Abstract
Background
The limited within-breed genetic heterogeneity and an enrichment of disease-predisposing alleles have made the dog a very suitable model for the identification of genes associated with risk for specific diseases. Canine mammary cancer is an example of such a disease. However, the underlying inherited risk factors for canine mammary tumours (CMTs) are still largely unknown. In this study, 52 single nucleotide polymorphisms (SNPs) in ten human cancer-associated genes were genotyped in two different datasets in order to identify genes/alleles associated with the development of CMTs. The first dataset consisted of English Springer Spaniel (ESS) CMT cases and controls. ESS is a dog breed known to be at increased risk of developing CMTs. In the second dataset, dogs from breeds known to have a high frequency of CMTs were compared to dogs from breeds with a lower occurrence of these tumours.
Results
We found significant associations to CMT for SNPs and haplotypes in the estrogen receptor 1 (ESR1) gene in the ESS material (best PBonf = 0.021). A large number of SNPs, among them several SNPs in ESR1, showed significantly different allele frequencies between the high and low risk breed groups (best PBonf = 8.8E-32, best PBPerm = 0.076).
Conclusions
The identification of CMT-associated SNPs in ESR1 in two independent datasets suggests that this gene might be involved in CMT development. These findings also support that CMT may serve as a good model for human breast cancer research.
Keywords: Dog, Single nucleotide polymorphism, Allele frequency, Risk, Association, Mammary tumour, Estrogen receptor
Background
The modern dog breeds are a result of vigorous line-breeding and often originate from a few founding ancestors. This has led to extreme phenotypic variation between breeds, but limited genetic variation within breeds [1]. Some breeds have a considerably higher susceptibility to certain diseases than others. This indicates an enrichment of risk alleles within these specific breeds. Canine cancer is an example of such a disease. Mammary tumours are among the most common canine cancer forms [2]. Elderly, intact bitches are primarily affected by these tumours [3,4], with a higher incidence in breeds such as the English Springer Spaniel, Boxer, Cocker Spaniel and Dachshund [3-8]. The differences in breed predisposition clearly indicate a genetic influence on canine mammary tumour (CMT) development. There are many similarities between CMTs and human breast cancer. Among the shared disease characteristics are a spontaneous occurrence of tumours, by which females are primarily affected, and a hormonal influence on tumour development (e.g. oestrogen and progesterone) (as reviewed by Queiroga et al. 2011 [9]). Regional lymph node metastasis seems to be less important as a prognostic factor in dogs than in humans, but metastatic spread is otherwise broadly equal [10]. There are also similarities regarding the histological features and classifications of human breast cancer and CMTs. But while epithelial tumours are by far the most common in humans, CMTs relatively frequently also contain myoepithelial and mesenchymal components. Furthermore studies indicate that CMT and human breast cancer have several mutual prognostic markers and genetic risk factors for disease (as reviewed by Queiroga et al. 2011 [9]). Still, the underlying inherited risk factors for CMTs are largely unknown. Previous studies of genotypes, gene and protein expression in CMTs have identified possible candidate genes and pathways, but the results need to be confirmed or are somewhat inconsistent. However, a study on BRCA1 and BRCA2, two genes well-known to be involved in human breast cancer, showed associations of these genes with mammary tumours in English Springer Spaniels [11]. Another CMT study found mutations in the cancer-associated gene TP53[12]. We have previously identified a considerable number of single nucleotide polymorphisms (SNPs) in ten cancer-associated genes known from studies in dogs and/or humans [13]. Some of these SNPs are likely to be associated with canine cancer, and the protein-changing SNPs are of particular interest due to their potential as functional disease-causing variants. In the present study, we aimed at exploring such CMT associations to identify genes involved in the development of mammary tumours in a case-control population of English Springer Spaniels (ESS) and in a second dataset of high and low risk breeds of CMT.
Results
After quality control, 165 ESS cases and 94 controls were left for analysis (Table 1). Of the 41 SNPs that passed quality control in the ESS case-control dataset, nominal single SNP association was found for two SNPs in ESR1 exon 2 (rs21960513) and intron 7–8 (ss244244344) (PRaw = 0.033 and 0.002, respectively, Table 2). Another SNP in ESR1 (ss244244343) showed borderline nominal significance (PRaw = 0.052). The risk alleles for the two SNPs ss244244344 and ss244244343 are extremely common in the ESS cohort (Table 2). The ss244244344 and rs21960513 SNPs were still significant after correcting for multiple testing using 10,000 permutations (PPerm = 0.018 and 0.042, Table 2). Ss244244344 also remained significant after applying Bonferroni correction. Of the classified cases, 32 malignant and 78 benign were left after quality control (Table 1). In the comparison of cases with malignant diagnosis vs. controls, ss244244344 showed nominal significance (PRaw = 0.016. Additional file 1: Table S3), but the result was not significant after multiple testing correction. We found no significant differences in allele frequencies between benign cases and controls or malignant and benign cases. All association results are provided in Additional file 1.
Table 1.
Datasets and samples
|
Dataset/Breed |
No. samples |
CMT grouping |
|
|---|---|---|---|
| Before QCa | After QC | ||
|
English Springer Spanielb |
|
|
|
| |
176 |
165 |
Cases |
| |
83/33 |
78/32 |
Benign/malignant casesc |
| |
122 |
94 |
Controls |
|
Canine Biobank dataset |
|
|
|
| Boxer |
50 |
46 |
High risk |
| Cocker Spaniel |
50 |
46 |
High risk |
| Dachshund |
50 |
48 |
High risk |
| English Setter |
50 |
50 |
High risk |
| Standard Poodle |
50 |
47 |
High risk |
| Beagle |
50 |
47 |
Low risk |
| Bernese Mountain Dog |
50 |
49 |
Low risk |
| Collie |
50 |
45 |
Low risk |
| Shetland Sheepdog | 50 | 50 | Low risk |
a QC: Quality control.
b Subset from the study by Rivera et al. [11].
c As identified by histopathology by a veterinary pathologist at the time of the genotyping analysis. See Methods for further description.
