Abstract
Background
Microbial transformation of steroids has been extensively used for the synthesis of steroidal drugs, that often yield novel analogues, not easy to obtain by chemical synthesis. We report here fungal transformation of a synthetic steroidal drug, exemestane, used for the treatment of breast cancer and function through inhibition of aromatase enzyme.
Results
Microbial transformation of anti-cancer steroid, exemestane (1), was investigated by using two filamentous fungi. Incubation of 1 with fungi Macrophomina phaseolina, and Fusarium lini afforded three new, 11α-hydroxy-6-methylene-androsta-1, 4-diene-3,17-dione (2), 16β, 17β-dihydroxy-6-methylene-androsta-1, 4-diene-3-one (3), and 17β-hydroxy-6-methylene-androsta-1, 4-diene-3, 16-dione (4), and one known metabolites, 17β-hydroxy-6-methylene-androsta-1, 4-diene-3-one (5). Their structures were deduced spectroscopically. Compared to 1 (steroidal aromatase inactivator), the transformed metabolites were also evaluated for cytotoxic activity by using a cell viability assay against cancer cell lines (HeLa and PC3). Metabolite 2 was found to be moderately active against both the cell lines.
Conclusions
Biotransformation of exemestane (1) provides an efficient method for the synthesis of new analogues of 1. The metabolites were obtained as a result of reduction of double bond and hydroxylation. The transformed product 2 exhibited a moderate activity against cancer cell lines (HeLa and PC3). These transformed products can be studied for their potential as drug candidates.
Keywords: Steroid, Exemestane, Anti-cancer activity, Cancer cell lines (HeLa, PC3), Fusarium lini, Macrophomina phaseolina, Microbial transformation
Background
Microbial transformation of steroids has been extensively employed for the synthesis of steroidal drugs, both at laboratory and industrial levels [1-7]. In modern drug discovery process, generation of libraries of bioactive compounds with diverse structures plays an important role [8].
Exemstane (trade name aromasin) is a steroidal irreversible aromatase inhibitor, used for the treatment of breast cancer. Breast cancers have estrogen receptors (ER-positive) and their growth depends on aromatase activity. Therefore, inhibition of aromatase enzyme reduces the estrogen levels and thus slows the growth of breast cancer [9-12].
Interestingly exemestane not only increases the testosterone level and lowers estrogen, but it also increases the levels of insulin-like growth factor (IGF) [10]. The large reduction in estrogen levels combined with a rise in IGF, makes exemestane an effective breast cancer medication [13-15]. Based on the importance of exemestane in the treatment of breast cancer, a number of exemestane derivatives were previously synthesized involving modification of C-6 methylene and reduction of C-17 keto group, and evaluated for their aromatase inhibitory potential [16-19].
During current study, we synthesized new analogues of 1 by biotransformation techniques. Screening experiments showed that Macrophomina phaseolina and Fusarium lini were able to efficiently transform 1 into several metabolites. Subsequent large scale fermentations produced three new metabolites 2-4 along with a known metabolite 5. The structures of metabolites were unambiguously established through detailed spectral analysis. The microbial transformed metabolites 2 and 4 of exemestane showed a moderate anti-cancer effect against PC3 and/or Hela cancer cell lines. This successful attempt to synthesize new derivatives of an anti-cancer steroid may lead to the discovery of new cancer therapeutic agents.
Results and discussion
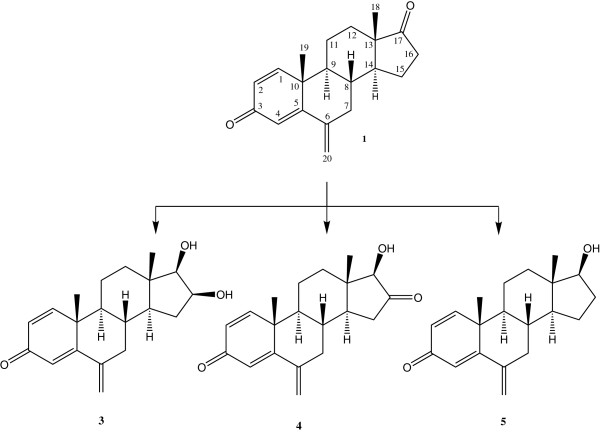
Four microbial metabolites were generated by the selected fungal strains, i.e. Macrophomina phaseolina and Fusarium lini (Figures 1 and 2). M. phaseolina is previously reported to catalyze the introduction of double bond between C-1 and C-2, hydroxyl groups at C-6, C-15, C-16 and C-17, and carbonyl group at C-17 of the steroidal skeleton [1,20]. F. lini is also reported to catalyze the oxidation at C-1, C-2, C-6, and C-11 of steroidal skeleton [21]. The chemical structures of the metabolites 2-4 are reported here for the first time along with their NMR data (Tables 1 and 2).
