Abstract
Nuclear receptors (NRs) regulate and coordinate multiple processes by integrating internal and external signals, thereby maintaining homeostasis in front of nutritional, behavioral and environment challenges. NRs exhibit strong similarities in their structure and mode of action: by selective transcriptional activation or repression of cognate target genes, which can either be controlled through a direct, DNA binding-dependent mechanism or through crosstalk with other transcriptional regulators, NRs modulate the expression of gene clusters thus achieving coordinated tissue responses. Additionally, non genomic effects of NR ligands appear mediated by ill-defined mechanisms at the plasma membrane. These effects mediate potential therapeutic effects as small lipophilic molecule targets, and many efforts have been put in elucidating their precise mechanism of action and pathophysiological roles. Currently, numerous nuclear receptor ligand analogs are used in therapy or are tested in clinical trials against various diseases such as hypertriglyceridemia, atherosclerosis, diabetes, allergies and cancer and others.
Keywords: Animals; Humans; Ligands; Molecular Biology; Receptors, Cytoplasmic and Nuclear; chemistry; genetics; metabolism
Keywords: transcriptional regulation, nuclear receptors, coactivators, corepressors, structure
INTRODUCTION
The nuclear receptor superfamily comprises evolutionarily related transcription factors fulfilling multiple regulatory functions in growth, development and homeostasis. Nuclear receptors share a common architecture and functional behavior. The effector function of nuclear receptors is to modulate transcription through several distinct mechanisms, which include both transactivation and transrepression activities upon receptor-specific ligand binding. Nuclear receptors can also be the targets of other signaling pathways that modify the receptor, or their transcriptional comodulators, post-translationally and affect their activity and functions. According to phylogenetic studies, nuclear receptors emerged long before the divergence of vertebrates and invertebrates, during the earliest metazoan evolution [1]. The first cloned human receptors were the glucocorticoid receptor (GR/NR3C1, [2,3]) together with the estrogen receptor (ER) [4,5] and the thyroid hormone receptor (T3R/NR1A1, [6,7]). Forty eight nuclear receptors have since been identified in the human [8].
Nuclear receptors share a common structural organization which defines this gene superfamily (Figure 1). The N-terminal domain is highly variable depending on the receptor and contains a ligand-independent transactivation domain termed Activation Function 1 (AF-1). The most conserved central region is the DNA-binding domain (DBD), which contains the P-box, a short motif responsible for direct DNA interaction and DNA-binding specificity. Additional sequences in the DBD are involved in the homo- or heterodimerization of nuclear receptors. Nuclear receptors bind to sequence-specific elements located not only in the vicinity of target gene promoters, but also in intronic and enhancer regions, either as monomers (Nor1/NR4A3), as homodimers such as the steroid receptors [GR/NR3C1, estrogen receptors (ERα/NR3A1 and ERβ/NR3A2), progesterone receptor (PR/NR3C3), mineralocorticoid receptor (MR/NR3C2), androgen receptor (AR/NR3C4)] and retinoid X receptors (RXRα/NR2B1, RXRβ/NR2B2, RXRγ/NR2B3), or as heterodimers with RXRs. The DBD and the C-terminal ligand-binding domain (LBD) are linked by the hinge region [9]. The C terminus of NRs harbors several functionally critical motifs, such as the activating function 2 (AF-2), conferring to many NRs a ligand-dependent transcriptional activity, a strong dimerization interface and a ligand binding pocket (LBP). The in-depth structural nuclear receptor architecture is delineated further in this review.
Figure 1. General structural organization of nuclear receptors.
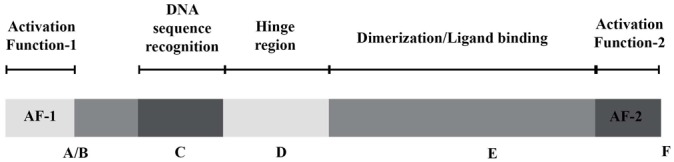
Letters from A to F represent nuclear receptor domains from N-terminus to C-terminus of the nuclear receptor respectively. The structure and functions of each domain is detailed in the text.
Nomenclatures of the nuclear receptor family have been proposed according to different criteria. Based on the sequence alignment of the two well-conserved domains (DBD and LBD) and phylogenetic tree construction, the nuclear receptor gene family has been divided into six subfamilies. Interestingly and importantly, a correlation exists between DNA-binding and dimerization abilities of each classified nuclear receptor and its phylogenetic position. Subfamily 1 comprises nuclear receptors forming heterodimers with RXR (T3Rs:NR1A; RARs: NR1B; VDR: NR1I1; PPARs: NR1C; RORs: NR1F: Rev-erbs: NR1D; CAR: NR1I3; PXR: NR1I2; LXRs: NR1H). Subfamily 2 is formed by HNF4s: NR2A1&2; COUP-TFs: NR2F; RXRs: NR1B. Subfamily 2 members can function in two configurations, either as homodimers and as heterodimers. Subfamily 3 includes the above mentioned steroid hormone receptors. Subfamily 4 contains the nerve growth factor-induced clone B group of orphan receptors NGFI-B/Nur77/NR4A1, Nurr1/NR4A2, and NOR1/NR4A3. The small subfamily 5 includes the steroidogenic factor 1 (SF1/NR5A1) and receptors related to Drosophila FTZ-F1 (LRH1/NR5A2). The sixth subfamily comprises only the GCNF1 receptor. Finally, subfamily 0 encompasses 2 atypical nuclear receptors lacking the DBD (Dax1/NR0B1 and SHP/NR0B2), thereby displaying constitutive dominant-negative activities [10].
Another functional classification according to the ligand-binding properties splits the superfamily of nuclear receptors into three groups. The most characterized subfamily called thyroid/steroid hormone receptor subfamily comprises ER, AR, PR, MR and GR and also includes the thyroid receptors T3Rs, VDR, and RARs. The second ‘orphan’ subfamily is composed by nuclear receptors for which regulatory molecules have not been identified so far. They are represented by NR4 receptors and COUP-TFs. The function and molecular mechanism of action for many ‘orphan’ receptors is only poorly investigated. The third subfamily of nuclear receptors is known as ‘adopted’ orphan receptors. Members of this subfamily were initially characterized as ‘orphans’ and afterwards, natural ligands have been identified that convey physiological functions. These nuclear receptors are sensors of the metabolic status of cells, organs and the whole body and trigger responses to xenobiotics, dietary signals, diatomic gases and metabolites. In this class are found Rev-erbα and β, PPARs, LXRs, FXRs, RORs, PXR and CAR.
The ability of nuclear receptors to be regulated by natural or synthetic molecules have led to intensive efforts to target nuclear receptors therapeutically. However, many currently available ligands have several deleterious side-effects, many of which seem to be related to their transactivating properties. It seems to be essential to determine the importance of positive and negative gene regulation in conferring the therapeutic benefits of nuclear receptor ligands in disease models. In this review we will discuss the relationship between the molecular structure and the molecular action of nuclear receptors.
A. STRUCTURAL FEATURES OF NUCLEAR RECEPTORS
Nuclear receptors reveal characteristic protein architecture that consists of five to six domains of homology designated A to F, starting from N-terminus to C-terminus of protein. The weakest conservancy is observed in the N-terminal A/B domain, D or hinge domain, and F region at the C-terminus which is not present in all nuclear receptors. The DBD and LBD are the most highly conserved domains (Figure 1). The most recent structural studies [11,12] of RXR heterodimers bound to DNA showed asymmetric complexes of 150–200Å, with LBDs being located on one side of the DNA, 5′ of the DNA response element (Figure 2). The hinge region plays an important structural role by specifying the relative orientation of the DBD with respect to the LBD.
Figure 2. Crystal structure of PPARγ-RXRα complex bound to a DR-1 response element.
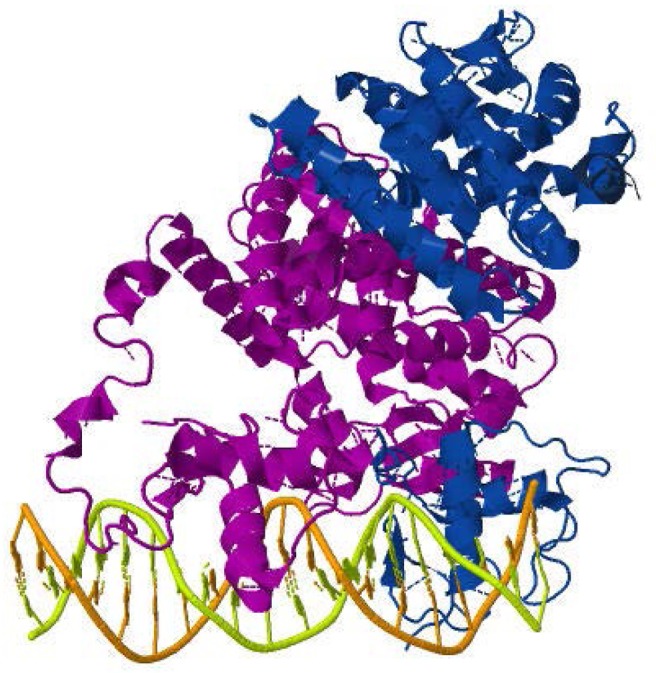
Crystallographic coordinates were obtained from the RCSB protein databank (PDB 3E00) and visualized using the Jmol software. PPARγ is purple and RXRα is blue.
1. A/B domain
The poorly structurally defined N-terminal A/B region reveals a strong diversity among nuclear receptors and because of its high mobility, its tertiary structure has not been elucidated so far. Isoform-specific differences in amino termini are observed for several NRs and these sequence variations may induce differential binding affinities to response elements and/or with members of the transcription initiation complex, distinct transcriptional activities and different in vivo roles (see for examples [13–18]).
The A/B domain contains the activation function 1 (AF-1) which is ligand-independent. Hydrogen/deuterium exchange mass spectrometry of PPARγ revealed that the ordering of A/B portion is not substantially changed upon ligand binding [11]. By contrast, the N-terminus of T3Rβ1 may transmit thyroid hormone-dependent signaling to the general transcriptional machinery by a direct interaction of the receptor with transcription factor IIB (TFIIB, [13,19]). Moreover, the N-terminal region is an interaction surface for multiple transcriptional coregulatory proteins: steroid receptor coactivator-1 (SRC-1/NCoA1), steroid receptor coactivator-2 (SRC-2/TIF2/NCoA2), p300 and CBP enable a functional synergism between AF-1 and AF-2 regions of steroid receptors, PPARγ or RARs and thus cooperatively enhances transactivation [20–23]. In addition, co-regulator-linked interactions with the N-terminal and C-terminal domains were found for AR, ER and PR [24]. Inter-domain communication also regulates ligand-independent transcriptional silencing: deletion of the PPARγ N-terminal domain prevents corepressor binding [25].
The A/B domains can be modified by phosphorylation and other post-translational, covalent modifications and confer distinct functional properties of nuclear receptors. In the case of ligand-activated receptors, AF-1 modifications have generally a tissue-specific modulatory effect on their transcriptional properties. For instance, the MR N-terminus harbors a serine/threonine-rich nuclear localization signal (NL0) that can be regulated by phosphorylation and influence receptor subcellular localization [26]. An elegant mechanism of regulation of the activity of RXR is provided by the piggyback nuclear exclusion of RXR upon association with Nur77, in a Nur77 AF-1 phosphorylation-dependent manner [27]. Similarly, MEK1-mediated phosphorylation of serine at position 84 inhibits PPARγ1 nuclear localization [28], although an alternative mechanism involving Pin1-mediated proteasomal degradation of PPARγ has been recently proposed [29]. Preventing phosphorylation at this residue in vivo generates mice with increased insulin sensitivity when fed a high fat diet [30]. Taken together, these and other data suggest that translocation of NRs to the nucleus is a property which can be very rapidly regulated by various signaling cascades.
Post-translational modifications also affect the intrinsic transactivating potential of NRs, i.e. by modulating their ability to recruit transcriptional comodulators, or by modifying the polypeptide half-life, both properties being in some instances intimately linked [31,32]. Very interestingly, phosphorylation of the A/B domain of GR by p38 MAPK was shown to induce stable tertiary structure formation in this domain, hence favoring its interaction with coregulatory proteins [33]. In turn, this tertiary structure may be stabilized by protein-protein interactions, as reported for the AR AF-1 [34]. More physiologically, the estrogenic effects of EGF are partially mediated by the phosphorylation of ER AF-1 by EGF-activated MAPKs [35]. In the case of orphan receptors, whose transcriptional activity is strongly dependent on AF-1 integrity, covalent modifications of this region have a very strong impact on their transcriptional output. Amino acid motifs in the A/B domain of Nurr1 mediating ERK5- or ERK2-mediated transcriptional activation have been identified [36,37]. Evidence for other post-translational modifications occurring in the N-terminus of NRs are scarce. Phosphorylation-dependent SUMOylation the AF-1 of ERRγ represses its transcriptional activity [38]. AR is SUMO-1ylated in its AF-1 domain at a SUMO consensus sequence found in all steroid receptors, thus inhibiting androgen-regulated signaling [39]. Conversely, N-terminal SUMO-1ylation of PPARγ strongly increases its transactivating potential [40]. However, as discussed below, SUMOylation in the C terminal AF-2 region is now viewed as a critical mechanism regulating the balance between transactivating and transrepressive functions of NRs.
2. The DNA-binding domain
The DNA-binding domain (DBD) or C domain is the most conserved domain within the nuclear receptor family. Its main function is to recognize and bind specific DNA regulatory sites called response elements (REs) [41] The core DBD region contains about 66 amino acids, but many nuclear receptors additionally contain a less conserved C-terminus, a poorly structured motif of about 25 amino acids called the C-terminal extension region (CTE). As the CTE is located in the so-called hinge region, its features will be detailed in the corresponding paragraph.
The DBD is a highly structured, very compact globular domain composed by a pair of perpendicular α-helices stabilized by two C4 zinc-binding domains each coordinating tetrahedrally a zinc atom, a short β-sheet, and a few stretches of amino acids [42,43]. Each receptor monomer establish specific DNA contact through the first N-terminal helix (helix 1) which directly interacts with the major groove of the DNA half-site. A motif called the P box is critical for the DNA-binding specificity of the receptor [43–47]. Three amino acids of the α-helical P box distinguish nuclear receptors that will bind to the core AGAACA half-element (the “GSV-P box” initially found in GR) or to the AGGTCA half-element (the “EGG-P box” initially found in ER). Structural studies revealed that V and E amino acids make direct and unique contacts with the DNA half-site [48,49].
Nuclear receptor homo- or heterodimers establish contacts with two DNA half-sites that can be arranged in different geometry and separated by a spacer of varying length (see below and [50]). The C-terminal helix (helix 2) contributes to stabilization of the overall DBD structure, establishes weak, non-specific contact with DNA. A 5-amino acid loop defines a strong dimerization interface (D box) for homodimer formation and contributes, to a much lesser extent, to heterodimer stabilization [8,51–53].
DNA also provides a template for dimer assembly, which in turn induces conformational changes of the DNA double helix, most notably by inducing distortion of the minor groove to facilitate sequence recognition by the CTE [54]. This phenomenon is correlated with increased DNA bending in vitro, which has been documented for a number of nuclear receptors [55–58]. The relevance of this phenomenon when response elements are in a chromatinized environment is not clear however, although intrinsic DNA bendability affects GR binding to nucleosomal response elements in vitro [59]. The important role of nucleosome assembly and of histone post-translational modifications on the DNA binding affinity and transcriptional activity of nuclear receptors was demonstrated in vitro [60–63] and in vivo [64].
Although being a domain poorly accessible when receptor dimers are bound to nucleosomal DNA, the DBD can be the target of post-translational modifications. Much attention has been paid to kinase-mediated regulation of nuclear receptor affinity, and consequently a wealth of data document the generally inhibitor role of DBD phosphorylation. Indeed, as expected from the introduction of a repulsive charge, phosphorylation of the DBD of HNF4 [65,66], T3R [67] and ER [68] decreases their DNA binding activity. In a possibly related fashion, phosphorylation of a number of nuclear receptors in this region alters their nuclear retention and decreases their transcriptional activity [65,69,70]. In contrast, phosphorylation of TR2 [71] and of FXR [72] increased their DNA binding activity and interaction with PGC-1α respectively. Other covalent modifications such as RARα methylation or 15d-PGJ(2) adduct formation on ERα favor or inhibit receptor activity, respectively [73,74].
3. The hinge region
The flexible hinge, or D region, also called the C-terminal extension of the DBD (CTE) links the C-domain to the multifunctional C-terminal E/F ligand-binding domain and displays very low amino acid identity and similarity between nuclear receptors. Being located between two functionally and structurally important domains, it seems likely that its functions, deduced mostly from deletion and/or site-directed mutagenesis, may also reflect structural and functional alterations of these neighboring domains. Nevertheless, hinge regions of numerous nuclear receptors have been extensively dissected from a molecular point of view and shown to contain motifs responsible for regulating the subcellular distribution of nuclear receptors. Such a function has been demonstrated for ER [75], AR [76], VDR [77] and Dax1 [78] and reflect the presence of conserved nuclear localization sequences (NLS). The hinge region is also involved in tethering activities. The GR hinge region interacts with GR corepressors HEXIM1 and Bag-1 [79,80]. A natural variant (V227A) in the PPARα hinge region is associated with dyslipidemia and this mutation increases PPARα interaction with the nuclear corepressor NCoR [81]. Quite similarly, natural hinge variants of T3R display impaired dissociation of NCoR and recruitment of the coactivator SRC-1 upon agonist binding [82]. Supporting its role as a flexible link between the DBD and the C terminal LBD, hinge domain mutations affect the synergy between the AF-1 and AF-2 domains of ER [83]. Furthermore, the conserved 3D structure of receptor heterodimers, irrespective of the geometry of the bound DNA response element, highlights this physical property [12]. The hinge domain integrity is also conditioning the DNA binding affinity: in vitro assays showed that alternative splice variants affecting the hinge region sequence of FXR display distinct DNA binding affinities [84].