Table 2.
CMT-associated ESR1 SNPs in the ESS cases and controls
|
SNP |
NCBI SNP ID |
Minor allele |
Frequency of minor allele |
Major allele |
PRawa |
PBonfb |
PPermc |
OR |
|
|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | ||||||||
|
ESR1 INT 7-8a |
ss244244344 |
A |
0.033 |
0.106 |
G |
0.002 |
0.021 |
0.018 |
0.3 |
|
ESR1 EX 2 |
rs21960513 |
C |
0.299 |
0.391 |
T |
0.033 |
0.362 |
0.042 |
0.7 |
| ESR1 EX 4 | ss244244343 | G | 0.006 | 0.027 | A | 0.052 | 0.568 | 0.155 | 0.2 |
(n = 165 and n = 94, respectively, after QC).
aP-value from chi-square test in PLINK.
b Bonferroni corrected for the number of LD blocks (11).
c EMP1 value after 10,000 permutations in PLINK.
We identified 11 LD blocks with D’ ~ 1 and LOD ≥ 2 in the ESS case-control dataset. One LD block was identified in each of the genes BRCA2, BRIP1, CDH1, CHEK2, EGFR and PTEN, whereas two and three blocks were found within ESR1 and ERBB2 (HER2), respectively. Only one SNP was genotyped in BRCA1 and STK11, making LD and haplotype analysis impossible. Nominal association was found for one and two haplotypes in each of two ESR1 LD blocks, respectively, and one haplotype showed borderline nominal significance (Table 3). No haplotypes remained significant after correction for multiple testing. As the total cancer risk of an individual is probably a result of risk alleles at multiple loci, we evaluated the combined effect of the risk haplotypes of the two ESR1 LD blocks compared to the protective haplotypes. A borderline Bonferroni-significant association was found for the combined risk haplotypes with an odds ratio of 3.3 (PBonf = 0.055) (Table 4).
Table 3.
CMT-associated ESR1 haplotypes in the ESS cases and controls
|
Haplotype |
Frequency |
PRawa |
PBonfb |
PPermc |
OR |
||
|---|---|---|---|---|---|---|---|
| Total | Cases | Controls | |||||
|
ESR1 EX 2 and ESR1 EX 4 (rs21960513 and ss244244343) | |||||||
|
TA |
0.668 |
0.701 |
0.610 |
0.035 |
0.381 |
0.549 |
1.5 |
|
CA |
0.318 |
0.293 |
0.363 |
0.098 |
1.000 |
0.941 |
0.7 |
|
CG |
0.014 |
0.006 |
0.027 |
0.052 |
0.572 |
0.741 |
0.2 |
|
ESR1 INT 7-8a, –b and ESR1 EX 8 (ss244244344, ss244244345 and ss244244346) | |||||||
|
GTG |
0.791 |
0.818 |
0.744 |
0.047 |
0.517 |
0.683 |
1.5 |
| ACA | 0.091 | 0.070 | 0.129 | 0.024 | 0.264 | 0.480 | 0.5 |
aP-value from chi-square test in Haploview.
b Corrected for the number of LD blocks available (11).
c Permutation P-value from Haploview, 10,000 permutations.
Table 4.
Combined effect of the haplotypes of the two ESR1 LD blocks in ESS cases and controls
|
Haplotype combination |
Frequency |
Haplotype comparison |
OR (95% CI) |
PRawa |
PBonfb |
|
|---|---|---|---|---|---|---|
| Cases | Controls | |||||
|
TA-GTG (risk) |
207 |
97 |
Risk TA-GTG vs. rest |
1.8 (1.2 - 2.8) |
0.007 |
0.077 |
| |
|
|
Risk TA-GTG vs. protective CA-ACA |
3.3 (1.4 - 7.9) |
0.005 |
0.055 |
|
CA-ACA (protective) |
9 |
14 |
Protective CA-ACA vs. rest |
0.3 (0.1 - 0.8) |
0.010 |
0.110 |
| Other c | 56 | 41 | ||||
a Fisher-Exact test applied due to low counts of the CG haplotype (less than five in both cases and controls).
b Corrected for the number of LD blocks (11).
c For 29 cases and 18 controls, the haplotype combinations could not be fully determined due to genotyping failure of one or more of the ESR1 SNPs.
In the second dataset, 237 dogs of high risk breeds, 191 dogs of low risk breeds and 43 SNPs passed quality control and were included in the study of allele frequencies (Table 1). Nineteen of the 31 SNPs with nominal single SNP associations were significant after 107 permutations, 23 after Bonferroni correction, and among them were the three ESR1 SNPs rs21960513, ss244244343 and ss244244344 (Table 5).
Table 5.
