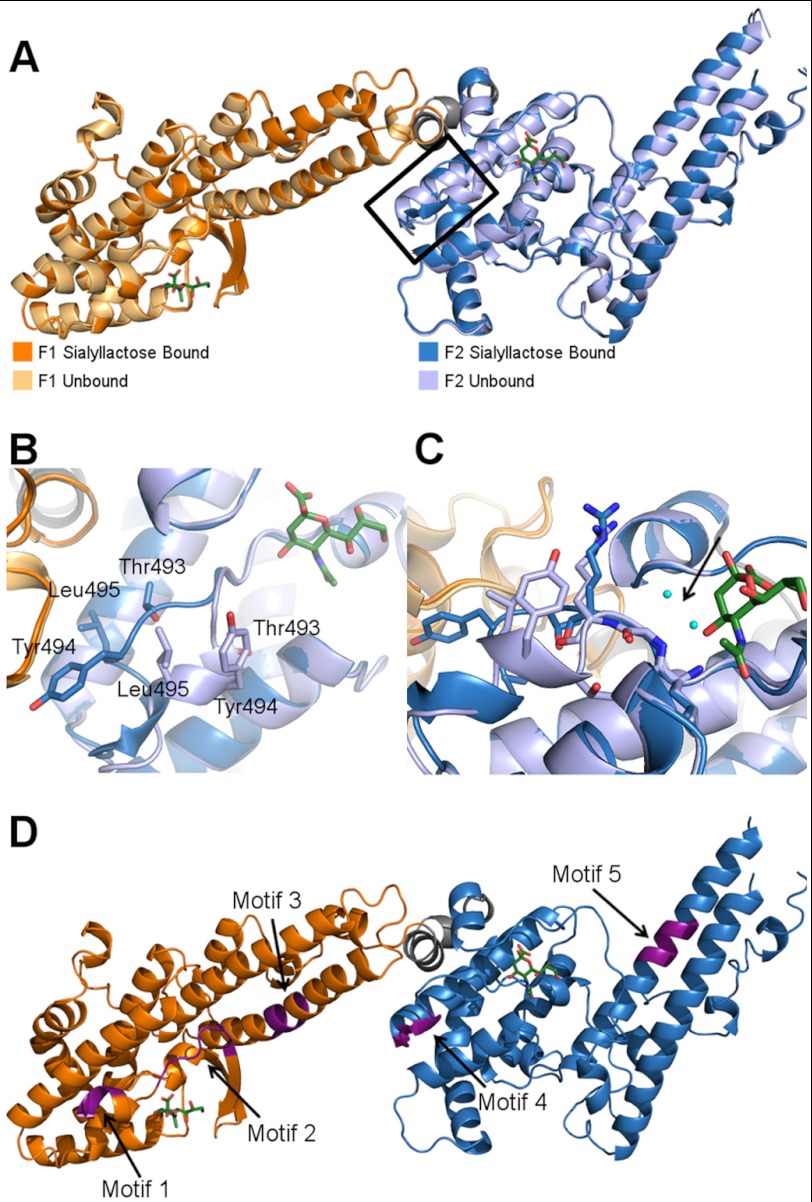
To identify receptor-binding pockets, we solved the crystal structure of RII PfEBA-140 in complex with α-2,3-sialyllactose (data collection and refinement statistics are presented in Table 1). The glycan, α-2,3-sialyllactose, is a trisaccharide composed of sialic acid, galactose, and glucose and thus contains the critical sugar groups present on GPC that are required for binding (Neu5Ac(α-2,3)-Gal). RII PfEBA-140 bound to sialyllactose is present as a monomer in the asymmetric unit, consistent with the solution and crystal structures of unbound RII PfEBA-140 (10). Clear density was observed for the sialic acid of two sialyllactose molecules in the co-crystal structure, with one molecule contacting each DBL domain. The sialic acid moieties were modeled into the two binding sites (Fig. 1, A–D). Average B-factors for the sialic acids in both pockets were comparable (64.42 and 64.04 Å2), suggesting similar occupancy and affinity. Additional density was observed for the galactose of each sialyllactose molecule, but due to inherent flexibility, galactose could not be adequately modeled. The galactose density extends out of both pockets and away from the contact surface (Fig. 1, B and C), supporting the importance of contacts between sialic acid and PfEBA-140. The two binding sites are present within a region spanning both DBL domains that contains a concentration of positively charged residues previously identified as a putative receptor-binding region (Fig. 1D) (10). The glycan-binding sites are in a structurally similar position on the F1 and F2 domains, with the sialic acid inserting into a valley at the interface of a helix and an extended loop (Fig. 1, E and F).
FIGURE 1.
The crystal structure of RII PfEBA-140 bound to sialyllactose reveals two receptor glycan-binding sites. A, RII PfEBA-140 bound to two sialyllactose molecules in the crystal structure. The F1 and F2 DBL domains each form a contact surface with one sialyllactose molecule. The F1 domain is shown in orange, and the F2 domain is in blue. A short linker connecting the two DBL domains is shown in gray. The modeled sialic acid molecules in each glycan contact site are shown in green and boxed in black. B, the bound sialic acid inserts into the F1- and F2-binding pockets, whereas the galactose extends away from the pocket surface. The sialic acid molecules and corresponding electron density observed in the F1 domain (left, orange) and F2 domain (right, blue) are displayed within their binding sites. The sialic acid is shown in red for clarity. Density for the galactose is shown to illustrate the location of this sugar moiety relative to the sialic acid-binding site. The Fo − Fc map prior to sialic acid modeling is shown in green and contoured at 3.0σ. The 2Fo − Fc map obtained following modeling of the sialic acid into the binding pocket is shown in blue and contoured at 1.0σ. C, close-up view of the electron density observed for the sialic acid and galactose of the bound sialyllactose in the F1 (left) and F2 (right) domains. The density maps are contoured and colored as in B. D, the two glycan-binding sites are located in a region of concentrated positively charged residues that may function as a high affinity interface for GPC engagement. Surface charge potential is colored from +3.5 eV (blue) to −3.5 eV (red). E, the sialic acid-binding pockets are in structurally similar locations on the F1 and F2 DBL domains. The glycan bound to the F1 domain is shown in green, and the glycan bound to the F2 domain is shown in yellow. F, the individual binding sites in each DBL domain are structurally similar. Specifically, the sialic acid inserts into a valley at the interface of a helix and an extended loop present on both domains. The glycan coloring is the same as in E.
