Abstract
The use of free-energy landscapes rationalizes a wide range of aspects of protein behavior by providing a clear illustration of the different states accessible to these molecules, as well as of their populations and pathways of interconversion. The determination of the free-energy landscapes of proteins by computational methods is, however, very challenging as it requires an extensive sampling of their conformational spaces. We describe here a technique to achieve this goal with relatively limited computational resources by incorporating nuclear magnetic resonance (NMR) chemical shifts as collective variables in metadynamics simulations. As in this approach the chemical shifts are not used as structural restraints, the resulting free-energy landscapes correspond to the force fields used in the simulations. We illustrate this approach in the case of the third Ig-binding domain of protein G from streptococcal bacteria (GB3). Our calculations reveal the existence of a folding intermediate of GB3 with nonnative structural elements. Furthermore, the availability of the free-energy landscape enables the folding mechanism of GB3 to be elucidated by analyzing the conformational ensembles corresponding to the native, intermediate, and unfolded states, as well as the transition states between them. Taken together, these results show that, by incorporating experimental data as collective variables in metadynamics simulations, it is possible to enhance the sampling efficiency by two or more orders of magnitude with respect to standard molecular dynamics simulations, and thus to estimate free-energy differences among the different states of a protein with a kBT accuracy by generating trajectories of just a few microseconds.
Keywords: NMR spectroscopy, protein folding, protein structure determination, bias-exchange metadynamics, enhanced sampling
In the past two decades, a series of experimental and theoretical advances has made it possible to obtain a detailed understanding of the molecular mechanisms underlying the folding process (1–6). With the increasing power of computers (7), as well as the improvements in force fields (8, 9), atomistic simulations are also becoming increasingly important because they can generate highly detailed descriptions of the motions of proteins (10–12). A supercomputer specifically designed to integrate Newton’s equations of motion of proteins (7) recently broke the millisecond time barrier. This achievement has allowed the direct calculation of repeated folding events for several fast-folding proteins (13) and the characterization of molecular mechanisms underlying protein dynamics and function (14). Reliable descriptions of the folding process have also been obtained by exploiting enhanced sampling techniques (15, 16), including replica-exchange molecular dynamics (17), metadynamics (18, 19), and distributed computing (20).
It has also been realized that by bringing together experimental measurements and computational methods, it is possible to expand the range of problems that may be addressed (4, 21–24). For example, by incorporating structural information relative to transition states (TSs; ϕ values) as structural restraints in molecular dynamics simulations, it is possible to obtain structural models of these transiently populated states (25, 26), as well as of native (27) and nonnative intermediates (28) explored during the folding process. By applying this strategy to structural parameters measured by NMR spectroscopy, one can determine the atomic-level structures and dynamics of proteins (29–32). In these approaches, the experimental information is exploited to create an additional term in the force field that penalizes the deviations from the measured values, thus restraining the sampling of the conformational space to regions close to those observed experimentally (25).
Here, we propose an alternative strategy to use experimental information to aid molecular dynamics simulations. In this approach, the measured parameters are not used as structural restraints in the simulations but rather to build collective variables (CVs) within metadynamics calculations. In metadynamics (18, 19), the conformational sampling is enhanced by constructing a time-dependent potential that discourages the explorations of regions already visited in terms of specific functions of the atomic coordinates called collective variables. In this work, we show that NMR chemical shifts may be used as collective variables to guide the sampling of conformational space in molecular dynamics simulations.
Because the method that we discuss here enables the conformational sampling to be enhanced without modifying the force field through the introduction of structural restraints, it provides the statistical weights corresponding to the force field used in the molecular dynamics simulations. In the present implementation, we used the bias-exchange metadynamics (BE-META) method (33), an enhanced sampling technique that allows the reconstruction of free energy as a simultaneous function of several variables. By using this approach, we computed the free-energy landscape in explicit solvent of the third Ig-binding domain of streptococcal protein G (GB3). Our calculations predict the native fold as the lowest free-energy minimum, also identifying the presence of an on-pathway compact intermediate with nonnative structural elements. In addition, we provide a detailed atomistic picture of the structure at the folding barrier, which shares with the native state a fraction of the secondary structure elements.
These results have been obtained using relatively limited computational resources. Through the advanced sampling method that we discuss, the total simulation time required to reach convergence in the free energy estimates was 380 ns on seven replicas, which is about three orders of magnitude less than the typical timescale required to fold similar proteins (34). We thus anticipate that the technique introduced here will allow the determination of the free-energy landscapes of a wide range of proteins in cases in which NMR chemical shifts are available.
Results and Discussion
We performed molecular dynamics simulations of GB3 at 330 K, using the Gromacs 4.5.3 package (35) and the AMBER99SB-ILDN force field (8). To enhance conformational sampling, we used the BE-META scheme (33) with seven replicas. We started the simulations from a structure at 5.7 Å from the reference structure [Protein Data Bank (PDB) ID code 2OED (36)] and ran them for a total of 380 × 7 ns. For each replica, we used a different metadynamics (18) history-dependent potential acting on a different CV (Methods and SI Text). Three CVs act at the secondary structure level by quantifying, respectively, the fraction of α-helical, antiparallel, and parallel β-sheet content of the protein. Three other CVs act at the tertiary structure level by biasing the number of hydrophobic contacts and the orientation of the side-chain dihedral angles χ1 and χ2 for hydrophobic and polar side chains. The seventh CV, called “CamShift,” measures the difference between the experimental and calculated chemical shifts, which were obtained using the CamShift method (37) (Methods and SI Text). Our results indicate that in the approach we present here, this last variable is essential to fold GB3 and reach convergence readily in the free-energy calculations.