Figure 1.
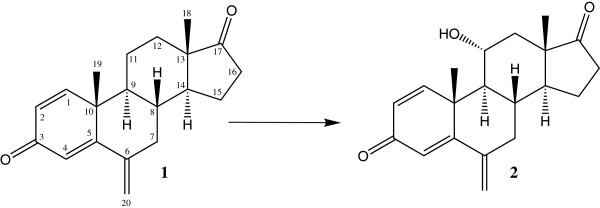
Biotransformation of exemestane (1) with Fusarium lini yielded metabolite 2.
Figure 2.
Biotransformation of exemestane (1) with Macrophomina phaseolina yielded metabolites 3-5.
Table 1.
1H-NMR data of compounds 1–5 in ppm, J in Hz
| COMPOUNDS | |||||
|---|---|---|---|---|---|
|
Carbon |
1a |
2a |
3b |
4b |
5b |
|
1 |
7.21 d (10.0) |
7.21 d (10.5) |
7.31 d (10.2) |
7.34 d (10.5) |
7.31 d (10.2) |
|
2 |
6.14 dd (10.5, 2.0) |
6.12 dd (10.5, 2.0) |
6.21 dd (10.2, 1.8) |
6.42, dd (10.5, 2.0) |
6.22 dd (10.2, 1.8) |
|
3 |
- |
- |
- |
- |
- |
|
4 |
5.99, d (2.0) |
6.00 d (2.0) |
6.08 d (1.8) |
6.11 d (2.0) |
6.09 d (1.8) |
|
5 |
- |
- |
- |
- |
- |
|
6 |
- |
- |
- |
- |
- |
|
7 |
2.69, 1.97 m |
2.13, 1.28 m |
2.56 d (9.0), 1.87 m |
2.57, 1.96 m |
2.66, 1.82 m |
|
8 |
2.03 m |
1.98 m |
1.86 m |
1.98 m |
1.83 m |
|
9 |
1.34 m |
1.62 m |
1.35 m |
1.48 m |
1.05 m |
|
10 |
- |
- |
- |
- |
- |
|
11 |
1.92, 1.76 m |
4.30 m |
1.91, 1.31 m, |
1.93, 1.84 m |
1.75, 1.83 m |
|
12 |
1.78, 1.28 m |
2.13, 1.28 m |
1.90, 1.16 m |
2.01, 1.45 m |
1.13, 1.92 m |
|
13 |
- |
- |
- |
- |
- |
|
14 |
1.43 m |
1.37 m |
0.93 m |
1.63 m |
1.27 m |
|
15 |
1.99, 1.67 m |
1.93, 1.80 m |
2.20, 1.30 m |
2.29, 1.95 m |
1.65, 1.38 m |
|
16 |
2.41, 1.95 m |
2.66, 1.95 m |
4.07 m |
- |
2.00, 1.51 m |
|
17 |
- |
- |
3.30 d (7.5) |
3.77 s |
3.55 t (8.7) |
|
18 |
0.92 s |
0.99 s |
0.99 s |
0.81 s |
0.81 s |
|
19 |
1.19 s |
1.18 s |
1.18 s |
1.21 s |
1.17 s |
| 20 | 5.03, 5.01 s | 5.04, 5.02 s | 5.02 t (1.9) | 5.06, 5.04 s | 4.99, 5.01 s |
a 500 MHz (CD3)2CO.
b 300 MHz CD3OD.
Table 2.