The CTE of monomeric receptor and of some dimeric receptors (also called T box and/or A box) adopts specific conformations which are context-dependent [85,86]. T and A boxes of dimeric receptors such as T3R, RARs, RXRs and VDR form an alpha-helical structure in solution and establish non-specific contacts with DNA [87–91] which can convert to an extended conformation favoring DNA binding in RXR homodimers and RXR-RAR heterodimers [92,93]. In contrast, the CTE of monomeric receptors such as rev-erbs, nur77 and ERRs establish specific contacts with DNA sequences located immediately 5′ of the NR response element through the A box, which adopts an extended loop conformation [94–96]. The CTE is the major determinant of heterodimer polarity on half-site DNA [11].
The other function of CTE via the T-box is to provide an additional dimerization interface with the second zinc finger helix of RXR. Recent crystallographic analysis of PPAR-RXR-DNA complexes revealed a previously unknown dimerization interface between the RXR CTE and the PPARγ LBD [11], although the relevance of this structure has been challenged [12]. In contrast, the PPARγ CTE makes extensive DNA interaction by binding to the AAACT DNA sequence upstream of the core response element. The interaction with this 5′ flanking sequence is similar to that observed with the Rev–Erb CTE [97,98].
As other domains, the hinge domain can be regulated by post-translational modifications such as methylation, acetylation, phosphorylation and sumoylation. p300-catalyzed acetylation of ERα hinge region regulates its transactivation properties and ligand sensitivity [99]. SUMOylation of RORalpha by both SUMO-1 and SUMO-2, as well as that of ERalpha has been reported, and mutations preventing SUMOylation generate transcription-defective receptors [100,101]. In contrast, hPPARα SUMOylation on lysine 185 increases the selective recruitment of NCoR and decreased transcriptional activity [102]. Phosphorylation of serine residues in RARα, RORα4 and Nur77 are detrimental for receptor-mediated transactivation, either by decreasing DNA recognition or by preventing receptor dimerization [103], while phosphorylation of the PPARα hinge domain favors transactivation over tethered transrepression [104]. No investigations were carried out to identify structural changes induced by these covalent modifications, nor are reports describing when such modifications occur on such a sterically hindered environment.
4. The E or ligand-binding domain
As the domain accommodating lipophilic ligands capable of activating or repressing the transcriptional activities of nuclear receptors, it has attracted considerable interest as a paradigm for a transcriptional molecular switch, and as a target for synthetic analogs since these receptors control signaling pathways involved in a wide range of pathophysiological processes. Since the first crystallization of the RXR LBD [105], more than 600 3D structures related to nuclear receptor LBD structures have been reported, and about 3000 publications relate to some aspects of LBD structure and function. For more details, readers may refer to recent reviews of this fascinating field, linking 3D structure determination and modeling to pharmacology and therapeutics [51,106].
The E domain (or LBD) of nuclear receptors is a multi-functional unit comprising, in addition to the ligand binding pocket, homo- and heterodimerization interfaces and a comodulator binding region. The LBD acts as a molecular switch by interpreting the ligand structure into conformational changes which will convert the receptor in a transcriptional activator or repressor. Although the ligand has been long considered as the sole conformational modifier, it is now recognized that DNA response elements also induce structural transitions (see below). Nevertheless, the LBD remains the main architectural feature triggering biological responses to very diverse lipophilic molecules.
X-ray crystallography established the E domain as organized as a three-layered antiparallel α-helical sandwich composed by 12 α-helices, including a β-sheet (s1–s2) which is part of the ligand binding pocket (LBP). The LBP is located inside of this structure and is composed of a group of surrounding helices [51]. The LBP of nuclear receptors is a highly variable region, both in volume, ranging from 300 to 1500Å3, and in structure. Such a diversity allows the binding of a variety of molecules ranging from phospholipids to heme, including steroid and fatty acid derivatives, xenobiotics..., and highlights the broad spectrum of physiological actions of nuclear receptors.
Ligand-LBP interactions involve amino acids located in most receptors in helices 3, 5 and 10/11. Additional interactions are brought into play as a function of the receptor and the chemical structure of the ligand. Hydrophobic interactions, hydrogen bonding networks and the steric size and shape of LBPs determine the strength and specificity of LBD-ligand complex [107]. This atomic network is variable according to receptor isoforms, allowing the design of isoform-selective agonists or antagonists [108].
Ligand binding causes conformational changes of nuclear receptors, which involve repositioning of H3, H4, L3-L4 and H12. Helix 12 (initially termed the AF-2 activating domain or AF-2 AD) is stabilized against the LBD core, generating a hydrophobic groove made of helices 12, 3, 4 and 5. This structure allows the LBD to interact with the LXXLL signature motif found in most if not all reported primary nuclear receptor coactivators [109]. This interaction is further stabilized by a charge clamp made in most cases of a lysine in H3 and a glutamic acid in H12, which is required for optimal binding of coactivator molecules [110–113]. Subtle changes in ligand structure seems to affect the coactivator binding interface, providing a molecular basis for the varying efficacy and potency of nuclear receptor agonists [112]. In a more extreme fashion, antagonist binding positions helix 12 to cause a steric obstruction of the LXXLL binding groove. Importantly, the helix 12 region contains a degenerated LXXLL motif allowing for this interaction. Alternatively, antagonism can be exerted by generating a structure favoring the recruitment of corepressor molecules such as SMRT and NCoR or by preventing H12 proper folding. [113–117]. Intriguingly, some nuclear receptors act, in the absence of ligand, as transcriptional repressors. While it is acknowleged that this repressive action is physiologically important, the structural basis for this ligand-independent repression was unknown until recently. Two reports described a specific structure in RARα and rev-erb-α in the LBD that forms an anti-parallel β-sheet with corepressor amino acids, identifying a novel interaction interface [118,119] and documenting a structural basis for the mechanism of derepression, which necessitates the active removal of corepressor molecules.
Finally, the LBD harbors a dimerization interface, the core of which mapping to H7, H9, H10, H11, loops L8-L9 and L9-L10. Although ligand binding has been long suspected to promote nuclear receptor dimerization [119–124], structural studies did not provide evidence for ligand-induced reshaping of this dimerization interface [106,125].
As other domains, the LBD is the target of posttranslational modifications. While it is beyond the scope of this review to provide an exhaustive list of identified covalent modifications (see also [126]), it is worth noting here SUMOylation plays an important role in channeling the transcriptional activity towards transactivation or tethered transrepression. SUMOylation of PPARγ at K365 is required for transrepression of the iNOS promoter in macrophages and targets PPARγ to the NCoR complex bound to NF-kappa-B regulated promoters [127]. This mechanism is detailed below. In an analogous manner, agonist-induced SUMOylation of LXRβ in the LBD promotes its interaction with GPS2 and binding to the NCoR complex associated to acute phase response genes [128]. PPARα also controls negatively hepatic gene expression in a sex-specific manner. Such a repression is exerted for example on Cyp7b1 expression, known to divert DHEA from the testosterone biosynthesis pathway. This occurs through the SUMOylation-dependent PPARα docking to the Cyp7b1 transactivating GA-binding protein, corepressor and HDAC recruitment to this promoter and DNMT3-catalyzed DNA methylation of a neighboring cis-activating SP-1 site [129]. Phosphorylatoin can exert opposite effects on NR activity through very diverse mechanisms. ATPase class 1 type 8B member [familial intrahepatic cholestasis 1 (FIC1) protein] activates FXR via PKCzeta-dependent phosphorylation of FXR at Thr-442. This covalent modification promotes the nuclear translocation of FXR and subsequent FXR target gene activation [130]. Through the combination of non genomic and genomic effects, retinoid acid activates the p38MAPK/MSK1 pathway, leading to phosphorylation of two serines in N-terminal domain and in RARα LBD and of histone H3. Phosphorylation of RARα increases the binding efficiency of cyclin H to the loop L8-L9 and promotes the right positioning of cdk7 and phosphorylation of RARα AF-1, to finally trigger RARα target genes activation [131]. This non-limitative set of examples thus point to the very complex integration of signaling events into nuclear receptor-mediated events.
5. The F domain
The F domain is located at the extreme C-terminus of NR. Because of its high variability in sequence, little is known about its structure and functional role. The length of the domain F can vary from no to 80 amino acids [132]. Crystal structure of progesterone receptor revealed that the F domain adopts an extended β-strand conformation [133] which may, in the case of RAR dimers, contact the dimerization partner [134]. Differences in ER isotype transcriptional activity is partly due to a variable F domain structure. Based on amino acid sequence, it is predicted that ERα F domain is an α-helical region followed by an extended β-strand-like region, separated by a random coil stretch. In contrast, ERβ domain F is more likely not to adopt an α-helical structure [135]. Mutagenesis and functional studies showed that domain F does not exert its activity independently and that it is dispensable for ligand binding or transcriptional activity. Nevertheless, deletion of the domain F or part of it may perturb NR activity and interactions with co-regulators. Deletion of the domain F eliminates the ability of human ERα to activate transcription via interaction with SP-1 [136]. HNF4, which harbors the longest domain F in its alternatively spliced isoform HNF4α2, is transcriptionally more active and is more responsive to overexpression of the co-activators NCoA2 and CBP [137]. The F domain of HNF4α1 interacts also with NCoR2/SMRT [138]. Interestingly, deletion of the F domain of RARα increased co-activator binding but decreased co-repressor binding [134]. Thus the F domain can be engaged in interactions with transcriptional co-regulators [139]. Moreover, different point mutations among domain F of ER suggested its involvement in ligand-receptor interaction, and impacts on the ligand responsiveness of ER tethered to an AP-1 response element [140]. Finally, the F domain can be covalently modified by phosphorylation and affect ER basal transcriptional activity. O-GlcNAcation of this domain leads to decreased ability of ER to bind to an estrogen response element in vitro [141].
B. DNA RESPONSE ELEMENTS GEOMETRY, ARCHITECTURE AND RECOGNITION BY NUCLEAR RECEPTORS
DNA sequence recognition and binding is the initial step of the transactivation process mediated by nuclear receptors. Consequently, NR monomers or dimers are positioned on RE which are made of one or two hexameric half-site motifs. Adopting a different geometry, they form palindromes, direct (DR), everted (ER) or inverted repeats (IR) separated by a spacer of varying length and sequence. Four conditions can be distinguished that determine the uniqueness of the response element. They are (i) the nucleotide sequence of the DNA-half sites, (ii) their relative orientation (iii) the sequence of the spacer and (iv) the length of the spacer.
Some NRs, mainly orphans, bind to DNA as monomers. The monomeric Nurr1 binds to a hormone response element 5′-AGGTCA-3′ flanked by a 5′ 1 to 6-bp long A/T-rich sequence [142]. This sequence referred to as an Nur77/NGFI-B response element (NBRE) [143] is also the target of Nur77 monomers [144]. Nurr1 can however dimerize with RXR, and in this configuration can display significant affinity for DR with spacing ranging from 10 to 27 bases [145]. A similar promiscuity in binding to naked DNA is observed for SF-1, FTZ-F1, rev-Erb-α and RORα which target a single copy of this extended core recognition sequence, although rev-Erb-α can also bind to a specific DR2 RE [146,147].
Receptors binding to DNA as homodimers, exemplified by the steroid hormone receptors GR, MR, AR and PR recognize two consensus half-sites 5′-AGAACA-3′ or in case of ER 5′-AGGTCA-3′ arranged as inverted repeats spaced by 3 bp (IR3) [148]. Formation of stable head-to-head homodimers is dependent on discrete dimerization interfaces located in both the DBD and the LBD (Figure 3).
Figure 3. Different architecture of selected response elements of nuclear receptors.
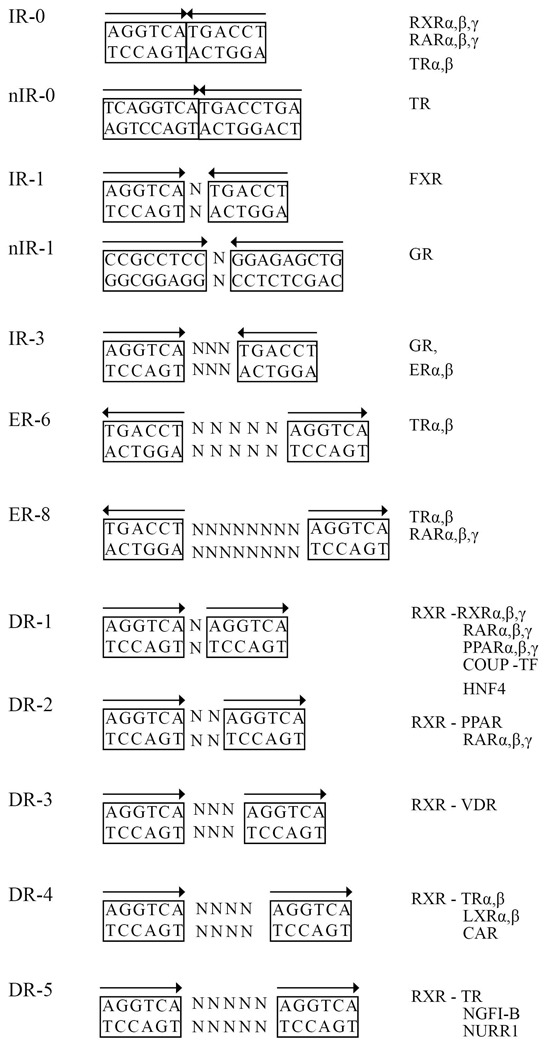
IR -inverted repeat, ER - everted repeat, DR - direct repeat, ‘N’ indicates any nucleotide, “n” indicates negative response elements.
Nuclear receptors that form heterodimers with RXRs recognize REs composed of two half-site motifs arranged as direct (DR), inverted (IR) or everted repeats (ER), the core consensus sequence being 5′-AGGTCA-3′. For instance PPARs, RARs, VDR and T3R recognize direct repeats following a specificity rule called the 1-2-3-4-5 rule [149–151]. Some RXR partners display a more relaxed specificity: PXR can bind to a variety of DNA response elements with various spacing, which includes direct repeats DR-3, DR-4, and DR-5, and everted repeats ER-6 and ER-8 [152,153]. FXR prefers binding to an inverted repeat of the ideal sequence 5′-AGGTCA-3′ separated by 1 bp (IR-1) [154], but several different response elements have been reported, including ER8 [155] and DR1 [156].
RXR partners can be divided into two groups depending on their functionality as heterodimers. Permissive RXR-containing heterodimers can be activated by RXR agonists in the absence of the agonist for the RXR partner. This group includes PPAR, LXR and FXR. Nonpermissive heterodimers formed by RXR and RAR, TR, VDR cannot be activated by RXR agonists and require agonists of the RXR partner to be activated.[157–159].
Heterodimers can adopt various polarities when bound to different REs, and RXR can be positioned either upstream or downstream of the heterodimer partner. This relative orientation and its impact on the transcriptional activity of receptors has been dissected for RAR-RXR heterodimers. On DR2 and DR5 elements, RXR occupies the 5′ hexameric motif, whereas the RAR partner occupies the 3′ motif. The polarity is reversed on DR1 response elements. This structural arrangement has dramatic consequences on the transactivation properties of RXR-RAR heterodimers, as RAR agonists are unable to activate transcription from a DR1 RE. This relates to the allosteric control of NCoR assembly on these various DR REs [157,160,161] whose geometry imposes an important structural adaptation of receptor domains. In support of this, DNA binding of RXR-VDR dimers was shown to alter VDR H12 structure [125]. Crystallographic structures of isolated GR DBD bound to DNA identified the so-called “lever arm”, located between the two GR zinc fingers, which adopts different conformations according to the RE geometry and influences coactivator recruitment [162]. Other heterodimers such as PPARα-RXRα bind to DNA similarly to RAR-RXRα and form a polar head-to-tail interaction with DR1, where RXRα binds exclusively to the 3′ site [11,92]. For VDR assembled on a DR3, TR and LXR on a DR4 and NGFI-B on a NBRE, the RXR DBD was found to bind to the 5′ upstream half-site [50,89,163].
Thus several structural features are brought into play to limit nuclear receptor DNA binding promiscuity, in addition to tissue- and cell-specific expression and limited ligand availability. It is worth noting that these rules have been defined using naked DNA templates. However, a genome-wide bioinformatic search for any of these consensus sequences will yield at least a hit every 500–1000 bp. This number is at odds with the number of actual NR binding sites determined by Chip-seq experiments (several thousands for ER and PPARγ, [164–167]) and the number of regulated genes determined in similar conditions (a few hundreds). Moreover, many of these sequences are located very distal to the transcriptional start site (TSS) when considering a linear sequence, either 5′ or 3′ to the TSS. Chromosomal conformational studies revealed that enhancer sequences act in cis with respect to promoter sequences, implying chromatin looping between TSS and enhancer sequences [165,168,169]. Quite intriguingly, the functionality of such an association is characterized by the induction of the so-called enhancer-templated non-coding RNA (eRNA) emanating from the distal binding site [170], a phenomenon whose functional significance has not yet been elucidated but which is not restricted to NR-mediated transcriptional control [171]. Genome-wide mapping of nuclear receptor binding sites also revealed the statistically- and biologically-significant association of a fraction of REs with other transcription factor binding sites. This led to the identification of cell-specific “pioneering factors” such as FoxA1, which act by priming NR DNA binding sites to bind their cognate NRs [172,173].
There are thus multiple mechanisms controlling the association of NRs with DNA, all of them having a significant impact on the assembly of NRs on chromatin templates and productive recruitment of the transcription machinery.
C. GENERAL MECHANISMS OF TRANSCRIPTIONAL REGULATION BY NUCLEAR RECEPTORS
As already mentioned above, nuclear receptors can control transcriptional events by exerting either a positive, direct effect or by imposing a repressed state to regulated promoters. They can also mediate, through protein-protein interaction, a repressive effect on a variety of other signaling pathways under the control of transcription factors such as AP-1, NF-kappa-B or C/EBP. Each of these aspects will be described below to provide a global view of the most recent concepts which have emerged in the field in the past years (Figure 4).
Figure 4. General mechanism of NR action.