SNPs significantly associated with CMT high and low risk breed groups after Bonferroni correction
|
SNP |
NCBI SNP ID |
Minor allele |
Frequency minor allele |
Major allele |
OR |
PRawa |
PBonfb |
PPermc |
PBPermd |
|
|---|---|---|---|---|---|---|---|---|---|---|
| High risk breeds | Low risk breeds | |||||||||
|
CHEK2 INT 8-9 |
ss244244336 |
G |
0.269 |
0.040 |
T |
8.8 |
8.6E-19 |
7.8E-18 |
<1.0E-07 |
0.076 |
|
ESR1 EX 2 |
rs21960513 |
T |
0.629 |
0.305 |
C |
3.9 |
5.0E-21 |
4.5E-20 |
<1.0E-07 |
0.107 |
|
ERBB2 EX 14 |
rs24537331 |
A |
0.253 |
0.511 |
G |
0.3 |
8.4E-15 |
7.6E-14 |
<1.0E-07 |
0.110 |
|
BRCA2 INT 18-19 |
ss244244323 |
G |
0.000 |
0.133 |
A |
_ e |
1.8E-16 |
1.6E-15 |
<1.0E-07 |
0.110 |
|
ERBB2 INT 3-4 |
ss244244348 |
G |
0.330 |
0.681 |
A |
0.2 |
2.4E-24 |
2.2E-23 |
<1.0E-07 |
0.112 |
|
BRCA2 EX 11 |
rs23244160 |
G |
0.174 |
0.346 |
T |
0.4 |
8.4E-09 |
7.6E-08 |
4.2E-05 |
0.151 |
|
CHEK2 INT 5-6 |
ss244244335 |
G |
0.051 |
0.000 |
A |
_ e |
9.6E-06 |
8.6E-05 |
0.005 |
0.182 |
|
ESR1 EX 4 |
ss244244343 |
G |
0.061 |
0.394 |
A |
0.1 |
9.8E-33 |
8.8E-32 |
<1.0E-07 |
0.204 |
|
PTEN EX 9 |
ss244244369 |
T |
0.112 |
0.346 |
C |
0.3 |
1.3E-16 |
1.2E-15 |
<1.0E-07 |
0.209 |
|
BRCA2 EX 5 |
rs23250374 |
C |
0.429 |
0.23 |
T |
2.5 |
1.1E-09 |
1.0E-08 |
1.0E-05 |
0.321 |
|
ERBB2 INT 12-13b |
rs24537327 |
A |
0.485 |
0.283 |
G |
2.4 |
2.2E-09 |
1.9E-08 |
2.0E-05 |
0.362 |
|
ERBB2 EX 13b |
rs24537329 |
T |
0.486 |
0.285 |
C |
2.4 |
2.4E-09 |
2.2E-08 |
2.1E-05 |
0.368 |
|
PTEN INT 7-8 |
ss244244368 |
A |
0.183 |
0.356 |
G |
0.5 |
8.0E-09 |
7.2E-08 |
4.0E-05 |
0.385 |
|
BRCA2 5UTR |
ss244244322 |
T |
0.343 |
0.506 |
C |
0.5 |
2.7E-06 |
2.4E-05 |
0.002 |
0.407 |
|
PTEN INT 3-4 |
ss244244367 |
G |
0.191 |
0.346 |
A |
0.6 |
2.8E-07 |
2.5E-06 |
5.0E-04 |
0.416 |
|
STK11 INT 1-2 |
rs22928814 |
T |
0.241 |
0.362 |
C |
0.6 |
1.1E-04 |
1.0E-03 |
0.031 |
0.476 |
|
BRIP1 EX 19b |
ss244244329 |
T |
0.538 |
0.386 |
T |
1.9 |
9.9E-06 |
8.9E-05 |
0.006 |
0.521 |
|
ERBB2 INT 8-9 |
ss244244349 |
G |
0.089 |
0.027 |
A |
3.2 |
0.001 |
5.8E-03 |
0.101 |
0.566 |
|
ESR1 INT 7-8a |
ss244244344 |
A |
0.124 |
0.223 |
G |
0.5 |
2.3E-04 |
2.2E-03 |
0.051 |
0.708 |
|
BRIP1 INT 8-9a |
ss244244325 |
G |
0.135 |
0.011 |
A |
14.5 |
3.2E-11 |
2.9E-10 |
<1.0E-07 |
0.817 |
|
ERBB2 INT 1-2 |
ss244244347 |
T |
0.021 |
0.000 |
C |
_ e |
0.005 |
0.045 |
0.340 |
0.845 |
|
BRIP1 INT 15-16 |
ss244244327 |
T |
0.131 |
0.011 |
A |
14.0 |
8.0E-11 |
7.2E-10 |
<1.0E-07 |
0.874 |
| ERBB2 EX13a | rs24616607 | G | 0.044 | 0.005 | C | 8.5 | 0.001 | 5.2E-03 | 0.095 | 0.983 |
(n = 237 and n = 191, respectively, after QC).
aP-value from chi-square test.
b Corrected for the number of across-breed LD blocks (9).
c EMP1 value after 107 permutations.
d EMP1 value after 10,000 breed permutations.
e OR could not be calculated as the minor allele was absent in either cases or controls.
However, there were considerable inter-breed variations in SNP allele frequencies for the high and low risk dataset also within the high and low risk breed groups (Table 6). Thus, the mean allele frequency of the group was often not representative for all the breeds included. The overall differences in allele frequency among breeds also caused general inflation of association P-values, complicating the interpretation. Breed permutation testing was therefore performed to correct for the inflation. None of the SNPs were significant after breed permutation testing (Table 5).
Table 6.
