Summary
Prevalence of etravirine genotypic resistance was assessed among 92 HIV-1C-infected patients failing nevirapine and efavirenz-based regimens from a cohort of 552 Indian patients. Overall prevalence of etravirine cross-resistance identified using the Tibotec weighted score was 41% (31.5% intermediately-resistant and 9.8% fully-resistant). The most frequently described NNRTI-associated mutations included Y181 (35.9%), K101(20.7%), G190(17.4%) and V108 (15.2%). The resistant group demonstrated higher viral load (p=0.01) and longer duration of antiretroviral treatment (p=0.03) compared to the susceptible group.
Keywords: Etravirine cross-resistance, HIV-1 subtype C, NNRTI experience, India
The low genetic barrier to development of resistance to first generation non-nucleoside reverse transcriptase inhibitors (NNRTI) is compounded by cross-resistance across the class which makes sequential therapy with the NNRTIs therapeutically inappropriate. Current first-line NNRTI in most resource-constrained regions includes nevirapine, except in cases of intolerance or potential drug interaction when efavirenz is used [1]. Etravirine, a new generation NNRTI (TMC-125, Intelence, Tibotec Pharmaceuticals Ltd) was approved by US FDA for use in ART-experienced adults with resistance to first-line NNRTIs. Etravirine resistance-associated mutations (RAMs) in reverse transcriptase (RT) gene were identified are as follows; V90I, A98G, L100I, K101E/P/H, V106I, V179D/F, Y181C/I/V, G190A/S, E138A, V179T, and M230L [2–4]. The Tibotec Weighted Score was proposed with 17 etravirine-RAMs and assigned differential weights based upon the impact on clinical response [5]. Alternatively, the Monogram Weighted (MW) Score included 30 etravirine-RAMs based on the genotypic and phenotypic inter-relationship [6]. Etravirine cross-resistance may be influenced by the prevailing HIV-1 subtype [7,8]. With a worldwide prevalence of 50% [9], and prevalence in India of 96%, HIV-1C undoubtedly has a significant impact on the evolution of the HIV epidemic globally. This study reports the selection of NNRTI RAMs and etravirine cross-resistance patterns among HIV-1C infected patients failing first-line ART.
Among a total of 552 participants participating in a 2-year longitudinal cohort study [10], 18% (n=101) with detectable viremia were assessed for presence of drug resistance-associated mutations during their baseline visit [11]. Drug resistance genotyping was successfully done from 92 plasma samples from failing patients (viral load >1000 copies/ml) using a validated in-house method [12]. Drug-resistant strains previously reported from India (n=429) from patients failing first-line ART were included as a second group in this study[13–19]. A third group of 1,122 global HIV-1C sequences were obtained from HIVseq Program (http://hivdb.stanford.edu/; accessed 13th August 2010) reported from patients worldwide with a history of treatment of NNRTI drugs. Indian sequences and duplicates were excluded from global subtype C sequences. NNRTI-DRMs in all these sequences were analyzed. Etravirine resistance was evaluated by Tibotec Etravirine Weighted Genotype Score [5]. Statistical analysis was performed in SPSS v11.5.
Plasma virus was successfully genotyped in 92 failing patients; their mean age was 39.6 years (SD 10.2yrs) and 67% were male, similar to the complete cohort. Among the 92 patients, 77% used nevirapine; 12% used efavirenz and 10% changed from an initial nevirapine-based regimen to an efavirenz-based regimen for clinical reasons. The mean duration of nevirapine and efavirenz exposure was 23 and 14 months, respectively.
The overall prevalence of etravirine resistance was 41% (38/92). Single etravirine-RAMs were seen in 13% and two etravirine-RAMs were seen in 33% strains. Eleven percent (10/92) strains harbored three or more etravirine-RAMs. The Tibotec Weighted Score identified 58.7% of the strains to be susceptible to etravirine whereas 31.5% and 9.8% strains displayed intermediate resistance and resistance respectively. Alternative scoring methods showed comparable patterns (39% of strains had an MW score ≥4) indicating that a significant percentage of isolates had reduced efficacy to etravirine.