Folding of GB3 Using Chemical Shifts as CVs.
The method that we introduce in this work makes it possible to visit efficiently a wide range of structures, ranging from extended to compact. Representative examples are shown in Fig. 1A. Native-like conformations are visited multiple times, reaching a backbone rmsd of 0.5 Å from the reference structure (PDB ID code 2OED). In these native-like structures, the internal packing of hydrophobic side chains is practically identical to that observed in the reference structure (Fig. 1C). In the calculations that we performed, this level of accuracy could be reached only by using a bias-exchange scheme in which the CamShift CV is included in the CV set (Methods and SI Text). To demonstrate this point, we performed another simulation with the same setup, using the six CVs discussed above that describe the secondary and tertiary structures, but not the CamShift CV. The difference between the two simulations is substantial. In the simulation without the CamShift CV, the closest configuration to the reference structure has an rmsd of 2.7 Å (Fig. 2, Inset B). After 50 ns, the rmsd starts increasing progressively (red line) and the folded state is not explored at all. By contrast, the simulation with the CamShift CV visits the folded state several times, with several unfolding–refolding events. During the first 50 ns, the latter simulation not only performed better, reaching an rmsd of 2.5 Å, but it also formed the correct secondary and tertiary contacts, particularly the ones involved in forming the first β-hairpin (Fig. 2, Inset A), which is critical for the folding of this protein (38, 39). The fraction of native contacts also was systematically higher in the simulation using the CamShift CV (Fig. 2, Inset C). These results indicate that the folding events observed later in the simulation are a result of the systematic bias induced by the CamShift CV toward the correct local topology in the native state.
Fig. 1.
(A) Representation of the conformational sampling achieved by the approach introduced in this work. The conformations visited are shown as a function of the CamShift collective variable (CV) and of the backbone rmsd from the reference structure (PDB ID code 2OED). (B) Structure with the lowest rmsd (0.5 Å) from the reference structure. (C) Detail of the side chain packing of the structure in B.
Fig. 2.
Time series of the trajectories that achieve the lowest rmsd value for the simulations with (black line) and without (red line) the CamShift CV. (Insets A and B) Lowest rmsd structures in the two simulations. (Inset C) Percentage of native contacts in each conformation in the first 50 ns in the two simulations.
Thermodynamics of GB3 Folding.
The molecular dynamics simulations that we performed using the approach presented in this work reached convergence after ∼240 ns, because at this point the bias potentials acting on all the replicas started to become stationary (40). We then continued the simulations for another 140 ns to reconstruct the free-energy landscape of the protein (Methods). In Fig. 3A, the free-energy landscape is represented as a function of three CVs: the fraction of antiparallel β-sheet, the fraction of parallel β-sheet, and the coordination number between the hydrophobic side chains (Fig. 3A). This representation reveals the organization of the free-energy landscape, with a deep minimum corresponding to native-like structures, separated by a relatively high barrier from other minima. The lowest free-energy minimum (Methods) includes configurations very similar to those of the reference structure (on average, at 1.3 Å rmsd). This result is confirmed by the analysis of the deviations of the calculated chemical shifts from the corresponding experimental values, both for the reference structure (PDB ID code 2OED) and for the structures belonging to the free-energy minimum (Fig. S1). The agreement is excellent in both cases, thus confirming that by our procedure we could find structures very close to the X-ray structure. These results also provide evidence of the excellent quality of the AMBER99SB-ILDN (8) force field that we used to model GB3.
Fig. 3.
