Abstract
The combinatorial architecture of cullin 1-RING ubiquitin ligases (CRL1s), in which multiple F-box containing substrate receptors (FBPs) compete for access to CUL1, poses special challenges to assembling CRL1 complexes through high affinity protein interactions while maintaining the flexibility to dynamically sample the entire FBP repertoire. Here, using highly quantitative mass spectrometry, we demonstrate that this problem is addressed by CAND1, a factor that controls the dynamics of the global CRL1 network by promoting the assembly of newly synthesized FBPs with CUL1-RBX1 core complexes. Our studies of in vivo CRL1 dynamics and in vitro biochemical findings showing that CAND1 can displace FBPs from Cul1p suggest that CAND1 functions in a cycle that serves to exchange FBPs on CUL1 cores. We propose that this cycle assures comprehensive sampling of the entire FBP repertoire in order to maintain the CRL1 landscape, a function that we show to be critical for substrate degradation and normal physiology.
Introduction
Cullin 1-RING ligases (CRL1s) are multifunctional ubiquitin ligases that uniquely exploit combinatorial diversity in order to achieve unparalleled versatility in substrate targeting and control of cell physiology 1–3. This combinatorial layout where multiple F-box containing substrate receptors (FBPs) compete for access to CUL1 poses special challenges to assembling CRL1 complexes through high affinity protein interactions while maintaining the flexibility to dynamically sample the entire FBP repertoire. Mounting evidence has implicated mechanisms related to the reversible modification of CUL1 with the ubiquitin-related peptide NEDD8 in this regulation 4, but no definitive model has been substantiated experimentally.
Conjugation of NEDD8 to CUL1 stimulates the ubiquitin ligase activity of CRL1s 5,6, and deneddylation by the COP9 signalosome resets CRL1s into an inactive state 7–9. Deneddylation has two important consequences: It prevents the autocatalytic destruction of FBPs 10–12 and it allows CUL1 to associate with CAND1, a highly conserved protein that inhibits CUL1 neddylation and hence CRL1 activity in vitro 13–18. This inhibition can be overcome by purified SKP1-FBP heterodimers which dissociate the CUL1-CAND1 complex in vitro 18,19. Paradoxically, however, CAND1 was also shown to promote CRL function in vivo 12,14,20–23.
In pursuit of this paradox, we previously showed that fission yeast cells deleted for knd1, the orthologue of human CAND1, display an imbalance in CRL complexes formed between Cul1p and two epitope-tagged FBPs, Pof1p-Myc and Pof3p-Myc 12. Whereas CRL1Pof1p-Myc complexes increased, CRL1Pof3p-Myc complexes were depleted in cells lacking knd1. The ~2-fold decrease in the CRL1Pof3p-Myc complex was sufficient to cause phenotypic defects that mirrored a Δpof3 deletion mutant 12,24, a finding that reinforced a positive role of CAND1/Knd1p in CRL1 control. Similar imbalances were reported for CUL1-TIR1 interactions in A. thaliana and for CUL3-KEAP1 interactions in human cells 25,26. The latter studies also demonstrated that substrate degradation by CRL1TIR1 and CRL3KEAP1 is compromised either in the absence of CAND1 or when CAND1 is overexpressed. Based on these findings, we proposed that a CAND1-mediated cycle of CRL1 complex disassembly and subsequent reassembly maintains the cellular balance of CRL1 complexes and optimal CRL1 activity 12. However, a subsequent study using siRNA-mediated knockdown in human cells achieved a partial reduction in CUL1-CAND1 complex but observed no significant effect on the recruitment of SKP1 (and presumably FBPs) to CUL1 and therefore relinquished a role of CAND1 in CRL1 assembly and remodeling 27.
We have used highly quantitative mass spectrometry to rigorously assess the impact of complete genetic depletion of CAND1/Knd1p on the global CRL1 repertoire and its assembly/disassembly dynamics. We demonstrate that CAND1/Knd1p plays a crucial role in maintaining a balanced repertoire through mechanisms that are consistent with our previously proposed CAND1 cycle 12.
Results
CAND1/Knd1p maintains the global CRL1 repertoire
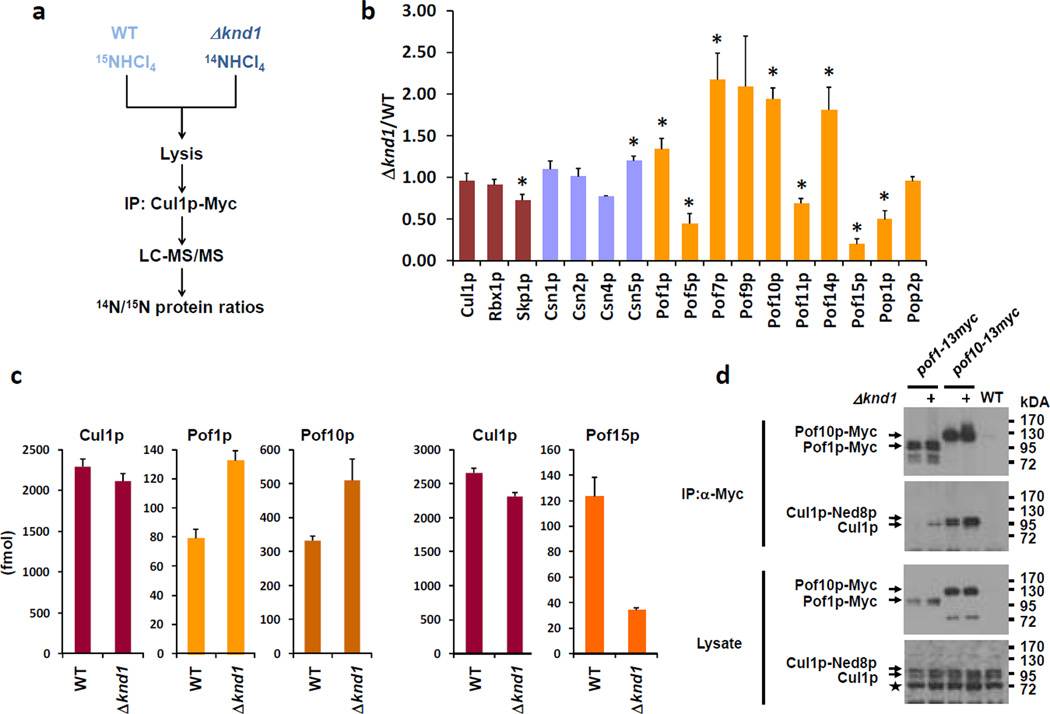
To test the effect of complete genetic ablation of CAND1 on the native CRL1 repertoire, we immunopurified Cul1p-associated proteins from wildtype and Δknd1 fission yeast cells differentially labeled with stable isotopes 28 and quantified them by liquid chromatography and tandem mass spectrometry (LC-MS/MS). Whereas Δknd1 cells were grown in medium containing regular ammonium-14N chloride as the nitrogen source, the wildtype cells were metabolically labeled with ammonium-15N chloride with an efficiency of > 98%. Cultures were mixed at a ratio of 1:1 and processed as a single sample for lysate preparation, Cul1p immunopurification, and LC-MS/MS to quantify the relative abundance of Cul1p-associated proteins in wildtype and Δknd1 cells based on averaged 14N/15N peptide ratios (Fig. 1a). Triplicate experiments revealed statistically significant (p ≤ 0.05) differences in Cul1p occupancy by various FBPs. While occupancy by Pof1p, Pof7p, Pof9p, Pof10p, and Pof14p increased by 1.3 – 2.2 fold, occupancy by Pof5p, Pof11p, Pof15p, and Pop1p decreased by factors of 1.5 – 5 fold (Fig. 1b, Supplementary Table S1, Supplementary Data 1). The modest amplitude of these changes is explained by the fact that ~50% of Cul1p is neddylated in cells 10. Since neddylated Cul1p cannot interact with CAND1, this fraction of CRL1 complexes is not responsive to the cellular CAND1 status.
