Abstract
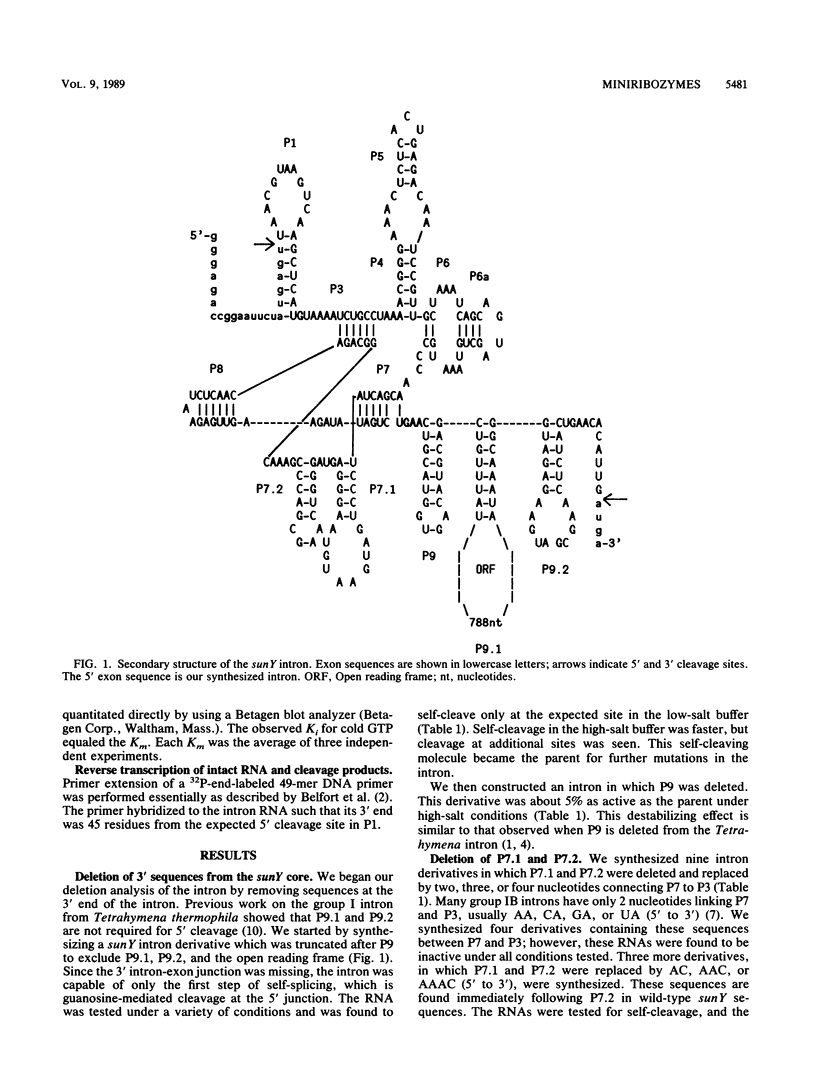
The self-splicing sunY intron from bacteriophage T4 has the smallest conserved core secondary structure of any of the active group I introns. Here we show that several nonconserved regions can be deleted from this intron without complete loss of catalytic activity. The 3' stems P9, P9.1, and P9.2 can be deleted while retaining 5' cleaving activity. Two base-paired stems (P7.1 and P7.2) that are peculiar to the group IA introns can also be deleted; however, the activities of the resulting derivatives depend greatly on the choice of replacement sequences and their lengths. The smallest active derivative is less than 180 nucleotides long. These experiments help to define the minimum structural requirements for catalysis.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barfod E. T., Cech T. R. Deletion of nonconserved helices near the 3' end of the rRNA intron of Tetrahymena thermophila alters self-splicing but not core catalytic activity. Genes Dev. 1988 Jun;2(6):652–663. doi: 10.1101/gad.2.6.652. [DOI] [PubMed] [Google Scholar]
- Belfort M., Pedersen-Lane J., West D., Ehrenman K., Maley G., Chu F., Maley F. Processing of the intron-containing thymidylate synthase (td) gene of phage T4 is at the RNA level. Cell. 1985 Jun;41(2):375–382. doi: 10.1016/s0092-8674(85)80010-6. [DOI] [PubMed] [Google Scholar]
- Cech T. R. The chemistry of self-splicing RNA and RNA enzymes. Science. 1987 Jun 19;236(4808):1532–1539. doi: 10.1126/science.2438771. [DOI] [PubMed] [Google Scholar]
- Doudna J. A., Gerber A. S., Cherry J. M., Szostak J. W. Genetic dissection of an RNA enzyme. Cold Spring Harb Symp Quant Biol. 1987;52:173–180. doi: 10.1101/sqb.1987.052.01.022. [DOI] [PubMed] [Google Scholar]
- Gardiner K. J., Marsh T. L., Pace N. R. Ion dependence of the Bacillus subtilis RNase P reaction. J Biol Chem. 1985 May 10;260(9):5415–5419. [PubMed] [Google Scholar]
- Kim S. H., Cech T. R. Three-dimensional model of the active site of the self-splicing rRNA precursor of Tetrahymena. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8788–8792. doi: 10.1073/pnas.84.24.8788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Dujon B. Conservation of RNA secondary structures in two intron families including mitochondrial-, chloroplast- and nuclear-encoded members. EMBO J. 1983;2(1):33–38. doi: 10.1002/j.1460-2075.1983.tb01376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shub D. A., Gott J. M., Xu M. Q., Lang B. F., Michel F., Tomaschewski J., Pedersen-Lane J., Belfort M. Structural conservation among three homologous introns of bacteriophage T4 and the group I introns of eukaryotes. Proc Natl Acad Sci U S A. 1988 Feb;85(4):1151–1155. doi: 10.1073/pnas.85.4.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W. Enzymatic activity of the conserved core of a group I self-splicing intron. Nature. 1986 Jul 3;322(6074):83–86. doi: 10.1038/322083a0. [DOI] [PubMed] [Google Scholar]