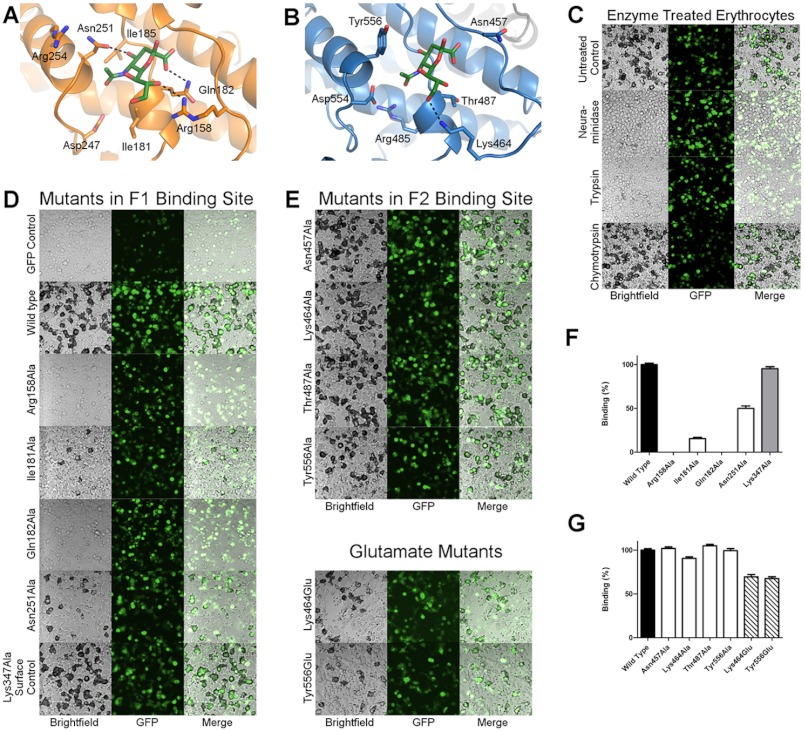
The sialic acid moiety contains a pyranose ring with single carboxyl, glycerol, and acetamido functional groups. The pyranose ring and functional groups of the bound sialic acid insert into the F1-binding pocket, and several contact residues form a cup-like interaction with the glycan (Fig. 2A). In the base of the pocket, Gln-182 hydrogen-bonds and forms van der Waal interactions with the sialic acid carboxyl. Contacts with the sialic acid glycerol chain are mediated by Arg-158, which forms a hydrogen bond and is involved in van der Waals contacts with hydroxyl groups. On the opposite face of the binding pocket, the sialic acid acetamido group is oriented toward a helix containing Asn-251. This helix places Asn-251 in proper orientation to hydrogen-bond with the 4-hydroxyl of the pyranose ring. In addition to the aforementioned contact surfaces, Ile-181 and Ile-185 form the base of the pocket and interact with hydrophobic regions of the sugar.
FIGURE 2.
Sialic acid modeling and erythrocyte binding studies provide a detailed molecular description of the RII PfEBA-140 erythrocyte invasion interface. A, close-up view of the F1-binding pocket. Four residues directly contact the sialic acid. Hydrogen bonds are designated with black lines. B, close-up view of the F2-binding pocket. Four residues directly contact the sialic acid. The single hydrogen bond interaction is shown with a black line. C, erythrocyte binding by RII PfEBA-140 expressed on the surface of HEK-293 cells is sensitive to trypsin and neuraminidase treatment of red blood cells, but chymotrypsin-resistant. Erythrocytes were treated with 0.1 mg/ml trypsin, 0.1 mg/ml chymotrypsin, or 5 milliunits of V. cholerae neuraminidase at 37 °C prior to the rosetting assay. Bound erythrocytes appear black around the transfected mammalian cells. The images are displayed at 20× magnification. D and E, RII PfEBA-140 mutants were expressed on the surface of mammalian cells and tested for erythrocyte binding. GFP was used to assess proper expression of each construct. Wild type RII PfEBA-140 extensively binds erythrocytes, and a construct containing GFP alone is not capable of binding. To identify critical binding interactions, individual glycan contact residues in the F1- and F2-binding pockets were mutated to alanine or glutamate and tested for erythrocyte binding by rosetting assay. Bound erythrocytes appear black around the transfected mammalian cells. The images are displayed at 20× magnification. F and G, the percentage of cells expressing point mutants of RII PfEBA-140 that bind erythrocytes relative to wild type RII. The binding phenotypes are representative of the binding percentage quantified for 30 fields of view per mutation. Lys-347, mutated to alanine as a surface control residue, displays wild type binding. The black bars indicate wild type RII PfEBA-140 binding. The white bars display binding percentages for single alanine mutants. The gray bar represents the surface control Lys-347. The hatched bars represent binding percentages for glutamate mutants in the F2-binding pocket. Error bars indicate S.E.
The F2-binding site contains a glycan contact surface that is similar to that observed in the F1 domain (Fig. 1F). However, the pocket residues are altered such that the observed glycan contacts are distinct (Fig. 2B). Asn-457 forms contacts not observed in the F1 pocket as a result of its location on a loop at the top of the binding site. This orientation allows Asn-457 to form van der Waal interactions with the glycan carboxyl. Interactions with the sialic acid glycerol chain are mediated by Lys-464, which hydrogen-bonds with a hydroxyl group and engages in van der Waal contacts with the sugar. Thr-487 is present in the base of the pocket and contacts hydrophobic regions of the glycan. A helix containing Tyr-556, analogous to the helix carrying Asn-251 in the F1 pocket, extends the side chain of Tyr-556 toward the glycan to engage in hydrophobic contacts with the sialic acid acetamido group.
