Abstract
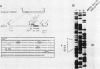
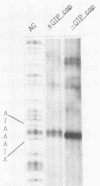
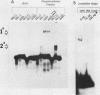
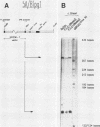
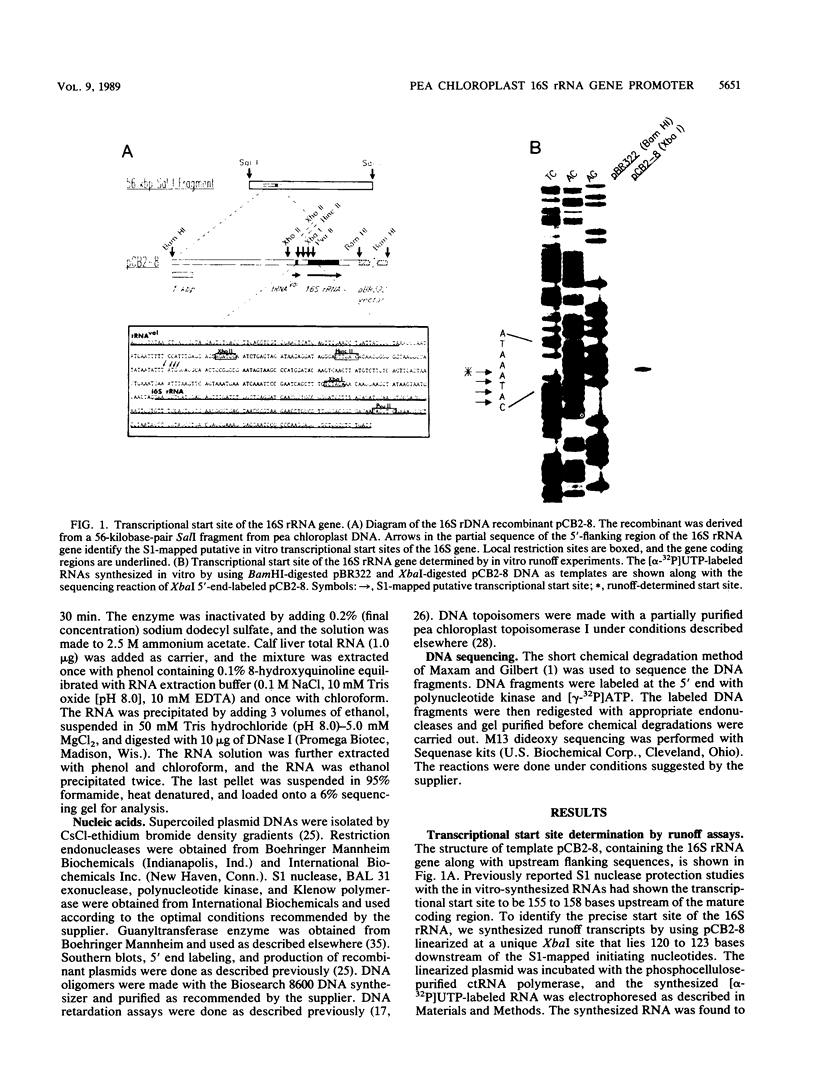
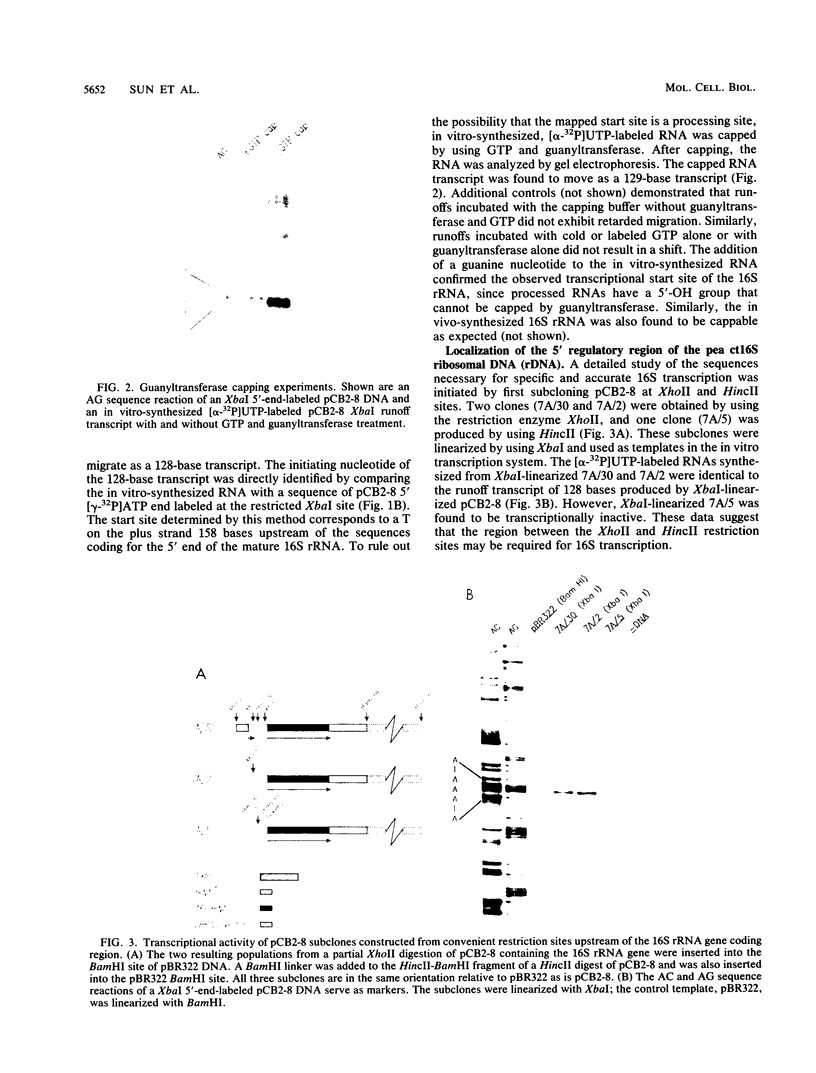
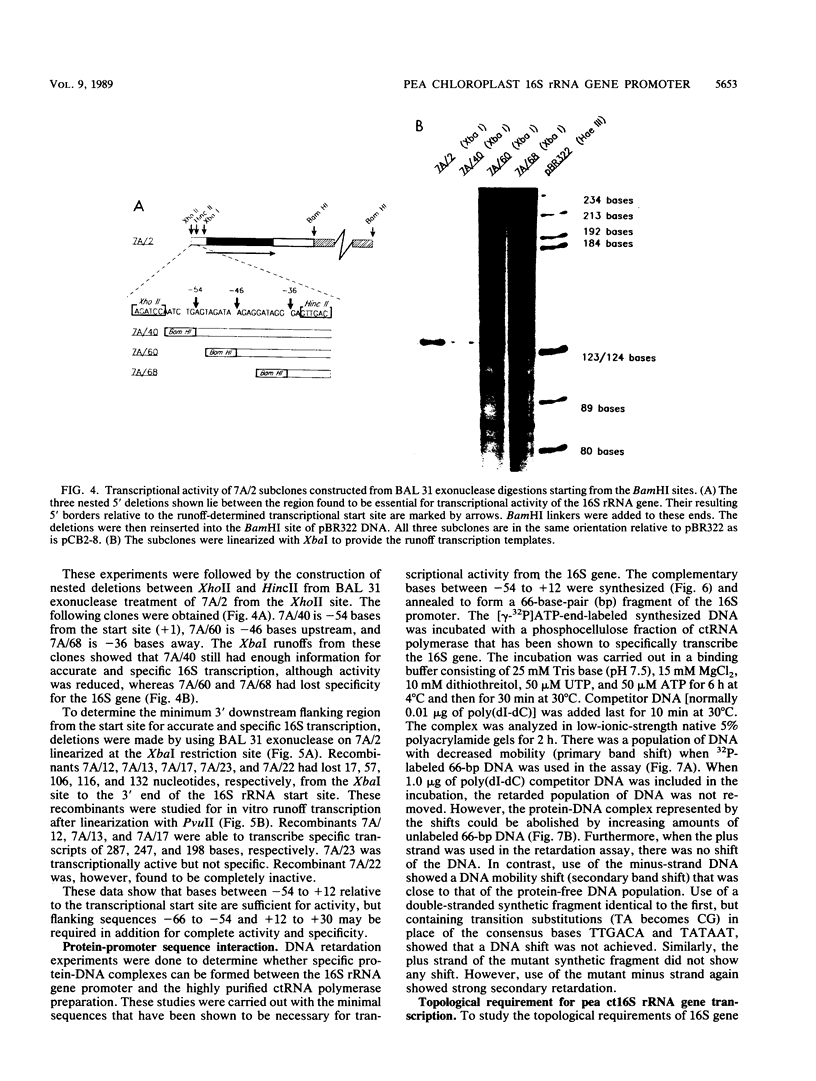
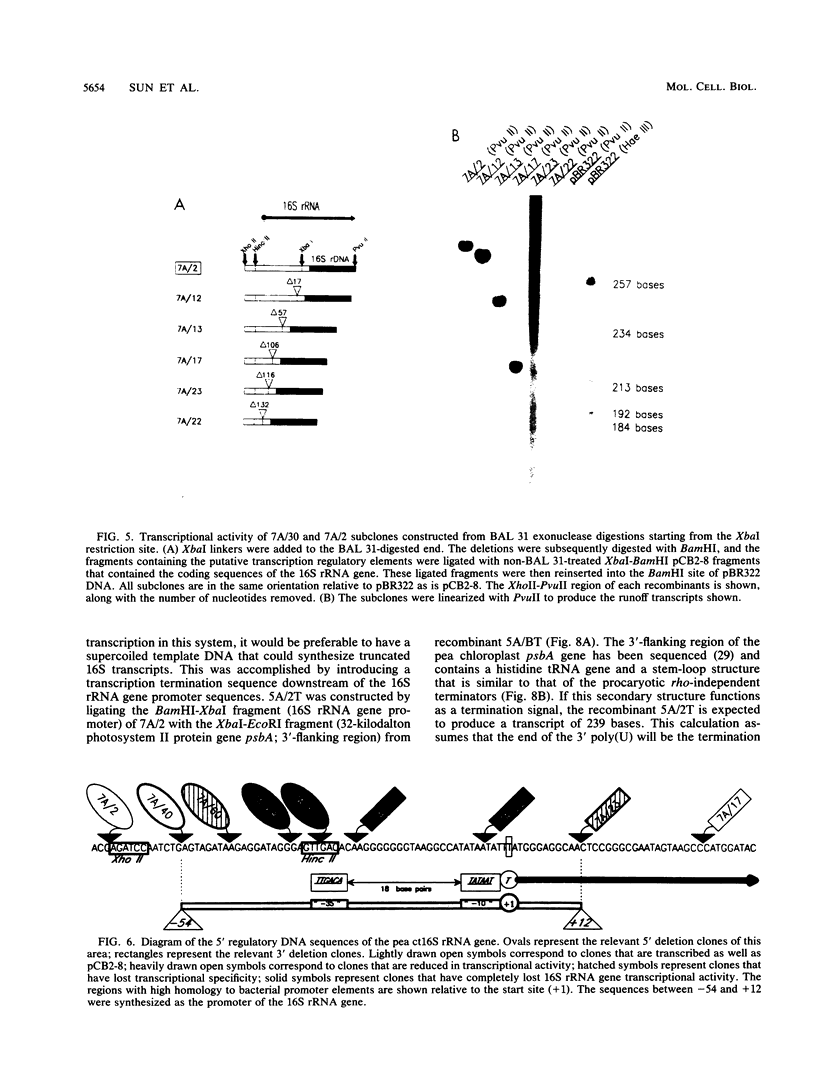
A cloned pea chloroplast 16S rRNA gene promoter has been characterized in detail by use of a homologous in vitro transcription system that contains a highly purified chloroplast RNA polymerase. The in vivo and in vitro 16S rRNA transcriptional start site has been identified to be a T on the plus strand, 158 bases upstream of the mature 5' end of the gene. BAL 31 deletions of the 16S rRNA leader region demonstrated that the bases between -66 to +30 relative to the transcriptional start site (+1) are necessary for specific 16S transcription. Disruption of canonical TTGACA or TATAAT elements within this region caused complete transcriptional inactivation and prevented protein binding. The topological requirement for 16S transcription was examined by using a construct that synthesized a transcript from the 16S promoter and released it from a pea plastid putative terminator sequence. This minigene was relaxed in vitro with a topoisomerase I from pea chloroplast. It was shown that the 16S promoter was most active when the minigene plasmid was supercoiled.