Figure 1. Effect of Knd1p on the CRL1 repertoire.
(a) Wildtype (WT) S. pombe cells and cells deleted for knd1 (Δknd1), which also carry a Myc-tagged allele of endogenous Cul1p, were grown in medium containing either ammonium-14N chloride or ammonium-15N chloride as the nitrogen sources. Equal numbers of cells were mixed at a ratio of 1:1, followed by the isolation of CRL1 complexes through immunopurification (IP) of Cul1p-Myc. The purified material was analyzed by LC-MS/MS to identify Cul1p-associated proteins and to quantify their relative abundance in WT and Δknd1 samples based on 14N/15N peptide/protein ratios.
(b) Relative abundance of Cul1p and Cul1p interacting proteins in WT versus Δknd1 cells. Triplicate datasets (Supplementary Data File 1) were averaged and standard deviations are indicated. Statistically significant changes (t-test, p ≤ 0.05) are indicated by asterisks.
(c) Absolute amounts of Cul1p, Pof1p, and Pof10p retrieved from a 1:1 mixture of 15N-labeled WT and 14N-labeled Δknd1 cells by Cul1p IP and mass spectrometry-based quantification by selected ion monitoring. The right hand panel shows the same measurements for Cul1p and Pof15p from an independent Cul1p IP sample.
(d) Lysate of WT and Δknd1 cells expressing Pof1p or Pof10p tagged with the c-Myc epitope were used in immunoprecipitation with anti-Myc antibodies, followed by detection of co-precipitated proteins with either anti-Myc or anti-Cul1p antibodies. The neddylated and unneddylated forms of Cul1p are indicated. Total cell lysate is shown in the bottom two panels. The asterisk denotes a cross-reactivity of the anti-Cul1p antibody.
To corroborate these data, we performed a control experiment to exclude that CRL1 complexes rearrange during the immunopurification from mixed 14N and 15N cell lysates. For this, we mixed equal parts of 15N-labeled lysate from wildtype cells carrying a Cul1p-Myc allele with unlabeled lysate of Δknd1 cells lacking a Cul1p-Myc allele. When Cul1p complexes were purified and analyzed by LC-MS/MS, CRL1 components were almost exclusively represented by 15N peptides (Supplementary Fig. S1a,b). This finding indicates that 14N-labeled subunits present in the lysate of untagged cells did not enter CRL1 complexes at an appreciable rate after cell lysis. In contrast, proteins that were unspecifically retrieved on the beads were represented by both 14N and 15N peptides at roughly equal parts (Supplementary Fig. S1b).
We subsequently applied LC-MS/MS-based absolute protein quantification technology using isotopically labeled reference peptides (AQUA; 29) to measure the exact amounts of FBPs retrieved in CRL1 complexes. In agreement with the relative quantification data, the absolute amounts of Pof1p and Pof10p were 1.5 – 1.7-fold higher in complexes isolated from Δknd1 cells than from wildtype cells (Fig. 1c, Supplementary Data 2). While 3.5% of Cul1p was occupied by Pof1p in wildtype cells, the fraction increased to 6.2% in Δknd1 cells (Supplementary Data 2). Likewise, Cul1p occupancy by Pof10p increased from 14.5 to 24% in Δknd1 cells. These increases were confirmed by reciprocal immunoprecipitation/immunoblotting experiments (Fig. 1d, Supplementary Fig. S1c). Conversely, Pof15p levels were 3.5-fold lower in Cul1p complexes isolated from Δknd1 cells (Fig. 1c) and the fractional occupancy for this FBP was reduced from 4.6% to 1.5% (Supplementary Data 2). Again these absolute measurements confirmed the relative quantifications (Fig. 1b).
The observed changes in Cul1p-FBP interactions are not due to concordant changes in steady-state FBP levels in Δknd1 cells (Fig. 2a; Ref. 12). Likewise, CRL1 core subunits Cul1p and Rbx1p were retrieved in equal amounts from both wildtype and Δknd1 cells (Fig. 1b). Skp1p protein levels were ~40% higher in the mutant as determined by immunoblotting and quantitative mass spectrometry (Fig. 2a,b). Nevertheless, binding of Skp1p to Cul1p was decreased by ~25% in Δknd1 cells (Fig. 1b). This finding, which contrasts with previous measurements in human cells showing that binding of SKP1 to CUL1 is slightly increased upon acute knockdown of CAND1 13,14,27, was highly reproducible across 179 individual Skp1p peptide measurements by LC-MS/MS (Supplementary Data 3). It therefore appears that S. pombe cells devoid of Knd1p activity might have a ~25% overall reduction in fully assembled CRL1 complexes. The complexes that are assembled, however, dramatically differ in FBP composition relative to wildtype cells.