Distinct Functional Roles for Each Glycan-binding Pocket
To examine the receptor specificity of the RII PfEBA-140 construct used for crystallization, we assessed the binding phenotype with enzyme-treated erythrocytes. The rosetting interaction is sensitive to neuraminidase and trypsin treatment, but resistant to chymotrypsin, consistent with previously observed results for RII PfEBA-140 (Fig. 2C) (12, 15, 21). These results confirm that the interaction is sialic acid-dependent and support the specificity of the recombinant construct for GPC on the erythrocyte surface.
To determine the role of specific sialic acid interactions identified in the crystal structure, individual glycan contact residues were mutated to alanine and the mutant constructs were tested for erythrocyte binding. Mutating individual contact residues in the F1 pocket severely diminished erythrocyte binding, suggesting that F1 glycan contacts are critical for receptor recognition (Fig. 2, D and F). In the absence of contacts mediated by Arg-158 or Gln-182, erythrocyte binding does not occur, suggesting that these residues are essential for binding interactions. The helix that presents the side chain of Asn-251 is supportive for binding, and it is likely that this helix plays an important role in maintaining the structure of the binding pocket. The hydrophobic base of the pocket, composed primarily of Ile-181, is crucial for erythrocyte binding. The observed binding deficiency upon mutating Ile-181 to alanine is likely due to its role in maintaining normal binding site conformation and engaging in strong hydrophobic contacts with the glycan.
In contrast to the severe phenotypes observed for alanine mutations in the F1-binding pocket, mutations in the F2-binding site resulted in a modest reduction in erythrocyte binding (Fig. 2, E and G). Mutation of Lys-464 to alanine resulted in a 9.3% decrease in binding, whereas mutation of other residues to alanine in the F2 pocket did not alter the erythrocyte binding phenotype. However, it is possible that a single mutation to alanine is insufficient to disrupt the function of this binding site. To further examine the functional role of glycan contacts in the F2-binding pocket, two residues were mutated to glutamate to induce charge repulsion with the sialic acid. Individually mutating Lys-464 or Tyr-556 to glutamate resulted in a 30.6 and 32.5% decrease in binding, respectively. The F2-binding pocket is also close to residues in a basic patch (Arg-485 and Asp-554) that when mutated individually to alanine drastically reduce binding to erythrocytes (Fig. 2B) (10). Together, these results suggest that the F2-binding site recognizes sialic acid in a functional, but nonessential, manner and that the F2 sialic acid-binding pocket and surrounding residues contact GPC. Reduced erythrocyte binding of alanine and glutamate mutants was not due to inefficient surface expression of RII PfEBA-140 as expression was monitored by GFP (Fig. 2, C–E). In addition, Lys-347 was mutated to alanine as a surface control and displays wild type binding.
PfEBA-140 Glycan-binding Contacts Are Unique among Sialic Acid-binding Proteins
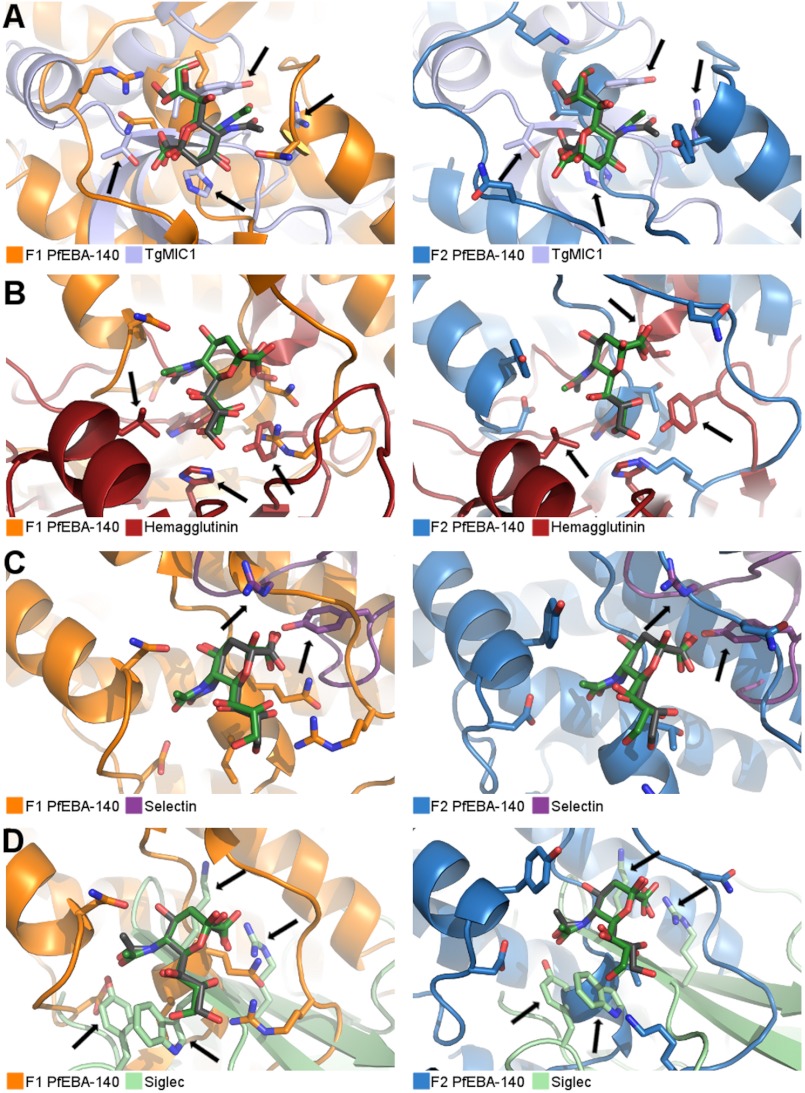
The glycan contacts observed in the two PfEBA-140-binding pockets are unique when compared with other proteins that require cell surface sialic acids for binding. In addition to Plasmodium species, other apicomplexan parasites, including Toxoplasma gondii, actively invade host cells during infection. Invasion by T. gondii is mediated by micronemal proteins (MICs). A member of this group, TgMIC1, recognizes sialic acid derivatives on the cell surface (31). Glycan recognition by TgMIC1 requires contact with the sialic acid carboxyl group mediated by a threonine conserved in the TgMIC1 protein fold (Fig. 3A). Recombinant TgMIC1 does not bind host cells when this threonine is mutated to alanine, confirming the essential role of this residue (31). In addition to the threonine, a histidine interacts with the carboxyl group, and a tyrosine forms the base of the sialic acid-binding pocket. The overall structures of the sialic acid-binding sites and the critical contact residues are thus distinct from both binding sites of PfEBA-140 (Fig. 3A).