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Briat J. F., Lescure A. M., Mache R. Transcription of the chloroplast DNA: a review. Biochimie. 1986 Jul-Aug;68(7-8):981–990. doi: 10.1016/s0300-9084(86)80041-4. [DOI] [PubMed] [Google Scholar]
- CHAMBERLIN M., BERG P. MECHANISM OF RNA POLYMERASE ACTION: CHARACTERIZATION OF THE DNA-DEPENDENT SYNTHESIS OF POLYADENYLIC ACID. J Mol Biol. 1964 May;8:708–726. doi: 10.1016/s0022-2836(64)80120-0. [DOI] [PubMed] [Google Scholar]
- Chen L. J., Orozco E. M., Jr Recognition of prokaryotic transcription terminators by spinach chloroplast RNA polymerase. Nucleic Acids Res. 1988 Sep 12;16(17):8411–8431. doi: 10.1093/nar/16.17.8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossland L. D., Rodermel S. R., Bogorad L. Single gene for the large subunit of ribulosebisphosphate carboxylase in maize yields two differentially regulated mRNAs. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4060–4064. doi: 10.1073/pnas.81.13.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delp G., Igloi G. L., Beck C. F., Kössel H. Functional in vivo verification in E. coli of promoter activities from the rDNA/tDNA(Val)(GAC) leader region of Zea mays chloroplasts. Curr Genet. 1987;12(4):241–246. doi: 10.1007/BF00435284. [DOI] [PubMed] [Google Scholar]
- Greenberg B. M., Narita J. O., DeLuca-Flaherty C., Gruissem W., Rushlow K. A., Hallick R. B. Evidence for two RNA polymerase activities in Euglena gracilis chloroplasts. J Biol Chem. 1984 Dec 10;259(23):14880–14887. [PubMed] [Google Scholar]