13C-NMR data of compounds 1–5 in ppm
| COMPOUNDS | |||||
|---|---|---|---|---|---|
|
Carbon |
1a |
2b |
3c |
4d |
5d |
|
1 |
155.0 |
154.9 |
158.1 |
157.6 |
158.2 |
|
2 |
128.0 |
128.0 |
127.8 |
127.9 |
127.7 |
|
3 |
185.8 |
185.8 |
188.7 |
188.6 |
188.7 |
|
4 |
122.9 |
122.9 |
122.7 |
122.8 |
122.7 |
|
5 |
167.9 |
167.9 |
171.6 |
171.2 |
171.7 |
|
6 |
147.1 |
147.0 |
147.7 |
147.3 |
147.8 |
|
7 |
39.87 |
32.1 |
41.4 |
41.2 |
41.3 |
|
8 |
36.0 |
39.6 |
36.6 |
35.9 |
37.2 |
|
9 |
50.9 |
48.6 |
51.9 |
51.4 |
51.7 |
|
10 |
44.3 |
44.3 |
45.6 |
45.5 |
45.6 |
|
11 |
22.63 |
71.5 |
23.0 |
23.2 |
23.6 |
|
12 |
32.0 |
32.0 |
38.1 |
36.6 |
37.5 |
|
13 |
48.1 |
48.1 |
43.7 |
43.5 |
44.2 |
|
14 |
51.3 |
50.8 |
48.2 |
45.4 |
51.8 |
|
15 |
22.3 |
22.3 |
35.9 |
36.6 |
24.3 |
|
16 |
35.8 |
39.7 |
70.6 |
217.7 |
30.5 |
|
17 |
218.8 |
218.0 |
81.7 |
86.8 |
82.1 |
|
18 |
13.9 |
14.5 |
12.5 |
11.9 |
11.6 |
|
19 |
20.1 |
20.1 |
20.1 |
20.1 |
20.1 |
| 20 | 112.2 | 112.2 | 112.6 | 112.9 | 112.4 |
a 125 MHz (CD3)2CO.
b 150 MHz (CD3)2CO.
c 125 MHz CD3OD.
d 75 MHz CD3OD.
The anti-cancer effect of exemestane (1) [2] and its synthetic analogues on HeLa and PC3 were determined by using MTT assay. Results obtained from these assays are presented in Table 3.
Table 3.
In vitro cytotoxicity of compounds 1–5
| Compound Codes | HeLa (Cervical cancer) (IC50± S.D.) μM | PC-3 (Prostate cancer) (IC50 ±S.D.) μM |
|---|---|---|
|
1 |
>50 |
>50 |
|
2 |
16.83±0.96 |
24.87±0.72 |
|
3 |
>50 |
>50 |
|
4 |
37.20 ± 0.88 |
>50 |
|
5 |
>50 |
>50 |
| Doxorubicin | 3.10 ± 0.20 | 0.91 ± 0.12 |
The molecular formula C20H24O3 [M+, m/z 312] of metabolite 2 was deduced from the HREI-MS (M+m/z 312.1705), suggested the addition of an oxygen in substrate 1. The 1H-NMR spectral analysis of 2 (Table 1) displayed a downfield methine signal, as compared to the starting material exemestane (1), resonating at δ 4.30 (m, W1/2 = 15.6 Hz), while its respective carbon signal was at δ 71.5 in 13C-NMR spectrum (Table 2). The HMBC spectrum (Figure 3) displayed long-range couplings of the hydroxyl-bearing methine proton (δ 4.30) with C-9 (δ 48.6), C-10 (δ 44.3), and C-13 (δ 48.1), which suggested the position of the hydroxyl-bearing methine at C-11. H-11 also showed COSY cross peaks with H-9 (δ 1.62) and H2-12 (δ 1.28, 2.13). The stereochemical assignments were based on NOESY interactions (Figure 3) between H-11 (δ 4.30), H-8 (δ 1.98), and Me-19 (δ 1.18). H-11 was thus deduced as β-oriented. Metabolite 2 was finally identified as 11α-hydroxy-6-methylene-androsta-1,4-diene-3,17-dione.
Figure 3.
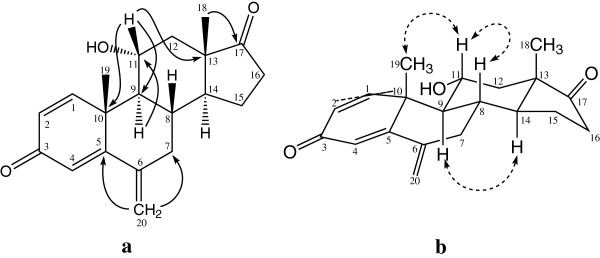
Important HMBC (a) and NOESY (b) correlations in metabolite 2.