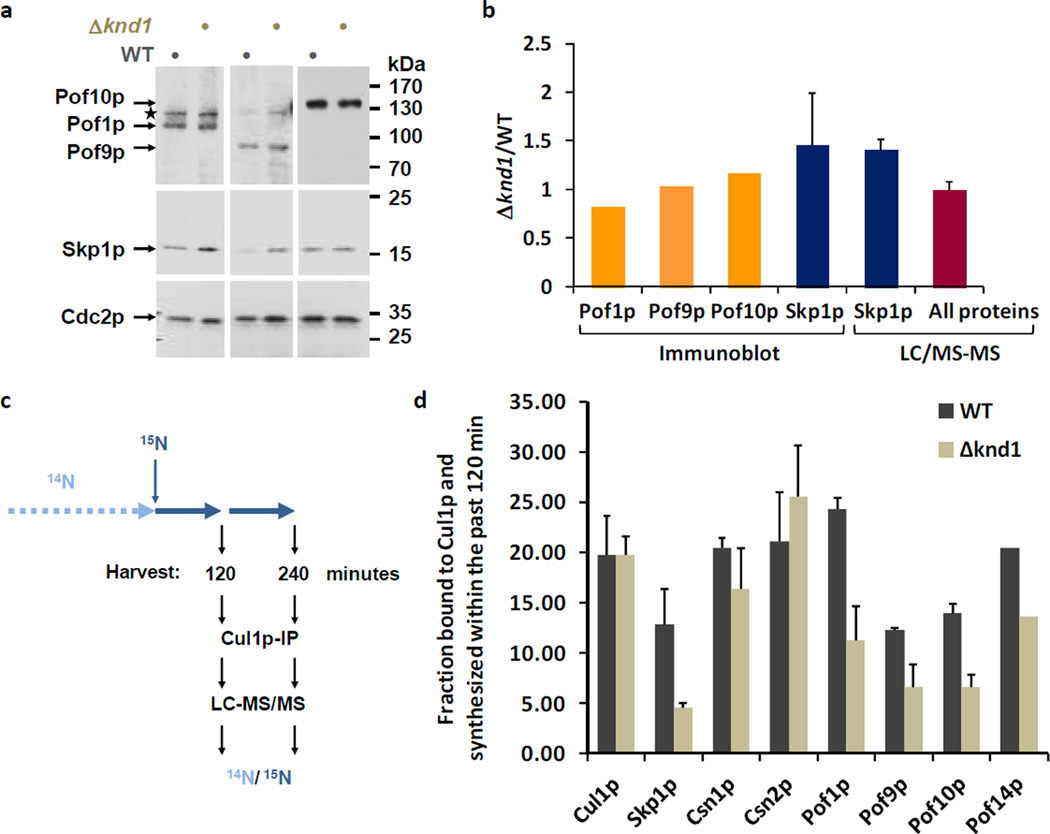
Figure 2. Effect of Knd1p on CRL1 dynamics.
(a) Steady-state levels of FBPs in WT and Δknd1 cells. The expression of Pof1p, Pof9p, and Pof10p modified with c-Myc epitopes at the endogenous genomic locus was determined by immunoblotting with Myc antibodies. Blots were reprobed with anti-Skp1p and anti-Cdc2p (PSTAIR) antibodies as a reference.
(b) The first 3 bars show a quantification of the FBP ratios in Δknd1 versus WT cells apparent from the immunoblots in (A) as determined by film densitometry. Ratios are from values normalized to Cdc2p. The fourth bar shows the average Skp1p ratio obtained from 4 independent immunoblot measurements including standard deviation. The fifth bar shows the Δknd1/WT ratio of Skp1p quantified by LC-MS/MS. Skp1p was immunopurified independently four times from 1:1 mixtures of 15N (WT) and 14N (Δknd1) labeled cells. The average ratio and the standard deviation were plotted. The sixth bar shows the average ratio of all proteins identified in the Skp1p purification. The ratio of these background proteins was close to 1.0 with a narrow standard deviation, supporting the significance of the ~1.5-fold increase in Skp1p levels in Δknd1 cells.
(c) Schematic outline of the 15N pulse labeling strategy. Cells maintained in routine 14N media were shifted to 15N media for 120 or 240 minutes prior to preparation of cell lysate, Cul1p-IP, and analysis by LC-MS/MS.
(d) Relative fractions of newly synthesized FBPs and CSN subunits associated with Cul1p in wildtype and Δknd1 cells. Data represent averages and standard deviations of three independent experiments (see Supplementary Data 4).
CAND1/Knd1p regulates CRL1 dynamics
To determine how the CRL1 repertoire of cells lacking CAND1/Knd1p could be disturbed in the presence of stable amounts of core components and FBPs, we developed a metabolic 15N pulse-labeling assay that allowed us to assess CRL1 complex dynamics. Wildtype and Δknd1 cells were maintained in 14N medium and then switched to 15N medium for 120 minutes, followed by lysate preparation, Cul1p immunopurification, and LC-MS/MS analysis of the retrieved proteins (Fig. 2c). Since we had previously shown that CAND1/Knd1p does not affect FBP stability 12, changes in the 14N/15N peptide ratios of CRL1 components inform primarily on the rate with which newly synthesized components become assembled into CRL1 complexes in wildtype cells relative to Δknd1 cells. Performing this experiment in triplicate, we found a statistically significant diminution in the incorporation of Skp1p and several FBPs into CRL1 complexes (Fig. 2D, Supplementary Data 4). The observation was confirmed in an independent experiment in which the pulse labeling was performed for 240 minutes (Supplementary Fig. S2, Supplementary Data 5).