- Gruissem W. Chloroplast gene expression: how plants turn their plastids on. Cell. 1989 Jan 27;56(2):161–170. doi: 10.1016/0092-8674(89)90889-1. [DOI] [PubMed] [Google Scholar]
- Gruissem W., Greenberg B. M., Zurawski G., Prescott D. M., Hallick R. B. Biosynthesis of chloroplast transfer RNA in a spinach chloroplast transcription system. Cell. 1983 Dec;35(3 Pt 2):815–828. doi: 10.1016/0092-8674(83)90114-9. [DOI] [PubMed] [Google Scholar]
- Gruissem W., Zurawski G. Analysis of promoter regions for the spinach chloroplast rbcL, atpB and psbA genes. EMBO J. 1985 Dec 16;4(13A):3375–3383. doi: 10.1002/j.1460-2075.1985.tb04093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruissem W., Zurawski G. Identification and mutational analysis of the promoter for a spinach chloroplast transfer RNA gene. EMBO J. 1985 Jul;4(7):1637–1644. doi: 10.1002/j.1460-2075.1985.tb03831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung S. D., Lin C. M. Chloroplast promoters from higher plants. Nucleic Acids Res. 1985 Nov 11;13(21):7543–7549. doi: 10.1093/nar/13.21.7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam E., Hanley-Bowdoin L., Chua N. H. Characterization of a chloroplast sequence-specific DNA binding factor. J Biol Chem. 1988 Jun 15;263(17):8288–8293. [PubMed] [Google Scholar]