Molecular composition of metabolite 3 was deduced to be C20H26O3 from the HREI-MS analysis (M+ = m/z 314.1933, calcd 314.1882). The 1H-NMR spectra μm (Table 1) of metabolite 3 showed two hydroxyl-bearing methine proton peaks at δ 3.30 (d, J17,16 = 7.5 Hz, H-17) and 4.07 (m, W1/2 = 20.0 Hz). The 13C-NMR spectrum of 3 lacks signal for C-17 carbonyl, whereas new methine carbon at δ 81.7 suggested the reduction of C-17 ketone into C-17 OH. The proton geminal to the –OH group (δ 4.07) was correlated with C-13 (δ 43.7), C-14 (δ 48.2) and C-17 (δ 81.7) in the HMBC spectrum. The methine C-17 (δ 81.7) showed HMBC correlations with H-14 (δ 0.93, m) and H-18 (δ 0.99, s). Based on the above observations, the hydroxyl-bearing methine carbon was identified as C-16. The H-16 (δ 4.07) showed NOESY cross peaks with H-14 (δ 0.93), but no interaction with H-18 (δ 0.99) (Figure 4). Therefore the C-16 proton was assigned to be α-oriented. The metabolite 3 was thus identified as 16β, 17β-dihydroxy-6-methylene-androsta-1, 4-diene-3-one.
Figure 4.
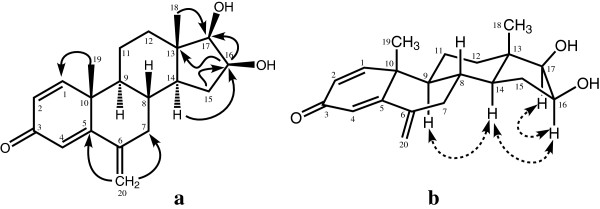
Important HMBC (a) and NOESY (b) correlations in metabolite 3.
Molecular formula C20H24O3 (M+m/z 312.1725, calcd 312.1720) was deduced from the HREI-MS of metabolite 4. A distinct downfield methine proton signal appeared at δ 3.77 (br. s, W1/2 = 9.3 Hz) in the 1H-NMR spectrum of 4. The 13C-NMR spectrum showed a saturated ketone carbon signal at δ 217.7. The rest of the spectrum was distinctly similar to metabolite 2. The deshielded methine proton was HMBC correlated with this ketonic carbon, while its corresponding methine carbon at δ 86.8 showed the HMBC correlations with H2-15 (δ 1.95, 2.29), and CH3-18 (δ 0.81). These interactions, along with appearance of a downfield proton (δ 3.77), indicated that the ketone at C-17 has been reduced into an –OH. Geminal H-17 (δ 3.77) showed NOESY correlations with H-14 (δ 1.63), indicating it to be axially (α-) oriented. The saturated ketone carbon (δ 217.7) was place at C-16, based on the above mentioned HMBC correlations (Figure 5). The structure of metabolite 4 was finally identified as 17β-hydroxy-6-methylene-androsta-1, 4-diene-3, 16-dione.
Figure 5.
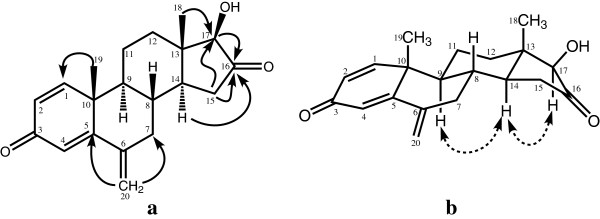
Important HMBC (a) and NOESY (b) correlations in metabolite 4.
Metabolite 5 has a molecular composition C20H26O2 (HREI-MS, M+m/z 298.1730, calcd 298.1733). Based on 1H- and 13C-NMR spectral data (Tables 1 and 2), compound 5 was identified as 17β-hydroxy-6-methylene-androsta-1, 4-diene-3-one. It has previously been reported as an in-vitro cytochrome P450-mediated transformed product of exemestane [22].
The cytotoxic effect of the compounds 1-5 against two tumor cell lines, PC-3 (prostate cancer cell) and Hela (cervical cancer cell), was evaluated (Table 3) using the MTT assay. Compound 2 showed a moderate cytotoxicity against both the cancer cell line with IC50 = 16.83 ± 0.96 and 24.87 ± 0.72 μM, respectively, as compared to the standard drug, doxorubicin. Compound 4 exhibited a moderate activity against HeLa cell line.