Breed-wise average allele frequencies in the high and low risk breed dataset
|
SNP |
Allele frequency (minor allele) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Minor allele | Boxer | Cocker Spaniel | Dachshund | English Setter | Standard Poodle | Beagle | Bernese Mountain Dog | Collie | Shetland Sheepdog | |
|
BRCA2 5UTR |
T |
0.138 |
0.589 |
0.682 |
0.177 |
0.167 |
0.244 |
0.904 |
0.524 |
0.330 |
|
BRCA2 EX 5 |
C |
0.130 |
0.848 |
0.292 |
0.160 |
0.720 |
0.141 |
0.684 |
0.055 |
0.011 |
|
BRCA2 EX 11 |
G |
0.202 |
0.141 |
0.417 |
0.020 |
0.100 |
0.160 |
0.245 |
0.556 |
0.436 |
|
BRCA2 INT 18-19 |
G |
0.000 |
0.000 |
0.000 |
0.000 |
0.000 |
0.000 |
0.000 |
0.463 |
0.117 |
|
BRIP1 INT 8-9a |
G |
0.660 |
0.000 |
0.021 |
0.000 |
0.010 |
0.021 |
0.020 |
0.000 |
0.000 |
|
BRIP1 INT 15-16 |
T |
0.652 |
0.000 |
0.021 |
0.000 |
0.010 |
0.021 |
0.020 |
0.000 |
0.000 |
|
BRIP1 EX 19b |
T |
0.872 |
0.705 |
0.187 |
0.120 |
0.837 |
0.489 |
0.204 |
0.033 |
0.809 |
|
CHEK2 INT 5-6 |
G |
0.033 |
0.159 |
0.073 |
0.000 |
0.000 |
0.000 |
0.000 |
0.000 |
0.000 |
|
CHEK2 INT 8-9 |
G |
0.670 |
0.120 |
0.271 |
0.270 |
0.050 |
0.064 |
0.073 |
0.000 |
0.021 |
|
ESR1 EX 2 |
T |
0.967 |
0.772 |
0.729 |
0.410 |
0.310 |
0.670 |
0.102 |
0.213 |
0.234 |
|
ESR1 EX 4 |
G |
0.011 |
0.043 |
0.000 |
0.000 |
0.240 |
0.053 |
0.000 |
0.791 |
0.766 |
|
ESR1 INT 7-8a |
A |
0.000 |
0.083 |
0.044 |
0.600 |
0.000 |
0.000 |
0.942 |
0.000 |
0.000 |
|
ERBB2 INT1-2 |
T |
0.106 |
0.000 |
0.000 |
0.000 |
0.000 |
0.000 |
0.000 |
0.000 |
0.000 |
|
ERBB2 INT 3-4 |
G |
0.033 |
0.326 |
0.365 |
0.720 |
0.173 |
0.223 |
0.918 |
0.637 |
0.936 |
|
ERBB2 INT 8-9 |
G |
0.043 |
0.318 |
0.052 |
0.020 |
0.000 |
0.109 |
0.000 |
0.000 |
0.000 |
|
ERBB2 INT 12-13b |
A |
0.915 |
0.533 |
0.542 |
0.240 |
0.230 |
0.798 |
0.071 |
0.225 |
0.043 |
|
ERBB2 EX 13a |
G |
0.223 |
0.000 |
0.000 |
0.000 |
0.000 |
0.000 |
0.020 |
0.000 |
0.000 |
|
ERBB2 EX 13b |
T |
0.926 |
0.533 |
0.531 |
0.240 |
0.230 |
0.798 |
0.071 |
0.233 |
0.043 |
|
ERBB2 EX 14 |
A |
0.021 |
0.250 |
0.260 |
0.460 |
0.260 |
0.170 |
0.561 |
0.451 |
0.870 |
|
PTEN INT 3-4 |
G |
0.021 |
0.413 |
0.031 |
0.450 |
0.040 |
0.128 |
0.000 |
0.538 |
0.734 |
|
PTEN INT 7-8 |
A |
0.000 |
0.076 |
0.000 |
0.470 |
0.340 |
0.128 |
0.010 |
0.544 |
0.766 |
|
PTEN EX 9 |
T |
0.000 |
0.076 |
0.000 |
0.450 |
0.020 |
0.128 |
0.000 |
0.538 |
0.734 |
| STK11 INT 1-2 | T | 0.394 | 0.348 | 0.156 | 0.010 | 0.310 | 0.213 | 0.745 | 0.467 | 0.011 |
To estimate the degree of association seen for the Bonferroni significant polymorphisms, correlation between disease risk and average breed allele frequency for each SNP was calculated. No statistically significant correlations were found.
We applied the LD block criteria D’ ~ 1 and LOD ≥ 2 to the second dataset to study if any of the blocks found in the ESS cases and controls could be re-identified in the high and low risk breeds. Not all blocks were present in all breeds, but a 9kilobase block of eight SNPs in ERBB2 (ss244244354, ss244244355, ss244244357, ss244244358, ss244244360, ss244244361, ss244244363 and ss244244364) was re-identified for all breeds but the Beagle.
When aligned in Sequencher® the canine ESR1 exons showed a high match percentage (≥84%) to the human exons, except for exon 1 and 8. We found no human cancer-associated polymorphisms in close proximity to the canine ESR1 exon 2 SNP (rs21960513). However, the canine SNP in exon 4 (ss244244343) was positioned one base pair (bp) next to the human rs1801132 SNP (Figure 1). Another human ESR1 polymorphism, rs2228480, aligned to the canine exon 8 at a position 207 bp downstream from the canine exon 8 SNP (ss244244346) (minimum match percentage of 84%) (Figure 1). The canine SNPs rs21960513 and ss244244346 were synonymous, while ss244244343 lead to an amino acid substitution from isoleucine to leucine. Also, ss244244343 is located in a gene region conserved across four species [13]. However, the substitution was predicted benign and tolerant by PolyPhen and SIFT, respectively [13,14].
Figure 1.
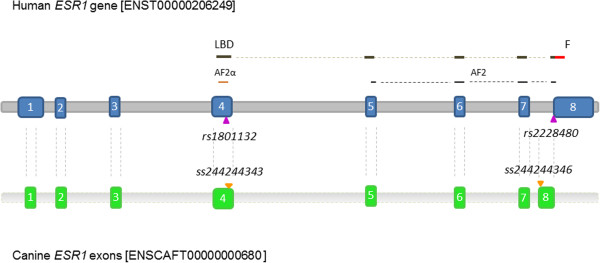
Schematic illustration of ESR1 alignments relative to the human ESR1 gene sequence. The human ESR1 gene sequence (grey) and exons (blue) with aligned canine ESR1 exons (green) and human and canine polymorphisms (purple and orange triangles, respectively). The exon numbers are in white letters. Bars above the human exon sequences are indicating the location of the ligand binding domain (LBD, dark brown), F domain (F, red bar), activation factor 2α domain (AF2α, light brown bar) and activation factor 2 domain (AF2, black bars). GATA interacts with the AF2 domain [15-19].