FIGURE 3.
RII PfEBA-140 glycan-binding sites are distinct from other classes of sialic acid-binding proteins. A–D, the PfEBA-140 F1 (orange) and F2 domains (blue) are shown overlaid with the TgMIC1 (light blue) (A), hemagglutinin (red) (B), selectin (purple) (C), and Siglec (light green) (D) sialic acid-binding sites. The sialic acid molecule bound to PfEBA-140 is shown in green, and the sialic acid bound to the protein of comparison is shown in gray. Arrows highlight structural differences in each case.
In addition to apicomplexan parasites, other pathogens recognize cell surface sialic acids during infection. All subtypes of the influenza virus bind to host cells in a sialic acid-dependent manner mediated by the surface protein hemagglutinin. The conserved hemagglutinin sialic acid-binding site utilizes essential glycerol chain contacts and hydrophobic interactions with the acetamido group to engage the glycan (Fig. 3B). Contacts with the carboxyl appear to be supportive (32–34). In comparison, although glycerol contacts are essential to PfEBA-140 binding, the binding site structures and interacting residues are distinct. In addition, interactions with the sialic acid carboxyl are critical for PfEBA-140, but not hemagglutinin binding.
Sialic acid is also required for a number of interactions involving proteins that modulate immune function, including selectins and Siglecs (35). Selectins interact most extensively with the sialic acid carboxylate and glycosidic oxygen and also contact the 4-hydroxyl of the pyranose ring (Fig. 3C). Contact with other functional groups does not appear to be important for selectin binding (36). This mode of contact sharply contrasts with that of PfEBA-140, which interacts with each functional group and requires both the carboxyl and the glycerol moieties to effectively engage the glycan. The PfEBA-140-binding sites are also distinct from that of the Siglecs. Siglec binding is dependent on a salt bridge formed with the sialic acid carboxyl group and van der Waal contacts with the acetamido methyl group, mediated by a conserved arginine and semiconserved tryptophan/tyrosine, respectively (Fig. 3D) (37, 38). Although other sialic acid-binding proteins utilize contacts that resemble those observed in RII PfEBA-140, the GPC-binding sites are distinct and represent a novel mode of glycan contact.
Structural Basis for Altered Receptor Binding Caused by Polymorphic Residues
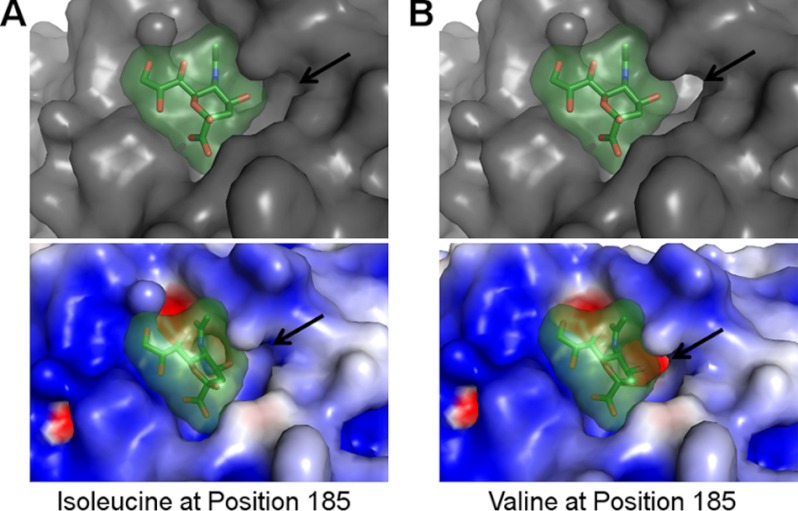
Polymorphisms in PfEBA-140 are thought to reduce affinity to GPC (21) and have been proposed to alter PfEBA-140 receptor specificity (11). One of the identified polymorphisms, Ile-185, is buried in the critical F1-binding pocket (Fig. 2A). Variants that contain a valine in place of isoleucine at position 185 display reduced binding to erythrocytes and GPC (21). The F1-binding pocket forms a tight fit with high shape complementarity to the sialic acid when isoleucine is present at position 185 (Fig. 4A). To examine the effect of a valine mutation, Ile-185 was mutated to valine in silico. This mutation opens a cavity at the base of the pocket adjacent to the 4-hydroxyl of the pyranose ring (Fig. 4B). Mutation to valine in vivo likely alters the pocket surface in this manner, reducing shape complementarity and increasing solvent accessibility to both the pocket and the glycan. This change would disrupt glycan binding and provides a structural basis for diminished affinity of polymorphic PfEBA-140 variants for erythrocytes (21). The structural alteration also elucidates a putative basis for altered receptor specificity.
FIGURE 4.
The F1-binding pocket surface is altered when Ile-185 is mutated to valine in silico. A, the glycan-binding surface containing Ile-185 (the residue present in the construct used for crystallization) exhibits strong shape complimentary with the sialic acid. The overall surface is shown in gray on the top, and the electrostatic surface is shown on the bottom (surface charge potential is colored from +3.5 eV (blue) to −3.5 eV (red)). The bound sialic acid is shown in green. B, the binding pocket surface is altered following in silico mutation of residue 185 to valine. The top displays the overall surface of the pocket in gray, and the bottom displays the electrostatic surface (surface charge potential is colored as in A). The altered surface cavity is identified with an arrow, and the sialic acid is shown in green.
