Abstract
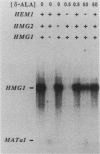
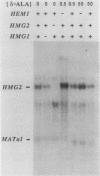
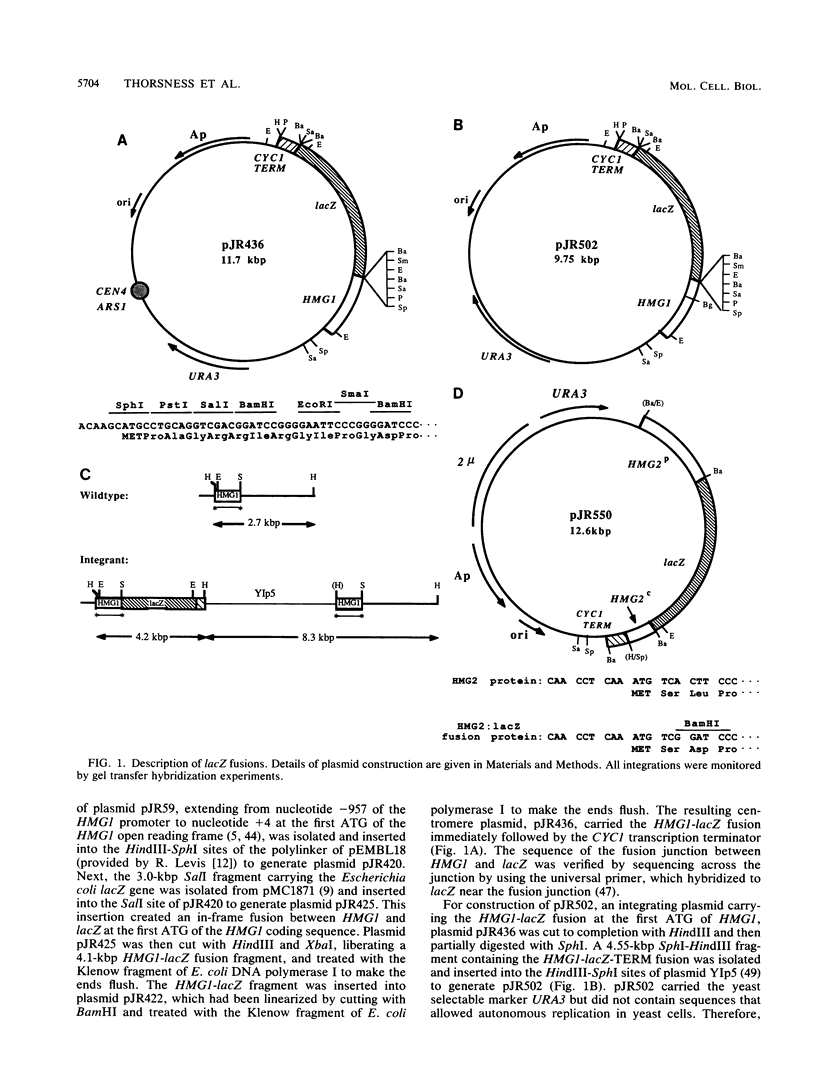
Responses of the yeast genes encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase, HMG1 and HMG2, to in vivo changes in heme concentrations were investigated. Expression of the genes was determined by direct measurement of the mRNA transcribed from each gene, by direct assay of the enzyme activity encoded by each gene, and by measurement of the expression of lacZ fusions to the control regions of each gene. These studies indicated that expression of HMG1 was stimulated by heme, whereas expression of HMG2 was repressed by heme. The effect of heme on HMG1 expression was mediated by the HAP1 transcriptional regulator and was independent of HAP2. Thus, the genes encoding the 3-hydroxy-3-methylglutaryl coenzyme A reductase isozymes join a growing list of gene pairs that are regulated by heme in opposite ways.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alberts A. W., Chen J., Kuron G., Hunt V., Huff J., Hoffman C., Rothrock J., Lopez M., Joshua H., Harris E. Mevinolin: a highly potent competitive inhibitor of hydroxymethylglutaryl-coenzyme A reductase and a cholesterol-lowering agent. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3957–3961. doi: 10.1073/pnas.77.7.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes G., Hansen W. J., Holcomb C. L., Rine J. Asparagine-linked glycosylation in Saccharomyces cerevisiae: genetic analysis of an early step. Mol Cell Biol. 1984 Nov;4(11):2381–2388. doi: 10.1128/mcb.4.11.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basson M. E., Moore R. L., O'Rear J., Rine J. Identifying mutations in duplicated functions in Saccharomyces cerevisiae: recessive mutations in HMG-CoA reductase genes. Genetics. 1987 Dec;117(4):645–655. doi: 10.1093/genetics/117.4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basson M. E., Thorsness M., Finer-Moore J., Stroud R. M., Rine J. Structural and functional conservation between yeast and human 3-hydroxy-3-methylglutaryl coenzyme A reductases, the rate-limiting enzyme of sterol biosynthesis. Mol Cell Biol. 1988 Sep;8(9):3797–3808. doi: 10.1128/mcb.8.9.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basson M. E., Thorsness M., Rine J. Saccharomyces cerevisiae contains two functional genes encoding 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5563–5567. doi: 10.1073/pnas.83.15.5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boll M., Löwel M., Still J., Berndt J. Sterol biosynthesis in yeast. 3-Hydorxy-3-methylglutaryl-Coenzyme A reductase as a regulatory enzyme. Eur J Biochem. 1975 Jun;54(2):435–444. doi: 10.1111/j.1432-1033.1975.tb04154.x. [DOI] [PubMed] [Google Scholar]
- Brown M. S., Goldstein J. L. Multivalent feedback regulation of HMG CoA reductase, a control mechanism coordinating isoprenoid synthesis and cell growth. J Lipid Res. 1980 Jul;21(5):505–517. [PubMed] [Google Scholar]
- Casadaban M. J., Martinez-Arias A., Shapira S. K., Chou J. Beta-galactosidase gene fusions for analyzing gene expression in escherichia coli and yeast. Methods Enzymol. 1983;100:293–308. doi: 10.1016/0076-6879(83)00063-4. [DOI] [PubMed] [Google Scholar]
- Chang T. Y., Limanek J. S. Regulation of cytosolic acetoacetyl coenzyme A thiolase, 3-hydroxy-3-methylglutaryl coenzyme A synthase, 3-hydroxy-3-methylglutaryl coenzyme A reductase, and mevalonate kinase by low density lipoprotein and by 25-hydroxycholesterol in Chinese hamster ovary cells. J Biol Chem. 1980 Aug 25;255(16):7787–7795. [PubMed] [Google Scholar]