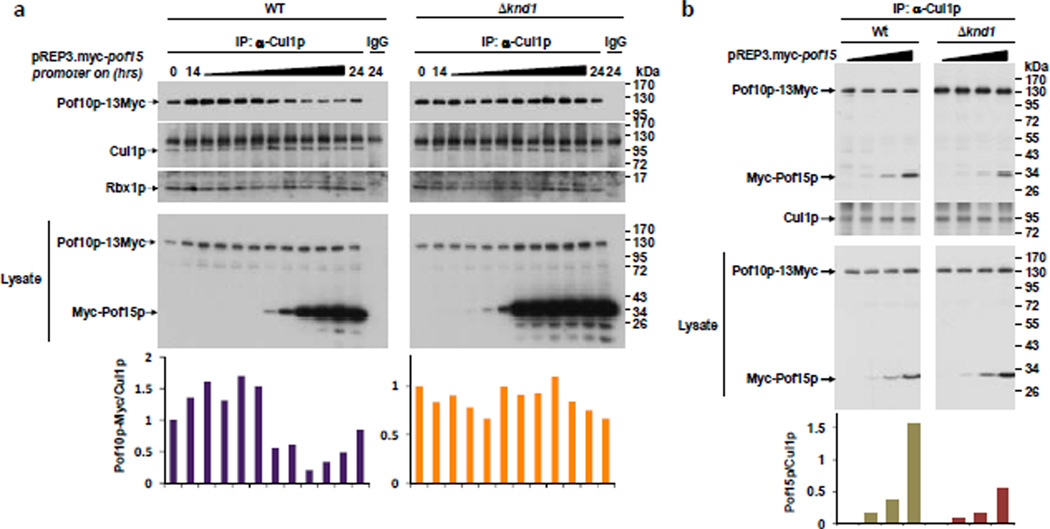
Slow incorporation of newly synthesized FBP may occur because preexisting CRL1 complexes are stabilized in the absence of knd1. We tested this prediction in an in vivo competition experiment. We perturbed steady-state CRL1 complexes by expressing from a plasmid Myc epitope-tagged Pof15p. This FBP is severely depleted from CRL complexes in Δknd1 cells (Fig. 1b, Supplementary Table S1) indicating that it requires Knd1p to successfully compete for CRL1 core subunits. Pof15p was expressed in a strain that harbors endogenously tagged Pof10p-Myc, an FPB that strongly accumulates in CRL1 complexes in Δknd1 cells (Fig. 1b, Supplementary Table S1). The strains were constructed in two backgrounds, wildtype and Δknd1. Myc-Pof15p expression was turned on and CRL1 complexes were immunoprecipitated after different times and monitored for the levels of co-precipitated Pof10p-Myc. We found that a ~10-fold excess of exogenous Pof15p efficiently displaced Pof10p-Myc from Cul1p (Fig. 3a). This competition was completely abolished by disrupting the F-box motif of Pof15p, confirming that it is not an indirect effect of overexpressing Pof15p (Supplementary Fig. S3). Importantly, the displacement of Pof10p-Myc from Cul1p by the competing Pof15p was drastically reduced in the absence of Knd1p even though Pof15p accumulated more readily upon overexpression in Δknd1 cells (Fig. 3a). The inefficiency of Pof15p in competing off Pof10p from Cul1p in the absence of Knd1p suggests that Pof15p is compromised in assembling into a CRL1 complex. Indeed, Pof15p expressed at low, non-competing levels in wildtype and Δknd1 cells was less efficiently recruited into a complex with Cul1p when Knd1p was missing (Fig. 3b). Thus, CAND1/Knd1p appears to assist in CRL1 assembly by destabilizing pre-existing Cul1p-FBP interactions thereby making Cul1p available for engagement in new CRL1 complexes.
Figure 3. CRL1 complexes are stabilized in the absence of Knd1p.
(a) Myc epitope-tagged Pof15p was expressed from a pREP3 plasmid containing an inducible promoter in strains that harbor endogenously tagged Pof10p-Myc. The strains were constructed in two backgrounds, wildtype (WT) and Δknd1. Myc-Pof15p expression was switched on by the removal of thiamine for 14 – 24 h as indicated. CRL1 complexes were immunoprecipitated with anti-Cul1p antibodies and monitored for the levels of co-precipitated Pof10p-Myc and Myc-Pof15p. As a specificity control, the 24 h lysate was used for immunoprecipitation with rabbit IgG. The level of Pof10p bound to Cul1p was quantified and plotted in a bar chart. The bottom panel shows total lysates.
(b) Same experiment as in (a) but with Myc-Pof15p expressed at low, non-competing levels. The level of Pof15p bound to Cul1p was quantified and plotted in a bar chart.
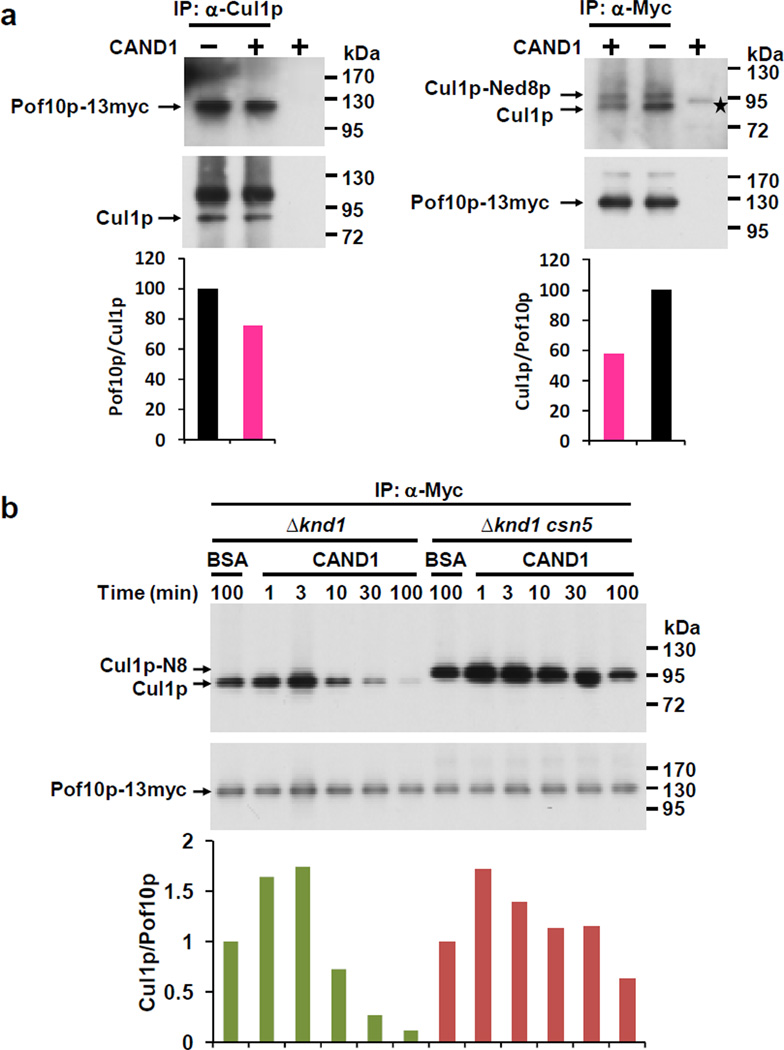
CAND1 displaces FBPs from Cul1p in vitro
To further test this idea, we asked whether recombinant CAND1 could displace FBPs from Cul1p. Cul1p complexes were immunopurified and incubated with recombinant human CAND1 (Supplementary Fig S4a) for 30 minutes followed by measuring the amount of Pof10p-Myc retained in the complex. In a reciprocal experiment, Pof10p-Myc complexes were purified and incubated with CAND1. In both experiments, CAND1 led to an apparent release of Pof10p from Cul1p. The same activity of CAND1 was observed for Pof1p-Myc complexes (Supplementary Fig. S4b). A kinetic experiment showed that the FBP releasing activity of CAND1 depended on its ability to bind Cul1p, since fully neddylated Cul1p complexes isolated from Δknd1 Δcsn5 cells were largely resistant to CAND1-mediated FBP release (Fig. 4B). The minor release of Pof10p that occurred after 100 minutes of incubation with CAND1 is consistent with the recent observation that neddylated Cul1p retains a minimal ability to interact with CAND1 30. Lack of the release activity of CAND1/Knd1p may thus cause the increased stability of Cul1p-FBP interactions observed in vivo.
Fig. 4. CAND1 displaces F-box proteins from Cul1p.