Discussion
Publications about the existence, frequency and importance of CMT-associated germline mutations and their role in the tumour development are sparse. In the present survey, we studied SNPs in known cancer-associated genes and observed significant differences in allele and haplotype frequencies for the ESR1 gene in the ESS material. These findings were supported by the high and low risk breed groups and suggested an association of ESR1 alleles with increased risk of CMTs. The ESR1 gene encodes an estrogen receptor which works as a ligand-activated transcription factor in the cell. Besides its normal role in e.g. sexual development and reproductive function, the estrogen receptor is involved in several pathological processes such as breast cancer (as reviewed by Dahlman-Wright et al. 2006 [20]). Previous studies on human breast cancer have suggested that ESR1 polymorphisms are associated with the development of these tumours (Additional file 2). The non-synonymous canine exon 4 SNP (ss244244343) is positioned one bp downstream compared to the position of the human rs1801132 SNP. While rs1801132 is in human codon 325, the canine SNP aligned to a position in human codon 326. Both codons encode amino acids in the hormone binding domain of the human estrogen receptor (Figure 1). This domain is related to receptor dimerization, chaperone binding and recruitment of co-regulators [21]. Studies on rs1801132 have shown an association with breast malignancies [22,23]. Its C allele has been associated with cancer, suggesting that it interferes with the binding of the GATA-1 and GATA-2 transcription factors to the estrogen receptor [21]. GATA transcription factors interact with the activation factor 2 (AF2) region of the ligand binding domain of the human estrogen receptor [15]. There are structural differences in the human and canine ERα proteins, but the major pocket sites seem to be very similar [24]. An association with breast cancer has also been suggested for the human rs2228480[25], which aligned in proximity of the synonymous canine exon 8 SNP (ss244244346). Rs2228480 is positioned near the F domain of the human estrogen receptor (Figure 1). This domain is believed to be important for the ability of estrogen receptor to distinguish between receptor agonist and antagonist binding [26]. Thus, the observed associations of the canine exon 4 and exon 8 SNPs in our two independent canine datasets are supported by similar effects of closely linked SNPs in the human gene. This suggests ESR1 as an interesting candidate gene for mammary tumours in dogs as well. Given the prior evidence that ESR1 plays a role in human breast cancer etiology, it seems probable that the described loci of the present study might be correlated with a causal variant affecting ESR1 function.
Correction for multiple testing is necessary to adjust for the multiple comparisons performed when testing several SNPs or haplotypes for association. However, SNPs within a gene are very closely linked and not independent observations. Correcting for the number of SNPs would therefore be too conservative. Moreover, this study comprises a selection of biologically important and previously cancer-associated genes where it is likely to find an association, rather than a random set of genes. To assure an appropriate correction for multiple testing of the single SNP and haplotype P-values, we therefore used the number of LD blocks. The fact that only one SNP in ESR1 was significant in the ESS material after Bonferroni correction was somewhat surprising. Possibly, the rest of the SNPs we assessed are not directly causative or in high LD with such variants, creating false negative gene association results. Further, a substantial number of sub-classifications of CMTs exists [27], potentially with different germline mutations contributing to the development of different tumour subtypes and grades of malignancy. Such heterogeneity might complicate the detection of truly cancer-associated mutations. Thus, our study of the ESS dataset might have insufficient power to prove SNP associations to CMT. Still, it has been indicated that about 100 cases and 100 controls should suffice to find loci with strong effects (fivefold) even for complex traits such as cancer [28]. As the one Bonferroni significant ESR1 SNP (ss244244344) is intronic, and we have tested only a limited set of SNPs, this SNP is more likely in LD with a functional mutation than being causative itself. It might also be a false positive. Thus, more studies on the ESR1 gene are required to establish its role in CMT development.
The considerable breed variations in allele frequencies for the high and low risk dataset might be expected due to between-breed genetic heterogeneity for risk alleles at different risk loci. Yet, the differences are interesting as it appears to be a significant breed-specific accumulation of certain coding variants also in genes that are vital for normal cell function. However, the large allele frequency differences between breeds within the same risk group complicated the interpretation of the results. There is potentially a genetic heterogeneity between breeds as to predominant CMT types and associated risk genes/alleles. If the candidate genes in this study are associated with cancer risk in the selected breeds, the variation in allele frequencies indicates that the associated genes/alleles differ between breeds. Another possibility is that cancer-driving mutations are in the regulatory parts of the genes, and the coding SNPs in the present study can be considered markers for functionally active regulatory sites. During breed differentiation there may have been recombinations between regulatory sites and coding parts resulting in different coding SNPs being linked to regulatory variation in different breeds. Consequently, even if a gene has an important role in cancer development, the associated coding SNP allele/haplotype might vary between breeds. Another challenge is the documentation of high and low risk breeds. As a result of e.g. ancestral patterns of geographical establishment, fluctuations in a dog breed’s popularity and extensive use of popular sires within a country, the genetic composition of a dog breed can change over time and between different geographical locations. Thus, breed predisposition of CMT might vary from subpopulation to subpopulation. We based the selection of high and low risk breeds in this study on previous publications, but not all of them were Norwegian, and some were up to twenty years old. This could possibly be a source of sample error in our study. Moreover, the dogs from the high and low risk breeds in our study are randomly selected without knowledge about the CMT phenotype. They would be a mixture of individuals with high and low risk corresponding to the population frequency of CMT. The frequency and effect of CMT-associated risk alleles would need to be relatively strong to be detected in such a material. It might be that our study is underpowered in that respect. However, we have documented large breed variations in allele frequencies of the coding SNPs in important cancer genes, and there are probably similar differences in the frequency of functionally active haplotypes between dog populations.