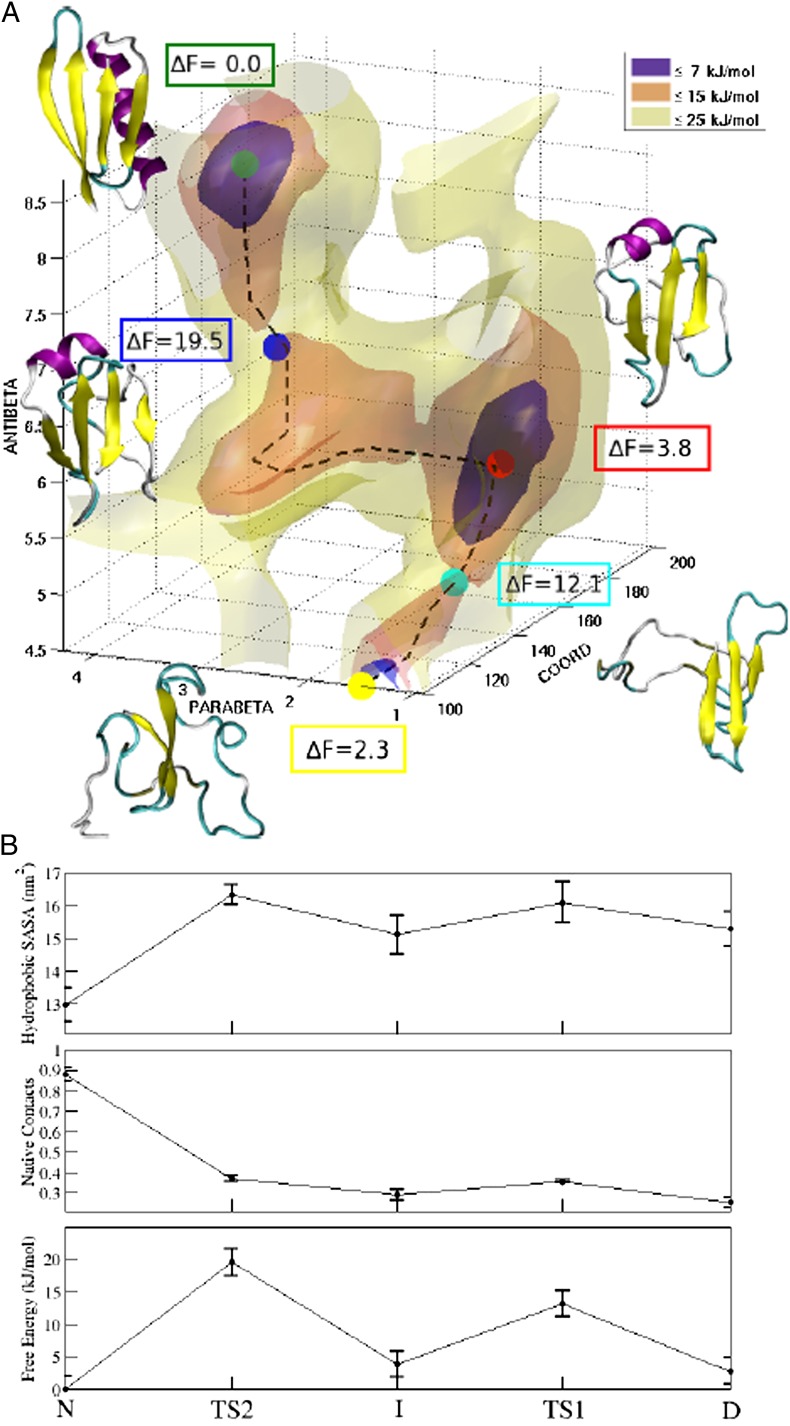
(A) Three-dimensional representation of the free-energy landscape of GB3 as a function of three CVs (see the main text). Along the folding pathway (black dashed line), the most relevant structures are reported with their relative free-energy values: the native state N is shown in green, the transition state TS2 in blue, the intermediate state I in red, the transition state TS1 in cyan, and the unfolded ensemble U in yellow. (B) Hydrophobic SASA, relative number of native contacts, and free energy along the folding pathway.
The shallow minimum immediately after the free-energy barrier separating the folded state from the rest of the conformational space includes compact structures with a high secondary structure content, but with a fold that is rather different from the native, as is discussed below. This second minimum is separated by another free-energy barrier from another minimum, which includes more disordered structures with a much lower secondary structure content. In these conformations, the native C-terminal β-hairpin appears to be present, confirming its high stability, whereas the α-helix and the N-terminal β-hairpin are completely disrupted (41–43). The folded-like and unfolded-like states have a free-energy difference of only 2.3 kJ/mol, which is comparable with the error of our free-energy estimates (40) (Methods). The relatively small difference in the free energies of the folded and unfolded states reflects the conformational properties of the protein at the temperature at which the simulation was performed (330 K), which is about 30 K below the experimental melting temperature of GB3 (34).
An Intermediate State in the Folding of GB3.
The free-energy landscape that we calculated illustrates explicitly the presence of three distinct states of GB3. In addition to the native (N, in dark green in Fig. 3A) and unfolded (U, in yellow in Fig. 3A) states, we identified the presence of an intermediate state (I, in red in Fig. 3A) with a free energy 3.8 kJ/mol higher than that of the N state. From the relative free energies, we calculated the populations of the three states at 330 K, which are 59% for N, 14% for I, and 26% for U. A control unbiased molecular dynamics simulation of 200 ns starting from a structure corresponding to the intermediate free-energy minimum remained extremely stable, with an average rmsd of 2.4 Å from the equilibrated initial structure. These results are consistent with the observation of the presence of an intermediate state of GB1 (44, 45), which shares 88% of the sequence identity of GB3. In particular, that work, which was based on the measurement of the kinetic folding constant as a function of the pH and denaturant concentration, reported a folding behavior consistent with the presence of an on-pathway intermediate and two different TSs (44, 45). However, the structure of the intermediate of GB1 is likely to be more native-like than the one that we find here. The ensemble of conformations making up the intermediate state characterized by our approach contains compact structures, which share specific secondary elements with the native state, including the C-terminal β-hairpin. The N-terminal extension is instead less structured, with only an incipient parallel pairing of the first β-strand (44) and the N-terminal region of the α-helix (residues 22–30). In addition, the C-terminal part of the α-helix exhibits a nonnative configuration by forming an antiparallel β-strand paired with the third β-strand of the protein (residues 41–47).