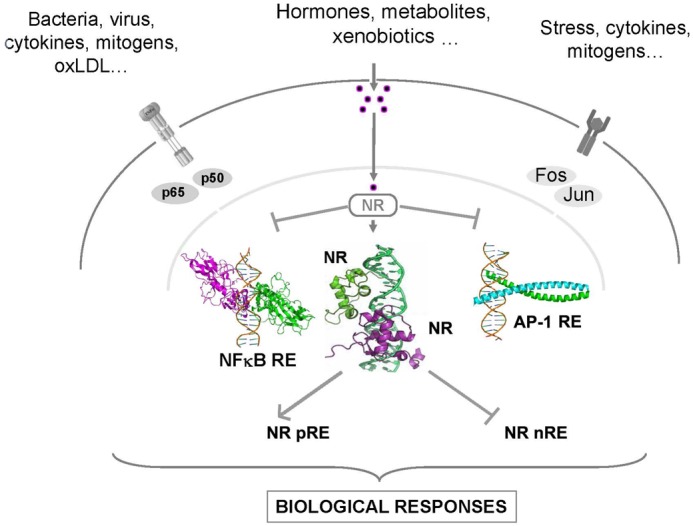
Nuclear receptors may act in two different ways. Upon ligand binding nuclear receptors forming heterodimers with RXR interact with a specific positive gene response element (pRE) and activate mRNA transcription of target genes. Alternatively, they may interact directly with repressive, negative response elements (nRE). The major suppressive effect of nuclear receptors is however thought to be mediated by monomers interaction with subunits of AP-1 and NF-kB transcription factors, and hamper the expression of inflammatory-related genes (see text for details).
1. Transcriptional activation
An important feature of steroid hormone receptors and of most of the heterodimeric nuclear receptors is the ability to activate transcription of target genes upon ligand binding. In general, this mechanism comprises ligand-dependent conformational changes of the nuclear receptor associated to chromatinized REs, that trigger co-repressor complex release and the sequential recruitment of co-activator complexes that modify chromatin structure and promote the assembly of the transcription initiation complex at regulated promoters. Various co-activators were identified for NRs and the repertoire is specific for certain cell types, genes and signals. Thus binding of agonists stimulate the exchange of co-repressors for co-activators necessary for transcriptional activation. Of note, the ligand-dependent association of NR corepressors such as LCoR and RIP140, through LXXLL motifs may play a significant role in transcription attenuation [174,175], however this mechanism has not been studied in great detail and will not be discussed here.
1a. Nuclear receptor corepressor binding
In the unliganded state, NRs are associated to corepressor complexes. These complexes are composed of a subunit (SMRT/NCoR2 or NCoR1) directly interacting with the receptor through a degenerated LXXLL motif, which harbor a consensus sequence L/I-X-X-I/V-I or LXXXI/LXXXI/L also called the CoRNR box [176,177]. This CoRNR box motif interacts, as the coactivator LXXLL motif, with amino acids from the LBD hydrophobic groove. This interaction interface is remodeled upon agonist binding and helix 12 positioning occludes part of the CoR binding interface. As mentioned above, additional CoR binding interfaces, as well as novel CoRNR boxes have been described [118,119,178], suggesting the use of alternative mechanisms for NR-corepressor interaction. Corepressor complexes are built around the SMRT or NCoR subunits, which harbor a conserved repression domain on which the core repressive machinery (including HDAC3, GPS2 and TBL1 or TBLR1) is assembled. Recent structural and functional studies highlighted a central role for TBL1 in assembling this very large complex (ca. 1–2 MDa) [179]. In some cases, ligand-binding is sufficient to inhibit co-repressor recruitment (e.g. for RXR and TR), but more generally the active removal of the co-repressor complex is required. This points again to the critical role of TBL1/TBLR1 which encompass a F-box domain interacting with the ubiquitin-conjugating enzyme H5 (UBCH5) and a 19S-proteasome complex, which mediates ubiquitination and proteosomal degradation of SMRT- or NCoR-GPS2-HDAC3 complexes [180].
1b. Nuclear receptor coactivator binding
Since the seminal discovery of SRC-1/NCoA1 as a progesterone receptor coactivator [181], more than 350 coactivators have been identified so far. This prodigious amount of polypeptides exhibit various enzymatic activities involved in the regulation of histone modification and chromatin remodeling, initiation of transcription, elongation of RNA transcripts, mRNA splicing and elongation, and proteasomal termination of nuclear receptor complexes. Their involvement and relative activity in nuclear receptor-controlled processes is modulated by their cell-specific expression levels and post-translational modifications, conditions which have been reviewed recently [182,183]. It is also nowadays accepted that many of these coregulators participate in molecular events driven by other transcription factors.
The coactivator family has been divided in two subfamilies. The first one defines coactivators which interact directly with NR AF-1&2 regions such as the SRC coactivators, CBP and p300. The second one includes other proteins which interact with primary coactivators such as CARM1, CoCoA, Fli-I... Primary and secondary coactivators are recruited to regulated promoters in an orchestrated fashion [184]. Since this issue is devoted to nuclear receptors involved in metabolism control, only coactivators associated to such an activity will be briefly described here.
b.1. The p160 and p300 families
Co-activators belonging to the p160 family [NCoA1/SRC-1, NCoA2/TIF2 (known as SRC-2 or GRIP1) and NCoA3/RAC3 (also known as SRC-3, ACTR, pCIP or TRAM-1)], p300 and the cAMP response element-binding protein (CBP) bind to the NR LBD via an alpha-helical LXXLL motif [185,186]. Co-activators such as CBP and p300 posses histone acetylase transferase (HAT) activity, which has a critical role in regulating NR-mediated transcription [187]. N-terminal tail acetylation of histone H4, which is likely to establish contacts with the histone H2A/H2B dimer, prevents this interaction and destabilizes chromatin compaction. Additionally, acetylation weakens the interaction of the histone tails with DNA [188]. Consequently, the chromatin is decondensated allowing the promoter initiation complex to bind at the promoter site.
Data emerging from studies of knockout animals suggest that the SRCs play critical and distinct roles in controlling energy homeostasis. SRC-1−/− mice have decreased energy expenditure and are prone to obesity. In opposition, SRC-2−/− mice are protected against high-fat diet-induced obesity, but can lead to a condition reminiscent of a glycogen storage disease type 1a. The ablation of SRC-3 generates mice highly resistant to high-fat diet-induced obesity. Collectively, these data and others point to a complex, but critical role of SRCs in metabolic regulation which has been in most instances related to the control of PPARγ transcriptional activity [189]. However, given the pleiotropic role of SRCs, it is very likely that other mechanisms contribute to these metabolic effects.
b.2. The ATP-dependent remodelling complex SWI/SNF
the SWI/SNF complex has a role in metabolic control, as it was identified in yeast to be essential for mating-type switching and growth on sucrose. The SWI/SNF family is evolutionary conserved and plays an important role in ATP-dependent chromatin remodeling [190] by catalyzing the disruption of DNA-histone interactions and sliding of the nucleosome along DNA [191]. The human homolog BAF complex is a multimeric entity of 1.2 MDa including BRG1/hBRM, BAF polypeptides (BAF155/170, BAF60, BAF57, BAF53a/b, BAF47, BAF250a/b, BAF200, BAF45a/b/c/d, Brd9, and Brd7) and actin. Several of these subunits harbor LXXLL motifs and have been identified not only as nuclear receptors coactivators for ER [192,193], AR [194], RAR [193,195], FXR [196] and GR [197], but also as corepressors of SHP [198], as SWI/SNF components can be integrated in corepressor complexes [199]. Interestingly, the BAF60a subunit displays a circadian expression in mouse liver and, acting as a coregulator of RORα, regulates the expression of clock and metabolic genes [200].
b.3. The mediator complex
Like the SWI/SNF complex, the Mediator complex has been originally identified in yeast and subsequently characterized in other eukaryotic cells. A number of studies described its role as a catalyzer of the transcription preinitiation complex (PIC) assembly at activated promoters. Through direct interaction with RNA polymerase II, general transcription factors (TFIID, TFIIH) and elongation factors, Mediator plays a key role in RNA polymerase II-controlled transcription [201]. Investigations about the role of Mediator in NR research gained momentum when it was realized that Mediator-like complexes bind directly to NRs [202–206]. Mediator is organized in four structural modules and includes more than 20 subunits, of which the Med1 subunit contains LXXLL motifs [207]. The liver-specific Med1 KO induces hepatic steatosis in a PPARγ-dependent manner [208], in agreement with its adipogenic [209] and PPARγ coactivator roles [210]. Skeletal muscle-specific KO of Med1 enhances insulin sensitivity and improves glucose tolerance and confers resistance to high-fat diet-induced obesity [211]. Thus given its broad and key roles in transcriptional regulation through a direct interaction with RNA polymerase II, Mediator is viewed as being the last complex recruited cyclically to NR-regulated promoters [184].
2. Transcriptional repression
2a. Transcriptional repression by unliganded receptors
Some nuclear receptors can actively repress transcription in the absence of ligand. This process is related to the recruitment of co-repressor complexes. There are several co-repressor complexes characterized, but the most commonly studied complex comprises nuclear receptor co-repressor (NCoR), silencing mediator for retinoid and thyroid hormone receptors (SMRT), histone deacetylase 3 (HDAC3), transducin-α-like 1 (TBL1), TBL-1-like related protein (TBLR1) and G-protein-pathway suppressor 2 (GPS2) [212,213]. HDACs posses a well-characterized role in transcriptional repression by deacetylating N-terminal lysines of histone proteins thus generating a condensed, transcriptional inactive chromatin structure. It was reported that SMRT and NCoR contain a deacetylase-activating domain which can trigger the enzymatic activity of HDAC3 [214].
In addition, other corepressor complexes have been described, such as SWI/SNF-containing complexes as mentioned above, PRC1&2 and CoRest complexes. Like the NCoR/SMRT complex, tethering these multiprotein entities to promoters leads to histone and DNA covalent modifications, followed by chromatin compaction and/or DNA masking. A critical step in NR-mediated transcriptional activation is the dismissal of corepressor complex from the DNA-bound receptor. In vitro assays have demonstrated that agonist-induced conformational changes are sufficient for SMRT or NCoR dissociation from the receptor, in agreement with crystal structure data. However, dynamic models of de-repression involving post-translational modifications of corepressor complex subunits leading either to their nuclear exclusion and/or degradation have been described [215]. The mechanism(s) by which such an active derepression take place is as of yet unknown.
2b. Direct transrepression by liganded receptors
Ligand-bound NRs repress the transcription of some genes by a mechanism called negative regulation. This process occurs with multiple NRs and genes and was detailed for GR and TR. It has been suggested that these NRs recognize and bind negative response elements and downregulate specific target genes. The analysis of specific DNA sites revealed that negative glucocorticoid response elements (nGRE) and negative thyroid response elements (nTRE) are different from positive response element that mediate transcriptional activation [216,217]. Overlapping binding sites for transcription factors such as Oct-1/Pbx, AP-1 and SP1 were found for negative response elements of GR and TR, and found to dictate the transcriptional cis effect of the response element [218–221]. These data thus posit that negative cis-acting glucocorticoid response elements exert such an activity by interacting with other transcription factors. However, a recent report described a novel class of negative glucocorticoid REs, organized as inverted repeats with a 1bp spacer, on which glucocorticoids promote the recruitment of GR-corepressor complexes [222]. Such a mechanistic principle does not seem to hold true for T3-mediated transcriptional repression. As detailed for the αTSH gene, the corepressor SMRT is recruited to the nTRE and promotes histone deacetylation. Upon agonist treatment, SMRT dismissal is correlated with histone acetylation and gene repression [223,224]. Furthermore, functional studies have shown a role for SRC-1 in transcriptional repression mediated by liganded TR [225,226]. The mechanistic basis for such a reversal of transcriptional activity is not known, but could be mediated by post-translational modifications such as phosphorylation, acetylation or SUMOylation of promoter-associated histones and/or of coregulatory proteins [227–229]. Thus direct repression occurs via distinct mechanisms which are receptor- and context-dependent. These studies also pinpoints to the versatility of coregulator complexes, which may exert either positive or negative effects on the transcriptional outcome following NR agonist stimulation.
2c. Tethered transrepression by liganded receptors
The mechanism referred to as tethered transrepression engages negative crosstalk of ligand-activated nuclear receptors with other signal-dependent transcription factors, including NF-kappa-B and activator protein-1 (AP-1). This process modulates inflammation in various cells of the central nervous system, the immune system as well as the liver, etc and interferes with cellular proliferation in various tissues.
Several mechanisms can be proposed to account for such a repression: (i) repression of PIC assembly on NF-kappa- or AP-1 regulated promoters [127,128,230,231]; (ii) inhibition of RNA polymerase II conversion towards an elongation-competent form [232,233]; (iii) upregulation of the expression of the inhibitor of NK-kappa-B [234]; (iv) interaction with upstream components of the NF-kappa-B or AP-1 activating cascade [235–237]; (v) coactivator exclusion by competition [238,239] and (vi) direct physical interaction with AP-1 or NF-kappa-B (mostly p65) subunits [240–243], although this process is much more complex and requires multiple factors in living cells [244].
Interestingly, inflammatory programs triggered by TLR-3, 4 or 9 activation in macrophages are only partially inhibited by GR, LXR and PPARγ agonists, each receptor inhibiting about one-third to one-half of the induced genes. Intriguingly, inhibited clusters of genes by each receptor were only partially overlapping [238].
NR co-repressors such as NCoR and SMRT play an important role in ligand-dependent tethered transrepression. NCoR-deficient macrophages display a derepressed expression of various AP-1 and NF-kappa-B-related genes, an effect linked to NCoR (or SMRT, [245]) association to these DNA-bound transcription factors [246]. Much like NR-mediated transcription, activation of signaling pathways leads to the transcription of NF-kappa-B-driven genes by removal of the corepressor complex through a proteasome-dependent pathway. NR activation upon ligand binding promotes tethering of sumoylated NR to NF-kappa-B complexes, which interrupts corepressor complexes clearance, hence maintaining the promoter in a repressed state [127]. More recently, sumoylated LXRs were found to be targeted at transrepressed promoters through interaction with a NCoR complex component, coronin A. This interaction prevents corepressor turnover by preventing oligomeric actin recruitment [247]. This very elaborate process has been described for LXR. in mouse macrophages, whereas transrepression of the acute phase reaction (APR) in mouse liver by LXR involves GPS2 rather than coronin A [128,247]. Thus, as suspected from many previous studies, tethered transrepression follows different mechanistic schemes which are receptor-, gene- and cell type-specific.
The structural features of NR specifically involved in transrepression are not clearly defined. Extensive mutagenesis studies of T3R, RAR, PPARγ, GR and ER (see for examples [239,248–261]) did not yield a clear-cut and unifying model for tethered transrepression. Taken as a whole, it clearly appeared that coactivator recruitment through the AF-2 domain is not required for this activity, as well as direct DNA binding. There are also strong evidences suggesting that homo- or hetero dimerization is not mandatory [239,262]. The lack of well-defined molecular structures involved in transrepression is an important pitfall in designing screening methods aiming at identifying dissociated ligands which would preferentially elicit tethered transrepression in inflammatory diseases.
D. NUCLEAR RECEPTORS AND NON-GENOMIC SIGNALING PATHWAYS
NR ligands regulate gene expression by genomic actions which are described above. Nevertheless, NR ligands also exhibit non-genomic effects manifested by the rapid and transient activation of several kinase cascades, which can be attributable to a subpopulation of NRs located at the cell membrane, although this point is still debated. Accordingly, conserved palmyltoylation sites have been identified in GR and ER [263–265], and together with MR, these receptors have been detected in lipid rafts [265–268].
This extranuclear localization provides a mean for steroid receptors to interact with various kinases. Estrogens trigger protein-protein interaction between ER and Src/p21ras/Erk and PI3K/Akt, through the SH2 domain of c-Src and the regulatory subunit of PI3K respectively. Estrogen-mediated induction of these kinase cascades play an important role in cell proliferation in breast cancer and vascular function [269–273]. Progestins can induce the Src/Erk1/2 pathway mediated by the interaction of two domains of the progesterone receptor (PR) with the LBD of ER. This crosstalk is essential for progestin induction of DNA synthesis and cell proliferation in breast cancer [274]. A complex of activated PR, ERK and its target kinase Msk1 is recruited to the promoter after hormone treatment and phosphorylates serine 10 of histone H3, where it induces the recruitment of SRC-1, RNA polymerase II and chromatin remodeling complex (hSnf2h and Brg1). This example constitutes a link between kinase cascade activation in the cytoplasm, chromatin remodeling, and transcriptional activation in the nucleus [275] which is possibly conserved for retinoid receptors [276]. It has been suggested that aldosterone can counteract vasoconstriction via stimulation of endothelial NO production. This occurs through a mechanism which engages PI3 kinase and its interaction with MR [277]. A similar mechanism seems to underlie the decreased vascular inflammation and reduced myocardial infarct size following ischemia and reperfusion injury induced by glucocorticoids [278].
Recent evidences show that dexamethasone, a synthetic GR agonist, reduces cPLA2 activation which releases arachidonic acid. This mechanism seems to be glucocorticoid receptor-dependent but transcription-independent [279,280]. Plasma membrane-bound GR [281] has indeed been described in a variety of cell types [282,283] and GR has been shown to associate to Src in lipid rafts [268].
Non-genomic effect events similar to those described for steroid hormones occur for retinoids. It has been reported that RAR is present in the cytoplasm and in membranes where it associates with PI3K or Src [284]. Retinoic acid (RA) rapidly activates mitogen-activated protein kinases (MAPKs) such as ERK and p38MAPK in fibroblasts, mouse embryocarcinoma cells, mammary breast tumor cells and leukemia cells [131,285,286]. A novel unexpected non-genomic activity has been demonstrated for RARα: RARα is transported to neuronal dendrites where associates with glutamate receptor 1 (GluR1) mRNA, via its C-terminal F region and, as a result, inhibits the translation of this mRNA. RA binding abrogates this translational repression. These effects have been correlated to the regulation of synaptic functions and neuronal plasticity controlled by RA [287–289].
Non-genomic effects were also observed for nuclear receptor ligands involved in metabolic control. Although there is no evidence for membrane-bound PPARγ, the synthetic agonist rosiglitazone (RGZ) as well as the natural agonist 15ΔPGJ2 regulate glucose and lipid metabolism and sperm activation in human spermatozoa by a rapid mechanism involving protein phosphorylation [290]. In human microvascular endothelial cells, RGZ interferes with pro-inflammatory actions of TNF and IFNγ by direct inhibition of ERK1/2 phosphorylation in a PPARγ-dependent manner [291]. RGZ-mediated ERK1/2 regulation and PI3K inhibition was observed in human adrenocortical cells and PC3 prostate cells [292,293]. Conversely, in vascular smooth muscle cells 15ΔPGJ2 and TZD activated the MEK/ERK pathway via PI3K [294]. Importantly, the energy-sensitive AMP kinase is activated by TZD-stimulated PPARγ, inducing acetyl CoA carboxylase phosphorylation, stimulation of glucose uptake and fatty acid oxidation in skeletal muscle, liver and adipose tissue [295,296].