(a) Cul1p- and Pof10p-associated protein complexes were immunopurified from Δknd1 cells and incubated with 1 µg of recombinant His-tagged human CAND1 for 30 minutes. 1 µg of bovine serum albumin was used as control instead of CAND1. The complexes were analyzed for the levels of Cul1p and Pof10p-Myc by immunoblotting with Cul1p and Myc antibodies, and signals were quantified. The asterisk denotes a band present in the CAND1 preparation that weakly cross-reacts with the α-Cul1p antiserum (possibly insect cell CUL1).
(b) Pof10p-Myc complexes were isolated from Δknd1 and Δknd1 csn5 cells as in (a) and incubated for the indicated times with 1 ug BSA or CAND1 as indicated. The complexes were analyzed for the levels of Cul1p and Pof10p-Myc by immunoblotting with Cul1p and Myc antibodies, and signals were quantified.
CAND1/Knd1p is required for efficient substrate degradation
Our data suggested an important role for CAND1/Knd1p in organizing the cellular CRL1 repertoire such as to optimize substrate degradation. To test this, we assessed the impact of Knd1p on the degradation of the CRL1Pof3p substrate Ams2p 31. Similar to what we describe for CRL1 complexes containing Pof5p, Pof11p, and Pof15p in Fig. 1b, the level of CRL1Pof3p is approximately 50% diminished in Δknd1 cells 12. Consistent with this downregulation, we found a ~2-fold upregulation of the steady state levels of Ams2p in Δknd1 cells, which coincided with an increase in half-life (Fig. 5a). Similar to Δpof3 cells which accumulate Ams2p 24, we found Δknd1 cells to be sensitive to the genotoxic replication inhibitor hydroxyurea (Fig. 5B). CAND1/Knd1p-mediated maintenance of the CRL1 landscape therefore appears critical for substrate degradation and normal physiology.
Fig. 5. Knd1p is required for efficient CRL1-mediated substrate degradation.
(a) Wildtype and Δknd1 cells were incubated with 100 ug/ml cycloheximide for the indicated times and Ams2p levels were determined by immunoblotting relative to the loading control Cdc2p. The graph on the right shows a quantification of the results. Exponential trend lines were fitted through the data points in Excel, and the resulting equations were used to calculate half-lives.
(b) Hydroxyurea sensitivity of Δknd1 cells. 5-fold serial dilutions of wildtype and Δknd1 cells were spotted onto media with (bottom) or without (top) 10 mM hydroxyurea (HU).
Discussion
Our data obtained by complete genetic ablation of CAND1 suggest a novel function of CAND1/Knd1p in controlling the in vivo dynamic exchange of FBPs from Cul1p. We propose that lack of this activity “freezes” FBP exchange. Therefore, in the presence of stable but limiting amounts of Cul1p, those CRL1 complexes that are formed by FBPs that have relatively higher intrinsic ratios of on-rate/off-rate for Cul1p would out-compete those formed by FBPs with lower ratios. In addition, the degradation rates of individual FBPs may vary and, in the absence of CAND1/Knd1p, FBPs that turn over more quickly would become progressively displaced by those that are more stable thus leading to the observed imbalances. These imbalances may augment the degradation of substrates of CRL1 complexes that accumulate in the absence of CAND1/Knd1p while at the same time curtailing the destruction of substrates of those CRL1 ligases that are depleted. The CRL1Pof3p substrate Ams2p is one such example. This interpretation can reconcile conflicting reports of simultaneous positive as well as negative effects of CAND1 depletion on CRL-mediated substrate degradation in vivo 32.
The proposition that CAND1/Knd1p functions transiently as a FBP exchange factor can rationalize our previous observation that only a very small fraction of Knd1p is in a stable complex with Cul1p 12, a finding that was subsequently confirmed in human cells 27. This finding combined with the data on CRL dynamics and our demonstration that CAND1 can displace FBPs from Cul1p in vitro strongly suggest that CAND1 drives a cycle of continuous and rapid FBP recruitment and displacement which we dubbed the “CAND1 cycle” 12. This cycle would provide the dynamicity that assures comprehensive sampling of the steady-state FBP repertoire. Our data therefore qualify the conclusion of a previous study that CRL network organization is driven by the abundance of FBPs but not by cycles of CAND1 binding and release 27. Rather we argue that CAND1/Knd1p accounts for the activity that assures that the CRL1 repertoire is a reflection of the steady-state abundance of FBPs. These considerations also resolve the surprising observation that the CRL1 repertoire is not greatly disturbed when CUL1 neddylation is pharmacologically inhibited by MLN4924 27,33 because CAND1-mediated equilibration of FBPs would continue to operate - and in fact would be predicted to be more efficient - in the absence of neddylation.
From our studies, the general principle arises that the problem of substrate receptor competition that is inherent to the vast combinatorial architecture of CRLs is solved through the coordinated interplay of the CAND1 cycle with the neddylation/deneddylation cycle. For a given molecule of CUL1, the CAND1-mediated continuous substrate receptor exchange cycle would be interrupted only upon neddylation. Since substrate can stimulate cullin neddylation 19,30,34, CRLs containing receptors fitting a particular substrate that is present at a certain point in time would be selectively removed from the CAND1 cycle and thus activated. Upon substrate consumption, substrate receptors likely succumb to autocatalytic destruction or be recycled 11,35. COP9 signalosome-mediated deneddylation would reenter these complexes into the CAND1 cycle. In this scenario, which is consistent with in vitro data 18,19 as well as our study on in vivo CRL dynamics, CAND1 would function as a substrate receptor exchange factor that accelerates the approach of the unneddylated CRL1 repertoire to equilibrium. Interestingly, CAND1 was originally identified as a factor that stimulates the integration of RNA polymerase II/TFIIF into the TFIID-TFIIB-DNA-complex 36, raising the intriguing possibility that CAND1 may also promote the dynamics of other large protein complexes that require the same high degree of dynamicity as CRLs.