Glycan Binding Induces a Helix Shift in the F2 Domain
The structure of RII PfEBA-140 bound to sialyllactose maintains the same overall architecture and hinge angle as unbound RII (Fig. 5A). Glycan binding did not result in any large structural changes in the F1 domain, but caused a helix containing Thr-493, Tyr-494, and Leu-495 to move away from the binding pocket of the F2 domain (Fig. 5B). The bound sialic acid in the F2 domain inserts into a cavity containing two water molecules in the unbound structure. Glycan binding blocks access of the water molecules at this position, disrupting main chain contacts and destabilizing the helix such that the helix movement is possible (Fig. 5C). The glycan-induced movement in the F2 domain may provide access to the highest affinity interface for GPC.
FIGURE 5.
The sialyllactose-bound structure illuminates a glycan-induced structural change and allows for localization of putative sulfate-binding sites. A, overlay of the sialyllactose bound (F1 domain in orange, F2 domain in blue) and unbound (F1 domain in light orange, F2 domain in light blue) structures of RII PfEBA-140. The bound sialic acid molecules are shown in green. The glycan-induced structural change is outlined in black. B, close-up view of residues Thr-493, Tyr-494, and Leu-495, which show a pronounced movement following glycan binding. C, the helix shift in the F2 domain observed upon glycan binding is propagated by sialic acid interference with main chain water molecule interactions. Glycan binding precludes access of two water molecules to the cavity indicated by the black arrow (the waters are present in the apo structure). In the absence of contacts mediated by these water molecules, the helix is destabilized, allowing the movement observed in the bound crystal structure. The water molecules are shown in cyan, and the sialic acid is in green. The bound F2 domain is shown in dark blue, and the unbound F2 domain is in light blue. D, three sulfate-binding motifs in RII PfEBA-140 are adjacent to the sialic acid-binding sites. Two other putative sulfate-binding motifs are distal to the binding sites. The putative sulfate-binding sites are shown in purple and identified with an arrow.
Putative Sulfate-binding Motifs Map Close to the Glycan-binding Pockets
In addition to binding GPC, RII PfEBA-140 is capable of interactions with heparin and potentially with cell surface GAGs. Heparitinase treatment of erythrocytes inhibits RII PfEBA-140 binding, providing support for a potential interaction with cell surface GAGs such as heparan sulfate (23). These observations suggest that GAGs on the erythrocyte surface interact with PfEBA-140 and may promote invasion. RII PfEBA-140 contains five putative sulfate-binding motifs that can potentially bind GAGs, three of which are adjacent to the identified sialic acid-binding sites in F1 and F2 (Fig. 5D). Mutation of the adjacent sulfate-binding motifs 1 and 4 greatly diminishes erythrocyte binding. The effect of mutating motif 2 has not been examined. In contrast to the adjacent motifs 1 and 4, mutation of sulfate-binding motifs 3 and 5, which are distal to the glycan-binding sites, has little or no effect on erythrocyte binding (23). Thus, it is plausible that sulfate-binding sites 1, 2, and 4 represent true interaction elements that may play a role in facilitating invasion. Although the putative sulfate-binding motifs are proposed to engage heparan sulfate (23), additional sulfate-containing molecules such as sulfatides, sulfated glycolipids, glycoprotein oligosaccharides, and chondroitin sulfate proteoglycans exist on erythrocytes. It is possible that putative sulfate-binding motifs 1, 2, and 4 may interact with any or all of these moieties.
DISCUSSION
The results presented here represent the first molecular description of specific interactions an EBL protein utilizes to recognize and bind sialic acid. Although DBL domains have been crystallized with sugars previously (40, 41), the crystal structure of RII PfEBA-140 presented here represents the first full modeling of sialic acid into a DBL domain sugar-binding site. This modeling allowed identification of two receptor-binding sites and elucidation of critical molecular interactions that the parasite utilizes to form the essential tight junction with the host erythrocyte membrane during invasion. Mapping molecular contacts demonstrated that PfEBA-140 recognizes sialic acid on receptor glycans utilizing unique interactions when compared with its paralog PfEBA-175 as well as compared with viral cell invasion and host immune proteins that also engage sialic acid. The bound structure also provided insight into the role of polymorphisms in altering receptor binding and illuminated the location of putative sulfate-binding motifs that may engage GAGs or other sulfate-containing surface molecules. The importance of these moieties during invasion is unclear, but localizing their binding sites within RII provides a basis for future studies examining their function during Plasmodium invasion.