Thus non genomic effects of NR ligands, mediated or not by an extranuclear subpopulation of NRs introduce a new layer of complexity in NR biology which must be determined when studying biological and pharmacological effects of NR ligand administration. Although impaired by technical limitations, the study of the subcellular localization of NRs in pathophysiological conditions may help deciphering mechanisms controlling the broad spectrum of biological responses controlled by NRs. Worth noting, the mitochondrial effects of some NRs such as SHP [297], GR [298] and Nur77 [299–301] which play an important role in apoptosis regulation through protein-protein interaction, deserve further investigations for other members of the NR family.
E. CONCLUSION
NRs are modular transcription involved in multiple pathophysiological processes. They can be viewed as an assembly platform on chromatin for multimeric coregulators which will dictate the cell-specific and even gene-selective transcriptional ouput of target cells. In addition to direct ligand binding, these multi-proteic complexes are integration modules of other signaling pathways which can additionally adjust NR-driven promoters response to their extracellular cues. With the advent of high throughput genomic, epigenetic and proteomic techniques, a NR system biology can now be elaborated to bring a global and detailed view of NR contribution to human biology and diseases.
Acknowledgments
This work was supported by grants from Institut National de la Santé et de la Recherche Médicale, Université de Lille 2, Région Nord-Pas de Calais/FEDER, COST (Action BM0602).
Footnotes
Conflict of interest: The authors declare no conflict of interest.
REFERENCE LIST
- 1.Escriva GH, Laudet V, Robinson-Rechavi M. Nuclear receptors are markers of animal genome evolution. J Struct Funct Genomics. 2003;3:177–184. [PubMed] [Google Scholar]
- 2.Weinberger C, Hollenberg SM, Rosenfeld MG, Evans RM. Domain structure of human glucocorticoid receptor and its relationship to the v-erb-A oncogene product. Nature. 1985;318:670–672. doi: 10.1038/318670a0. [DOI] [PubMed] [Google Scholar]
- 3.Hollenberg SM, Weinberger C, Ong ES, Cerelli G, Oro A, Lebo R, Thompson EB, Rosenfeld MG, Evans RM. Primary structure and expression of a functional human glucocorticoid receptor cDNA. Nature. 1985;318:635–641. doi: 10.1038/318635a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greene GL, Gilna P, Waterfield M, Baker A, Hort Y, Shine J. Sequence and expression of human estrogen receptor complementary DNA. Science. 1986;231:1150–1154. doi: 10.1126/science.3753802. [DOI] [PubMed] [Google Scholar]
- 5.Walter P, Green S, Greene G, Krust A, Bornert JM, Jeltsch JM, Staub A, Jensen E, Scrace G, Waterfield M. Cloning of the human estrogen receptor cDNA. Proc Natl Acad Sci U S A. 1985;82:7889–7893. doi: 10.1073/pnas.82.23.7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sap J, Munoz A, Damm K, Goldberg Y, Ghysdael J, Leutz A, Beug H, Vennstrom B. The c-erb-A protein is a high-affinity receptor for thyroid hormone. Nature. 1986;324:635–640. doi: 10.1038/324635a0. [DOI] [PubMed] [Google Scholar]
- 7.Weinberger C, Thompson CC, Ong ES, Lebo R, Gruol DJ, Evans RM. The c- erb-A gene encodes a thyroid hormone receptor. Nature. 1986;324:641–646. doi: 10.1038/324641a0. [DOI] [PubMed] [Google Scholar]
- 8.Germain P, Staels B, Dacquet C, Spedding M, Laudet V. Overview of nomenclature of nuclear receptors. Pharmacol Rev. 2006;58:685–704. doi: 10.1124/pr.58.4.2. [DOI] [PubMed] [Google Scholar]
- 9.Giguere V, Hollenberg SM, Rosenfeld MG, Evans RM. Functional domains of the human glucocorticoid receptor. Cell. 1986;46:645–652. doi: 10.1016/0092-8674(86)90339-9. [DOI] [PubMed] [Google Scholar]
- 10.A unified nomenclature system for the nuclear receptor superfamily. Cell. 1999;97:161–163. doi: 10.1016/s0092-8674(00)80726-6. [DOI] [PubMed] [Google Scholar]
- 11.Chandra V, Huang P, Hamuro Y, Raghuram S, Wang Y, Burris TP, Rastinejad F. Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA. Nature. 2008;456:350–356. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rochel N, Ciesielski F, Godet J, Moman E, Roessle M, Peluso-Iltis C, Moulin M, Haertlein M, Callow P, Mely Y, Svergun DI, Moras D. Common architecture of nuclear receptor heterodimers on DNA direct repeat elements with different spacings. Nat Struct Mol Biol. 2011;18:564–570. doi: 10.1038/nsmb.2054. [DOI] [PubMed] [Google Scholar]
- 13.Hollenberg AN, Monden T, Madura JP, Lee K, Wondisford FE. Function of nuclear co-repressor protein on thyroid hormone response elements is regulated by the receptor A/B domain. J Biol Chem. 1996;271:28516–28520. doi: 10.1074/jbc.271.45.28516. [DOI] [PubMed] [Google Scholar]
- 14.Borjesson AE, Windahl SH, Lagerquist MK, Engdahl C, Frenkel B, Moverare-Skrtic S, Sjogren K, Kindblom JM, Stubelius A, Islander U, Antal MC, Krust A, Chambon P, Ohlsson C. Roles of transactivating functions 1 and 2 of estrogen receptor-alpha in bone. Proc Natl Acad Sci U S A. 2011;108:6288–6293. doi: 10.1073/pnas.1100454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Billon-Gales A, Fontaine C, Filipe C, Douin-Echinard V, Fouque MJ, Flouriot G, Gourdy P, Lenfant F, Laurell H, Krust A, Chambon P, Arnal JF. The transactivating function 1 of estrogen receptor alpha is dispensable for the vasculoprotective actions of 17beta-estradiol. Proc Natl Acad Sci U S A. 2009;106:2053–2058. doi: 10.1073/pnas.0808742106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Briancon N, Weiss MC. In vivo role of the HNF4alpha AF-1 activation domain revealed by exon swapping. EMBO J. 2006;25:1253–1262. doi: 10.1038/sj.emboj.7601021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bour G, Plassat JL, Bauer A, Lalevee S, Rochette-Egly C. Vinexin beta interacts with the non-phosphorylated AF-1 domain of retinoid receptor gamma (RARgamma) and represses RARgamma-mediated transcription. J Biol Chem. 2005;280:17027–17037. doi: 10.1074/jbc.M501344200. [DOI] [PubMed] [Google Scholar]
- 18.Mascrez B, Mark M, Krezel W, Dupe V, LeMeur M, Ghyselinck NB, Chambon P. Differential contributions of AF-1 and AF-2 activities to the developmental functions of RXR alpha. Development. 2001;128:2049–2062. doi: 10.1242/dev.128.11.2049. [DOI] [PubMed] [Google Scholar]
- 19.Tomura H, Lazar J, Phyillaier M, Nikodem VM. The N-terminal region (A/B) of rat thyroid hormone receptors alpha 1, beta 1, but not beta 2 contains a strong thyroid hormone-dependent transactivation function. Proc Natl Acad Sci U S A. 1995;92:5600–5604. doi: 10.1073/pnas.92.12.5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bommer M, Benecke A, Gronemeyer H, Rochette-Egly C. TIF2 mediates the synergy between RARalpha 1 activation functions AF-1 and AF-2. J Biol Chem. 2002;277:37961–37966. doi: 10.1074/jbc.M206001200. [DOI] [PubMed] [Google Scholar]
- 21.Gianni M, Tarrade A, Nigro EA, Garattini E, Rochette-Egly C. The AF-1 and AF-2 domains of RAR gamma 2 and RXR alpha cooperate for triggering the transactivation and the degradation of RAR gamma 2/RXR alpha heterodimers. J Biol Chem. 2003;278:34458–34466. doi: 10.1074/jbc.M304952200. [DOI] [PubMed] [Google Scholar]
- 22.Bugge A, Grontved L, Aagaard MM, Borup R, Mandrup S. The PPARgamma2 A/B-domain plays a gene-specific role in transactivation and cofactor recruitment. Mol Endocrinol. 2009;23:794–808. doi: 10.1210/me.2008-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Onate SA, Boonyaratanakornkit V, Spencer TE, Tsai SY, Tsai MJ, Edwards DP, O’Malley BW. The steroid receptor coactivator-1 contains multiple receptor interacting and activation domains that cooperatively enhance the activation function 1 (AF1) and AF2 domains of steroid receptors. J Biol Chem. 1998;273:12101–12108. doi: 10.1074/jbc.273.20.12101. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi Y, Kitamoto T, Masuhiro Y, Watanabe M, Kase T, Metzger D, Yanagisawa J, Kato S. p300 mediates functional synergism between AF-1 and AF-2 of estrogen receptor alpha and beta by interacting directly with the N-terminal A/B domains. J Biol Chem. 2000;275:15645–15651. doi: 10.1074/jbc.M000042200. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki S, Sasaki S, Morita H, Oki Y, Turiya D, Ito T, Misawa H, Ishizuka K, Nakamura H. The role of the amino-terminal domain in the interaction of unliganded peroxisome proliferator-activated receptor gamma-2 with nuclear receptor co-repressor. J Mol Endocrinol. 2010;45:133–145. doi: 10.1677/JME-10-0007. [DOI] [PubMed] [Google Scholar]
- 26.Walther RF, Atlas E, Carrigan A, Rouleau Y, Edgecombe A, Visentin L, Lamprecht C, Addicks GC, Hache RJ, Lefebvre YA. A serine/threonine-rich motif is one of three nuclear localization signals that determine unidirectional transport of the mineralocorticoid receptor to the nucleus. J Biol Chem. 2005;280:17549–17561. doi: 10.1074/jbc.M501548200. [DOI] [PubMed] [Google Scholar]
- 27.Katagiri Y, Takeda K, Yu ZX, Ferrans VJ, Ozato K, Guroff G. Modulation of retinoid signalling through NGF-induced nuclear export of NGFI-B. Nat Cell Biol. 2000;2:435–440. doi: 10.1038/35017072. [DOI] [PubMed] [Google Scholar]
- 28.Burgermeister E, Chuderland D, Hanoch T, Meyer M, Liscovitch M, Seger R. Interaction with MEK causes nuclear export and downregulation of peroxisome proliferator-activated receptor gamma. Mol Cell Biol. 2007;27:803–817. doi: 10.1128/MCB.00601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujimoto Y, Shiraki T, Horiuchi Y, Waku T, Shigenaga A, Otaka A, Ikura T, Igarashi K, Aimoto S, Tate S, Morikawa K. Proline cis/trans-isomerase Pin1 regulates peroxisome proliferator-activated receptor gamma activity through the direct binding to the activation function-1 domain. J Biol Chem. 2010;285:3126–3132. doi: 10.1074/jbc.M109.055095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rangwala SM, Rhoades B, Shapiro JS, Rich AS, Kim JK, Shulman GI, Kaestner KH, Lazar MA. Genetic modulation of PPARgamma phosphorylation regulates insulin sensitivity. Dev Cell. 2003;5:657–663. doi: 10.1016/s1534-5807(03)00274-0. [DOI] [PubMed] [Google Scholar]
- 31.Picard N, Charbonneau C, Sanchez M, Licznar A, Busson M, Lazennec G, Tremblay A. Phosphorylation of activation function-1 regulates proteasome-dependent nuclear mobility and E6-associated protein ubiquitin ligase recruitment to the estrogen receptor beta. Mol Endocrinol. 2008;22:317–330. doi: 10.1210/me.2007-0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gianni M, Bauer A, Garattini E, Chambon P, Rochette-Egly C. Phosphorylation by p38MAPK and recruitment of SUG-1 are required for RA-induced RAR gamma degradation and transactivation. EMBO J. 2002;21:3760–3769. doi: 10.1093/emboj/cdf374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garza AM, Khan SH, Kumar R. Site-specific phosphorylation induces functionally active conformation in the intrinsically disordered N-terminal activation function (AF1) domain of the glucocorticoid receptor. Mol Cell Biol. 2010;30:220–230. doi: 10.1128/MCB.00552-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar R, Betney R, Li J, Thompson EB, McEwan IJ. Induced alpha-helix structure in AF1 of the androgen receptor upon binding transcription factor TFIIF. Biochemistry. 2004;43:3008–3013. doi: 10.1021/bi035934p. [DOI] [PubMed] [Google Scholar]
- 35.Kato S, Endoh H, Masuhiro Y, Kitamoto T, Uchiyama S, Sasaki H, Masushige S, Gotoh Y, Nishida E, Kawashima H, Metzger D, Chambon P. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science. 1995;270:1491–1494. doi: 10.1126/science.270.5241.1491. [DOI] [PubMed] [Google Scholar]
- 36.Sacchetti P, Carpentier R, Segard P, Olive-Cren C, Lefebvre P. Multiple signaling pathways regulate the transcriptional activity of the orphan nuclear receptor Nurr1. Nucleic Acids Res. 2006;34:5515–5527. doi: 10.1093/nar/gkl712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang T, Jia N, Fei E, Wang P, Liao Z, Ding L, Yan M, Nukina N, Zhou J, Wang G. Nurr1 is phosphorylated by ERK2 in vitro and its phosphorylation upregulates tyrosine hydroxylase expression in SH-SY5Y cells. Neurosci Lett. 2007;423:118–122. doi: 10.1016/j.neulet.2007.06.041. [DOI] [PubMed] [Google Scholar]
- 38.Hentschke M, Susens U, Borgmeyer U. Transcriptional ERRgamma2-mediated activation is regulated by sentrin-specific proteases. Biochem J. 2009;419:167–176. doi: 10.1042/BJ20081556. [DOI] [PubMed] [Google Scholar]
- 39.Poukka H, Karvonen U, Janne OA, Palvimo JJ. Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1) Proc Natl Acad Sci U S A. 2000;97:14145–14150. doi: 10.1073/pnas.97.26.14145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohshima T, Koga H, Shimotohno K. Transcriptional activity of peroxisome proliferator-activated receptor gamma is modulated by SUMO-1 modification. J Biol Chem. 2004;279:29551–29557. doi: 10.1074/jbc.M403866200. [DOI] [PubMed] [Google Scholar]
- 41.Rastinejad F, Perlmann T, Evans RM, Sigler PB. Structural determinants of nuclear receptor assembly on DNA direct repeats. Nature. 1995;375:203–211. doi: 10.1038/375203a0. [DOI] [PubMed] [Google Scholar]
- 42.Hard T, Kellenbach E, Boelens R, Maler BA, Dahlman K, Freedman LP, Carlstedt-Duke J, Yamamoto KR, Gustafsson JA, Kaptein R. Solution structure of the glucocorticoid receptor DNA-binding domain. Science. 1990;249:157–160. doi: 10.1126/science.2115209. [DOI] [PubMed] [Google Scholar]
- 43.Freedman LP, Luisi BF, Korszun ZR, Basavappa R, Sigler PB, Yamamoto KR. The function and structure of the metal coordination sites within the glucocorticoid receptor DNA binding domain. Nature. 1988;334:543–546. doi: 10.1038/334543a0. [DOI] [PubMed] [Google Scholar]
- 44.Nelson CC, Faris JS, Hendy SC, Romaniuk PJ. Functional analysis of the amino acids in the DNA recognition alpha-helix of the human thyroid hormone receptor. Mol Endocrinol. 1993;7:1185–1195. doi: 10.1210/mend.7.9.8247021. [DOI] [PubMed] [Google Scholar]
- 45.Danielsen M, Hinck L, Ringold GM. Two amino acids within the knuckle of the first zinc finger specify DNA response element activation by the glucocorticoid receptor. Cell. 1989;57:1131–1138. doi: 10.1016/0092-8674(89)90050-0. [DOI] [PubMed] [Google Scholar]
- 46.Mader S, Kumar V, de VH, Chambon P. Three amino acids of the oestrogen receptor are essential to its ability to distinguish an oestrogen from a glucocorticoid-responsive element. Nature. 1989;338:271–274. doi: 10.1038/338271a0. [DOI] [PubMed] [Google Scholar]
- 47.Smit-McBride Z, Privalsky ML. DNA sequence specificity of the v-erb A oncoprotein/thyroid hormone receptor: role of the P-box and its interaction with more N-terminal determinants of DNA recognition. Mol Endocrinol. 1994;8:819–828. doi: 10.1210/mend.8.7.7984144. [DOI] [PubMed] [Google Scholar]
- 48.Remerowski ML, Kellenbach E, Boelens R, van der Marel GA, van Boom JH, Maler BA, Yamamoto KR, Kaptein R. 1H NMR studies of DNA recognition by the glucocorticoid receptor: complex of the DNA binding domain with a half-site response element. Biochemistry. 1991;30:11620–11624. doi: 10.1021/bi00114a003. [DOI] [PubMed] [Google Scholar]
- 49.Luisi BF, Xu WX, Otwinowski Z, Freedman LP, Yamamoto KR, Sigler PB. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991;352:497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- 50.Chen Y, Young MA. Structure of a thyroid hormone receptor DNA-binding domain homodimer bound to an inverted palindrome DNA response element. Mol Endocrinol. 2010;24:1650–1664. doi: 10.1210/me.2010-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bain DL, Heneghan AF, Connaghan-Jones KD, Miura MT. Nuclear receptor structure: implications for function. Annu Rev Physiol. 2007;69:201–220. doi: 10.1146/annurev.physiol.69.031905.160308. [DOI] [PubMed] [Google Scholar]
- 52.Baumann H, Paulsen K, Kovacs H, Berglund H, Wright AP, Gustafsson JA, Hard T. Refined solution structure of the glucocorticoid receptor DNA-binding domain. Biochemistry. 1993;32:13463–13471. doi: 10.1021/bi00212a011. [DOI] [PubMed] [Google Scholar]
- 53.Zechel C, Shen XQ, Chambon P, Gronemeyer H. Dimerization interfaces formed between the DNA binding domains determine the cooperative binding of RXR/RAR and RXR/TR heterodimers to DR5 and DR4 elements. EMBO J. 1994;13:1414–1424. doi: 10.1002/j.1460-2075.1994.tb06395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sierk ML, Zhao Q, Rastinejad F. DNA deformability as a recognition feature in the reverb response element. Biochemistry. 2001;40:12833–12843. doi: 10.1021/bi011086r. [DOI] [PubMed] [Google Scholar]