Identification and Characterization of the TSs.
To better characterize the folding mechanism of GB3, we simulated by a kinetic Monte Carlo approach (46) the dynamics on the multidimensional free-energy landscape reconstructed by our procedure (Methods). All the trajectories connecting the folded and unfolded states go through the intermediate state, confirming that it is an on-pathway intermediate, like the one observed for GB1 (45). The black dashed line in Fig. 3A represents the 3D projection of the trajectory of highest probability connecting the folded and unfolded states. Consistent with this topology, the trajectory crosses two TSs: TS1 between the unfolded and intermediate states (in cyan in Fig. 3A) and TS2 between the intermediate and native states (in blue in Fig. 3A). The rate-limiting step is represented by TS2, with a barrier of 19.5 kJ/mol from the native state, whereas TS1 is at a free energy of 12 kJ/mol.
The hydrophobic solvent-accessible surface area (SASA) reveals how the two TSs are less compact than the N and I states but still quite structured (Fig. 3B). A similar conclusion was reached by the experimental Tanford β-values for the two transition states of GB1: βTS1 = 0.76 ± 0.04 and βTS2 = 0.93 ± 0.04 (45). These values are consistent with those computed by the ratio of the total SASA between N and the corresponding TS obtained in the present study for GB3, βTS1 = 0.82 ± 0.03, and βTS2 = 0.91 ± 0.03.
We found that TS2 of GB3 is more compact than TS1 (Fig. 3A), at least in part because of the presence of a native salt bridge between Lys-10 and Glu-56 that is missing in TS1. This aspect also was suggested in the case of GB1 (45) to explain the differences in the pH dependence for the unfolding rate constant of the two TSs. Indeed, an inspection of the TS1, I, and TS2 structures reveals how this salt bridge may trigger the correct arrangement between the C terminus and the first β-strand (residues 1–10). The formation of the salt bridge, which is absent in TS1, acts in I as an anchor that may allow the parallel pairing of the first β-strand, increasing the fraction of native contacts from 29% in I to 37% in TS2. On this view, the second β-hairpin represents the initial native element in the folding process, followed by the formation of the N terminus of the native helix and the parallel pairing of the first β-strand to the C terminus β-hairpin, to then stabilize the formation of the first β-hairpin.
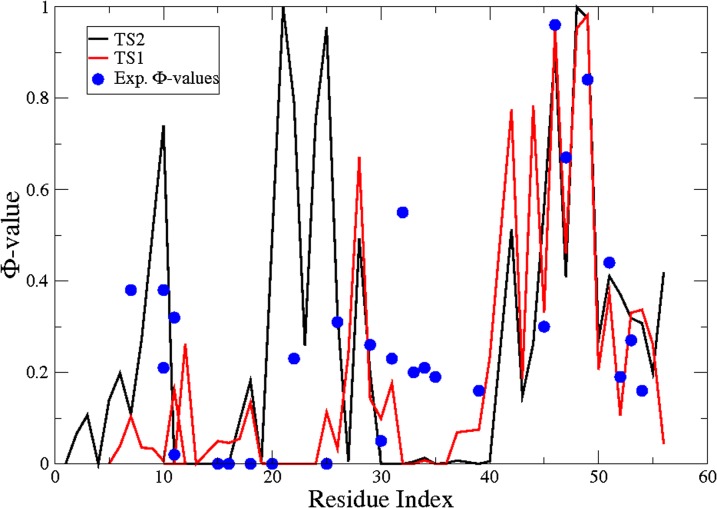
These findings are consistent with the ϕ-values measured for GB1 (38). A comparison between the experimental ϕ-values of GB1 and those calculated for GB3 for TS1 and TS2 is presented in Fig. 4 through the fraction of native contacts of amino acid side chains (25, 26). Despite the differences in sequence between GB1 and GB3, the structure of the TS2 of GB3 exhibits a pattern approximately consistent with experimental ϕ-values of the TS of GB1 (Fig. 4), especially in the two β-hairpin regions. These results, which are consistent with previous conclusions (38), indicate that in the TS the C-terminal hairpin is completely formed as well as the parallel pairing of the first β-strand. Instead, the ϕ-values in the α-helical region show a more complex behavior compatible with a variety of conformations in the transition ensemble.
Fig. 4.
Comparison of the experimental ϕ-values (blue circles) of GB1 (38) with the ϕ-values for GB3 calculated from the TS2 (black line) and TS1 (red line) structures determined in this work.
Conclusions
We have introduced a method for calculating the free-energy landscapes of proteins based on the incorporation of experimental NMR chemical shifts as collective variables in bias-exchange metadynamics simulations (18, 33). To this end, we have defined a collective variable that measures the difference between experimental and calculated chemical shifts, and helps the simulations find a route to the folded state by exploiting the capability of the chemical shifts to characterize in detail the local configuration of a protein molecule. We have found that this procedure facilitates the formation of the correct native contacts and, consequently, the identification of structures effectively indistinguishable from the native-state conformation determined experimentally.