- Lescure A. M., Bisanz-Seyer C., Pesey H., Mache R. In vitro transcription initiation of the spinach chloroplast 16S rRNA gene at two tandem promoters. Nucleic Acids Res. 1985 Dec 20;13(24):8787–8796. doi: 10.1093/nar/13.24.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin J. R., Krummel B., Chamberlin M. J. Isolation and properties of transcribing ternary complexes of Escherichia coli RNA polymerase positioned at a single template base. J Mol Biol. 1987 Jul 5;196(1):85–100. doi: 10.1016/0022-2836(87)90512-2. [DOI] [PubMed] [Google Scholar]
- Link G. DNA sequence requirements for the accurate transcription of a protein-coding plastid gene in a plastid in vitro system from mustard (Sinapis alba L.). EMBO J. 1984 Aug;3(8):1697–1704. doi: 10.1002/j.1460-2075.1984.tb02034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M. T. Binding of the herpes simplex virus immediate-early gene product ICP4 to its own transcription start site. J Virol. 1987 Mar;61(3):858–865. doi: 10.1128/jvi.61.3.858-865.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K. K., Shapiro D. R., Tewari K. K. Sequence organization of a pea chloroplast DNA gene coding for a 34,500-dalton protein. Mol Cell Biol. 1984 Nov;4(11):2556–2563. doi: 10.1128/mcb.4.11.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco E. M., Jr, Mullet J. E., Chua N. H. An in vitro system for accurate transcription initiation of chloroplast protein genes. Nucleic Acids Res. 1985 Feb 25;13(4):1283–1302. doi: 10.1093/nar/13.4.