- 55.Elferink CJ, Whitlock JP., Jr 2,3,7,8-Tetrachlorodibenzo-p-dioxin-inducible, Ah receptor-mediated bending of enhancer DNA. J Biol Chem. 1990;265:5718–5721. [PubMed] [Google Scholar]
- 56.Nardulli AM, Shapiro DJ. Binding of the estrogen receptor DNA-binding domain to the estrogen response element induces DNA bending. Mol Cell Biol. 1992;12:2037–2042. doi: 10.1128/mcb.12.5.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu XP, Eberhardt NL, Pfahl M. DNA bending by retinoid X receptor-containing retinoid and thyroid hormone receptor complexes. Mol Cell Biol. 1993;13:6509–6519. doi: 10.1128/mcb.13.10.6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McBroom LD, Flock G, Giguere V. The nonconserved hinge region and distinct amino-terminal domains of the ROR alpha orphan nuclear receptor isoforms are required for proper DNA bending and ROR alpha-DNA interactions. Mol Cell Biol. 1995;15:796–808. doi: 10.1128/mcb.15.2.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Q, Wrange O. Translational positioning of a nucleosomal glucocorticoid response element modulates glucocorticoid receptor affinity. Genes Dev. 1993;7:2471–2482. doi: 10.1101/gad.7.12a.2471. [DOI] [PubMed] [Google Scholar]
- 60.Lefebvre P, Mouchon A, Lefebvre B, Formstecher P. Binding of retinoic acid receptor heterodimers to DNA. A role for histones NH2 termini. J Biol Chem. 1998;273:12288–12295. doi: 10.1074/jbc.273.20.12288. [DOI] [PubMed] [Google Scholar]
- 61.Dilworth FJ, Fromental-Ramain C, Remboutsika E, Benecke A, Chambon P. Ligand-dependent activation of transcription in vitro by retinoic acid receptor alpha/retinoid X receptor alpha heterodimers that mimics transactivation by retinoids in vivo. Proc Natl Acad Sci U S A. 1999;96:1995–2000. doi: 10.1073/pnas.96.5.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dilworth FJ, Fromental-Ramain C, Yamamoto K, Chambon P. ATP-driven chromatin remodeling activity and histone acetyltransferases act sequentially during transactivation by RAR/RXR In vitro. Mol Cell. 2000;6:1049–1058. doi: 10.1016/s1097-2765(00)00103-9. [DOI] [PubMed] [Google Scholar]
- 63.Lefebvre B, Brand C, Lefebvre P, Ozato K. Chromosomal integration of retinoic acid response elements prevents cooperative transcriptional activation by retinoic acid receptor and retinoid X receptor. Mol Cell Biol. 2002;22:1446–1459. doi: 10.1128/mcb.22.5.1446-1459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Garcia-Bassets I, Kwon YS, Telese F, Prefontaine GG, Hutt KR, Cheng CS, Ju BG, Ohgi KA, Wang J, Escoubet-Lozach L, Rose DW, Glass CK, Fu XD, Rosenfeld MG. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128:505–518. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun K, Montana V, Chellappa K, Brelivet Y, Moras D, Maeda Y, Parpura V, Paschal BM, Sladek FM. Phosphorylation of a conserved serine in the deoxyribonucleic acid binding domain of nuclear receptors alters intracellular localization. Mol Endocrinol. 2007;21:1297–1311. doi: 10.1210/me.2006-0300. [DOI] [PubMed] [Google Scholar]
- 66.Viollet B, Kahn A, Raymondjean M. Protein kinase A-dependent phosphorylation modulates DNA-binding activity of hepatocyte nuclear factor 4. Mol Cell Biol. 1997;17:4208–4219. doi: 10.1128/mcb.17.8.4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tzagarakis-Foster C, Privalsky ML. Phosphorylation of thyroid hormone receptors by protein kinase A regulates DNA recognition by specific inhibition of receptor monomer binding. J Biol Chem. 1998;273:10926–10932. doi: 10.1074/jbc.273.18.10926. [DOI] [PubMed] [Google Scholar]
- 68.Chen D, Pace PE, Coombes RC, Ali S. Phosphorylation of human estrogen receptor alpha by protein kinase A regulates dimerization. Mol Cell Biol. 1999;19:1002–1015. doi: 10.1128/mcb.19.2.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ponguta LA, Gregory CW, French FS, Wilson EM. Site-specific androgen receptor serine phosphorylation linked to epidermal growth factor-dependent growth of castration-recurrent prostate cancer. J Biol Chem. 2008;283:20989–21001. doi: 10.1074/jbc.M802392200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Iwamoto F, Umemoto T, Motojima K, Fujiki Y. Nuclear transport of peroxisome-proliferator activated receptor alpha. J Biochem. 2011;149:311–319. doi: 10.1093/jb/mvq144. [DOI] [PubMed] [Google Scholar]
- 71.Khan SA, Park SW, Huq MD, Wei LN. Ligand-independent orphan receptor TR2 activation by phosphorylation at the DNA-binding domain. Proteomics. 2006;6:123–130. doi: 10.1002/pmic.200500068. [DOI] [PubMed] [Google Scholar]
- 72.Gineste R, Sirvent A, Paumelle R, Helleboid S, Aquilina A, Darteil R, Hum DW, Fruchart JC, Staels B. Phosphorylation of farnesoid X receptor by protein kinase C promotes its transcriptional activity. Mol Endocrinol. 2008;22:2433–2447. doi: 10.1210/me.2008-0092. [DOI] [PubMed] [Google Scholar]
- 73.Huq MD, Ha SG, Wei LN. Modulation of retinoic acid receptor alpha activity by lysine methylation in the DNA binding domain. J Proteome Res. 2008;7:4538–4545. doi: 10.1021/pr800375z. [DOI] [PubMed] [Google Scholar]
- 74.Kim HJ, Kim JY, Meng Z, Wang LH, Liu F, Conrads TP, Burke TR, Veenstra TD, Farrar WL. 15-deoxy-Delta12,14-prostaglandin J2 inhibits transcriptional activity of estrogen receptor-alpha via covalent modification of DNA-binding domain. Cancer Res. 2007;67:2595–2602. doi: 10.1158/0008-5472.CAN-06-3043. [DOI] [PubMed] [Google Scholar]
- 75.Burns KA, Li Y, Arao Y, Petrovich RM, Korach KS. Selective mutations in estrogen receptor alpha D-domain alters nuclear translocation and non-estrogen response element gene regulatory mechanisms. J Biol Chem. 2011;286:12640–12649. doi: 10.1074/jbc.M110.187773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tanner TM, Denayer S, Geverts B, Van TN, Kerkhofs S, Helsen C, Spans L, Dubois V, Houtsmuller AB, Claessens F, Haelens A. A 629RKLKK633 motif in the hinge region controls the androgen receptor at multiple levels. Cell Mol Life Sci. 2010;67:1919–1927. doi: 10.1007/s00018-010-0302-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shaffer PL, McDonnell DP, Gewirth DT. Characterization of transcriptional activation and DNA-binding functions in the hinge region of the vitamin D receptor. Biochemistry. 2005;44:2678–2685. doi: 10.1021/bi0477182. [DOI] [PubMed] [Google Scholar]
- 78.Bernard P, Ludbrook L, Queipo G, Dinulos MB, Kletter GB, Zhang YH, Phelan JK, McCabe ER, Harley VR, Vilain E. A familial missense mutation in the hinge region of DAX1 associated with late-onset AHC in a prepubertal female. Mol Genet Metab. 2006;88:272–279. doi: 10.1016/j.ymgme.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 79.Hong W, Baniahmad A, Liu Y, Li H. Bag-1M is a component of the in vivo DNA-glucocorticoid receptor complex at hormone-regulated promoter. J Mol Biol. 2008;384:22–30. doi: 10.1016/j.jmb.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 80.Yoshikawa N, Shimizu N, Sano M, Ohnuma K, Iwata S, Hosono O, Fukuda K, Morimoto C, Tanaka H. Role of the hinge region of glucocorticoid receptor for HEXIM1-mediated transcriptional repression. Biochem Biophys Res Commun. 2008;371:44–49. doi: 10.1016/j.bbrc.2008.03.155. [DOI] [PubMed] [Google Scholar]
- 81.Liu MH, Li J, Shen P, Husna B, Tai ES, Yong EL. A natural polymorphism in peroxisome proliferator-activated receptor-alpha hinge region attenuates transcription due to defective release of nuclear receptor corepressor from chromatin. Mol Endocrinol. 2008;22:1078–1092. doi: 10.1210/me.2007-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Safer JD, Cohen RN, Hollenberg AN, Wondisford FE. Defective release of corepressor by hinge mutants of the thyroid hormone receptor found in patients with resistance to thyroid hormone. J Biol Chem. 1998;273:30175–30182. doi: 10.1074/jbc.273.46.30175. [DOI] [PubMed] [Google Scholar]
- 83.Zwart W, de LR, Rondaij M, Neefjes J, Mancini MA, Michalides R. The hinge region of the human estrogen receptor determines functional synergy between AF-1 and AF-2 in the quantitative response to estradiol and tamoxifen. J Cell Sci. 2010;123:1253–1261. doi: 10.1242/jcs.061135. [DOI] [PubMed] [Google Scholar]
- 84.Zhang Y, Kast-Woelbern HR, Edwards PA. Natural structural variants of the nuclear receptor farnesoid X receptor affect transcriptional activation. J Biol Chem. 2003;278:104–110. doi: 10.1074/jbc.M209505200. [DOI] [PubMed] [Google Scholar]
- 85.Holmbeck SM, Dyson HJ, Wright PE. DNA-induced conformational changes are the basis for cooperative dimerization by the DNA binding domain of the retinoid X receptor. J Mol Biol. 1998;284:533–539. doi: 10.1006/jmbi.1998.2207. [DOI] [PubMed] [Google Scholar]
- 86.van Tilborg PJ, Mulder FA, de Backer MM, Nair M, van Heerde EC, Folkers G, van der Saag PT, Karimi-Nejad Y, Boelens R, Kaptein R. Millisecond to microsecond time scale dynamics of the retinoid X and retinoic acid receptor DNA-binding domains and dimeric complex formation. Biochemistry. 1999;38:1951–1956. doi: 10.1021/bi982526q. [DOI] [PubMed] [Google Scholar]
- 87.Lee MS, Kliewer SA, Provencal J, Wright PE, Evans RM. Structure of the retinoid X receptor alpha DNA binding domain: a helix required for homodimeric DNA binding. Science. 1993;260:1117–1121. doi: 10.1126/science.8388124. [DOI] [PubMed] [Google Scholar]
- 88.Parker KL, Schimmer BP. Transcriptional regulation of the adrenal steroidogenic enzymes. Trends Endocrinol Metab. 1993;4:46–50. doi: 10.1016/s1043-2760(05)80014-1. [DOI] [PubMed] [Google Scholar]
- 89.Hsu MH, Palmer CN, Song W, Griffin KJ, Johnson EF. A carboxyl-terminal extension of the zinc finger domain contributes to the specificity and polarity of peroxisome proliferator-activated receptor DNA binding. J Biol Chem. 1998;273:27988–27997. doi: 10.1074/jbc.273.43.27988. [DOI] [PubMed] [Google Scholar]
- 90.Hsieh JC, Whitfield GK, Oza AK, Dang HT, Price JN, Galligan MA, Jurutka PW, Thompson PD, Haussler CA, Haussler MR. Characterization of unique DNA-binding and transcriptional-activation functions in the carboxyl-terminal extension of the zinc finger region in the human vitamin D receptor. Biochemistry. 1999;38:16347–16358. doi: 10.1021/bi9916574. [DOI] [PubMed] [Google Scholar]
- 91.Roemer SC, Donham DC, Sherman L, Pon VH, Edwards DP, Churchill ME. Structure of the progesterone receptor-deoxyribonucleic acid complex: novel interactions required for binding to half-site response elements. Mol Endocrinol. 2006;20:3042–3052. doi: 10.1210/me.2005-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rastinejad F, Wagner T, Zhao Q, Khorasanizadeh S. Structure of the RXR-RAR DNA-binding complex on the retinoic acid response element DR1. EMBO J. 2000;19:1045–1054. doi: 10.1093/emboj/19.5.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhao Q, Chasse SA, Devarakonda S, Sierk ML, Ahvazi B, Rastinejad F. Structural basis of RXR-DNA interactions. J Mol Biol. 2000;296:509–520. doi: 10.1006/jmbi.1999.3457. [DOI] [PubMed] [Google Scholar]
- 94.Sem DS, Casimiro DR, Kliewer SA, Provencal J, Evans RM, Wright PE. NMR spectroscopic studies of the DNA-binding domain of the monomer-binding nuclear orphan receptor, human estrogen related receptor-2. The carboxyl-terminal extension to the zinc-finger region is unstructured in the free form of the protein. J Biol Chem. 1997;272:18038–18043. doi: 10.1074/jbc.272.29.18038. [DOI] [PubMed] [Google Scholar]
- 95.Wilson TE, Paulsen RE, Padgett KA, Milbrandt J. Participation of non-zinc finger residues in DNA binding by two nuclear orphan receptors. Science. 1992;256:107–110. doi: 10.1126/science.1314418. [DOI] [PubMed] [Google Scholar]
- 96.Gearhart MD, Holmbeck SM, Evans RM, Dyson HJ, Wright PE. Monomeric complex of human orphan estrogen related receptor-2 with DNA: a pseudo-dimer interface mediates extended half-site recognition. J Mol Biol. 2003;327:819–832. doi: 10.1016/s0022-2836(03)00183-9. [DOI] [PubMed] [Google Scholar]
- 97.Zhao Q, Khorasanizadeh S, Miyoshi Y, Lazar MA, Rastinejad F. Structural elements of an orphan nuclear receptor-DNA complex. Mol Cell. 1998;1:849–861. doi: 10.1016/s1097-2765(00)80084-2. [DOI] [PubMed] [Google Scholar]
- 98.Ijpenberg A, Jeannin E, Wahli W, Desvergne B. Polarity and specific sequence requirements of peroxisome proliferator-activated receptor (PPAR)/retinoid X receptor heterodimer binding to DNA. A functional analysis of the malic enzyme gene PPAR response element. J Biol Chem. 1997;272:20108–20117. doi: 10.1074/jbc.272.32.20108. [DOI] [PubMed] [Google Scholar]
- 99.Wang C, Fu M, Angeletti RH, Siconolfi-Baez L, Reutens AT, Albanese C, Lisanti MP, Katzenellenbogen BS, Kato S, Hopp T, Fuqua SA, Lopez GN, Kushner PJ, Pestell RG. Direct acetylation of the estrogen receptor alpha hinge region by p300 regulates transactivation and hormone sensitivity. J Biol Chem. 2001;276:18375–18383. doi: 10.1074/jbc.M100800200. [DOI] [PubMed] [Google Scholar]
- 100.Hwang EJ, Lee JM, Jeong J, Park JH, Yang Y, Lim JS, Kim JH, Baek SH, Kim KI. SUMOylation of RORalpha potentiates transcriptional activation function. Biochem Biophys Res Commun. 2009;378:513–517. doi: 10.1016/j.bbrc.2008.11.072. [DOI] [PubMed] [Google Scholar]
- 101.Sentis S, Le RM, Bianchin C, Rostan MC, Corbo L. Sumoylation of the estrogen receptor alpha hinge region regulates its transcriptional activity. Mol Endocrinol. 2005;19:2671–2684. doi: 10.1210/me.2005-0042. [DOI] [PubMed] [Google Scholar]
- 102.Pourcet B, Pineda-Torra I, Derudas B, Staels B, Glineur C. SUMOylation of human peroxisome proliferator-activated receptor alpha inhibits its trans-activity through the recruitment of the nuclear corepressor NCoR. J Biol Chem. 2010;285:5983–5992. doi: 10.1074/jbc.M109.078311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Delmotte MH, Tahayato A, Formstecher P, Lefebvre P. Serine 157, a retinoic acid receptor alpha residue phosphorylated by protein kinase C in vitro, is involved in RXR.RARalpha heterodimerization and transcriptional activity. J Biol Chem. 1999;274:38225–38231. doi: 10.1074/jbc.274.53.38225. [DOI] [PubMed] [Google Scholar]
- 104.Blanquart C, Mansouri R, Fruchart JC, Paumelle R, Staels B, Glineur C. The Protein Kinase C Signaling Pathway Regulates a Molecular Switch Between Transactivation and Transrepression Activity of the Peroxisome Proliferator-Activated Receptor Alpha. 2004. pp. 663–670. [DOI] [PubMed] [Google Scholar]
- 105.Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D. Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-alpha. Nature. 1995;375:377–382. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- 106.Huang P, Chandra V, Rastinejad F. Structural overview of the nuclear receptor superfamily: insights into physiology and therapeutics. Annu Rev Physiol. 2010;72:247–272. doi: 10.1146/annurev-physiol-021909-135917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Germain P, Kammerer S, Perez E, Peluso-Iltis C, Tortolani D, Zusi FC, Starrett J, Lapointe P, Daris JP, Marinier A, de Lera AR, Rochel N, Gronemeyer H. Rational design of RAR-selective ligands revealed by RARbeta crystal stucture. EMBO Rep. 2004;5:877–882. doi: 10.1038/sj.embor.7400235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.de Lera AR, Bourguet W, Altucci L, Gronemeyer H. Design of selective nuclear receptor modulators: RAR and RXR as a case study. Nat Rev Drug Discov. 2007;6:811–820. doi: 10.1038/nrd2398. [DOI] [PubMed] [Google Scholar]
- 109.Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 110.Perissi V, Staszewski LM, McInerney EM, Kurokawa R, Krones A, Rose DW, Lambert MH, Milburn MV, Glass CK, Rosenfeld MG. Molecular determinants of nuclear receptor-corepressor interaction. Genes Dev. 1999;13:3198–3208. doi: 10.1101/gad.13.24.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bledsoe RK, Montana VG, Stanley TB, Delves CJ, Apolito CJ, McKee DD, Consler TG, Parks DJ, Stewart EL, Willson TM, Lambert MH, Moore JT, Pearce KH, Xu HE. Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell. 2002;110:93–105. doi: 10.1016/s0092-8674(02)00817-6. [DOI] [PubMed] [Google Scholar]
- 112.Wu Y, Chin WW, Wang Y, Burris TP. Ligand and coactivator identity determines the requirement of the charge clamp for coactivation of the peroxisome proliferator-activated receptor gamma. J Biol Chem. 2003;278:8637–8644. doi: 10.1074/jbc.M210910200. [DOI] [PubMed] [Google Scholar]