A distinctive aspect of the approach that we have presented is that it uses the chemical shifts only to define a reaction coordinate, without modifying the underlying force field used in the molecular dynamics simulations. Hence, the resulting free-energy landscape derives from the Boltzmann distribution of the system for the force field used in the simulations.
This procedure allows the free-energy landscapes of proteins to be determined with relatively limited computational resources. In the case of GB3 discussed here, only 380-ns simulations for seven replicas were required. Our calculations have revealed the presence of an on-pathway, partially nonnative intermediate state and have enabled us to estimate accurately the free-energy differences between the different states populated by this protein.
Because the approach that we have described is based on the use of chemical shifts as collective variables in metadynamics simulations, it also may be adopted if only incomplete data are available. It thus will be interesting to explore its applicability to larger proteins and to intrinsically disordered proteins (IDPs) to generate ensembles of structures and free-energy landscapes consistent with chemical shift data. Furthermore, this kind of approach may be generalized by incorporating other experimental data in a metadynamics framework, including NOEs, J-couplings, and residual dipolar couplings, or data from other experimental techniques, such as Small-Angle X-ray Scattering (SAXS) or Fluorescence Resonance Energy Transfer (FRET) methods. We anticipate that these developments will provide molecular dynamics descriptions of the behavior of a variety of proteins for which only sparse experimental data are available.
Methods
BE-META.
Bias-exchange metadynamics (BE-META) is a technique that may be used for enhancing the conformational search and for reconstructing the free energy of complex biological systems. This method involves a combination of replica exchange (17) and metadynamics (18), in which a set of CVs is chosen and several metadynamics simulations are performed in parallel on different replicas of the system at the same temperature, each replica biasing a different CV (SI Text). Exchanges between the replicas are attempted periodically according to a replica-exchange scheme, and this process is repeated until convergence of the free energy profiles is obtained. As a consequence of the effectively multidimensional nature of the bias, this procedure allows complex free-energy landscapes to be explored with great efficiency, and it may be very useful to study the folding process (33). To reach this goal, the choice of CVs is crucial, as inappropriate CVs do not enable one to reach convergence. Here, together with other variables for folding used in other works (47), we introduced a CV based on experimental NMR chemical shifts as the driving force for folding. This procedure represents an alternative way to incorporate experimental data in molecular dynamics simulations, not as restraints (30) but in a metadynamics framework.
CamShift CV.
To predict the NMR chemical shifts corresponding to a given structure, we used the CamShift method (37), which is based on an approximation of the chemical shifts as polynomial functions of interatomic distances (SI Text). Unlike other methods for the semiempirical calculation of protein backbone chemical shifts (48–50), the functions used in CamShift are differentiable, thus allowing the forces to be computed and the CV to be defined as a penalty function based on the differences between the experimentally measured and the calculated backbone chemical shifts (1Hα, 13Cα, 13Cβ,13C’, 1HN, 15N) (SI Text and Fig. S2) (30, 32). Because the chemical shifts are extremely sensitive to the details of the local configuration and environment of the atoms, the aim of this CV is to reproduce the local rearrangement of the protein compatible with the experimental data, especially when it approaches low values, which correspond to a better overlap between the predicted and the experimental chemical shifts. Even if the calculation of the chemical shifts is restricted to the backbone atoms, some contributions also depend on the orientation of the side chains (37). Therefore, the forces applied by metadynamics to all the atoms involved in the calculation of the CV help the slow transition of side-chain dihedral angles in finding the correct arrangement crucial to avoid a bad steric hindrance and to reach the correct fold. The CamShift CV has been implemented in a modified version of the freely available plug-in PLUMED (51) for Gromacs (35).
Simulation Details.
We performed a BE-META simulation of GB3 at 330 K, using seven replicas, one for each of the CVs (SI Text):
CamShift (see above): This CV was used as a local folding-driving force. Parameters: Gaussian width σ = 1.
AlphaRMSD, ParaBetaRMSD, and AntiBetaRMSD: These CVs were used to measure the fractions of α-helix and parallel and antiparallel β-sheet of the protein conformation (47). Parameters: for AlphaRMSD, m = 4, n = 2, R0 = 0.08, and σ = 0.2; for ParaBetaRMSD, m = 12, n = 8, R0 = 0.08, and σ = 0.1; for AntiBetaRMSD, m = 12, n = 8, R0 = 0.08, and σ = 0.2.
Coordination Number: This CV was used to measure the number of hydrophobic contacts. Parameters: m = 8, n = 4, R0 = 0.4, and σ = 10.
Two AlphaBeta Similarities: These CVs were applied to the dihedral angles χ1 and χ2, respectively, for hydrophobic and polar amino acids to enhance the side-chain packing search. Parameters: σ = 0.5 for both replicas.
The functional forms of the CVs are defined in ref. 51 (SI Text).