Conclusion
In conclusion, the biotransformation of exemestane (1) with F. lini and M. phaseolina were investigated for the first time which provided an efficient route towards the synthesis of several new metabolites 2–5. Metabolite 2 was found to be moderately active against both cancer cell lines (HeLa and PC3). The work presented here can be helpful for the study of in vivo metabolism of exemestane (1), as well as for the discovery of new anticancer drugs
Experimental
Substrate and chemicals
Exemestane (1) was purchased from local market as drug (Pfizer Canada Inc., Brand name Aromasin), extracted and further purified by flash chromatography. Thin layer chromatography (TLC) was carried out on silica gel precoated plates (PF254; Merck). Column chromatography (CC) was performed by using silica gel (E. Merck, Germany). Optical rotations were measured in methanol with a JASCO P-2000 polarimeter. 1H- and 13C-NMR spectra were recorded in (CD3)2CO and CD3OD on Bruker Avance spectrometers. The chemical shifts (δ values) are presented in ppm and the coupling constants (J) are in Hz. For 1D- and 2D-NMR experiments, standard Bruker pulse sequences were used. UV Spectra (in nm) were recorded in methanol with a Hitachi U-3200 spectrophotometer. Infrared (IR) spectra (in cm-1) were recorded with an FT-IR-8900 spectrophotometer. JEOL (Japan) JMS-600H mass spectrometer was used for recording of EI-MS and high-resolution mass spectra (HREI-MS) in m/z (rel. %).
The anticancer activity of compounds 2-5 was evaluated in 96-well flat-bottomed micro-titer plates [Iwaki, Japan] by using the standard dye MTT (3-[4, 5-dimethylthiazole-2-yl]-2, 5-diphenyl-tetrazolium bromide) [Sigma-Aldrich Chemicals, St. Louis, USA] through colorimetric analysis. For this purpose, the cells were cultured in Minimum Essential Medium (MEM), supplemented with 10% Fetal Calf Serum (FCS), 1 mmol/l sodium pyruvate, 1% (v/v) antibiotic / antimycotic and passaged weekly, using 0.25% trypsin / EDTA [Sigma-Aldrich Chemicals, St. Louis, USA] in tissue culture flasks T-75 [Iwaki, Japan]. Absorbance was taken at 540 nm wavelength by using microplate reader (Spectra Max plus, Molecular Devices, USA) using software SoftMax Pro 340 [Molecular Devices, CA, USA]. Dimethyl sulfoxide (DMSO) and doxorubicin (standard inhibitor) were purchased from Sigma-Aldrich Chemicals, [St. Louis, USA].
Microorganisms and culture medium
The fungi were purchased from the Northern Regional Research Laboratories (NRRL), or obtained as gift from the Karachi University Culture Collection (KUCC).
Fusarium lini (NRRL 2204), and Macrophomina phaseolina (KUCC 730) were grown in a culture medium prepared by mixing glucose (40.0 g), glycerol (40.0 mL), peptone (20.0 g), yeast extract (20.0 g), KH2PO4 (20.0 g), and NaCl (20.0 g) in distilled H2O (4.0 L).
Cell lines
PC3 (prostate cancer) and HeLa (cervical cancer) cell lines were purchased from the American Type Culture Collection (ATCC) for anticancer activity.
General Fermentation and Extraction Conditions
4 Liters fungal media was prepared and distributed into 40 conical flasks (100 mL in each flask). All flasks were then autoclaved at 121°C. The fungal cultures were then inoculated into each flask containing media and incubated at room temperature on shaker for three days. Compound 1 was dissolved in 40 mL methanol and distributed equally to all 40 flasks. All experimental flasks were then kept for fermentation. Two control experiments, i.e. media + compound 1 and media + fungus were also conducted. The transformation was then checked on TLC. After the detection of transformation on TLC, fungal culture from all 40 flasks was filtered and extracted with CH2Cl2 (12 L) by using liquid-liquid chromatography. The dichloromethane layer was evaporated in vaccue. The obtained gum was analyzed by thin-layer chromatography.
Fermentation and Purification of Exemestane (1) with Fusarium lini
Exemestane (1; 1.0 g) was dissolved in 40 mL methanol, and incubated with culture of F. Lini. The obtained gum (2.3 g) was fractionated (ARC 1-3) by using silica gel column chromatography. The mobile phase was composed of petroleum ether and acetone with a gradient of 10%. Fraction ARC-2 yielded metabolite 2 (4 mg, pet. ether: acetone = 8:2) after elution through silica gel column.