- 113.Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, Greene GL. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 114.Brzozowski AM, Pike AC, Dauter Z, Hubbard RE, Bonn T, Engstrom O, Ohman L, Greene GL, Gustafsson JA, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 115.Baxter JD, Funder JW, Apriletti JW, Webb P. Towards selectively modulating mineralocorticoid receptor function: lessons from other systems. Mol Cell Endocrinol. 2004;217:151–165. doi: 10.1016/j.mce.2003.10.044. [DOI] [PubMed] [Google Scholar]
- 116.Heldring N, Pawson T, McDonnell D, Treuter E, Gustafsson JA, Pike AC. Structural insights into corepressor recognition by antagonist-bound estrogen receptors. J Biol Chem. 2007;282:10449–10455. doi: 10.1074/jbc.M611424200. [DOI] [PubMed] [Google Scholar]
- 117.Graham JD, Bain DL, Richer JK, Jackson TA, Tung L, Horwitz KB. Nuclear receptor conformation, coregulators, and tamoxifen-resistant breast cancer. Steroids. 2000;65:579–584. doi: 10.1016/s0039-128x(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 118.le MA, Teyssier C, Erb C, Grimaldi M, Alvarez S, de Lera AR, Balaguer P, Gronemeyer H, Royer CA, Germain P, Bourguet W. A unique secondary-structure switch controls constitutive gene repression by retinoic acid receptor. Nat Struct Mol Biol. 2010;17:801–807. doi: 10.1038/nsmb.1855. [DOI] [PubMed] [Google Scholar]
- 119.Phelan CA, Gampe RT, Jr, Lambert MH, Parks DJ, Montana V, Bynum J, Broderick TM, Hu X, Williams SP, Nolte RT, Lazar MA. Structure of Rev-erbalpha bound to N-CoR reveals a unique mechanism of nuclear receptor-co-repressor interaction. Nat Struct Mol Biol. 2010;17:808–814. doi: 10.1038/nsmb.1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fawell SE, Lees JA, White R, Parker MG. Characterization and colocalization of steroid binding and dimerization activities in the mouse estrogen receptor. Cell. 1990;60:953–962. doi: 10.1016/0092-8674(90)90343-d. [DOI] [PubMed] [Google Scholar]
- 121.Depoix C, Delmotte MH, Formstecher P, Lefebvre P. Control of retinoic acid receptor heterodimerization by ligand-induced structural transitions. A novel mechanism of action for retinoid antagonists. J Biol Chem. 2001;276:9452–9459. doi: 10.1074/jbc.m008004200. [DOI] [PubMed] [Google Scholar]
- 122.Quack M, Carlberg C. Ligand-triggered stabilization of vitamin D receptor/retinoid X receptor heterodimer conformations on DR4-type response elements. J Mol Biol. 2000;296:743–756. doi: 10.1006/jmbi.2000.3499. [DOI] [PubMed] [Google Scholar]
- 123.Cheskis B, Freedman LP. Modulation of nuclear receptor interactions by ligands: kinetic analysis using surface plasmon resonance. Biochemistry. 1996;35:3309–3318. doi: 10.1021/bi952283r. [DOI] [PubMed] [Google Scholar]
- 124.Cheskis B, Freedman LP. Ligand modulates the conversion of DNA-bound vitamin D3 receptor (VDR) homodimers into VDR-retinoid X receptor heterodimers. Mol Cell Biol. 1994;14:3329–3338. doi: 10.1128/mcb.14.5.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhang J, Chalmers MJ, Stayrook KR, Burris LL, Wang Y, Busby SA, Pascal BD, Garcia-Ordonez RD, Bruning JB, Istrate MA, Kojetin DJ, Dodge JA, Burris TP, Griffin PR. DNA binding alters coactivator interaction surfaces of the intact VDR-RXR complex. Nat Struct Mol Biol. 2011;18:556–563. doi: 10.1038/nsmb.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Berrabah W, Aumercier P, Lefebvre P, Staels B. Control of nuclear receptor activities in metabolism by post-translational modifications. FEBS Lett. 2011;585:1640–1650. doi: 10.1016/j.febslet.2011.03.066. [DOI] [PubMed] [Google Scholar]
- 127.Pascual G, Fong AL, Ogawa S, Gamliel A, Li AC, Perissi V, Rose DW, Willson TM, Rosenfeld MG, Glass CK. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature. 2005;437:759–763. doi: 10.1038/nature03988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Venteclef N, Jakobsson T, Ehrlund A, Damdimopoulos A, Mikkonen L, Ellis E, Nilsson LM, Parini P, Janne OA, Gustafsson JA, Steffensen KR, Treuter E. GPS2-dependent corepressor/SUMO pathways govern anti-inflammatory actions of LRH-1 LXRbeta in the hepatic acute phase response. Genes Dev. 2010;24:381–395. doi: 10.1101/gad.545110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Leuenberger N, Pradervand S, Wahli W. Sumoylated PPARalpha Mediates Sex-Specific Gene Repression and Protects the Liver From Estrogen-Induced Toxicity in Mice. 2009. pp. 3188–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Frankenberg T, Miloh T, Chen FY, Ananthanarayanan M, Sun AQ, Balasubramaniyan N, Arias I, Setchell KD, Suchy FJ, Sheider BL. The Membrane Protein ATPase Class I Type 8B Member 1 Signals Through Protein Kinase C Zeta to Activate the Farnesoid X Receptor. 2008. pp. 1896–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bruck N, Vitoux D, Ferry C, Duong V, Bauer A, de TH, Rochette-Egly C. A coordinated phosphorylation cascade initiated by p38MAPK/MSK1 directs RARalpha to target promoters. EMBO J. 2009;28:34–47. doi: 10.1038/emboj.2008.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kumar V, Green S, Stack G, Berry M, Jin JR, Chambon P. Functional domains of the human estrogen receptor. Cell. 1987;51:941–951. doi: 10.1016/0092-8674(87)90581-2. [DOI] [PubMed] [Google Scholar]
- 133.Williams SP, Sigler PB. Atomic structure of progesterone complexed with its receptor. Nature. 1998;393:392–396. doi: 10.1038/30775. [DOI] [PubMed] [Google Scholar]
- 134.Farboud B, Privalsky ML. Retinoic acid receptor-alpha is stabilized in a repressive state by its C-terminal, isotype-specific F domain. Mol Endocrinol. 2004;18:2839–2853. doi: 10.1210/me.2004-0236. [DOI] [PubMed] [Google Scholar]
- 135.Schwartz JA, Zhong L, ighton-Collins S, Zhao C, Skafar DF. Mutations targeted to a predicted helix in the extreme carboxyl-terminal region of the human estrogen receptor-alpha alter its response to estradiol and 4-hydroxytamoxifen. J Biol Chem. 2002;277:13202–13209. doi: 10.1074/jbc.M112215200. [DOI] [PubMed] [Google Scholar]
- 136.Kim K, Thu N, Saville B, Safe S. Domains of estrogen receptor alpha (ERalpha) required for ERalpha/Sp1-mediated activation of GC-rich promoters by estrogens and antiestrogens in breast cancer cells. Mol Endocrinol. 2003;17:804–817. doi: 10.1210/me.2002-0406. [DOI] [PubMed] [Google Scholar]
- 137.Sladek FM, Ruse MD, Jr, Nepomuceno L, Huang SM, Stallcup MR. Modulation of transcriptional activation coactivator interaction by a splicing variation in the F domain of nuclear receptor hepatocyte nuclear factor 4alpha1. Mol Cell Biol. 1999;19:6509–6522. doi: 10.1128/mcb.19.10.6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ruse MD, Jr, Privalsky ML, Sladek FM. Competitive cofactor recruitment by orphan receptor hepatocyte nuclear factor 4alpha1: modulation by the F domain. Mol Cell Biol. 2002;22:1626–1638. doi: 10.1128/MCB.22.6.1626-1638.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Warnmark A, Almlof T, Leers J, Gustafsson JA, Treuter E. Differential recruitment of the mammalian mediator subunit TRAP220 by estrogen receptors ERalpha and ERbeta. J Biol Chem. 2001;276:23397–23404. doi: 10.1074/jbc.M011651200. [DOI] [PubMed] [Google Scholar]
- 140.Weatherman RV, Scanlan TS. Unique protein determinants of the subtype-selective ligand responses of the estrogen receptors (ERalpha and ERbeta ) at AP-1 sites. J Biol Chem. 2001;276:3827–3832. doi: 10.1074/jbc.M005414200. [DOI] [PubMed] [Google Scholar]
- 141.Jiang MS, Hart GW. A subpopulation of estrogen receptors are modified by O- linked N-acetylglucosamine. J Biol Chem. 1997;272:2421–2428. doi: 10.1074/jbc.272.4.2421. [DOI] [PubMed] [Google Scholar]
- 142.Meinke G, Sigler PB. DNA-binding mechanism of the monomeric orphan nuclear receptor NGFI-B. Nat Struct Biol. 1999;6:471–477. doi: 10.1038/8276. [DOI] [PubMed] [Google Scholar]
- 143.Pirih FQ, Tang A, Ozkurt IC, Nervina JM, Tetradis S. Nuclear orphan receptor Nurr1 directly transactivates the osteocalcin gene in osteoblasts. J Biol Chem. 2004;279:53167–53174. doi: 10.1074/jbc.M405677200. [DOI] [PubMed] [Google Scholar]
- 144.Wilson TE, Fahrner TJ, Milbrandt J. The orphan receptors NGFI-B and steroidogenic factor 1 establish monomer binding as a third paradigm of nuclear receptor-DNA interaction. Mol Cell Biol. 1993;13:5794–5804. doi: 10.1128/mcb.13.9.5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sacchetti P, Dwornik H, Formstecher P, Rachez C, Lefebvre P. Requirements for heterodimerization between the orphan nuclear receptor Nurr1 and retinoid X receptors. J Biol Chem. 2002;277:35088–35096. doi: 10.1074/jbc.M205816200. [DOI] [PubMed] [Google Scholar]
- 146.Giguere V, McBroom LD, Flock G. Determinants of target gene specificity for ROR alpha 1: monomeric DNA binding by an orphan nuclear receptor. Mol Cell Biol. 1995;15:2517–2526. doi: 10.1128/mcb.15.5.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Harding HP, Lazar MA. The monomer-binding orphan receptor Rev-Erb represses transcription as a dimer on a novel direct repeat. Mol Cell Biol. 1995;15:4791–4802. doi: 10.1128/mcb.15.9.4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Schwabe JW, Chapman L, Finch JT, Rhodes D. The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements. Cell. 1993;75:567–578. doi: 10.1016/0092-8674(93)90390-c. [DOI] [PubMed] [Google Scholar]
- 149.Umesono K, Murakami KK, Thompson CC, Evans RM. Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell. 1991;65:1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Naar AM, Boutin JM, Lipkin SM, Yu VC, Holloway JM, Glass CK, Rosenfeld MG. The orientation and spacing of core DNA-binding motifs dictate selective transcriptional responses to three nuclear receptors. Cell. 1991;65:1267–1279. doi: 10.1016/0092-8674(91)90021-p. [DOI] [PubMed] [Google Scholar]
- 151.Kliewer SA, Umesono K, Noonan DJ, Heyman RA, Evans RM. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature. 1992;358:771–774. doi: 10.1038/358771a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ihunnah CA, Jiang M, Xie W. Nuclear receptor PXR, transcriptional circuits and metabolic relevance. Biochim Biophys Acta. 2011;1812:956–963. doi: 10.1016/j.bbadis.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Orans J, Teotico DG, Redinbo MR. The nuclear xenobiotic receptor pregnane X receptor: recent insights and new challenges. Mol Endocrinol. 2005;19:2891–2900. doi: 10.1210/me.2005-0156. [DOI] [PubMed] [Google Scholar]
- 154.Chong HK, Infante AM, Seo YK, Jeon TI, Zhang Y, Edwards PA, Xie X, Osborne TF. Genome-wide interrogation of hepatic FXR reveals an asymmetric IR-1 motif and synergy with LRH-1. Nucleic Acids Res. 2010;38:6007–6017. doi: 10.1093/nar/gkq397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Renga B, Mencarelli A, Cipriani S, D’Amore C, Zampella A, Monti MC, Distrutti E, Fiorucci S. The nuclear receptor FXR regulates hepatic transport and metabolism of glutamine and glutamate. Biochim Biophys Acta. 2011;1812:1522–1531. doi: 10.1016/j.bbadis.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 156.Anisfeld AM, Kast-Woelbern HR, Meyer ME, Jones SA, Zhang Y, Williams KJ, Willson T, Edwards PA. Syndecan-1 expression is regulated in an isoform-specific manner by the farnesoid-X receptor. J Biol Chem. 2003;278:20420–20428. doi: 10.1074/jbc.M302505200. [DOI] [PubMed] [Google Scholar]
- 157.Kurokawa R, Soderstrom M, Horlein A, Halachmi S, Brown M, Rosenfeld MG, Glass CK. Polarity-specific activities of retinoic acid receptors determined by a co-repressor. Nature. 1995;377:451–454. doi: 10.1038/377451a0. [DOI] [PubMed] [Google Scholar]
- 158.Vivat V, Zechel C, Wurtz JM, Bourguet W, Kagechika H, Umemiya H, Moras D, Gronemeyer H, Chambon P. A mutation mimicking ligand-induced conformational change yields a constitutive RXR that senses allosteric effects in heterodimers. EMBO J. 1997;15:5697–5709. doi: 10.1093/emboj/16.18.5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Westin S, Kurokawa R, Nolte RT, Wisely GB, McInerney EM, Rose DW, Milburn MV, Rosenfeld MG, Glass CK. Interactions controlling the assembly of nuclear-receptor heterodimers and co-activators. Nature. 1998;395:199–202. doi: 10.1038/26040. [DOI] [PubMed] [Google Scholar]
- 160.Kurokawa R, DiRenzo J, Boehm M, Sugarman J, Gloss B, Rosenfeld MG, Heyman RA, Glass CK. Regulation of retinoid signalling by receptor polarity and allosteric control of ligand binding. Nature. 1994;371:528–531. doi: 10.1038/371528a0. [DOI] [PubMed] [Google Scholar]
- 161.Mouchon A, Delmotte MH, Formstecher P, Lefebvre P. Allosteric regulation of the discriminative responsiveness of retinoic acid receptor to natural and synthetic ligands by retinoid X receptor and DNA. Mol Cell Biol. 1999;19:3073–3085. doi: 10.1128/mcb.19.4.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324:407–410. doi: 10.1126/science.1164265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Svensson S, Ostberg T, Jacobsson M, Norstrom C, Stefansson K, Hallen D, Johansson IC, Zachrisson K, Ogg D, Jendeberg L. Crystal structure of the heterodimeric complex of LXRalpha and RXRbeta ligand-binding domains in a fully agonistic conformation. EMBO J. 2003;22:4625–4633. doi: 10.1093/emboj/cdg456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 165.Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, Chew EG, Huang PY, Welboren WJ, Han Y, Ooi HS, Ariyaratne PN, Vega VB, Luo Y, Tan PY, Choy PY, Wansa KD, Zhao B, Lim KS, Leow SC, Yow JS, Joseph R, Li H, Desai KV, Thomsen JS, Lee YK, Karuturi RK, Herve T, Bourque G, Stunnenberg HG, Ruan X, Cacheux-Rataboul V, Sung WK, Liu ET, Wei CL, Cheung E, Ruan Y. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, Denissov S, Borgesen M, Francoijs KJ, Mandrup S, Stunnenberg HG. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 2008;22:2953–2967. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lefterova MI, Zhang Y, Steger DJ, Schupp M, Schug J, Cristancho A, Feng D, Zhuo D, Stoeckert CJ, Jr, Liu XS, Lazar MA. PPARgamma C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008;22:2941–2952. doi: 10.1101/gad.1709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Li G, Thomas AM, Hart SN, Zhong X, Wu D, Guo GL. Farnesoid X receptor activation mediates head-to-tail chromatin looping in the Nr0b2 gene encoding small heterodimer partner. Mol Endocrinol. 2010;24:1404–1412. doi: 10.1210/me.2010-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hsu PY, Hsu HK, Singer GA, Yan PS, Rodriguez BA, Liu JC, Weng YI, Deatherage DE, Chen Z, Pereira JS, Lopez R, Russo J, Wang Q, Lamartiniere CA, Nephew KP, Huang TH. Estrogen-mediated epigenetic repression of large chromosomal regions through DNA looping. Genome Res. 2010;20:733–744. doi: 10.1101/gr.101923.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, Glass CK, Rosenfeld MG, Fu XD. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Eeckhoute J, Carroll JS, Geistlinger TR, Torres-Arzayus MI, Brown M. A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev. 2006;20:2513–2526. doi: 10.1101/gad.1446006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lee CH, Chinpaisal C, Wei LN. Cloning and characterization of mouse RIP140, a corepressor for nuclear orphan receptor TR2. Mol Cell Biol. 1998;18:6745–6755. doi: 10.1128/mcb.18.11.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Fernandes I, Bastien Y, Wai T, Nygard K, Lin R, Cormier O, Lee HS, Eng F, Bertos NR, Pelletier N, Mader S, Han VK, Yang XJ, White JH. Ligand-dependent nuclear receptor corepressor LCoR functions by histone deacetylase-dependent and -independent mechanisms. Mol Cell. 2003;11:139–150. doi: 10.1016/s1097-2765(03)00014-5. [DOI] [PubMed] [Google Scholar]
- 176.Hu X, Li Y, Lazar MA. Determinants of CoRNR-dependent repression complex assembly on nuclear hormone receptors. Mol Cell Biol. 2001;21:1747–1758. doi: 10.1128/MCB.21.5.1747-1758.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hu X, Lazar MA. The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature. 1999;402:93–96. doi: 10.1038/47069. [DOI] [PubMed] [Google Scholar]