Starting from an unfolded state at 5.7 Å from the reference structure [PDB ID code 2OED (36)], obtained by a simulated annealing procedure, we ran 380 ns for each replica using the Gromacs 4.5.3 package (35) with the AMBER99SB-ILDN force field (8) and the TIP3P water model (52). The protein was solvated by 6,524 water molecules in a 212-nm3 periodic box. The particle-mesh Ewald method (53) was used for long-range electrostatic interactions with a short-range cutoff of 1 nm. A cutoff was used for Lennard–Jones interactions at 1.2 nm. All bond lengths were constrained to their equilibrium length with the LINCS (LINear Costraint Solver) algorithm (54). The time step for the molecular dynamics simulation was set at 2.0 fs, and the Nosé–Hoover thermostat (55, 56) with a relaxation time of 1ps was used. The atomic coordinates and the energy were saved every 1 ps. Concerning the metadynamics setup, 1D Gaussian functions of height w = 0.30 kJ/mol were added every 4 ps, and exchanges of the bias potentials were attempted every 20 ps.
After 120 ns of simulation, in which very wide regions of the CVs were explored, we introduced loose upper boundaries to help the convergence of the bias potentials (57). At this time, we also reduced to 0.5 the Gaussian width of the CamShift CV and doubled the σ of the AlphaRMSD CV (SI Text and Table S1). To run the BE-META, we used a modified version of the PLUMED plugin (51) for Gromacs, which will be made publicly available in a future release. A second BE-META simulation of 300 ns was run with six replicas, excluding the CamShift CV, to benchmark the importance and the power of this CV in folding the protein. Finally, a standard molecular dynamics simulation of 200 ns was performed to evaluate the stability of the intermediate state.
Free-Energy Reconstruction in the CV Space.
BE-META allows the free energy of a system to be reconstructed once the bias potentials become stable (33) (SI Text). This happens in our case after an equilibration time teq = 240 ns. After selecting the CVs that are most effective in discriminating different states of the system, the CV space is divided in hypercubes and each simulation frame is assigned to the corresponding microstate according to its CV value (SI Text). The structures within each hypercube must be consistent to define a proper microstate of the system; otherwise, its size must be reduced. Then, a free-energy value is computed for the microstate, according to the corresponding bias potentials and the populations observed after the teq. In our study, we have chosen the CamShift, Coordination Number, and Anti- and Para-BetaRMSD CVs; the relative free-energy profiles are reported in Fig. S3. The error on the free-energy difference of the microstates corresponding to the three local free-energy minima in Fig. 3 is approximately 3 kJ/mol. All the analyses have been performed as previously described (40) (SI Text), using METAGUI (58), a Visual Molecular Dynamics (VMD) (59) interface for analyzing metadynamics and molecular dynamics simulations.
Supplementary Material
Acknowledgments
We acknowledge Standard High Performance Computing (HPC) Grant 2011 from CASPUR Supercomputing Center for computational resources and Associazione Italiana per la Ricerca sul Cancro 5 per Mille Grant Rif.12214. C.C. was supported by a Marie Curie Intra-European Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218350110/-/DCSupplemental.
References
- 1.Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254(5038):1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 2.Fersht A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding. San Francisco: Freeman; 1998. [Google Scholar]
- 3.Muñoz V, Eaton WA. A simple model for calculating the kinetics of protein folding from three-dimensional structures. Proc Natl Acad Sci USA. 1999;96(20):11311–11316. doi: 10.1073/pnas.96.20.11311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dobson C, Šali A, Karplus M. Protein folding: A perspective from theory and experiment. Angew Chem Int Ed. 1998;37(7):868–893. doi: 10.1002/(SICI)1521-3773(19980420)37:7<868::AID-ANIE868>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 5.Wolynes PG, Onuchic JN, Thirumalai D. Navigating the folding routes. Science. 1995;267(5204):1619–1620. doi: 10.1126/science.7886447. [DOI] [PubMed] [Google Scholar]
- 6.Dill KA, Chan HS. From Levinthal to pathways to funnels. Nat Struct Biol. 1997;4(1):10–19. doi: 10.1038/nsb0197-10. [DOI] [PubMed] [Google Scholar]
- 7.Shaw DE, et al. Anton, a special-purpose machine for molecular dynamics simulation. Commun ACM. 2008;51(7):91–97. [Google Scholar]
- 8.Piana S, Lindorff-Larsen K, Shaw DE. How robust are protein folding simulations with respect to force field parameterization? Biophys J. 2011;100(9):L47–L49. doi: 10.1016/j.bpj.2011.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MacKerell AD, Jr, Banavali N, Foloppe N. Development and current status of the CHARMM force field for nucleic acids. Biopolymers. 2000-2001;56(4):257–265. doi: 10.1002/1097-0282(2000)56:4<257::AID-BIP10029>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 10.Karplus M, McCammon JA. Molecular dynamics simulations of biomolecules. Nat Struct Biol. 2002;9(9):646–652. doi: 10.1038/nsb0902-646. [DOI] [PubMed] [Google Scholar]