- Dente L., Cesareni G., Cortese R. pEMBL: a new family of single stranded plasmids. Nucleic Acids Res. 1983 Mar 25;11(6):1645–1655. doi: 10.1093/nar/11.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollub E. G., Liu K. P., Dayan J., Adlersberg M., Sprinson D. B. Yeast mutants deficient in heme biosynthesis and a heme mutant additionally blocked in cyclization of 2,3-oxidosqualene. J Biol Chem. 1977 May 10;252(9):2846–2854. [PubMed] [Google Scholar]
- Guarente L., Lalonde B., Gifford P., Alani E. Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene of S. cerevisiae. Cell. 1984 Feb;36(2):503–511. doi: 10.1016/0092-8674(84)90243-5. [DOI] [PubMed] [Google Scholar]
- Guarente L., Mason T. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell. 1983 Apr;32(4):1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- Hagen D. C., Sprague G. F., Jr Induction of the yeast alpha-specific STE3 gene by the peptide pheromone a-factor. J Mol Biol. 1984 Oct 5;178(4):835–852. doi: 10.1016/0022-2836(84)90314-0. [DOI] [PubMed] [Google Scholar]
- Hahn S., Guarente L. Yeast HAP2 and HAP3: transcriptional activators in a heteromeric complex. Science. 1988 Apr 15;240(4850):317–321. doi: 10.1126/science.2832951. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hata S., Nishino T., Komori M., Katsuki H. Involvement of cytochrome P-450 in delta 22-desaturation in ergosterol biosynthesis of yeast. Biochem Biophys Res Commun. 1981 Nov 16;103(1):272–277. doi: 10.1016/0006-291x(81)91689-2. [DOI] [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R. Transformation of yeast. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge M. R., Kim G., Singh K., Cumsky M. G. Inverse regulation of the yeast COX5 genes by oxygen and heme. Mol Cell Biol. 1989 May;9(5):1958–1964. doi: 10.1128/mcb.9.5.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm C., Meeks-Wagner D. W., Fangman W. L., Botstein D. A rapid, efficient method for isolating DNA from yeast. Gene. 1986;42(2):169–173. doi: 10.1016/0378-1119(86)90293-3. [DOI] [PubMed] [Google Scholar]
- Ishitani K., Niitsu Y., Listowsky I. Characterization of the different polypeptide components and analysis of subunit assembly in ferritin. J Biol Chem. 1975 Apr 25;250(8):3124–3128. [PubMed] [Google Scholar]
- Keyhani J., Keyhani E. Mevalonic acid as a precursor of the alkyl sidechain of heme a of cytochrome c oxidase in yeast Saccharomyces cerevisiae. FEBS Lett. 1978 Sep 15;93(2):271–274. doi: 10.1016/0014-5793(78)81119-3. [DOI] [PubMed] [Google Scholar]
- Kozak M. How do eucaryotic ribosomes select initiation regions in messenger RNA? Cell. 1978 Dec;15(4):1109–1123. doi: 10.1016/0092-8674(78)90039-9. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Limanek J. S., Chin J., Chang T. Y. Mammalian cell mutant requiring cholesterol and unsaturated fatty acid for growth. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5452–5456. doi: 10.1073/pnas.75.11.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R. T., Parks L. W. Regulation of ergosterol biosynthesis and sterol uptake in a sterol-auxotrophic yeast. J Bacteriol. 1987 Aug;169(8):3707–3711. doi: 10.1128/jb.169.8.3707-3711.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry C. V., Lieber R. H. Negative regulation of the Saccharomyces cerevisiae ANB1 gene by heme, as mediated by the ROX1 gene product. Mol Cell Biol. 1986 Dec;6(12):4145–4148. doi: 10.1128/mcb.6.12.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry C. V., Zitomer R. S. Oxygen regulation of anaerobic and aerobic genes mediated by a common factor in yeast. Proc Natl Acad Sci U S A. 1984 Oct;81(19):6129–6133. doi: 10.1073/pnas.81.19.6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry C. V., Zitomer R. S. ROX1 encodes a heme-induced repression factor regulating ANB1 and CYC7 of Saccharomyces cerevisiae. Mol Cell Biol. 1988 Nov;8(11):4651–4658. doi: 10.1128/mcb.8.11.4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent S. A., Fenimore C. M., Bostian K. A. Vector systems for the expression, analysis and cloning of DNA sequences in S. cerevisiae. Yeast. 1985 Dec;1(2):83–138. doi: 10.1002/yea.320010202. [DOI] [PubMed] [Google Scholar]