- 178.Varlakhanova N, Snyder C, Jose S, Hahm JB, Privalsky ML. Estrogen receptors recruit SMRT and N-CoR corepressors through newly recognized contacts between the corepressor N terminus and the receptor DNA binding domain. Mol Cell Biol. 2010;30:1434–1445. doi: 10.1128/MCB.01002-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Oberoi J, Fairall L, Watson PJ, Yang JC, Czimmerer Z, Kampmann T, Goult BT, Greenwood JA, Gooch JT, Kallenberger BC, Nagy L, Neuhaus D, Schwabe JW. Structural basis for the assembly of the SMRT/NCoR core transcriptional repression machinery. Nat Struct Mol Biol. 2011;18:177–184. doi: 10.1038/nsmb.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116:511–526. doi: 10.1016/s0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- 181.Onate SA, Tsai SY, Tsai MJ, O’Malley BW. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 182.Han SJ, Lonard DM, O’Malley BW. Multi-modulation of nuclear receptor coactivators through posttranslational modifications. Trends Endocrinol Metab. 2009;20:8–15. doi: 10.1016/j.tem.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Lonard DM, Kumar R, O’Malley BW. Minireview: the SRC family of coactivators: an entree to understanding a subset of polygenic diseases? Mol Endocrinol. 2010;24:279–285. doi: 10.1210/me.2009-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 185.Chen D, Huang SM, Stallcup MR. Synergistic, p160 coactivator-dependent enhancement of estrogen receptor function by CARM1 and p300. J Biol Chem. 2000;275:40810–40816. doi: 10.1074/jbc.M005459200. [DOI] [PubMed] [Google Scholar]
- 186.McKenna NJ, O’Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 187.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 188.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 189.York B, O’Malley BW. Steroid receptor coactivator (SRC) family: masters of systems biology. J Biol Chem. 2010;285:38743–38750. doi: 10.1074/jbc.R110.193367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Liu N, Hayes JJ. When push comes to shove: SWI/SNF uses a nucleosome to get rid of a nucleosome. Mol Cell. 2010;38:484–486. doi: 10.1016/j.molcel.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Belandia B, Orford RL, Hurst HC, Parker MG. Targeting of SWI/SNF chromatin remodelling complexes to estrogen-responsive genes. EMBO J. 2002;21:4094–4103. doi: 10.1093/emboj/cdf412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Chiba H, Muramatsu M, Nomoto A, Kato H. Two human homologues of Saccharomyces cerevisiae SWI2/SNF2 and Drosophila brahma are transcriptional coactivators cooperating with the estrogen receptor and the retinoic acid receptor. Nucleic Acids Res. 1994;22:1815–1820. doi: 10.1093/nar/22.10.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Link KA, Burd CJ, Williams E, Marshall T, Rosson G, Henry E, Weissman B, Knudsen KE. BAF57 governs androgen receptor action and androgen-dependent proliferation through SWI/SNF. Mol Cell Biol. 2005;25:2200–2215. doi: 10.1128/MCB.25.6.2200-2215.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Flajollet S, Lefebvre B, Cudejko C, Staels B, Lefebvre P. The core component of the mammalian SWI/SNF complex SMARCD3/BAF60c is a coactivator for the nuclear retinoic acid receptor. Mol Cell Endocrinol. 2007;270:23–32. doi: 10.1016/j.mce.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 196.Miao J, Fang S, Lee J, Comstock C, Knudsen KE, Kemper JK. Functional specificities of Brm and Brg-1 Swi/Snf ATPases in the feedback regulation of hepatic bile acid biosynthesis. Mol Cell Biol. 2009;29:6170–6181. doi: 10.1128/MCB.00825-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Hsiao PW, Fryer CJ, Trotter KW, Wang W, Archer TK. BAF60a mediates critical interactions between nuclear receptors and the BRG1 chromatin-remodeling complex for transactivation. Mol Cell Biol. 2003;23:6210–6220. doi: 10.1128/MCB.23.17.6210-6220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Kemper JK, Kim H, Miao J, Bhalla S, Bae Y. Role of an mSin3A-Swi/Snf chromatin remodeling complex in the feedback repression of bile acid biosynthesis by SHP. Mol Cell Biol. 2004;24:7707–7719. doi: 10.1128/MCB.24.17.7707-7719.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Underhill C, Qutob MS, Yee SP, Torchia J. A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J Biol Chem. 2000;275:40463–40470. doi: 10.1074/jbc.M007864200. [DOI] [PubMed] [Google Scholar]
- 200.Tao W, Chen S, Shi G, Guo J, Xu Y, Liu C. The SWI/SNF complex subunit BAF60a integrates hepatic circadian clock and energy metabolism. Hepatology. 2011 doi: 10.1002/hep.24514. [DOI] [PubMed] [Google Scholar]
- 201.Conaway RC, Conaway JW. Origins and activity of the Mediator complex. Semin Cell Dev Biol. 2011 doi: 10.1016/j.semcdb.2011.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Fondell JD, Guermah M, Malik S, Roeder RG. Thyroid hormone receptor-associated proteins and general positive cofactors mediate thyroid hormone receptor function in the absence of the TATA box-binding protein-associated factors of TFIID. Proc Natl Acad Sci U S A. 1999;96:1959–1964. doi: 10.1073/pnas.96.5.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Gu W, Malik S, Ito M, Yuan CX, Fondell JD, Zhang X, Martinez E, Qin J, Roeder RG. A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol Cell. 1999;3:97–108. doi: 10.1016/s1097-2765(00)80178-1. [DOI] [PubMed] [Google Scholar]
- 204.Ito M, Yuan CX, Malik S, Gu W, Fondell JD, Yamamura S, Fu ZY, Zhang X, Qin J, Roeder RG. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol Cell. 1999;3:361–370. doi: 10.1016/s1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- 205.Rachez C, Lemon BD, Suldan Z, Bromleigh V, Gamble M, Naar AM, Erdjument-Bromage H, Tempst P, Freedman LP. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 206.Naar AM, Beaurang PA, Zhou S, Abraham S, Solomon W, Tjian R. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature. 1999;398:828–832. doi: 10.1038/19789. [DOI] [PubMed] [Google Scholar]
- 207.Taatjes DJ. The human Mediator complex: a versatile, genome-wide regulator of transcription. Trends Biochem Sci. 2010;35:315–322. doi: 10.1016/j.tibs.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Bai L, Jia Y, Viswakarma N, Huang J, Vluggens A, Wolins NE, Jafari N, Rao MS, Borensztajn J, Yang G, Reddy JK. Transcription coactivator mediator subunit MED1 is required for the development of fatty liver in the mouse. Hepatology. 2011;53:1164–1174. doi: 10.1002/hep.24155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Ge K, Cho YW, Guo H, Hong TB, Guermah M, Ito M, Yu H, Kalkum M, Roeder RG. Alternative mechanisms by which mediator subunit MED1/TRAP220 regulates peroxisome proliferator-activated receptor gamma-stimulated adipogenesis and target gene expression. Mol Cell Biol. 2008;28:1081–1091. doi: 10.1128/MCB.00967-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Ge K, Guermah M, Yuan CX, Ito M, Wallberg AE, Spiegelman BM, Roeder RG. Transcription coactivator TRAP220 is required for PPAR gamma 2-stimulated adipogenesis. Nature. 2002;417:563–567. doi: 10.1038/417563a. [DOI] [PubMed] [Google Scholar]
- 211.Chen W, Zhang X, Birsoy K, Roeder RG. A muscle-specific knockout implicates nuclear receptor coactivator MED1 in the regulation of glucose and energy metabolism. Proc Natl Acad Sci U S A. 2010;107:10196–10201. doi: 10.1073/pnas.1005626107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Zhang J, Kalkum M, Chait BT, Roeder RG. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol Cell. 2002;9:611–623. doi: 10.1016/s1097-2765(02)00468-9. [DOI] [PubMed] [Google Scholar]
- 213.Yoon HG, Chan DW, Huang ZQ, Li J, Fondell JD, Qin J, Wong J. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO J. 2003;22:1336–1346. doi: 10.1093/emboj/cdg120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Li J, Wang J, Wang J, Nawaz Z, Liu JM, Qin J, Wong J. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Perissi V, Jepsen K, Glass CK, Rosenfeld MG. Deconstructing repression: evolving models of co-repressor action. Nat Rev Genet. 2010;11:109–123. doi: 10.1038/nrg2736. [DOI] [PubMed] [Google Scholar]
- 216.Sakai DD, Helms S, Carlstedt-Duke J, Gustafsson JA, Rottman FM, Yamamoto KR. Hormone-mediated repression: a negative glucocorticoid response element from the bovine prolactin gene. Genes Dev. 1988;2:1144–1154. doi: 10.1101/gad.2.9.1144. [DOI] [PubMed] [Google Scholar]
- 217.Chatterjee VK, Lee JK, Rentoumis A, Jameson JL. Negative regulation of the thyroid-stimulating hormone alpha gene by thyroid hormone: receptor interaction adjacent to the TATA box. Proc Natl Acad Sci U S A. 1989;86:9114–9118. doi: 10.1073/pnas.86.23.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Diamond MI, Miner JN, Yoshinaga SK, Yamamoto KR. Transcription factor interactions: selectors of positive or negative regulation from a single DNA element. Science. 1990;249:1266–1272. doi: 10.1126/science.2119054. [DOI] [PubMed] [Google Scholar]
- 219.Subramaniam N, Cairns W, Okret S. Glucocorticoids repress transcription from a negative glucocorticoid response element recognized by two homeodomain-containing proteins, Pbx and Oct-1. J Biol Chem. 1998;273:23567–23574. doi: 10.1074/jbc.273.36.23567. [DOI] [PubMed] [Google Scholar]
- 220.Villa A, Santiago J, Belandia B, Pascual A. A response unit in the first exon of the beta-amyloid precursor protein gene containing thyroid hormone receptor and Sp1 binding sites mediates negative regulation by 3,5,3′-triiodothyronine. Mol Endocrinol. 2004;18:863–873. doi: 10.1210/me.2003-0260. [DOI] [PubMed] [Google Scholar]
- 221.Miner JN, Yamamoto KR. The basic region of AP-1 specifies glucocorticoid receptor activity at a composite response element. Genes Dev. 1992;6:2491–2501. doi: 10.1101/gad.6.12b.2491. [DOI] [PubMed] [Google Scholar]
- 222.Surjit M, Ganti KP, Mukherji A, Ye T, Hua G, Metzger D, Li M, Chambon P. Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell. 2011;145:224–241. doi: 10.1016/j.cell.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 223.Berghagen H, Ragnhildstveit E, Krogsrud K, Thuestad G, Apriletti J, Saatcioglu F. Corepressor SMRT functions as a coactivator for thyroid hormone receptor T3Ralpha from a negative hormone response element. J Biol Chem. 2002;277:49517–49522. doi: 10.1074/jbc.M209546200. [DOI] [PubMed] [Google Scholar]
- 224.Tagami T, Park Y, Jameson JL. Mechanisms that mediate negative regulation of the thyroid-stimulating hormone alpha gene by the thyroid hormone receptor. J Biol Chem. 1999;274:22345–22353. doi: 10.1074/jbc.274.32.22345. [DOI] [PubMed] [Google Scholar]
- 225.Takeuchi Y, Murata Y, Sadow P, Hayashi Y, Seo H, Xu J, O’Malley BW, Weiss RE, Refetoff S. Steroid receptor coactivator-1 deficiency causes variable alterations in the modulation of T(3)-regulated transcription of genes in vivo. Endocrinology. 2002;143:1346–1352. doi: 10.1210/endo.143.4.8730. [DOI] [PubMed] [Google Scholar]
- 226.Weiss RE, Xu J, Ning G, Pohlenz J, O’Malley BW, Refetoff S. Mice deficient in the steroid receptor co-activator 1 (SRC-1) are resistant to thyroid hormone. EMBO J. 1999;18:1900–1904. doi: 10.1093/emboj/18.7.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Wang D, Xia X, Weiss RE, Refetoff S, Yen PM. Distinct and histone-specific modifications mediate positive versus negative transcriptional regulation of TSHalpha promoter. PLoS One. 2010;5:e9853. doi: 10.1371/journal.pone.0009853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Metzger E, Imhof A, Patel D, Kahl P, Hoffmeyer K, Friedrichs N, Muller JM, Greschik H, Kirfel J, Ji S, Kunowska N, Beisenherz-Huss C, Gunther T, Buettner R, Schule R. Phosphorylation of histone H3T6 by PKCbeta(I) controls demethylation at histone H3K4. Nature. 2010;464:792–796. doi: 10.1038/nature08839. [DOI] [PubMed] [Google Scholar]
- 229.Han SJ, Lonard DM, O’Malley BW. Multi-modulation of nuclear receptor coactivators through posttranslational modifications. Trends Endocrinol Metab. 2009;20:8–15. doi: 10.1016/j.tem.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.De BK, Vanden BW, Vermeulen L, Plaisance S, Boone E, Haegeman G. Glucocorticoids repress NF-kappaB-driven genes by disturbing the interaction of p65 with the basal transcription machinery, irrespective of coactivator levels in the cell. Proc Natl Acad Sci U S A. 2000;97:3919–3924. doi: 10.1073/pnas.97.8.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Delerive P, Martin-Nizard F, Chinetti G, Trottein F, Fruchart JC, Najib J, Duriez P, Staels B. Peroxisome proliferator-activated receptor activators inhibit thrombin-induced endothelin-1 production in human vascular endothelial cells by inhibiting the activator protein-1 signaling pathway. Circ Res. 1999;85:394–402. doi: 10.1161/01.res.85.5.394. [DOI] [PubMed] [Google Scholar]
- 232.Luecke HF, Yamamoto KR. The glucocorticoid receptor blocks P-TEFb recruitment by NFkappaB to effect promoter-specific transcriptional repression. Genes Dev. 2005;19:1116–1127. doi: 10.1101/gad.1297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Nissen RM, Yamamoto KR. The glucocorticoid receptor inhibits NFkappaB by interfering with serine-2 phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 2000;14:2314–2329. doi: 10.1101/gad.827900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Auphan N, DiDonato JA, Rosette C, Helmberg A, Karin M. Immunosuppression by glucocorticoids: inhibition of NF-kappa B activity through induction of I kappa B synthesis. Science. 1995;270:286–290. doi: 10.1126/science.270.5234.286. [DOI] [PubMed] [Google Scholar]
- 235.Dedieu S, Lefebvre P. Retinoids interfere with the AP1 signalling pathway in human breast cancer cells. Cell Signal. 2006;18:889–898. doi: 10.1016/j.cellsig.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 236.Bruna A, Nicolas M, Munoz A, Kyriakis JM, Caelles C. Glucocorticoid receptor-JNK interaction mediates inhibition of the JNK pathway by glucocorticoids. EMBO J. 2003;22:6035–6044. doi: 10.1093/emboj/cdg590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Gonzalez MV, Jimenez B, Berciano MT, Gonzalez-Sancho JM, Caelles C, Lafarga M, Munoz A. Glucocorticoids antagonize AP-1 by inhibiting the Activation/phosphorylation of JNK without affecting its subcellular distribution. J Cell Biol. 2000;150:1199–1208. doi: 10.1083/jcb.150.5.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Ogawa S, Lozach J, Benner C, Pascual G, Tangirala RK, Westin S, Hoffmann A, Subramaniam S, David M, Rosenfeld MG, Glass CK. Molecular determinants of crosstalk between nuclear receptors and toll-like receptors. Cell. 2005;122:707–721. doi: 10.1016/j.cell.2005.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Benkoussa M, Brand C, Delmotte MH, Formstecher P, Lefebvre P. Retinoic acid receptors inhibit AP1 activation by regulating extracellular signal-regulated kinase and CBP recruitment to an AP1-responsive promoter. Mol Cell Biol. 2002;22:4522–4534. doi: 10.1128/MCB.22.13.4522-4534.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Saijo K, Collier JG, Li AC, Katzenellenbogen JA, Glass CK. An ADIOL-ERbeta-CtBP transrepression pathway negatively regulates microglia-mediated inflammation. Cell. 2011;145:584–595. doi: 10.1016/j.cell.2011.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Glass CK, Saijo K. Nuclear receptor transrepression pathways that regulate inflammation in macrophages and T cells. Nat Rev Immunol. 2010;10:365–376. doi: 10.1038/nri2748. [DOI] [PubMed] [Google Scholar]
- 242.Saijo K, Winner B, Carson CT, Collier JG, Boyer L, Rosenfeld MG, Gage FH, Glass CK. A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell. 2009;137:47–59. doi: 10.1016/j.cell.2009.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Stein B, Yang MX. Repression of the interleukin-6 promoter by estrogen receptor is mediated by NF-kappa B and C/EBP beta. Mol Cell Biol. 1995;15:4971–4979. doi: 10.1128/mcb.15.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Beck IM, Vanden BW, Vermeulen L, Yamamoto KR, Haegeman G, De BK. Crosstalk in inflammation: the interplay of glucocorticoid receptor-based mechanisms and kinases and phosphatases. Endocr Rev. 2009;30:830–882. doi: 10.1210/er.2009-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Ghisletti S, Huang W, Jepsen K, Benner C, Hardiman G, Rosenfeld MG, Glass CK. Cooperative NCoR/SMRT interactions establish a corepressor-based strategy for integration of inflammatory and anti-inflammatory signaling pathways. Genes Dev. 2009;23:681–693. doi: 10.1101/gad.1773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Ghisletti S, Huang W, Ogawa S, Pascual G, Lin ME, Willson TM, Rosenfeld MG, Glass CK. Parallel SUMOylation-dependent pathways mediate gene- and signal-specific transrepression by LXRs and PPARgamma. Mol Cell. 2007;25:57–70. doi: 10.1016/j.molcel.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Huang W, Ghisletti S, Saijo K, Gandhi M, Aouadi M, Tesz GJ, Zhang DX, Yao J, Czech MP, Goode BL, Rosenfeld MG, Glass CK. Coronin 2A mediates actin-dependent de-repression of inflammatory response genes. Nature. 2011;470:414–418. doi: 10.1038/nature09703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Sanchez-Pacheco A, Martinez-Iglesias O, Mendez-Pertuz M, Aranda A. Residues K128, 132, and 134 in the thyroid hormone receptor-alpha are essential for receptor acetylation and activity. Endocrinology. 2009;150:5143–5152. doi: 10.1210/en.2009-0117. [DOI] [PubMed] [Google Scholar]