Genotypic analysis predicted that 41.6% (30/72) of samples from nevirapine-experienced and 9.1%(1/11) from efavirenz-experienced patients were cross-resistant to etravirine. The maximum level of cross-resistance (77.8%, 7/9) was observed in those patients who had exposure of both the drugs. The most frequently described RAMs included amino acid substitutions at positions Y181 (35.9%), K101(20.7%), G190(17.4%) and V108(15.2%).. Similar trends were observed in sequences reported previously from India (n=429); however among global subtype C sequences, K103N was the most frequent RAM (Figure 1).
Figure 1. Selection of NNRTI mutations in ART experienced patients harboring HIV-1 subtype C viruses.
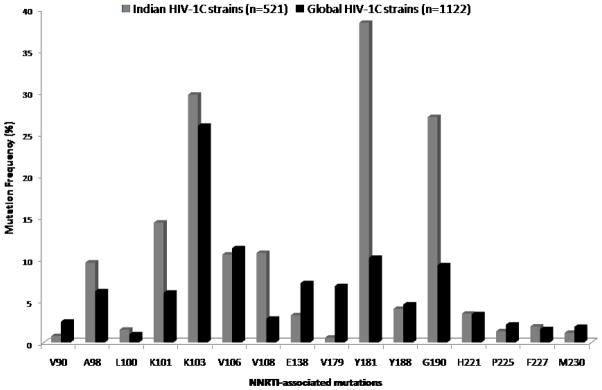
Higher frequencies of NNRTI drug resistance mutations are present in residues Y181, K101, G190 and V108 in Indian sequences (n=521, 92 primary isolates and 429 previously reported sequences) compared to global subtype C sequences (n=1122) obtained from HIVseq Program from Stanford University HIV Drug resistance database (http://hivdb.stanford.edu/; accessed on 13th August 2010).
Compared to patients with susceptible virus, those who harbored etravirine-resistant virus were more likely to have been on ART for a longer duration (p=0.03) and to have higher viral load (p=0.01) (Supplementary digital content 1). There was no significant difference in age, CD4 count, time since diagnosis or self-reported adherence in the last month measured by Visual Analog Scale between the two groups.
Our report highlights the high prevalence of etravirine cross-resistance (41%) among the patients infected with HIV-1C viruses and failing first generation NNRTI-based regimens in India. Etravirine RAMs has also been described in ART-naïve patients from France, Mali and India [20, 21]. Our finding of etravirine resistance is higher than among HIV-infected patients harboring subtype B in UK (11.5%) and Spain (18.7%). A similar study from Thailand found 56% etravirine cross-resistance in HIV-1 CRF01_AE strains [24].
The high prevalence of Y181 and K101 found in our setting is also seen in other places where nevirapine is widely used as first-line NNRTI. Similar trends have been observed in patients with CRF01_AE strains from Thailand (50% Y181C/I/V and 18.7% K101E/H/P) [24] and UK (17% Y181C in those failing efavirenz and 40.5% Y181C were in those failing nevirapine) [25], thus lending credence to the conclusion that Y181C is particularly selected during prolonged exposure to a failing nevirapine-containing regimen [11].
The association between etravirine resistance and higher viral loads in the study cohort may be reflective of the longer duration on poorly suppressive regimens experienced by these patients [26]. In settings like India where routine viral load monitoring is not a part of standard of care, the second-line antiretroviral therapy regimens have to be designed with caution when including NNRTI drugs. As over 50% of failing isolates are susceptible to etravirine, it can be used as salvage therapy among those patients failing first generation NNRTI-based regimens. Patients with high level of etravirine-RAMs were also more likely to have tenofovir-associated mutations [27] which may raise challenges in designing an effective second-line regimen in resource-constrained settings like India. The presence of cross-resistance also highlights the need for developing effective and sustainable adherence interventions that target local adherence patterns and barriers in order to keep the limited first-line ART agents effective for as long as possible [10].
In summary our study highlights the high level of etravirine cross-resistance in a cohort of ART-experienced patients failing NNRTI-containing first-line therapy in India. The pattern of NNRTI-mutations in nevirapine exposed patients also suggests the possible benefit of reconsidering the use of nevirapine in favor of efavirenz as first-line NNRTI choice in resource-constrained settings.