11α-Hydroxy-6-methylene-androsta-1,4-diene-3,17-dione (2)
Amorphous material; [α]25D: +81.4 (c = 0.096, MeOH); IR (KBr): νmax 3408, 1657 cm-1; UV (MeOH): λmax nm (log ε) 247 (3.78); 1H- and 13C-NMR: see Tables 1 and 2 (Additional file 1).
Fermentation and Purification of Exemestane (1) with Macrophomina phaseolina
Incubation of 1 (1.0 g / 40 mL methanol) with 3 days old culture of M. phaseolina in 40 flasks for 12 days produced the metabolites 3 (10 mg), 4 (5 mg) and 5 (6 mg).
16β, 17β-Dihydroxy-6-methylene-androsta-1,4-diene-3-one (3)
Amorphous material; [α]25D: +181.6 (c = 0.032, MeOH); IR (KBr): νmax 3388, 1658 cm-1; UV (MeOH): λmax nm (log ε) 249 (4.03); 1H- and 13C-NMR: see Tables 1, and 2 (Additional file 2).
17β-Hydroxy-6-methylene-androsta-1,4-diene-3,16-dione (4)
Amorphous material; [α]25D: -56.0 (c = 0.043, MeOH); IR (KBr): νmax 3411, 1749, 1658 cm-1; UV (MeOH): λmax nm (log ε) 247 (4.04); 1H- and 13C-NMR: see Tables 1 and 2 (Additional file 3).
17β-Hydroxy-6-methylene-androsta-1,4-diene-3-one (5)
Amorphous material; [α]25D: +174.5 (c = 0.046, MeOH); IR (KBr): νmax 3421, 1657, cm-1; UV (MeOH): λmax nm (log ε) 248 (4.24); 1H- and 13C-NMR: see Tables 1 and 2 (Additional file 4).
Cell Viability Assay
The cytotoxicity of metabolites 1-5 were determined by using MTT-based colorimetric assay in 96-well plate [23]. Both cell lines (PC-3 and HeLa) were cultured in DMEM and MEM media, respectively, in 25 cm3 tissue culture flasks. The media were supplemented with FBS (5%), pencillin (100 IU/mL) and streptomycin (100 mg/mL). The flasks were then incubated at 37°C in an incubator containing 5% CO2. The flask (80% confluence) was processed for MTT-based cytotoxicity assay. The percent viability of the cells was monitored by trypan blue dye. The cells with clear cytoplasm were considered viable. For the assay, the cells (1 × 105) were loaded onto 96-well tissue culture treated plate. The plate was incubated for 24 hours at 37°C. After incubation, the cells were treated with different concentrations (1.56-50 μM dissolved in DMSO) of compounds 1-5 and kept in an incubator for 48 hours at 37°C. At the end of the incubation, the MTT dye (50 μL, 2 mg/mL) was added to each well and the plate was incubated for 4 hours at 37°C in an incubator. Following incubation, the insoluble formazan crystals were dissolved by adding DMSO (100 μL).
The following formula was used to analyze the cytotoxic effects of the compounds.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
EB, MIC, AR, DF, and CM participated in experimental strategy design, supervision and manuscript writing. MB and MAI carried out the experiments. AW performed NMR experiments, while SAS carried out the biological screenings. All authors read and approved the final manuscript.
Supplementary Material
Spectroscopic data of metabolite 2. Include spectra of EI-MS, UV, IR, 1H-NMR, 13C-NMR (BB, DEPT-135°, DEPT-90°), HMQC, HMBC, COSY-45°, NOESY experiments.
Spectroscopic data of metabolite 3. Include spectra of EI-MS, UV, IR, 1H-NMR, 13C-NMR (BB, DEPT-135°, DEPT-90°), HSQC, HMBC, COSY-45°, NOESY experiments.
Spectroscopic data of metabolite 4. Include spectra of EI-MS, UV, IR, 1H-NMR, 13C-NMR (BB, DEPT-135°, DEPT-90°), HMQC, HMBC, COSY-45°, NOESY, experiments.
Spectroscopic data of metabolite 5. Include spectra of EI-MS, UV, IR, 1H-NMR, 13C-NMR (BB, DEPT-135°, DEPT-90°), HSQC, HMBC, COSY-45°, NOESY experiments.
Contributor Information
Elias Baydoun, Email: eliasbay1@yahoo.com.
Marium Bibi, Email: mariumhej@yahoo.com.
Muhammad Asif Iqbal, Email: asif_90pk@yahoo.com.
Atia-tul Wahab, Email: tulwahab@yahoo.com.