Methods
15N stable isotope labeling and immunopurification of CRL1
The yeast strains used in this study are summarized in Table 1. For stable isotope labeling, cells lacking any nutritional markers were grown at 30 °C for 10 generations on Edinburgh Minimal Medium (EMM) plates containing either ammonium-14N chloride (strain SDW542/2, cul1-13myc:kan knd1::ura4) or ammonium-15N chloride (strain SDW542/1, cul1-13myc:kan) as the nitrogen sources. 15N labeling efficiency was at least 98% as determined by mass spectrometry. For immunopurification of CRL1 complexes, cells were grown to OD600nm ~1.0 in 1000 ml EMM supplemented with the respective light or heavy ammonium chloride. Cells were harvested by centrifugation and washed with STOP buffer (150 mM NaCl, 10 mM EDTA, 50 mM NaF, 1 mM NaN3) once and flash frozen in liquid nitrogen. Equal amounts of wildtype and ∆knd1 mutant cells were mixed and disrupted by bead lysis in 14 ml immunoprecipitation buffer (25 mM Tris-HCl pH7.5, 50 mM NaCl, 0.1% Triton X100, 1 mM PMSF, 5 µg/ml aprotinin, 10 µg/ml leupeptin, 10 µg/ml pepstatin). Cell lysates were cleared by centrifugation at 18000 rpm and subjected to immunoprecipitation with 300 ul pre-equilibrated anti-c-Myc agarose (Clontech, cat#631208) for 2 hours. Beads were washed and eluted with 200 ul of 100 mM glycine pH 2.0 and neutralized with 20 ul of 2M Tris-HCl, pH 9.0. 5% of the eluate was loaded onto a gel to estimate the protein amount. The remainder was analyzed by LC-MS/MS.
Pulse labeling with 15N
Wildtype (sdw542/1) and ∆knd1 mutant cells (sdw542/2) were grown to OD600nm ~1.0 in 1000 ml EMM containing ammonium-14N chloride (light medium) at 30 °C. Cells were harvested by centrifugation and transferred to 1000 ml of EMM containing ammonium-15N chloride (heavy media). After labeling periods of 120 or 240 minutes, cells were harvested by centrifugation and washed with STOP buffer once and frozen in liquid nitrogen. Cell lysis and affinity purification of CRL1 complexes was as described above.
1D liquid chromatography and tandem mass spectrometry
Immunopurified protein complexes were first reduced and alkylated using TCEP and iodoacetamide, then digested using Trypsin as previously described in detail 37. After desalting, the peptides were analyzed by reverse-phase LC-MS/MS, using a Michrom Paradigm HPLC with a Magic C18 column and a LTQ-Orbitrap XL mass spectrometer (Thermo-Fisher). The peptides were separated on a 120 min gradient of 10% – 30% buffer B (100% acetonitrile/ 0.1% formic acid). Tandem mass spectrometry (MS/MS) spectra were collected during the LC-MS runs. Each scan was set to acquire a full MS scan, followed by MS/MS scans on the four most intense ions from the preceding MS scan. To maximize coverage, each IP sample was analyzed by LC-MS/MS three times (technical replicates).
Data analysis and protein quantification
Both protein identification and quantification of isotopically labeled peptides/proteins were performed by IP2, a comprehensive program package including SEQUEST, ProLuCID, DTASelect, and Census (Integrated Proteomics Applications, Inc, San Diego, CA). First, MS1 and MS2 files were extracted by RawExtract (The Scripps Research Institute, La Jolla, CA) from the raw LC-MS/MS spectra obtained on the LTQ-Orbitrap XL. Then a Swissprot S. pombe protein database was uploaded into IP2, which automatically generated a reverse decoy protein database. The MS1 and MS2 files of the three technical replicates of each sample were uploaded to IP2 server, and the combined MS1 and MS2 files were searched using ProLuCID and the decoy database. Differential modification included 16 Da on methionine for oxidation. The identification results from ProLuCID were filtered and organized by DTASelect. The DTASelect parameters were carefully set so that the false positive rate of protein identification was below 1.5%. The heavy and light spectral counts were obtained for each protein after ProLuCID/ DTASelect analysis. Finally the quantification analysis was selected for 15N. Protein ratios and standard deviations (SDs) were calculated by Census (based on peptides with a factor of determination of ≥ 0.5. 15N enrichment of each sample was calculated by IP2 as well and found to be at least 98%. Protein quantifications with high standard deviations for individual peptides or low peptide numbers were manually examined, and outliers were removed if necessary. To adjust for slight differences in 15N labeling efficiency in short term experiments, the protein ratios in the pulse labeling samples were normalized to the average ratio of all proteins quantified in the corresponding sample from wildtype cells. Average protein ratios were averaged over multiple replicates and standard deviations and p-values were calculated. To obtain robust datasets, quantifications with a relative standard deviation > 35% were excluded from the final data.
Absolute quantification of CRL1 components by LC-MS/MS
Based on our LC-MS/MS analysis of tryptic digests, we selected peptides TFFETNFIENTK, ELADDDVIWHR, TGVSLQSFQFR, and LSFLDEYSLR as AQUA standard peptides for Cul1p, Pof1p,Pof10p, and Pof15p, respectively. The absolute quantification (AQUA) peptides were purchased from Thermo-Fisher and Sigma-Aldrich, with stable isotopes (13C, 15N) in the C-terminal lysine or arginine. Since the AQUA reference peptides are isotopically distinct from their corresponding endogenous 14N and 15N peptides, we can quantify both “light” and “heavy” proteins in a single analysis of immunopurified CRL1 complexes isolated from 15N or 14N-labeled cells. An LTQ-Orbitrap method was developed that performs selected ion monitoring (SIM) followed by MS/MS confirmation of the peptides. 100 fmol of the standard peptides were spiked into the digests of CRL1 complexes immunopurified purified from a 1:1 mixture of wildtype and Δknd1 cell lysate. The peptides were separated by the same 120 min gradient LC method on the same LC-LTQ Orbitrap XL system as used for peptide identification. During the AQUA analysis, SIM spectra were first collected in the 630–760 m/z range with 60,000 FTMS resolution, followed by MS/MS scans of the nine peptide ions with known m/z. Peak integrations were obtained through application of extracted ion chromatograms over 10-ppm mass intervals on Qual Browser (Thermo-Fisher). No contaminating peptides were found to co-elute with the three peptides chosen for Cul1p, Pof1p, Pof10p, and Pof15p. The analysis was repeated five times. Raw data for a representative AQUA experiment are shown in Supplementary Fig. 5.
Hydroxyurea sensitivity assay
Five-fold serial dilutions of wildtype (SDW542/1) and ∆knd1 mutant (SDW542/2) cells were spotted onto control YES plates or plates containing 10 mM hydroxyurea. Plates were incubated at 30 °C for 5 days.
FBP competition experiment
The pof15 (SPAPB1A10.14) gene was cloned into the vector pREP3-6His.Myc, which drives the expression of N-terminal 6His-Myc tagged protein from the thiamine repressible nmt1 promoter. A Pof15p mutant lacking residues 34 – 40 (LPVEVID) within the F-box motif was generated in the same expression vector. The plasmid were confirmed by sequencing and transformed into wildtype and ∆knd1 mutant cells harboring a Myc-tagged allele of pof10 at the endogenous genomic locus (pof10-13myc). The expression of pof15 was induced by removal of thiamine from the culture medium for increasing periods of time followed by preparation of cell lysates by bead lysis in immunoprecipitation buffer (25 mM Tris-HCl pH7.5, 50 mM NaCl, 0.1% Triton X100, 1 mM PMSF, 5 µg/ml aprotinin, 10 µg/ml leupeptin, 10 µg/ml pepstatin). Lysates were subjected to immunoprecipitation with rabbit anti-Cul1p antibody 38, followed by collection of immunocomplexes on protein A Sepharose beads (Rockland, cat#PA50-00-0005). The immunoprecipitated complexes were analyzed by immunoblotting. Myc-tagged FBPs were detected with a monoclonal anti-Myc antibody (9E10, cat#13-2500, Invitrogen).