Supplementary Material
Acknowledgments
The authors would like to thank the Prerana study team for their generous help with field work, data collection and Karthika Arumugam, St. John’s National Academy of Health Sciences, Bangalore for her help with statistical analysis and Dr Prabhakar of Bowring and Lady Curzon Hospital for his help with patient recruitment. The study was approved by the Committee for Human Research at University of California, San Francisco, USA and the Institutional Ethical Review Board St. John’s Medical College and Hospital, Bangalore, India.
References
- 1.Joly V, Yeni P. Non-nucleoside reverse transcriptase inhibitors. Ann Med Interne (Paris) 2000;151:260–267. [PubMed] [Google Scholar]
- 2.Madruga JV, Cahn P, Grinsztejn B, Haubrich R, Lalezari J, Mills A, et al. Efficacy and safety of TMC125 (etravirine) in treatment-experienced HIV-1-infected patients in DUET-1: 24-week results from a randomised, double-blind, placebo-controlled trial. Lancet. 2007;370:29–38. doi: 10.1016/S0140-6736(07)61047-2. [DOI] [PubMed] [Google Scholar]
- 3.Lazzarin A, Campbell T, Clotet B, Johnson M, Katlama C, Moll A, et al. Efficacy and safety of TMC125 (etravirine) in treatment-experienced HIV-1-infected patients in DUET-2: 24-week results from a randomised, double-blind, placebo-controlled trial. Lancet. 2007;370:39–48. doi: 10.1016/S0140-6736(07)61048-4. [DOI] [PubMed] [Google Scholar]
- 4.Johnson VA, Brun-Vezinet F, Clotet B, Gunthard HF, Kuritzkes DR, et al. Update of the Drug Resistance Mutations in HIV-1. Top HIV Med. 2008;16:138–45. [PubMed] [Google Scholar]
- 5.Vingerhoets J, Tambuyzer L, Azijn H, Hoogstoel A, Nijs S, Peeters M, et al. Resistance profile of etravirine: combined analysis of baseline genotypic and phenotypic data from the randomized, controlled Phase III clinical studies. AIDS. 2010;24:503–14. doi: 10.1097/QAD.0b013e32833677ac. [DOI] [PubMed] [Google Scholar]
- 6.Benhamida J, Chappey C, Coakley E, Parkin NT. HIV-1 genotype algorithms for prediction of etravirine susceptibility: novel mutations and weighting factors identified through correlations to phenotype. Antivir Ther. 2008;13:A142. [Google Scholar]
- 7.Martinez-Cajas JL, Pant-Pai N, Klein MB, Wainberg MA. Role of genetic diversity amongst HIV-1 non-B subtypes in drug resistance: A systematic review of virologic and biochemical evidence. AIDS Rev. 2008;10:212–223. [PubMed] [Google Scholar]
- 8.Kosakovsky Pond SL, Smith DM. Are all subtypes created equal? The effectiveness of antiretroviral therapy against non-subtype B HIV-1. Clin Infect Dis. 2009;48:1306–1309. doi: 10.1086/598503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor BS, Sobieszczyk ME, McCutchan FE, Hammer SM. The challenge of HIV-1 subtype diversity. N Engl J Med. 2008;358:1590–1602. doi: 10.1056/NEJMra0706737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ekstrand ML, Chandy S, Heylen E, Steward W, Singh G. Developing useful highly active antiretroviral therapy adherence measures for India: the Prerana study. J Acquir Immune Defic Syndr. 2010;53:415–416. doi: 10.1097/QAI.0b013e3181ba3e4e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ekstrand ML, Shet A, Chandy S, Singh G, Shamsundar R, Madhavan V, Saravanan S, Kumarasamy N. Suboptimal adherence associated with virologic failure and resistance mutations among patients on 1st line HAART in Bangalore, India[Abstract]. XVIII International AIDS Conference; Vienna, Austria. July 18–23 2010; p. Abstract no. WEAB0202. [Google Scholar]