- 11.Pande VS, et al. Atomistic protein folding simulations on the submillisecond time scale using worldwide distributed computing. Biopolymers. 2003;68(1):91–109. doi: 10.1002/bip.10219. [DOI] [PubMed] [Google Scholar]
- 12.Best RB. Atomistic molecular simulations of protein folding. Curr Opin Struct Biol. 2012;22(1):52–61. doi: 10.1016/j.sbi.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Lindorff-Larsen K, Piana S, Dror RO, Shaw DE. How fast-folding proteins fold. Science. 2011;334(6055):517–520. doi: 10.1126/science.1208351. [DOI] [PubMed] [Google Scholar]
- 14.Shaw DE, et al. Atomic-level characterization of the structural dynamics of proteins. Science. 2010;330(6002):341–346. doi: 10.1126/science.1187409. [DOI] [PubMed] [Google Scholar]
- 15.Christen M, van Gunsteren WF. On searching in, sampling of, and dynamically moving through conformational space of biomolecular systems: A review. J Comput Chem. 2008;29(2):157–166. doi: 10.1002/jcc.20725. [DOI] [PubMed] [Google Scholar]
- 16.Bolhuis PG, Chandler D, Dellago C, Geissler PL. Transition path sampling: Throwing ropes over rough mountain passes, in the dark. Annu Rev Phys Chem. 2002;53:291–318. doi: 10.1146/annurev.physchem.53.082301.113146. [DOI] [PubMed] [Google Scholar]
- 17.Sugita Y, Okamoto Y. Replica-exchange molecular dynamics method for protein folding. Chem Phys Lett. 1999;314(1-2):141–151. [Google Scholar]
- 18.Laio A, Parrinello M. Escaping free-energy minima. Proc Natl Acad Sci USA. 2002;99(20):12562–12566. doi: 10.1073/pnas.202427399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laio A, Gervasio F. Metadynamics: A method to simulate rare events and reconstruct the free energy in biophysics, chemistry and material science. Rep Prog Phys. 2008;71(12):126601. [Google Scholar]
- 20.Shirts M, Pande VS. Screen savers of the world unite! Science. 2000;290(5498):1903–1904. doi: 10.1126/science.290.5498.1903. [DOI] [PubMed] [Google Scholar]
- 21.Fersht AR, Daggett V. Protein folding and unfolding at atomic resolution. Cell. 2002;108(4):573–582. doi: 10.1016/s0092-8674(02)00620-7. [DOI] [PubMed] [Google Scholar]
- 22.Sułkowska JI, Morcos F, Weigt M, Hwa T, Onuchic JN. Genomics-aided structure prediction. Proc Natl Acad Sci USA. 2012;109(26):10340–10345. doi: 10.1073/pnas.1207864109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ratje AH, et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468(7324):713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Velázquez-Muriel J, et al. Assembly of macromolecular complexes by satisfaction of spatial restraints from electron microscopy images. Proc Natl Acad Sci USA. 2012;109(46):18821–18826. doi: 10.1073/pnas.1216549109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vendruscolo M, Paci E, Dobson CM, Karplus M. Three key residues form a critical contact network in a protein folding transition state. Nature. 2001;409(6820):641–645. doi: 10.1038/35054591. [DOI] [PubMed] [Google Scholar]
- 26.Paci E, Vendruscolo M, Dobson CM, Karplus M. Determination of a transition state at atomic resolution from protein engineering data. J Mol Biol. 2002;324(1):151–163. doi: 10.1016/s0022-2836(02)00944-0. [DOI] [PubMed] [Google Scholar]
- 27.Gsponer J, Caflisch A. Molecular dynamics simulations of protein folding from the transition state. Proc Natl Acad Sci USA. 2002;99(10):6719–6724. doi: 10.1073/pnas.092686399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Simone A, Montalvao RW, Vendruscolo M. Determination of conformational equilibria in proteins using residual dipolar couplings. J Chem Theory Comput. 2011;7(12):4189–4195. doi: 10.1021/ct200361b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindorff-Larsen K, Best RB, Depristo MA, Dobson CM, Vendruscolo M. Simultaneous determination of protein structure and dynamics. Nature. 2005;433(7022):128–132. doi: 10.1038/nature03199. [DOI] [PubMed] [Google Scholar]
- 30.Robustelli P, Kohlhoff K, Cavalli A, Vendruscolo M. Using NMR chemical shifts as structural restraints in molecular dynamics simulations of proteins. Structure. 2010;18(8):923–933. doi: 10.1016/j.str.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 31.Neudecker P, et al. Structure of an intermediate state in protein folding and aggregation. Science. 2012;336(6079):362–366. doi: 10.1126/science.1214203. [DOI] [PubMed] [Google Scholar]
- 32.Camilloni C, Robustelli P, De Simone A, Cavalli A, Vendruscolo M. Characterization of the conformational equilibrium between the two major substates of RNase A using NMR chemical shifts. J Am Chem Soc. 2012;134(9):3968–3971. doi: 10.1021/ja210951z. [DOI] [PubMed] [Google Scholar]
- 33.Piana S, Laio A. A bias-exchange approach to protein folding. J Phys Chem B. 2007;111(17):4553–4559. doi: 10.1021/jp067873l. [DOI] [PubMed] [Google Scholar]