Expression and purification of recombinant human CAND1
The CAND1 isoform 1 open reading frame was PCR amplified from the MegaMan Human Transcriptome library (Agilent) with 3' sequences encoding a hexahistidine tag immediately 5' of the stop codon and used to generate a recombinant baculovirus using the FastBac system (Invitrogen). After 48 hours of infecting Hi5 insect cell cultures (10e8 cells) at a multiplicity of infection greater than 10, CAND1 was purified by Ni-NTA chromatography and desalted into 25 mM HEPES pH 7.6, 100 mM NaCl, and 10% glycerol.
FBP displacement assay with recombinant CAND1
Cells harboring endogenous Pof10p modified with Myc epitopes were grown to an OD600nm of ~1.0 in 200 ml of YES. Cell lysates were prepared by beads lysis in immunoprecipitation buffer (25 mM Tris-HCl pH7.5, 50 mM NaCl, 0.1% Triton X100, 1 mM PMSF, 5 µg/ml aprotinin, 10 µg/ml leupeptin, 10 µg/ml pepstatin). Pof10p was immunopurified on pre-equilibrated anti-c-Myc agarose beads (Clontech, cat#631208). In a reciprocal experiment, Cul1p complexes were immunoprecipitated with Cul1p antibodies. The complexes were washed and divided into two aliquots. 1 µg of recombinant His-tagged human CAND1 expressed in and purified from insect cells was added to one aliquot and incubated at 30 °C for 30 minutes. 1 µg of bovine serum albumin was used as control instead of CAND1. After washing, the immunoprecipitated complexes were analyzed by immunoblot. Myc tagged protein was detected with monoclonal anti-c-Myc antibody (9E10, cat#13-2500, Invitrogen), and Cul1p with rabbit anti-Cul1p antibody.
Immunoprecipitation and immunoblotting of CRL1 complexes
Cell lysates for immunoprecipitation of Myc-tagged FBPs were prepared as described in a buffer containing 50 mM Tris, pH 7.4, 50 mM NaCl, and 0.5% Triton X-100 12. Lysates were cleared by centrifugation, and proteins were precipitated with the respective antisera (anti-Myc, anti-Rbx1p). Immunocomplexes were collected by binding to protein A beads, washed, and analyzed by immunoblotting. Affinity-purified rabbit antisera against Cul1p, Skp1p, and Rbx1p were described previously 38,39. Ams2p antisera were kindly provided by Y. Takayama 31. For loading controls Cdc2 (PSTAIRE, Santa Cruz) antibodies were used at a dilution of 1:500.
Supplementary Material
Acknowledgements
We thank Y. Takayama for Ams2p antibodies and Z. Ronai for review of the manuscript. This work was supported by NIH grants GM59780, P20 CA132386-03, and P50 GM085764-03.
Footnotes
Author Contributions
S.W. prepared yeast strains and all samples for MS analysis, contributed to MS data analysis, and performed immunoblots. W. Z. performed LC-MS/MS and MS data analysis. T.N. prepared yeast strains and performed immunoblot analyses and the hydroxyurea sensitivity experiment. J.I.T and M.D.P. expressed and purified human CAND1. D.A.W. conceived the study, directed the experimental design, performed immunoprecipitation experiments, assisted with data analysis, and drafted the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
REFERENCES
- 1.Petroski MD, Deshaies RJ. Function and regulation of cullin–RING ubiquitin ligases. Nature Reviews Molecular Cell Biology. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 2.Bosu DR, Kipreos ET. Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div. 2008;3:7. doi: 10.1186/1747-1028-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hua Z, Vierstra RD. The Cullin-RING Ubiquitin-Protein Ligases. Annu Rev Plant Biol. 2010;62:299–334. doi: 10.1146/annurev-arplant-042809-112256. [DOI] [PubMed] [Google Scholar]
- 4.Wolf DA, Zhou C, Wee S. The COP9 signalosome: an assembly and maintenance platform for cullin ubiquitin ligases? Nature Cell Biology. 2003;5:1029–1033. doi: 10.1038/ncb1203-1029. [DOI] [PubMed] [Google Scholar]
- 5.Duda D, et al. Structural Insights into NEDD8 Activation of Cullin-RING Ligases: Conformational Control of Conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saha A, Deshaies R. Multimodal Activation of the Ubiquitin Ligase SCF by Nedd8 Conjugation. Molecular Cell. 2008;32:21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lyapina S, et al. Promotion of NEDD8-CUL1 conjugate cleavage by COP9 signalosome. Science. 2001;292:1382. doi: 10.1126/science.1059780. [DOI] [PubMed] [Google Scholar]
- 8.Schwechheimer C, et al. Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIR1 in mediating auxin response. Science. 2001;292:1379. doi: 10.1126/science.1059776. [DOI] [PubMed] [Google Scholar]
- 9.Zhou C, et al. The fission yeast COP9/signalosome is involved in cullin modification by ubiquitin-related Ned8p. BMC Biochem. 2001;2:7. doi: 10.1186/1471-2091-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou C, et al. Fission yeast COP9/signalosome suppresses cullin activity through recruitment of the deubiquitylating enzyme Ubp12p. Molecular Cell. 2003;11:927–938. doi: 10.1016/s1097-2765(03)00136-9. [DOI] [PubMed] [Google Scholar]
- 11.Cope GA, Deshaies RJ. Targeted silencing of Jab1/Csn5 in human cells downregulates SCF activity through reduction of F-box protein levels. BMC biochemistry. 2006;7:1. doi: 10.1186/1471-2091-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt MW, McQuary PR, Wee S, Hofmann K, Wolf DA. F-Box-Directed CRL Complex Assembly and Regulation by the CSN and CAND1. Molecular Cell. 2009;35:586–597. doi: 10.1016/j.molcel.2009.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J, Furukawa M, Matsumoto T, Xiong Y. NEDD8 modification of CUL1 dissociates p120CAND1, an inhibitor of CUL1-SKP1 binding and SCF ligases. Molecular Cell. 2002;10:1511–1518. doi: 10.1016/s1097-2765(02)00783-9. [DOI] [PubMed] [Google Scholar]
- 14.Zheng J, et al. CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Mol. Cell. 2002;10:1519–1526. doi: 10.1016/s1097-2765(02)00784-0. [DOI] [PubMed] [Google Scholar]
- 15.Hwang J-W, Min K-W, Tamura T, Yoon J-B. TIP120A associates with unneddylated cullin 1 and regulates its neddylation. FEBS Lett. 2003;541:102–108. doi: 10.1016/s0014-5793(03)00321-1. [DOI] [PubMed] [Google Scholar]