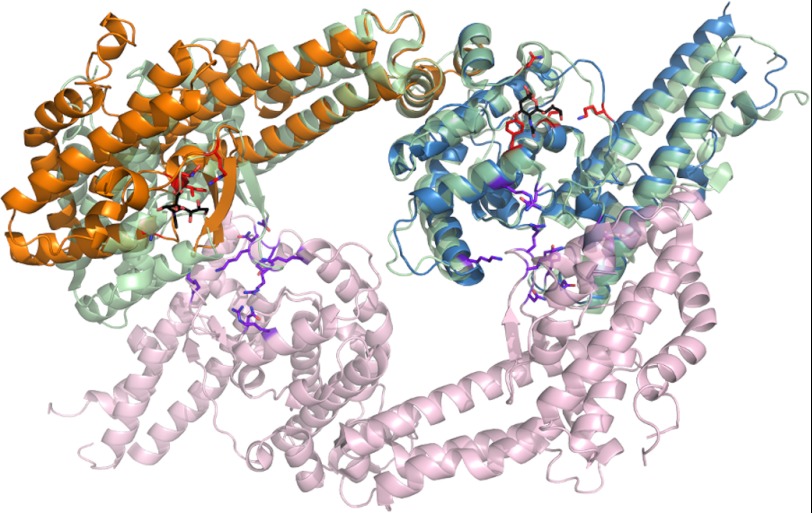
The identification and description of receptor-binding sites within PfEBA-140 allow for direct comparison with the glycan-binding sites of other DBL domains. DBL domains function in a number of settings including erythrocyte invasion and cytoadherence of infected erythrocytes mediated by erythrocyte membrane protein 1 (PfEMP1) (39). The two glycan-binding regions of RII PfEBA-140 are structurally distinct from receptor-binding regions of PfEBA-175, P. vivax Duffy-binding protein, PfEMP1 DBL3X, and the PfEMP1 VarO head region and thus represent novel receptor-binding sites for the DBL fold (40–43). Specifically, the glycan interactions observed in RII PfEBA-175 are limited to the dimer interface (41). In contrast, formation of the glycan-binding sites in RII PfEBA-140 does not require dimerization (Fig. 6). We previously demonstrated that RII PfEBA-140 is monomeric in solution in the absence of receptor (10). RII PfEBA-140 has been observed only as a monomer in solution and crystal forms, and the lack of dimerization in the presence of sialyllactose suggests that PfEBA-140 may engage GPC as a monomer. Studies examining the oligomeric state of PfEBA-140 in the presence of GPC are required to fully understand the mechanism of receptor engagement and specificity. In addition to the different oligomeric states observed upon sialic acid binding for PfEBA-175 and PfEBA-140, the location within the DBL domain and overall structure of the binding pockets is distinct (Fig. 6). Furthermore, PfEBA-175 contains six glycan-binding sites per dimer, in contrast to the two observed for the monomer of PfEBA-140. It is likely that the novel modes of contact observed for PfEBA-140 confer specificity for sialic acid moieties present on GPC over glycophorin A or B, and this suggests how structurally similar DBL domains specifically recognize diverse receptors.
FIGURE 6.
PfEBA-140 sialic acid recognition is distinct from that of PfEBA-175. Shown here is an overlay of the RII PfEBA-140 monomer with the dimer of RII PfEBA-175. The PfEBA-140 F1 domain is shown in orange, and the F2 domain is shown in blue. The RII PfEBA-175 monomer overlaid with RII PfEBA-140 is shown in light green. The second monomer of RII PfEBA-175 is shown in light purple. Residues of PfEBA-140 involved in sialic acid binding are shown in red. The sialic acid molecules bound to RII PfEBA-140 are shown in black for clarity. Residues involved in PfEBA-175 sialic acid binding are shown in purple.
In addition to structural and molecular contrasts with the glycan-binding sites of DBL domains, this work allows for comparison with other apicomplexan invasion proteins that recognize cell surface sialic acid. As shown in Fig. 3A, sialic acid recognition by TgMIC1 is quite distinct when compared with PfEBA-140. In addition, there is no structural similarity in the overall protein fold of the minimal binding regions of TgMIC1 and PfEBA-140 (31). However, both proteins contain two tandem, conserved domains comprising the minimal binding region, with each domain containing a single sialic acid-binding site on the same face of the protein. This resemblance suggests some functional similarities in their mode of glycan engagement (31). The Plasmodium merozoite is specifically marked for the erythrocyte, unlike T. gondii, which is capable of invading any nucleated cell. It thus appears that TgMIC1 has developed a sialic acid binding mode that allows recognition of a broad range of glycans during cell binding, sharply contrasting the proposed erythrocyte sialic acid glycan specificity of PfEBA-140. PfEBA-140 receptor specificity may thus be conferred by a unique glycan composition on the erythrocyte surface combined with essential interactions with the GPC protein backbone.
The erythrocyte binding phenotypes observed upon mutation of contact residues in each glycan pocket suggest distinct roles for the individual DBL domains of RII PfEBA-140 during erythrocyte binding. GPC contains several O-linked glycans and a single N-linked glycan, each with the essential sugar moiety Neu5Ac(α-2,3)-Gal. Recognition of the N-linked glycan is essential to the interaction (16). Thus, interactions with the N-linked glycan may be mediated solely by the F1 sialic acid-binding site. The null and severely deficient binding phenotypes observed upon mutation of contact residues in the F1 pocket could be the result of losing the critical interaction with the N-linked glycan.
Receptor contacts with the F2 domain are also required for erythrocyte binding as neither individual DBL domain is sufficient to bind erythrocytes independently (10, 11). The observation that only mutations to glutamate in the F2-binding pocket reduced binding efficiency may be explained by compensatory interactions between the glycan-binding site and the sialic acid. In addition to glycan contacts, PfEBA-140 erythrocyte binding is dependent on the GPC protein backbone. It is thus also plausible that F2 engages in strong contacts with additional regions of GPC and that glycan interactions are supportive but not essential in maintaining this interaction.
In addition to invading via GPC, it has been proposed that polymorphic variants of PfEBA-140 are capable of invading using an alternative receptor. The identification of Ile-185 in the F1-binding pocket suggests that polymorphism at this residue may play an important role in determining receptor affinity and possibly specificity. Our studies suggest a model to explain the altered binding profile of polymorphic variants. The proximity of the glycan-binding pockets and sulfate-binding motifs 1, 2, and 4 suggests that GPC binding precludes GAGs or sulfate-containing molecules from accessing these sugar-binding sites. This proposal is supported by the fact that the helix containing sulfate-binding motif 4 is shifted upon sialyllactose binding (Fig. 5, A and B). It is plausible that a valine mutation at position 185 destabilizes the F1-binding pocket, reducing affinity for sialic acid and opening sulfate-binding motifs 1 and 2 for functional GAG binding. The altered surface observed following in silico mutation of Ile-185 to valine supports the destabilization hypothesis (Fig. 4). It is equally possible that a secondary sialylated receptor is recognized by specific variants of PfEBA-140 during invasion.
The structural and functional data described here have provided a detailed molecular description of sialic acid recognition by an EBL family erythrocyte invasion ligand, PfEBA-140. Preventing erythrocyte receptor engagement provides an excellent opportunity to inhibit merozoite invasion and blood stage growth of the parasite. The identification of critical interactions at the invasion interface will thus aid in the design of rational therapeutics and vaccines that target the EBL ligands.