- 34.Alexander P, Orban J, Bryan P. Kinetic analysis of folding and unfolding the 56 amino acid IgG-binding domain of streptococcal protein G. Biochemistry. 1992;31(32):7243–7248. doi: 10.1021/bi00147a006. [DOI] [PubMed] [Google Scholar]
- 35.Hess B, Kutzner C, Van Der Spoel D, Lindahl E. Gromacs 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4(3):435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 36.Ulmer TS, Ramirez BE, Delaglio F, Bax A. Evaluation of backbone proton positions and dynamics in a small protein by liquid crystal NMR spectroscopy. J Am Chem Soc. 2003;125(30):9179–9191. doi: 10.1021/ja0350684. [DOI] [PubMed] [Google Scholar]
- 37.Kohlhoff KJ, Robustelli P, Cavalli A, Salvatella X, Vendruscolo M. Fast and accurate predictions of protein NMR chemical shifts from interatomic distances. J Am Chem Soc. 2009;131(39):13894–13895. doi: 10.1021/ja903772t. [DOI] [PubMed] [Google Scholar]
- 38.McCallister EL, Alm E, Baker D. Critical role of beta-hairpin formation in protein G folding. Nat Struct Biol. 2000;7(8):669–673. doi: 10.1038/77971. [DOI] [PubMed] [Google Scholar]
- 39.Camilloni C, Broglia RA, Tiana G. Hierarchy of folding and unfolding events of protein G, CI2, and ACBP from explicit-solvent simulations. J Chem Phys. 2011;134(4):045105. doi: 10.1063/1.3523345. [DOI] [PubMed] [Google Scholar]
- 40.Marinelli F, Pietrucci F, Laio A, Piana S. A kinetic model of trp-cage folding from multiple biased molecular dynamics simulations. PLOS Comput Biol. 2009;5(8):e1000452. doi: 10.1371/journal.pcbi.1000452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blanco FJ, Serrano L. Folding of protein G B1 domain studied by the conformational characterization of fragments comprising its secondary structure elements. Eur J Biochem. 1995;230(2):634–649. doi: 10.1111/j.1432-1033.1995.tb20605.x. [DOI] [PubMed] [Google Scholar]
- 42.Bussi G, Laio A, Parrinello M. Equilibrium free energies from nonequilibrium metadynamics. Phys Rev Lett. 2006;96(9):090601. doi: 10.1103/PhysRevLett.96.090601. [DOI] [PubMed] [Google Scholar]
- 43.Camilloni C, Provasi D, Tiana G, Broglia RA. Exploring the protein G helix free-energy surface by solute tempering metadynamics. Proteins. 2008;71(4):1647–1654. doi: 10.1002/prot.21852. [DOI] [PubMed] [Google Scholar]
- 44.Park SH, O’Neil KT, Roder H. An early intermediate in the folding reaction of the B1 domain of protein G contains a native-like core. Biochemistry. 1997;36(47):14277–14283. doi: 10.1021/bi971914+. [DOI] [PubMed] [Google Scholar]
- 45.Morrone A, et al. GB1 is not a two-state folder: identification and characterization of an on-pathway intermediate. Biophys J. 2011;101(8):2053–2060. doi: 10.1016/j.bpj.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bortz A, Kalos M, Lebowitz J. A new algorithm for monte carlo simulation of ising spin systems. J Comput Phys. 1975;17(1):10–18. [Google Scholar]
- 47.Pietrucci F, Laio A. A collective variable for the efficient exploration of protein beta-sheet structures: Application to sh3 and gb1. J Chem Theory Comput. 2009;5(9):2197–2201. doi: 10.1021/ct900202f. [DOI] [PubMed] [Google Scholar]
- 48.Han B, Liu Y, Ginzinger SW, Wishart DS. SHIFTX2: Significantly improved protein chemical shift prediction. J Biomol NMR. 2011;50(1):43–57. doi: 10.1007/s10858-011-9478-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44(4):213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen Y, Bax A. SPARTA+: A modest improvement in empirical NMR chemical shift prediction by means of an artificial neural network. J Biomol NMR. 2010;48(1):13–22. doi: 10.1007/s10858-010-9433-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bonomi M, et al. Plumed: A portable plugin for free-energy calculations with molecular dynamics. Comput Phys Commun. 2009;180(10):1961–1972. [Google Scholar]
- 52.Jorgensen W, Chandrasekhar J, Madura J, Impey R, Klein M. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 53.Essmann U, et al. A smooth particle mesh ewald method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 54.Hess B, et al. Lincs: A linear constraint solver for molecular simulations. J Comput Chem. 1997;18(12):1463–1472. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- 55.Nosé S. A molecular dynamics method for simulations in the canonical ensemble. Mol Phys. 1984;52(2):255–268. [Google Scholar]
- 56.Hoover WG. Canonical dynamics: Equilibrium phase-space distributions. Phys Rev A. 1985;31(3):1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 57.Baftizadeh F, Cossio P, Pietrucci F, Laio A. Protein folding and ligand-enzyme binding from bias-exchange metadynamics simulations. Curr Phys Chem. 2012;2:79–91. [Google Scholar]
- 58.Biarnés X, Pietrucci F, Marinelli F, Laio A. Metagui. A vmd interface for analyzing metadynamics and molecular dynamics simulations. Comput Phys Commun. 2012;183(1):203–211. [Google Scholar]
- 59.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14(1):33–38, 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.