- 249.Jennewein C, Kuhn AM, Schmidt MV, Meilladec-Jullig V, von KA, Gonzalez FJ, Brune B. Sumoylation of peroxisome proliferator-activated receptor gamma by apoptotic cells prevents lipopolysaccharide-induced NCoR removal from kappaB binding sites mediating transrepression of proinflammatory cytokines. J Immunol. 2008;181:5646–5652. doi: 10.4049/jimmunol.181.8.5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Jeninga EH, van BO, van Dijk AD, Hamers N, Hendriks-Stegeman BI, Bonvin AM, Berger R, Kalkhoven E. Impaired peroxisome proliferator-activated receptor gamma function through mutation of a conserved salt bridge (R425C) in familial partial lipodystrophy. Mol Endocrinol. 2007;21:1049–1065. doi: 10.1210/me.2006-0485. [DOI] [PubMed] [Google Scholar]
- 251.Wu J, Li Y, Dietz J, Lala DS. Repression of p65 transcriptional activation by the glucocorticoid receptor in the absence of receptor-coactivator interactions. Mol Endocrinol. 2004;18:53–62. doi: 10.1210/me.2002-0373. [DOI] [PubMed] [Google Scholar]
- 252.Reichardt HM, Tuckermann JP, Gottlicher M, Vujic M, Weih F, Angel P, Herrlich P, Schutz G. Repression of inflammatory responses in the absence of DNA binding by the glucocorticoid receptor. EMBO J. 2001;20:7168–7173. doi: 10.1093/emboj/20.24.7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Whitacre DC, Karnas KJ, Miesfeld RL. Analysis of glucocorticoid and androgen receptor gene fusions delineates domains required for transcriptional specificity. Endocrine. 2001;15:111–118. doi: 10.1385/ENDO:15:1:111. [DOI] [PubMed] [Google Scholar]
- 254.Jakacka M, Ito M, Weiss J, Chien PY, Gehm BD, Jameson JL. Estrogen receptor binding to DNA is not required for its activity through the nonclassical AP1 pathway. J Biol Chem. 2001;276:13615–13621. doi: 10.1074/jbc.M008384200. [DOI] [PubMed] [Google Scholar]
- 255.Valentine JE, Kalkhoven E, White R, Hoare S, Parker MG. Mutations in the estrogen receptor ligand binding domain discriminate between hormone-dependent transactivation and transrepression. J Biol Chem. 2000;275:25322–25329. doi: 10.1074/jbc.M002497200. [DOI] [PubMed] [Google Scholar]
- 256.De BK, Vanden BW, Vermeulen L, Plaisance S, Boone E, Haegeman G. Glucocorticoids repress NF-kappaB-driven genes by disturbing the interaction of p65 with the basal transcription machinery, irrespective of coactivator levels in the cell. Proc Natl Acad Sci U S A. 2000;97:3919–3924. doi: 10.1073/pnas.97.8.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.DiSepio D, Sutter M, Johnson AT, Chandraratna RA, Nagpal S. Identification of the AP1-antagonism domain of retinoic acid receptors. Mol Cell Biol Res Commun. 1999;1:7–13. doi: 10.1006/mcbr.1999.0101. [DOI] [PubMed] [Google Scholar]
- 258.Reichardt HM, Kaestner KH, Wessely O, Gass P, Schmid W, Schutz G. Analysis of glucocorticoid signalling by gene targeting. J Steroid Biochem Mol Biol. 1998;65:111–115. doi: 10.1016/s0960-0760(97)00181-7. [DOI] [PubMed] [Google Scholar]
- 259.Lefebvre B, Mouchon A, Formstecher P, Lefebvre P. H11-H12 loop retinoic acid receptor mutants exhibit distinct trans-activating and trans-repressing activities in the presence of natural or synthetic retinoids. Biochemistry. 1998;37:9240–9249. doi: 10.1021/bi9804840. [DOI] [PubMed] [Google Scholar]
- 260.Reichardt HM, Kaestner KH, Tuckermann J, Kretz O, Wessely O, Bock R, Gass P, Schmid W, Herrlich P, Angel P, Schutz G. DNA binding of the glucocorticoid receptor is not essential for survival. Cell. 1998;93:531–541. doi: 10.1016/s0092-8674(00)81183-6. [DOI] [PubMed] [Google Scholar]
- 261.Saatcioglu F, Lopez G, West BL, Zandi E, Feng W, Lu H, Esmaili A, Apriletti JW, Kushner PJ, Baxter JD, Karin M. Mutations in the conserved C-terminal sequence in thyroid hormone receptor dissociate hormone-dependent activation from interference with AP-1 activity. Mol Cell Biol. 1997;17:4687–4695. doi: 10.1128/mcb.17.8.4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Rauch A, Seitz S, Baschant U, Schilling AF, Illing A, Stride B, Kirilov M, Mandic V, Takacz A, Schmidt-Ullrich R, Ostermay S, Schinke T, Spanbroek R, Zaiss MM, Angel PE, Lerner UH, David JP, Reichardt HM, Amling M, Schutz G, Tuckermann JP. Glucocorticoids suppress bone formation by attenuating osteoblast differentiation via the monomeric glucocorticoid receptor. Cell Metab. 2010;11:517–531. doi: 10.1016/j.cmet.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 263.Acconcia F, Ascenzi P, Bocedi A, Spisni E, Tomasi V, Trentalance A, Visca P, Marino M. Palmitoylation-dependent estrogen receptor alpha membrane localization: regulation by 17beta-estradiol. Mol Biol Cell. 2005;16:231–237. doi: 10.1091/mbc.E04-07-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Pedram A, Razandi M, Sainson RC, Kim JK, Hughes CC, Levin ER. A conserved mechanism for steroid receptor translocation to the plasma membrane. J Biol Chem. 2007;282:22278–22288. doi: 10.1074/jbc.M611877200. [DOI] [PubMed] [Google Scholar]
- 265.Razandi M, Pedram A, Levin ER. Heat shock protein 27 is required for sex steroid receptor trafficking to and functioning at the plasma membrane. Mol Cell Biol. 2010;30:3249–3261. doi: 10.1128/MCB.01354-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Kim HP, Lee JY, Jeong JK, Bae SW, Lee HK, Jo I. Nongenomic stimulation of nitric oxide release by estrogen is mediated by estrogen receptor alpha localized in caveolae. Biochem Biophys Res Commun. 1999;263:257–262. doi: 10.1006/bbrc.1999.1348. [DOI] [PubMed] [Google Scholar]
- 267.Razandi M, Oh P, Pedram A, Schnitzer J, Levin ER. ERs associate with and regulate the production of caveolin: implications for signaling and cellular actions. Mol Endocrinol. 2002;16:100–115. doi: 10.1210/mend.16.1.0757. [DOI] [PubMed] [Google Scholar]
- 268.Matthews L, Berry A, Ohanian V, Ohanian J, Garside H, Ray D. Caveolin mediates rapid glucocorticoid effects and couples glucocorticoid action to the antiproliferative program. Mol Endocrinol. 2008;22:1320–1330. doi: 10.1210/me.2007-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Pedram A, Razandi M, Aitkenhead M, Hughes CC, Levin ER. Integration of the non-genomic and genomic actions of estrogen. Membrane-initiated signaling by steroid to transcription and cell biology. J Biol Chem. 2002;277:50768–50775. doi: 10.1074/jbc.M210106200. [DOI] [PubMed] [Google Scholar]
- 270.Castoria G, Migliaccio A, Bilancio A, Di DM, de FA, Lombardi M, Fiorentino R, Varricchio L, Barone MV, Auricchio F. PI3-kinase in concert with Src promotes the S-phase entry of oestradiol-stimulated MCF-7 cells. EMBO J. 2001;20:6050–6059. doi: 10.1093/emboj/20.21.6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Migliaccio A, Di DM, Castoria G, de FA, Bontempo P, Nola E, Auricchio F. Tyrosine kinase/p21ras/MAP-kinase pathway activation by estradiol-receptor complex in MCF-7 cells. EMBO J. 1996;15:1292–1300. [PMC free article] [PubMed] [Google Scholar]
- 272.Meyer MR, Haas E, Prossnitz ER, Barton M. Non-genomic regulation of vascular cell function and growth by estrogen. Mol Cell Endocrinol. 2009;308:9–16. doi: 10.1016/j.mce.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Geffroy N, Guedin A, Dacquet C, Lefebvre P. Cell cycle regulation of breast cancer cells through estrogen-induced activities of ERK and Akt protein kinases. Mol Cell Endocrinol. 2005;237:11–23. doi: 10.1016/j.mce.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 274.Ballare C, Uhrig M, Bechtold T, Sancho E, Di DM, Migliaccio A, Auricchio F, Beato M. Two domains of the progesterone receptor interact with the estrogen receptor and are required for progesterone activation of the c-Src/Erk pathway in mammalian cells. Mol Cell Biol. 2003;23:1994–2008. doi: 10.1128/MCB.23.6.1994-2008.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Vicent GP, Ballare C, Nacht AS, Clausell J, Subtil-Rodriguez A, Quiles I, Jordan A, Beato M. Convergence on chromatin of non-genomic and genomic pathways of hormone signaling. J Steroid Biochem Mol Biol. 2008;109:344–349. doi: 10.1016/j.jsbmb.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 276.Lefebvre B, Ozato K, Lefebvre P. Phosphorylation of histone H3 is functionally linked to retinoic acid receptor beta promoter activation. EMBO Rep. 2002;3:335–340. doi: 10.1093/embo-reports/kvf066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Uhrenholt TR, Schjerning J, Rasmussen LE, Hansen PB, Norregaard R, Jensen BL, Skott O. Rapid non-genomic effects of aldosterone on rodent vascular function. Acta Physiol Scand. 2004;181:415–419. doi: 10.1111/j.1365-201X.2004.01313.x. [DOI] [PubMed] [Google Scholar]
- 278.Hafezi-Moghadam A, Simoncini T, Yang Z, Limbourg FP, Plumier JC, Rebsamen MC, Hsieh CM, Chui DS, Thomas KL, Prorock AJ, Laubach VE, Moskowitz MA, French BA, Ley K, Liao JK. Acute cardiovascular protective effects of corticosteroids are mediated by non-transcriptional activation of endothelial nitric oxide synthase. Nat Med. 2002;8:473–479. doi: 10.1038/nm0502-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Croxtall JD, Choudhury Q, Flower RJ. Glucocorticoids act within minutes to inhibit recruitment of signalling factors to activated EGF receptors through a receptor-dependent, transcription-independent mechanism. Br J Pharmacol. 2000;130:289–298. doi: 10.1038/sj.bjp.0703272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Croxtall JD, Choudhury Q, Newman S, Flower RJ. Lipocortin 1 and the control of cPLA2 activity in A549 cells. Glucocorticoids block EGF stimulation of cPLA2 phosphorylation. Biochem Pharmacol. 1996;52:351–356. doi: 10.1016/0006-2952(95)02442-5. [DOI] [PubMed] [Google Scholar]
- 281.Bartholome B, Spies CM, Gaber T, Schuchmann S, Berki T, Kunkel D, Bienert M, Radbruch A, Burmester GR, Lauster R, Scheffold A, Buttgereit F. Membrane glucocorticoid receptors (mGCR) are expressed in normal human peripheral blood mononuclear cells and up-regulated after in vitro stimulation and in patients with rheumatoid arthritis. FASEB J. 2004;18:70–80. doi: 10.1096/fj.03-0328com. [DOI] [PubMed] [Google Scholar]
- 282.Buttgereit F, Straub RH, Wehling M, Burmester GR. Glucocorticoids in the treatment of rheumatic diseases: an update on the mechanisms of action. Arthritis Rheum. 2004;50:3408–3417. doi: 10.1002/art.20583. [DOI] [PubMed] [Google Scholar]
- 283.Song IH, Gold R, Straub RH, Burmester GR, Buttgereit F. New Glucocorticoids on the Horizon: Repress, Don’t Activate! J Rheumatol. 2005;32:1199–1207. [PubMed] [Google Scholar]
- 284.Rochette-Egly C, Germain P. Dynamic and combinatorial control of gene expression by nuclear retinoic acid receptors (RARs) Nucl Recept Signal. 2009;7:e005. doi: 10.1621/nrs.07005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Alsayed Y, Uddin S, Mahmud N, Lekmine F, Kalvakolanu DV, Minucci S, Bokoch G, Platanias LC. Activation of Rac1 and the p38 mitogen-activated protein kinase pathway in response to all-trans-retinoic acid. J Biol Chem. 2001;276:4012–4019. doi: 10.1074/jbc.M007431200. [DOI] [PubMed] [Google Scholar]
- 286.Gianni M, Parrella E, Raska I, Jr, Gaillard E, Nigro EA, Gaudon C, Garattini E, Rochette-Egly C. P38MAPK-dependent phosphorylation and degradation of SRC-3/AIB1 and RARalpha-mediated transcription. EMBO J. 2006;25:739–751. doi: 10.1038/sj.emboj.7600981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Poon MM, Chen L. Retinoic acid-gated sequence-specific translational control by RARalpha. Proc Natl Acad Sci U S A. 2008;105:20303–20308. doi: 10.1073/pnas.0807740105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Aoto J, Nam CI, Poon MM, Ting P, Chen L. Synaptic signaling by all-trans retinoic acid in homeostatic synaptic plasticity. Neuron. 2008;60:308–320. doi: 10.1016/j.neuron.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Chen N, Onisko B, Napoli JL. The nuclear transcription factor RARalpha associates with neuronal RNA granules and suppresses translation. J Biol Chem. 2008;283:20841–20847. doi: 10.1074/jbc.M802314200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Aquila S, Bonofiglio D, Gentile M, Middea E, Gabriele S, Belmonte M, Catalano S, Pellegrino M, Ando S. Peroxisome proliferator-activated receptor (PPAR)gamma is expressed by human spermatozoa: its potential role on the sperm physiology. J Cell Physiol. 2006;209:977–986. doi: 10.1002/jcp.20807. [DOI] [PubMed] [Google Scholar]
- 291.Lombardi A, Cantini G, Piscitelli E, Gelmini S, Francalanci M, Mello T, Ceni E, Varano G, Forti G, Rotondi M, Galli A, Serio M, Luconi M. A new mechanism involving ERK contributes to rosiglitazone inhibition of tumor necrosis factor-alpha and interferon-gamma inflammatory effects in human endothelial cells. Arterioscler Thromb Vasc Biol. 2008;28:718–724. doi: 10.1161/ATVBAHA.107.160713. [DOI] [PubMed] [Google Scholar]
- 292.Cantini G, Lombardi A, Piscitelli E, Poli G, Ceni E, Marchiani S, Ercolino T, Galli A, Serio M, Mannelli M, Luconi M. Rosiglitazone inhibits adrenocortical cancer cell proliferation by interfering with the IGF-IR intracellular signaling. PPAR Res. 2008;2008:904041. doi: 10.1155/2008/904041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Papageorgiou E, Pitulis N, Manoussakis M, Lembessis P, Koutsilieris M. Rosiglitazone attenuates insulin-like growth factor 1 receptor survival signaling in PC-3 cells. Mol Med. 2008;14:403–411. doi: 10.2119/2008-00021.Papageorgiou. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Ichiki T, Tokunou T, Fukuyama K, Iino N, Masuda S, Takeshita A. 15-Deoxy- Delta12,14-prostaglandin J2 and thiazolidinediones transactivate epidermal growth factor and platelet-derived growth factor receptors in vascular smooth muscle cells. Biochem Biophys Res Commun. 2004;323:402–408. doi: 10.1016/j.bbrc.2004.08.101. [DOI] [PubMed] [Google Scholar]
- 295.Fryer LG, Parbu-Patel A, Carling D. The Anti-diabetic drugs rosiglitazone and metformin stimulate AMP-activated protein kinase through distinct signaling pathways. J Biol Chem. 2002;277:25226–25232. doi: 10.1074/jbc.M202489200. [DOI] [PubMed] [Google Scholar]
- 296.Fediuc S, Pimenta AS, Gaidhu MP, Ceddia RB. Activation of AMP-activated protein kinase, inhibition of pyruvate dehydrogenase activity, and redistribution of substrate partitioning mediate the acute insulin-sensitizing effects of troglitazone in skeletal muscle cells. J Cell Physiol. 2008;215:392–400. doi: 10.1002/jcp.21321. [DOI] [PubMed] [Google Scholar]
- 297.Zhang Y, Soto J, Park K, Viswanath G, Kuwada S, Abel ED, Wang L. Nuclear receptor SHP, a death receptor that targets mitochondria, induces apoptosis and inhibits tumor growth. Mol Cell Biol. 2010;30:1341–1356. doi: 10.1128/MCB.01076-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Du J, McEwen B, Manji HK. Glucocorticoid receptors modulate mitochondrial function: A novel mechanism for neuroprotection. Commun Integr Biol. 2009;2:350–352. doi: 10.4161/cib.2.4.8554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Chen L, Hu L, Chan TH, Tsao GS, Xie D, Huo KK, Fu L, Ma S, Zheng BJ, Guan XY. Chromodomain helicase/adenosine triphosphatase DNA binding protein 1-like (CHD1l) gene suppresses the nucleus-to-mitochondria translocation of nur77 to sustain hepatocellular carcinoma cell survival. Hepatology. 2009;50:122–129. doi: 10.1002/hep.22933. [DOI] [PubMed] [Google Scholar]
- 300.Thompson J, Winoto A. During negative selection, Nur77 family proteins translocate to mitochondria where they associate with Bcl-2 and expose its proapoptotic BH3 domain. J Exp Med. 2008;205:1029–1036. doi: 10.1084/jem.20080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Cao X, Liu W, Lin F, Li H, Kolluri SK, Lin B, Han YH, Dawson MI, Zhang XK. Retinoid X receptor regulates Nur77/TR3-dependent apoptosis [corrected] by modulating its nuclear export and mitochondrial targeting. Mol Cell Biol. 2004;24:9705–9725. doi: 10.1128/MCB.24.22.9705-9725.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]