Acknowledgments
We thank Nichole Salinas for assistance with experiments, Wandy Beatty for help with confocal imaging, David Sibley for constructive comments on the manuscript, the Structural Biology Center (SBC) at Argonne National Laboratory for use of beamline 19-ID at the Advanced Photon Source, and Stephen L. Ginell and SBC beamline scientists for assistance during data collection. Argonne is operated by UChicago Argonne, LLC, for the United States Department of Energy, Office of Biological and Environmental Research under Contract DE-AC02-06CH11357.
This work was supported, in whole or in part, by National Institutes of Health Grant AI080792 from the National Institutes of Health and by a grant from the Edward Mallinckrodt, Jr. Foundation (to N. H. T.). This work was also supported by a National Science Foundation Graduate Research Fellowship (to B. M. M.) under Grant DGE-1143954.
The atomic coordinates and structure factors (code 4JNO) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- EBL
- erythrocyte binding-like
- PfEBA-140
- P. falciparum erythrocyte-binding antigen-140
- DBL
- Duffy binding-like
- RII
- region II
- GPC
- glycophorin C
- GAG
- glycosaminoglycan
- MIC
- micronemal protein
- PfEMP1
- P. falciparum erythrocyte membrane protein 1
- Siglec
- sialic acid-binding immunoglobulin-type lectin.
REFERENCES
- 1. Sim B. K., Orlandi P. A., Haynes J. D., Klotz F. W., Carter J. M., Camus D., Zegans M. E., Chulay J. D. (1990) Primary structure of the 175K Plasmodium falciparum erythrocyte binding antigen and identification of a peptide which elicits antibodies that inhibit malaria merozoite invasion. J. Cell Biol. 111, 1877–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Camus D., Hadley T. J. (1985) A Plasmodium falciparum antigen that binds to host erythrocytes and merozoites. Science 230, 553–556 [DOI] [PubMed] [Google Scholar]
- 3. Adams J. H., Sim B. K., Dolan S. A., Fang X., Kaslow D. C., Miller L. H. (1992) A family of erythrocyte binding proteins of malaria parasites. Proc. Natl. Acad. Sci. U.S.A. 89, 7085–7089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dvorak J. A., Miller L. H., Whitehouse W. C., Shiroishi T. (1975) Invasion of erythrocytes by malaria merozoites. Science 187, 748–750 [DOI] [PubMed] [Google Scholar]
- 5. Aikawa M., Miller L. H., Johnson J., Rabbege J. (1978) Erythrocyte entry by malarial parasites. A moving junction between erythrocyte and parasite. J. Cell Biol. 77, 72–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cowman A. F., Crabb B. S. (2006) Invasion of red blood cells by malaria parasites. Cell 124, 755–766 [DOI] [PubMed] [Google Scholar]
- 7. Aravind L., Iyer L. M., Wellems T. E., Miller L. H. (2003) Plasmodium biology: genomic gleanings. Cell 115, 771–785 [DOI] [PubMed] [Google Scholar]
- 8. Mayor A., Bir N., Sawhney R., Singh S., Pattnaik P., Singh S. K., Sharma A., Chitnis C. E. (2005) Receptor-binding residues lie in central regions of Duffy-binding-like domains involved in red cell invasion and cytoadherence by malaria parasites. Blood 105, 2557–2563 [DOI] [PubMed] [Google Scholar]
- 9. Lobo C. A., Rodriguez M., Reid M., Lustigman S. (2003) Glycophorin C is the receptor for the Plasmodium falciparum erythrocyte binding ligand PfEBP-2 (baebl). Blood 101, 4628–4631 [DOI] [PubMed] [Google Scholar]
- 10. Lin D. H., Malpede B. M., Batchelor J. D., Tolia N. H. (2012) Crystal and solution structures of Plasmodium falciparum erythrocyte-binding antigen 140 reveal determinants of receptor specificity during erythrocyte invasion. J. Biol. Chem. 287, 36830–36836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mayer D. C., Mu J. B., Feng X., Su X. Z., Miller L. H. (2002) Polymorphism in a Plasmodium falciparum erythrocyte-binding ligand changes its receptor specificity. J. Exp. Med. 196, 1523–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mayer D. C., Kaneko O., Hudson-Taylor D. E., Reid M. E., Miller L. H. (2001) Characterization of a Plasmodium falciparum erythrocyte-binding protein paralogous to EBA-175. Proc. Natl. Acad. Sci. U.S.A. 98, 5222–5227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Miller L. H., McAuliffe F. M., Mason S. J. (1977) Erythrocyte receptors for malaria merozoites. Am. J. Trop. Med. Hyg. 26, 204–208 [DOI] [PubMed] [Google Scholar]
- 14. Jiang L., Duriseti S., Sun P., Miller L. H. (2009) Molecular basis of binding of the Plasmodium falciparum receptor BAEBL to erythrocyte receptor glycophorin C. Mol. Biochem. Parasitol. 168, 49–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Narum D. L., Fuhrmann S. R., Luu T., Sim B. K. (2002) A novel Plasmodium falciparum erythrocyte binding protein-2 (EBP2/BAEBL) involved in erythrocyte receptor binding. Mol. Biochem. Parasitol. 119, 159–168 [DOI] [PubMed] [Google Scholar]
- 16. Mayer D. C., Jiang L., Achur R. N., Kakizaki I., Gowda D. C., Miller L. H. (2006) The glycophorin C N-linked glycan is a critical component of the ligand for the Plasmodium falciparum erythrocyte receptor BAEBL. Proc. Natl. Acad. Sci. U.S.A. 103, 2358–2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gosi P., Khusmith S., Khalambaheti T., Lanar D. E., Schaecher K. E., Fukuda M. M., Miller S. R. (2008) Polymorphism patterns in Duffy-binding protein among Thai Plasmodium vivax isolates. Malar. J. 7, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xainli J., Adams J. H., King C. L. (2000) The erythrocyte binding motif of Plasmodium vivax Duffy binding protein is highly polymorphic and functionally conserved in isolates from Papua New Guinea. Mol. Biochem. Parasitol. 111, 253–260 [DOI] [PubMed] [Google Scholar]