Conclusions
Cancer is a very complex disease. As in human breast malignancies, it is likely that the development of CMTs is influenced by several genes. The identified association of ESR1 to CMT in the present survey supports the power of the canine model for human breast cancer and the fact that combined studies within and between breeds can add power to the detection of risk alleles also for complex traits. However, the increased risk of CMTs in ESS and other high risk breeds might be due to other SNPs and/or genes than those selected in the present study. There is also a chance that predisposing CMT variants are undetected in our study due to limited power. Nevertheless, this is to our knowledge the first reported association of ESR1 polymorphisms to CMT and supports ESR1 as a candidate gene for canine cancer that should be further studied.
Methods
Samples
Two separate datasets were included in this study. The first consisted of blood DNA from English Springer Spaniel CMT cases and controls [11] (Table 1). These were privately owned female dogs registered in the Swedish Kennel Club. Approximately half of the dogs were confirmed unrelated at the parental level, while the rest could be as closely related as siblings. A subset of the cases had been classified as malignant (n = 33) or benign (n = 83) CMTs by histopathology by a veterinary pathologist at the time of the genotyping analysis. The rest of the cases were selected based on a veterinary clinical examination confirming the presence of single or multiple nodules within the mammary glands, but they had not (yet) had their mammary tumours surgically removed or histopathologically evaluated. The control dogs were older than eight years with confirmed absence of CMT after palpation of the mammary glands by a veterinarian. However, some of the samples from the material by Rivera et al. were not available for the present study. The same ESS cohort has been genotyped in a parallel study using Illumina 170 K canine HD SNP array, and multidimensional scaling plots were used to evaluate population stratification (data not shown). In this analysis, an outlier group of 29 dogs was identified. These dogs were consequently removed from further analysis in the present study.
The second dataset consisted of EDTA blood samples from the Canine Biobank at the Norwegian School of Veterinary Science (NSVS). In total, this dataset comprised samples from 450 individuals of nine dog breeds (Table 1). The selected breeds were known to be at either high or low risk of developing CMTs according to previous studies. However, the CMT status of the individual dogs from the Canine Biobank was unknown. But according to the higher genetic risk for some of the breeds, an increased allele frequency of associated risk alleles would be expected. Representing breeds at high risk, the Boxer, Cocker Spaniel, Dachshund, English Setter and Standard Poodle were selected. Assumed low risk breeds included in the study were the Beagle, Bernese Mountain Dog, Collie and Shetland Sheepdog [3-8]. Genomic DNA was extracted from the EDTA blood samples using E.Z.N.A Blood Kit according to the manufacturer’s protocol (Omega ®, VWR International, West Chester, Pennsylvania, USA). The DNA was analysed for quality and quantity using NanoDrop (Thermo Fisher Scientific, Wilmington, Pennsylvania, USA).
Single nucleotide polymorphism selection and genotyping
All samples were genotyped for 52 previously described canine SNPs [13] (Additional file 3). These SNPs were located in ten genes previously reported to be cancer-associated in humans; BRCA1, BRCA2, BRIP1, CDH1, CHEK2, EGFR, ERBB2 (HER2), ESR1, PTEN and STK11 (Table 7). Twenty of the SNPs were found in coding regions, including 11 synonymous and 9 non-synonymous SNPs. The SNPs were distributed into two pools and genotyped using the Sequenom iPLEX Gold Mass ARRAY® according to the manufacturer’s protocol (Sequenom®, San Diego, California, USA). The genotyping was performed at Broad Institute, Cambridge, Massachusetts.
Table 7.
Genes evaluated for association to CMT
| Gene | Human chromosome | Canine chromosome | No. SNPs genotyped | Location of SNPs (Mb) (CanFam 2.0) |
|---|---|---|---|---|
|
BRCA1 |
17 |
9 |
1 |
23.306 |
|
BRCA2 |
13 |
25 |
5 |
10.782-10.732 |
|
BRIP1 |
17 |
9 |
5 |
38.191-38.304 |
|
CDH1 |
16 |
5 |
5 |
83.792-83.771 |
|
CHEK2 |
22 |
26 |
3 |
25.104-25.086 |
|
EGFR |
7 |
18 |
5 |
8.982-9.027 |
|
ERBB2 (HER2) |
17 |
9 |
18 |
26.106-26.088 |
|
ESR1 |
6 |
1 |
5 |
45.129-45.409 |
|
PTEN |
10 |
26 |
4 |
40.921-40.978 |
| STK11 | 19 | 20 | 1 | 60.709 |
Single SNP and haplotype association analysis
Single SNP and haplotype analysis were performed separately for the two different study datasets; the ESS cases were compared to the controls, and the high risk breeds from the Canine Biobank were all compared to the low risk breeds. Single SNP association analysis was also performed for the subset of ESS cases with benign tumours vs. controls, malignant tumours vs. controls and benign vs. malignant tumours. Only samples and SNPs with a genotyping success rate of ≥75% and SNPs with a minor allele frequency (MAF) ≥1% were included in the single SNP and haplotype association analysis. The PLINK software [29,30] was used for analysing allele frequencies, single χ2 SNP association and SNP odds ratios. Haploview [31,32] was used to identify LD blocks with a D’ ~ 1 and LOD ≥ 2 for each dataset and to generate haplotypes and haplotype association statistics. Odds ratios for haplotypes at each specific locus were estimated using calculators at VassarStats [33]. The nominal (raw) χ2P-values from the single SNP and haplotype analysis were Bonferroni corrected using the number of LD blocks to adjust for the problem of multiple comparisons that arises from evaluating several SNPs or haplotypes. Multiple testing correction using 10,000 permutations for the ESS dataset and 107 permutations for the high and low risk breed dataset was also performed. Further, we did permutation testing by permuting the high/low risk labels simultaneously for all dogs in each breed in combination with PLINK analysis of association, using 10,000 permutations. A P-value of less than 0.05 after correction for multiple testing was reckoned statistically significant.