Dina Farran, Email: dkf01@aub.edu.lb.
Colon Smith, Email: cs10@aub.edu.lb.
Samina A Sattar, Email: sam_asattar@yahoo.com.
Atta-ur Rahman, Email: aurahman786@gmail.com.
M Iqbal Choudhary, Email: iqbal.choudhary@iccs.edu.
Acknowledgements
We would like to acknowledge the Lebanese CNRS (Lebanese Council for Scientific Research) for a research grant.
References
- Choudhary MI, Zafar S, Khan NT, Ahmad S, Noreen S, Marasini BP, Al-Khedhairy AA. Atta-ur-Rahman: Biotransformation of dehydroepiandrosterone with Macrophomina phaseolina and β-glucuronidase inhibitory activity of transformed products. J Enzyme Inhib Med Chem. 2012;27:348–355. doi: 10.3109/14756366.2011.590804. [DOI] [PubMed] [Google Scholar]
- Choudhary MI, Erum S, Atif M, Malik R, Khan NT, Atta-ur-Rahman. Biotransformation of (20S)-20-hydroxymethylpregna-1,4-dien-3-one by four filamentous fungi. Steroids. 2011;76:1288–1296. doi: 10.1016/j.steroids.2011.06.007. [DOI] [PubMed] [Google Scholar]
- Choudhary MI, Shah SAA, Atta-ur-Rahman, Khan SN, Khan MTH. α-Glucosidase and tyrosinase inhibitors from fungal hydroxylation of tibolone and hydroxytibolones. Steroids. 2010;75:956–966. doi: 10.1016/j.steroids.2010.05.017. [DOI] [PubMed] [Google Scholar]
- Al-Aboudi A, Mohammad MY, Haddad S, Al-Far R, Choudhary MI, Atta-ur-Rahman. Biotransformation of methyl cholate by Aspergillus niger. Steroids. 2009;74:483–486. doi: 10.1016/j.steroids.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Choudhary MI, Mohammad MY, Musharraf SG, Parvez M, Al-Aboudi A, Atta-ur-Rahman. New oxandrolone derivatives by biotransformation using Rhizopus stolonifer. Steroids. 2009;74:1040–1044. doi: 10.1016/j.steroids.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Choudhary MI, Khan NT, Musharraf SG, Anjum S, Atta-ur-Rahman. Biotransformation of adrenosterone by filamentous fungus, Cunninghamella elegans. Steroids. 2007;72:923–929. doi: 10.1016/j.steroids.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Devkota KP, Choudhary MI, Nawaz SA, Lannang AM, Lenta BN, Fokou PA, Sewald N. Microbial transformation of the steroidal alkaloid dictyophlebine by Rhizopus stolonifer. Chem Pharm Bull. 2007;55:682–684. doi: 10.1248/cpb.55.682. [DOI] [PubMed] [Google Scholar]
- Tong WY, Dong X. Microbial biotransformation: Recent development on steroid drugs. Recent Pat Biotechnol. 2009;3(2):141–153. doi: 10.2174/187220809788700157. [DOI] [PubMed] [Google Scholar]
- Henry NL, Azzouz F, Desta Z, Li L, Nguyen AT, Lemler S, Hayden J, Tarpinian K, Yakim E, Flockhart DA, Stearns V, Hayes DF, Storniolo AM. Predictors of aromatase inhibitor discontinuation as a result of treatment-Emergent symptoms in early-stage breast cancer. J Clin Oncol. 2012;30(9):936–942. doi: 10.1200/JCO.2011.38.0261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrozek E, Layman R, Ramaswamy B, Schaaf L, Li X, Ottman S, Shapiro CL. Phase II trial of exemestane in combination with fulvestrant in postmenopausal women with advanced, Hormone-Responsive Breast Cancer. Clin Breast Cancer. 2012;12(2):151–156. doi: 10.1016/j.clbc.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debled M, Le Loarer F, Callonnec F, Soubeyran I, Cambon-Michot C, Dujardin F, Italiano A. Complete response to exemestane in a patient with a desmoid tumor. Futur Oncol. 2012;8(4):483–486. doi: 10.2217/fon.12.24. [DOI] [PubMed] [Google Scholar]
- Hille U, Soergel P, Laenger F, Schippert C, Makowski L, Hillemanns P. Aromatase inhibitors as solely treatment in postmenopausal breast cancer patients. The Breast Journal. 2012;18(2):145–150. doi: 10.1111/j.1524-4741.2011.01203.x. [DOI] [PubMed] [Google Scholar]
- Long B, Groothuis PG, Hicklin D. IGF1R Inhibitor based treatment of prostrate cancer. International application number: PCT/US2010/055608 Publication number: WO/2011/05706, Filing date: Nov 05, 2010.