- 19. Baum J., Thomas A. W., Conway D. J. (2003) Evidence for diversifying selection on erythrocyte-binding antigens of Plasmodium falciparum and P. vivax. Genetics 163, 1327–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thompson J. K., Triglia T., Reed M. B., Cowman A. F. (2001) A novel ligand from Plasmodium falciparum that binds to a sialic acid-containing receptor on the surface of human erythrocytes. Mol. Microbiol. 41, 47–58 [DOI] [PubMed] [Google Scholar]
- 21. Maier A. G., Baum J., Smith B., Conway D. J., Cowman A. F. (2009) Polymorphisms in erythrocyte binding antigens 140 and 181 affect function and binding but not receptor specificity in Plasmodium falciparum. Infect. Immun. 77, 1689–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Boyle M. J., Richards J. S., Gilson P. R., Chai W., Beeson J. G. (2010) Interactions with heparin-like molecules during erythrocyte invasion by Plasmodium falciparum merozoites. Blood 115, 4559–4568 [DOI] [PubMed] [Google Scholar]
- 23. Kobayashi K., Kato K., Sugi T., Takemae H., Pandey K., Gong H., Tohya Y., Akashi H. (2010) Plasmodium falciparum BAEBL binds to heparan sulfate proteoglycans on the human erythrocyte surface. J. Biol. Chem. 285, 1716–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ford L., Lobo C. A., Rodriguez M., Zalis M. G., Machado R. L., Rossit A. R., Cavasini C. E., Couto A. A., Enyong P. A., Lustigman S. (2007) Differential antibody responses to Plasmodium falciparum invasion ligand proteins in individuals living in malaria-endemic areas in Brazil and Cameroon. Am. J. Trop. Med. Hyg. 77, 977–983 [PubMed] [Google Scholar]
- 25. Lopaticki S., Maier A. G., Thompson J., Wilson D. W., Tham W. H., Triglia T., Gout A., Speed T. P., Beeson J. G., Healer J., Cowman A. F. (2011) Reticulocyte and erythrocyte binding-like proteins function cooperatively in invasion of human erythrocytes by malaria parasites. Infect. Immun. 79, 1107–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maier A. G., Duraisingh M. T., Reeder J. C., Patel S. S., Kazura J. W., Zimmerman P. A., Cowman A. F. (2003) Plasmodium falciparum erythrocyte invasion through glycophorin C and selection for Gerbich negativity in human populations. Nat. Med. 9, 87–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kabsch W. (2010) XDS. Acta. Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 30. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 31. Blumenschein T. M. A., Friedrich N., Childs R. A., Saouros S., Carpenter E. P., Campanero-Rhodes M. A., Simpson P., Chai W. G., Koutroukides T., Blackman M. J., Feizi T., Soldati-Favre D., Matthews S. (2007) Atomic resolution insight into host cell recognition by Toxoplasma gondii. EMBO J. 26, 2808–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Skehel J. J., Wiley D. C. (2000) Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69, 531–569 [DOI] [PubMed] [Google Scholar]
- 33. Ha Y., Stevens D. J., Skehel J. J., Wiley D. C. (2001) X-ray structures of H5 avian and H9 swine influenza virus hemagglutinins bound to avian and human receptor analogs. Proc. Natl. Acad. Sci. U.S.A. 98, 11181–11186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Watowich S. J., Skehel J. J., Wiley D. C. (1994) Crystal structures of influenza virus hemagglutinin in complex with high-affinity receptor analogs. Structure 2, 719–731 [DOI] [PubMed] [Google Scholar]
- 35. Varki N. M., Varki A. (2007) Diversity in cell surface sialic acid presentations: implications for biology and disease. Lab. Invest. 87, 851–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Somers W. S., Tang J., Shaw G. D., Camphausen R. T. (2000) Insights into the molecular basis of leukocyte tethering and rolling revealed by structures of P- and E-selectin bound to SLeX and PSGL-1. Cell 103, 467–479 [DOI] [PubMed] [Google Scholar]
- 37. Crocker P. R., Paulson J. C., Varki A. (2007) Siglecs and their roles in the immune system. Nat. Rev. Immunol. 7, 255–266 [DOI] [PubMed] [Google Scholar]
- 38. Attrill H., Takazawa H., Witt S., Kelm S., Isecke R., Brossmer R., Ando T., Ishida H., Kiso M., Crocker P. R., van Aalten D. M. (2006) The structure of siglec-7 in complex with sialosides: leads for rational structure-based inhibitor design. Biochem. J. 397, 271–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kraemer S. M., Smith J. D. (2006) A family affair: var genes, PfEMP1 binding, and malaria disease. Curr. Opin. Microbiol. 9, 374–380 [DOI] [PubMed] [Google Scholar]
- 40. Singh K., Gittis A. G., Nguyen P., Gowda D. C., Miller L. H., Garboczi D. N. (2008) Structure of the DBL3x domain of pregnancy-associated malaria protein VAR2CSA complexed with chondroitin sulfate A. Nat. Struct. Mol. Biol. 15, 932–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tolia N. H., Enemark E. J., Sim B. K., Joshua-Tor L. (2005) Structural basis for the EBA-175 erythrocyte invasion pathway of the malaria parasite Plasmodium falciparum. Cell 122, 183–193 [DOI] [PubMed] [Google Scholar]
- 42. Vigan-Womas I., Guillotte M., Juillerat A., Hessel A., Raynal B., England P., Cohen J. H., Bertrand O., Peyrard T., Bentley G. A., Lewit-Bentley A., Mercereau-Puijalon O. (2012) Structural basis for the ABO blood-group dependence of Plasmodium falciparum rosetting. PLoS Pathog. 8, e1002781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Batchelor J. D., Zahm J. A., Tolia N. H. (2011) Dimerization of Plasmodium vivax DBP is induced upon receptor binding and drives recognition of DARC. Nat. Struct. Mol. Biol. 18, 908–914 [DOI] [PMC free article] [PubMed] [Google Scholar]