Considering each individual breed as the study unit rather than the individual dog, we performed pairwise correlation analysis for the high and low risk breed dataset using JMP 8 (SAS Institute Inc., North Carolina, USA). The correlation between the disease risk (high or low) and average allele frequency for each breed for each SNP was estimated, regarding a P-value of less than 0.05 statistically significant. Bonferroni correction was performed using the number of LD blocks across breeds (9).
Known human breast cancer-associated ESR1 polymorphisms (Additional file 2) were compared to the canine SNPs in this study by alignment using the Sequencher software (Gene Codes Corporation, Ann Arbor, MI, USA). The full genomic human ESR1 gene sequence [ENST00000206249] was aligned to the canine [ENSCAFT00000000680, (CanFam2.0)] [16] and human exon sequences and polymorphisms with flanking sequence.
Abbreviations
BRCA1: Breast cancer 1, early onset; BRCA2: Breast cancer 2, early onset; BRIP1: BRCA1 interacting protein C-terminal helicase 1; CDH1: Cadherin 1, type 1, E-cadherin (epithelial); CHEK2: Checkpoint kinase 2; CMT: Canine mammary tumour; D’: Normalized measure of linkage disequilibrium; EGFR: Epidermal growth factor receptor; ESR1: Estrogen receptor 1; ERBB2 (HER2): v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian); ESS: English Springer Spaniel; MAF: Minor allele frequency; LD: Linkage disequilibrium; NSVS: Norwegian School of Veterinary Science; PTEN: Phosphatase and tensin homolog; SNP: Single nucleotide polymorphism; nsSNP: Non-synonymous SNP; STK11: Serine/threonine kinase 11.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
KSB helped conceive the study and its design, carried out the DNA isolation, SNP and haplotype association analysis, sequence alignments and drafted the manuscript. MM contributed to the SNP and haplotype association analysis and revised the manuscript. PR and HVE collected the ESS case and control samples and planned CMT analysis. SIT was responsible for the collection of samples for the Canine Biobank and revised the manuscript. MTW helped with statistical analysis and performed the breed permutations. KLT contributed to the SNP and haplotype association analysis and revised the manuscript. FL conceived the study and its design, contributed to the SNP and haplotype association analysis and revised the manuscript. All authors read and approved the final manuscript.
Supplementary Material
All results from ESS single SNP and haplotype association analysis.
Cancer-associated human ESR1 polymorphisms that were compared to the canine SNPs.
Genotyped single nucleotide polymorphisms, genome position, reference ID and alleles.
Contributor Information
Kaja Sverdrup Borge, Email: KajaSverdrup.Borge@nvh.no.
Malin Melin, Email: Malin.Melin@imbim.uu.se.
Patricio Rivera, Email: patricio.rivera@kv.slu.se.
Stein Istre Thoresen, Email: Stein.Thoresen@nvh.no.
Matthew Thomas Webster, Email: matthew.webster@imbim.uu.se.
Henrik von Euler, Email: Henrik.von.Euler@slu.se.
Kerstin Lindblad-Toh, Email: kersli@broadinstitute.org.
Frode Lingaas, Email: Frode.Lingaas@nvh.no.
Acknowledgements
This work was (partly) funded by the European Commission (LUPA, GA-201370). We are also grateful to the “The research fund; Cancer in the dog” for the financial contribution to this study. KLT is the recipient for a EURYI award from the ESF.
References
- Patterson DF. Companion animal medicine in the age of medical genetics. J Vet Intern Med. 2000;14:1–9. doi: 10.1111/j.1939-1676.2000.tb01492.x. [DOI] [PubMed] [Google Scholar]
- Bronden LB, Nielsen SS, Toft N, Kristensen AT. Data from the Danish veterinary cancer registry on the occurrence and distribution of neoplasms in dogs in Denmark. Vet Rec. 2010;166:586–590. doi: 10.1136/vr.b4808. [DOI] [PubMed] [Google Scholar]
- Egenvall A, Bonnett BN, Ohagen P, Olson P, Hedhammar A, von Euler H. Incidence of and survival after mammary tumors in a population of over 80,000 insured female dogs in Sweden from 1995 to 2002. Prev Vet Med. 2005;69:109–127. doi: 10.1016/j.prevetmed.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Boldizsar H, Szenci O, Muray T, Csenki J. Studies on canine mammary tumours. I. Age, seasonal and breed distribution. Acta Vet Hung. 1992;40:75–87. [PubMed] [Google Scholar]
- Arnesen K, Gamlem H, Glattre E, Grøndalem J, Moe L, Nordstoga K. The Norwegian canine cancer register 1990–1998. Report from the project "Cancer in the dog". EJCAP. 2001;11:159–169. [Google Scholar]
- Dahl K, Moe L, Indrebø I, Gamlem H. Forekomst av mammatumor hos beslektede boxere [Occurence of mammary tumor in related Boxers] Nor Vet Tidsskr. 2002;114:615–622. [Google Scholar]
- Priester WA. Occurrence of mammary neoplasms in bitches in relation to breed, age, tumour type, and geographical region from which reported. J Small Anim Pract. 1979;20:1–11. doi: 10.1111/j.1748-5827.1979.tb07014.x. [DOI] [PubMed] [Google Scholar]