- Pfeifer K., Arcangioli B., Guarente L. Yeast HAP1 activator competes with the factor RC2 for binding to the upstream activation site UAS1 of the CYC1 gene. Cell. 1987 Apr 10;49(1):9–18. doi: 10.1016/0092-8674(87)90750-1. [DOI] [PubMed] [Google Scholar]
- Pfeifer K., Prezant T., Guarente L. Yeast HAP1 activator binds to two upstream activation sites of different sequence. Cell. 1987 Apr 10;49(1):19–27. doi: 10.1016/0092-8674(87)90751-3. [DOI] [PubMed] [Google Scholar]
- Pinkham J. L., Olesen J. T., Guarente L. P. Sequence and nuclear localization of the Saccharomyces cerevisiae HAP2 protein, a transcriptional activator. Mol Cell Biol. 1987 Feb;7(2):578–585. doi: 10.1128/mcb.7.2.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prezant T., Pfeifer K., Guarente L. Organization of the regulatory region of the yeast CYC7 gene: multiple factors are involved in regulation. Mol Cell Biol. 1987 Sep;7(9):3252–3259. doi: 10.1128/mcb.7.9.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rine J., Hansen W., Hardeman E., Davis R. W. Targeted selection of recombinant clones through gene dosage effects. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6750–6754. doi: 10.1073/pnas.80.22.6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M., Botstein D. Construction and use of gene fusions to lacZ (beta-galactosidase) that are expressed in yeast. Methods Enzymol. 1983;101:167–180. doi: 10.1016/0076-6879(83)01012-5. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servouse M., Karst F. Regulation of early enzymes of ergosterol biosynthesis in Saccharomyces cerevisiae. Biochem J. 1986 Dec 1;240(2):541–547. doi: 10.1042/bj2400541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueblood C. E., Poyton R. O. Identification of REO1, a gene involved in negative regulation of COX5b and ANB1 in aerobically grown Saccharomyces cerevisiae. Genetics. 1988 Nov;120(3):671–680. doi: 10.1093/genetics/120.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueblood C. E., Wright R. M., Poyton R. O. Differential regulation of the two genes encoding Saccharomyces cerevisiae cytochrome c oxidase subunit V by heme and the HAP2 and REO1 genes. Mol Cell Biol. 1988 Oct;8(10):4537–4540. doi: 10.1128/mcb.8.10.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Arsdell S. W., Stetler G. L., Thorner J. The yeast repeated element sigma contains a hormone-inducible promoter. Mol Cell Biol. 1987 Feb;7(2):749–759. doi: 10.1128/mcb.7.2.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdière J., Creusot F., Guarente L., Slonimski P. P. The overproducing CYP1 and the underproducing hap1 mutations are alleles of the same gene which regulates in trans the expression of the structural genes encoding iso-cytochromes c. Curr Genet. 1986;10(5):339–342. doi: 10.1007/BF00418404. [DOI] [PubMed] [Google Scholar]
- Wang S. S., Brandriss M. C. Proline utilization in Saccharomyces cerevisiae: sequence, regulation, and mitochondrial localization of the PUT1 gene product. Mol Cell Biol. 1987 Dec;7(12):4431–4440. doi: 10.1128/mcb.7.12.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitomer R. S., Sellers J. W., McCarter D. W., Hastings G. A., Wick P., Lowry C. V. Elements involved in oxygen regulation of the Saccharomyces cerevisiae CYC7 gene. Mol Cell Biol. 1987 Jun;7(6):2212–2220. doi: 10.1128/mcb.7.6.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]