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco E. M., Jr, Mullet J. E., Hanley-Bowdoin L., Chua N. H. In vitro transcription of chloroplast protein genes. Methods Enzymol. 1986;118:232–253. doi: 10.1016/0076-6879(86)18076-1. [DOI] [PubMed] [Google Scholar]
- Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- Stern D. B., Gruissem W. Control of plastid gene expression: 3' inverted repeats act as mRNA processing and stabilizing elements, but do not terminate transcription. Cell. 1987 Dec 24;51(6):1145–1157. doi: 10.1016/0092-8674(87)90600-3. [DOI] [PubMed] [Google Scholar]
- Strittmatter G., Gozdzicka-Jozefiak A., Kössel H. Identification of an rRNA operon promoter from Zea mays chloroplasts which excludes the proximal tRNAValGAC from the primary transcript. EMBO J. 1985 Mar;4(3):599–604. doi: 10.1002/j.1460-2075.1985.tb03672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R. J., Mosig G. An ATP-dependent supercoiling topoisomerase of Chlamydomonas reinhardtii affects accumulation of specific chloroplast transcripts. Nucleic Acids Res. 1985 Feb 11;13(3):873–891. doi: 10.1093/nar/13.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zaitlin D., Hu J., Bogorad L. Binding and transcription of relaxed DNA templates by fractions of maize chloroplast extracts. Proc Natl Acad Sci U S A. 1989 Feb;86(3):876–880. doi: 10.1073/pnas.86.3.876. [DOI] [PMC free article] [PubMed] [Google Scholar]