- Moe L. Population-based incidence of mammary tumours in some dog breeds. J Reprod Fertil Suppl. 2001;57:439–443. [PubMed] [Google Scholar]
- Queiroga FL, Raposo T, Carvalho MI, Prada J, Pires I. Canine mammary tumours as a model to study human breast cancer: most recent findings. In Vivo. 2011;25:455–465. [PubMed] [Google Scholar]
- Owen LN. A comparative study of canine and human breast cancer. Invest Cell Pathol. 1979;2:257–275. [PubMed] [Google Scholar]
- Rivera P, Melin M, Biagi T, Fall T, Haggstrom J, Lindblad-Toh K, von Euler H. Mammary tumor development in dogs is associated with BRCA1 and BRCA2. Cancer Research. 2009;69:8770–8774. doi: 10.1158/0008-5472.CAN-09-1725. [DOI] [PubMed] [Google Scholar]
- Veldhoen N, Watterson J, Brash M, Milner J. Identification of tumour-associated and germ line p53 mutations in canine mammary cancer. Br J Cancer. 1999;81:409–415. doi: 10.1038/sj.bjc.6690709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borge KS, Borresen-Dale AL, Lingaas F. Identification of genetic variation in 11 candidate genes of canine mammary tumour. Vet Comp Oncol. 2011;9:241–250. doi: 10.1111/j.1476-5829.2010.00250.x. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DR, Schmidt RJ, Pickard RT, Foxworthy PS, Eacho PI. Estrogen receptor-mediated repression of human hepatic lipase gene transcription. J Lipid Res. 2002;43:383–391. [PubMed] [Google Scholar]
- Ensembl release 67. [ http://www.ensembl.org/index.html]
- Norris JD, Fan D, Kerner SA, McDonnell DP. Identification of a third autonomous activation domain within the human estrogen receptor. Mol Endocrinol. 1997;11:747–754. doi: 10.1210/me.11.6.747. [DOI] [PubMed] [Google Scholar]
- Herynk MH, Fuqua SA. Estrogen receptor mutations in human disease. Endocr Rev. 2004;25:869–898. doi: 10.1210/er.2003-0010. [DOI] [PubMed] [Google Scholar]
- Kumar R, Zakharov MN, Khan SH, Miki R, Jang H, Toraldo G, Singh R, Bhasin S, Jasuja R. The dynamic structure of the estrogen receptor. J Amino Acids. 2011;2011:812540. doi: 10.4061/2011/812540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlman-Wright K, Cavailles V, Fuqua SA, Jordan VC, Katzenellenbogen JA, Korach KS, Maggi A, Muramatsu M, Parker MG, Gustafsson JA. International Union of Pharmacology. LXIV. Estrogen receptors. Pharmacol Rev. 2006;58:773–781. doi: 10.1124/pr.58.4.8. [DOI] [PubMed] [Google Scholar]
- Anghel A, Raica M, Narita D, Seclaman E, Nicola T, Ursoniu S, Anghel M, Popovici E. Estrogen receptor alpha polymorphisms: correlation with clinicopathological parameters in breast cancer. Neoplasma. 2010;57:306–315. doi: 10.4149/neo_2010_04_306. [DOI] [PubMed] [Google Scholar]
- Vasconcelos A, Medeiros R, Veiga I, Pereira D, Carrilho S, Palmeira C, Azevedo C, Lopes CS. Analysis of estrogen receptor polymorphism in codon 325 by PCR-SSCP in breast cancer: association with lymph node metastasis. Breast J. 2002;8:226–229. doi: 10.1046/j.1524-4741.2002.08407.x. [DOI] [PubMed] [Google Scholar]
- Kallel I, Rebai M, Khabir A, Farid NR, Rebai A. Genetic polymorphisms in the EGFR (R521K) and estrogen receptor (T594T) genes, EGFR and ErbB-2 protein expression, and breast cancer risk in Tunisia. J Biomed Biotechnol. 2009;2009:753683. doi: 10.1155/2009/753683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toniti W, Suthiyotha N, Puchadapirom P, Jenwitheesuk E. Binding capacity of ER-alpha ligands and SERMs: comparison of the human, dog and cat. Asian Pac J Cancer Prev. 2011;12:2875–2879. [PubMed] [Google Scholar]
- Tapper W, Hammond V, Gerty S, Ennis S, Simmonds P, Collins A, Eccles D. The influence of genetic variation in 30 selected genes on the clinical characteristics of early onset breast cancer. Breast Cancer Res. 2008;10:R108. doi: 10.1186/bcr2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montano MM, Muller V, Trobaugh A, Katzenellenbogen BS. The carboxy-terminal F domain of the human estrogen receptor: role in the transcriptional activity of the receptor and the effectiveness of antiestrogens as estrogen antagonists. Mol Endocrinol. 1995;9:814–825. doi: 10.1210/me.9.7.814. [DOI] [PubMed] [Google Scholar]
- Goldschmidt M, Pena L, Rasotto R, Zappulli V. Classification and grading of canine mammary tumors. Vet Pathol. 2011;48:117–131. doi: 10.1177/0300985810393258. [DOI] [PubMed] [Google Scholar]
- Karlsson EK, Lindblad-Toh K. Leader of the pack: gene mapping in dogs and other model organisms. Nat Rev Genet. 2008;9:713–725. doi: 10.1038/nrg2382. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PLINK (version 1.07) [ http://pngu.mgh.harvard.edu/purcell/plink/]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Haploview. [ http://www.broadinstitute.org/haploview]
- VassarStats:Website for statistical computation. [ http://vassarstats.net]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All results from ESS single SNP and haplotype association analysis.
Cancer-associated human ESR1 polymorphisms that were compared to the canine SNPs.
Genotyped single nucleotide polymorphisms, genome position, reference ID and alleles.