- Van-Gool SA, Wit JM, De-Schutter T, De-Clerck N, Postnov AA, Hovinga SK, Van-Doorn J, Veiga SJ, Garcia-Segura LM, Karperien M. Marginal growth increase, altered bone quality and polycystic ovaries in female prepubertal rats after treatment with the aromatase inhibitor exemestane. Horm Res Paediatr. 2010;73(1):49–60. doi: 10.1159/000271916. [DOI] [PubMed] [Google Scholar]
- Yamashita H, Takahashi S, Ito Y, Yamashita T, Ando Y, Toyama T, Sugiura H, Yoshimoto N, Kobayashi S, Fujii Y, Hirotaka I. Predictors of response to exemestane as primary endocrine therapy in estrogen receptor-positive breast cancer. Cancer Sci. 2009;100(11):2028–2033. doi: 10.1111/j.1349-7006.2009.01274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pariza RJ, Yarger JG. (S)-6-Methyloxaalkyl exemestane compounds and related methods of use. 2010. US patent application number: 11/541,987 Publication number: US 2007/0088013 A1 Filing date: Oct 2, 2006 Issued patent: US7846918 (Issue date Dec 7, 2010)
- Ariazi EA, Leitão A, Oprea TI, Chen B, Louis T, Bertucci AM, Sharma CGN, Gill SD, Kim HR, Shupp HA, Pyle JR, Madrack A, Donato AL, Cheng D, Paige JR, Jordan VC. Exemestane's 17-hydroxylated metabolite exerts biological effects as an androgen. Mol Cancer Ther. 2007;6(11):2817–2827. doi: 10.1158/1535-7163.MCT-07-0312. [DOI] [PubMed] [Google Scholar]
- Buzzetti F, Di Salle E, Longo A, Briatico G. Synthesis and aromatase inhibition by potential metabolites of exemestane (6-methylenandrosta-1,4-diene-3,17-dione) Steroids. 1993;58(11):527–532. doi: 10.1016/0039-128X(93)90029-M. [DOI] [PubMed] [Google Scholar]
- Gorlitzer K, Bonnekessel C, Jones PG, Palusczak A, Hartmann RW. Exemestan-derivate – Synthese und prüfung auf aromatase-hemmung. Die Pharmazie. 2006;61:575–581. [PubMed] [Google Scholar]
- Zafar S, Bibi M, Yousuf S, Choudhary MI. New metabolites from fungal biotransformation of an oral contraceptive agent: Methyloestrenolone. Steroids. 2013;78(4):418–425. doi: 10.1016/j.steroids.2013.01.002. [DOI] [PubMed] [Google Scholar]
- Al-Maruf MA, Khan NT, Sakil MAA, Choudhary MI, Ali MU, Islam MA. Biotransformations of 11-ketoprogesterone by filamentous fungus, Fusarium lini. J Sci Res. 2011;3(2):347–356. [Google Scholar]
- Kamdem LK, Flockhart DA, Desta Z. In Vitro cytochrome P450-mediated metabolism of exemestane. Drug Metab Disposition. 2011;39(1):98–105. doi: 10.1124/dmd.110.032276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Spectroscopic data of metabolite 2. Include spectra of EI-MS, UV, IR, 1H-NMR, 13C-NMR (BB, DEPT-135°, DEPT-90°), HMQC, HMBC, COSY-45°, NOESY experiments.
Spectroscopic data of metabolite 3. Include spectra of EI-MS, UV, IR, 1H-NMR, 13C-NMR (BB, DEPT-135°, DEPT-90°), HSQC, HMBC, COSY-45°, NOESY experiments.
Spectroscopic data of metabolite 4. Include spectra of EI-MS, UV, IR, 1H-NMR, 13C-NMR (BB, DEPT-135°, DEPT-90°), HMQC, HMBC, COSY-45°, NOESY, experiments.
Spectroscopic data of metabolite 5. Include spectra of EI-MS, UV, IR, 1H-NMR, 13C-NMR (BB, DEPT-135°, DEPT-90°), HSQC, HMBC, COSY-45°, NOESY experiments.