- 16.Min K-W. TIP120A Associates with Cullins and Modulates Ubiquitin Ligase Activity. Journal of Biological Chemistry. 2003;278:15905–15910. doi: 10.1074/jbc.M213070200. [DOI] [PubMed] [Google Scholar]
- 17.Oshikawa K, et al. Preferential interaction of TIP120A with Cul1 that is not modified by NEDD8 and not associated with Skp1. Biochemical and biophysical research communications. 2003;303:1209–1216. doi: 10.1016/s0006-291x(03)00501-1. [DOI] [PubMed] [Google Scholar]
- 18.Siergiejuk E, et al. Cullin neddylation and substrate-adaptors counteract SCF inhibition by the CAND1-like protein Lag2 in Saccharomyces cerevisiae. The EMBO Journal. 2009;28:3845–3856. doi: 10.1038/emboj.2009.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bornstein G, Ganoth D, Hershko A. Regulation of neddylation and deneddylation of cullin1 in SCFSkp2 ubiquitin ligase by F-box protein and substrate. Proceedings of the National Academy of Sciences. 2006;103:11515–11520. doi: 10.1073/pnas.0603921103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chuang H, Zhang W, Gray WM. Arabidopsis ETA2, an apparent ortholog of the human cullin-interacting protein CAND1, is required for auxin responses mediated by the SCFTIR1 ubiquitin ligase. The Plant Cell Online. 2004;16:1883. doi: 10.1105/tpc.021923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng S, et al. Arabidopsis CAND1, an unmodified CUL1-interacting protein, is involved in multiple developmental pathways controlled by ubiquitin/proteasome-mediated protein degradation. The Plant Cell. 2004;16:1870. doi: 10.1105/tpc.021949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bosu DR, et al. C. elegans CAND-1 regulates cullin neddylation, cell proliferation and morphogenesis in specific tissues. Developmental Biology. 2010 doi: 10.1016/j.ydbio.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helmstaedt K, et al. Recruitment of the inhibitor Cand1 to the cullin substrate adaptor site mediates interaction to the neddylation site. Molecular Biology of the Cell. 2011;22:153–164. doi: 10.1091/mbc.E10-08-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katayama S, Kitamura K, Lehmann A, Nikaido O, Toda T. Fission Yeast F-Box Protein Pof3 Is Required for Genome Integrity and Telomere Function. Mol. Biol. Cell. 2002;13:211–224. doi: 10.1091/mbc.01-07-0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lo SC, Hannink M. CAND1-mediated substrate adaptor recycling is required for efficient repression of Nrf2 by Keap1. Molecular and cellular biology. 2006;26:1235. doi: 10.1128/MCB.26.4.1235-1244.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang W, et al. Genetic analysis of CAND1–CUL1 interactions in Arabidopsis supports a role for CAND1-mediated cycling of the SCFTIR1 complex. Proceedings of the National Academy of Sciences. 2008;105:8470–8375. doi: 10.1073/pnas.0804144105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bennett EJ, Rush J, Gygi SP, Harper JW. Dynamics of Cullin-RING Ubiquitin Ligase Network Revealed by Systematic Quantitative Proteomics. Cell. 2010;143:951–965. doi: 10.1016/j.cell.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Washburn MP, Ulaszek R, Deciu C, Schieltz DM, Yates III JR. Analysis of quantitative proteomic data generated via multidimensional protein identification technology. Analytical chemistry. 2002;74:1650–1657. doi: 10.1021/ac015704l. [DOI] [PubMed] [Google Scholar]
- 29.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. SciU.SA. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emberley ED, Mosadeghi R, Deshaies RJ. Deconjugation of Nedd8 from Cul1 is directly regulated by Skp1-Fbox and substrate, and CSN inhibits deneddylated SCF by a non-catalytic mechanism. J. Biol. Chem. 2012;287:29679–29689. doi: 10.1074/jbc.M112.352484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takayama Y, et al. Hsk1- and SCFPof3-Dependent Proteolysis of Spombe Ams2 Ensures Histone Homeostasis and Centromere Function. Developmental Cell. 2010;18:385–396. doi: 10.1016/j.devcel.2009.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim S-H, Kim H-J, Kim S, Yim J. Drosophila Cand1 regulates Cullin3-dependent E3 ligases by affecting the neddylation of Cullin3 and by controlling the stability of Cullin3 and adaptor protein. Developmental Biology. 2010 doi: 10.1016/j.ydbio.2010.07.031. [DOI] [PubMed] [Google Scholar]
- 33.Lee JE, et al. The steady-state repertoire of human SCF Ubiquitin ligase complexes does not require ongoing Nedd8 conjugation. Mol Cell Proteomics. 2010 doi: 10.1074/mcp.M110.006460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chew E-H, Hagen T. Substrate-mediated Regulation of Cullin Neddylation. Journal of Biological Chemistry. 2007;282:17032–17040. doi: 10.1074/jbc.M701153200. [DOI] [PubMed] [Google Scholar]
- 35.Wee S, Geyer RK, Toda T, Wolf DA. CSN facilitates Cullin–RING ubiquitin ligase function by counteracting autocatalytic adapter instability. Nature Cell Biology. 2005;7:387–391. doi: 10.1038/ncb1241. [DOI] [PubMed] [Google Scholar]
- 36.Makino Y, et al. TATA-Binding Protein-Interacting Protein 120, TIP120, Stimulates Three Classes of Eukaryotic Transcription Via a Unique Mechanism. Mol. Cell. Biol. 1999;19:7951–7960. doi: 10.1128/mcb.19.12.7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brill LM, Motamedchaboki K, Wu S, Wolf DA. Comprehensive proteomic analysis of Schizosaccharomyces pombe by two-dimensional HPLC-tandem mass spectrometry. Methods. 2009;48:311–319. doi: 10.1016/j.ymeth.2009.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seibert V, et al. Combinatorial diversity of fission yeast SCF ubiquitin ligases by homo- and heterooligomeric assemblies of the F-box proteins Pop1p and Pop2p. BMC Biochem. 2002;3:22. doi: 10.1186/1471-2091-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geyer R, Wee S, Anderson S, Yates J, Wolf DA. BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol. Cell. 2003;12:783–790. doi: 10.1016/s1097-2765(03)00341-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.