- 12.Saravanan S, Vidya M, Balakrishnan P, Kumarasamy N, Solomon SS, Solomon S, et al. Evaluation of two human immunodeficiency virus-1 genotyping systems: ViroSeqTM 2. 0 and in-house methods. J Virol Methods. 2009;159:211–216. doi: 10.1016/j.jviromet.2009.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sen S, Tripathy SP, Patil AA, Chimanpure VM, Paranjape RP. High prevalence of Human Immunodeficiency Virus type 1 drug resistance mutations in antiretroviral treatment-experienced patients from Pune, India. AIDS Res Hum Retroviruses. 2007a;23:1303–1308. doi: 10.1089/aid.2007.0090. [DOI] [PubMed] [Google Scholar]
- 14.Sen S, Tripathy SP, Chimanpure VM, Patil AA, Bagul RD, Paranjape RP. Human Immunodeficiency Virus type 1 drug resistance mutations in peripheral blood mononuclear cell proviral DNA among antiretroviral treatment-naive and treatment-experienced patients from Pune, India. AIDS Res Hum Retroviruses. 2007b;23:489–497. doi: 10.1089/aid.2006.0221. [DOI] [PubMed] [Google Scholar]
- 15.Deshpande A, Jauvin V, Magnin N, Pinson P, Faure M, Masquelier B, et al. Resistance mutations in subtype C HIV type 1 isolates from Indian patients of Mumbai receiving NRTIs Plus NNRTIs and experiencing a treatment failure: resistance to AR. AIDS Res Hum Retroviruses. 2007;23:335–340. doi: 10.1089/aid.2006.0183. [DOI] [PubMed] [Google Scholar]
- 16.Vidya M, Saravanan S, Uma S, Kumarasamy N, Sunil SS, Kantor R, et al. Genotypic HIV type-1 drug resistance among patients with immunological failure to first-line antiretroviral therapy in south India. Antivir Ther. 2009;14:1005–1009. doi: 10.3851/IMP1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kandathil AJ, Kannangai R, Verghese VP, Pulimood SA, Rupali P, Sridharan G, et al. Drug resistant mutations detected by genotypic drug resistance testing in patients failing therapy in clade C HIV-1 infected individuals from India. Indian J Med Microbiol. 2009;27:231–236. doi: 10.4103/0255-0857.53205. [DOI] [PubMed] [Google Scholar]
- 18.Rajesh L, Karunaianantham R, Narayanan PR, Swaminathan S. Antiretroviral Drug-Resistant Mutations at Baseline and at Time of Failure of Antiretroviral Therapy in HIV Type 1-Coinfected TB Patients. AIDS Res Hum Retroviruses. 2009;25:1179–85. doi: 10.1089/aid.2009.0110. [DOI] [PubMed] [Google Scholar]
- 19.Choudhury SD, Choudhury AK, Kalra R, Andrabi R, Wig N, Biswas A, et al. Anti retroviral drug resistant mutations in the reverse transcriptase gene of HIV-1 isolates from northern Indian patients: A follow up study. Arch Virol. 2010;155:563–569. doi: 10.1007/s00705-010-0605-4. [DOI] [PubMed] [Google Scholar]
- 20.Maiga AI, Descamps D, Morand-Joubert L, Morand-Joubert L, Malet I, Derache A, et al. Resistance-associated mutations to etravirine (TMC-125) in antiretroviral-naive patients infected with non-B HIV-1 subtypes. Antimicrob Agents Chemother. 2010;54:728–33. doi: 10.1128/AAC.01335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neogi U, Prarthana BS, Gupta S, D’souza G, De Costa A, Kuttiatt VS, et al. Naturally occurring polymorphisms and primary drug resistance profile among antiretroviral-naïve individuals in Bangalore, India. AIDS Res Hum Retroviruses. 2010;26:1097–1101. doi: 10.1089/aid.2010.0092. [DOI] [PubMed] [Google Scholar]
- 22.Scott C, Grover D, Nelson M. Is there a role for etravirine in patients with Nonnucleoside reverse transcriptase inhibitor resistance? AIDS. 2008;22:989–992. doi: 10.1097/QAD.0b013e3282fa75df. [DOI] [PubMed] [Google Scholar]
- 23.Poveda E, Anta L, Blanco JL, Perez-Elias, Gracia F, Leal M, et al. Etravirine resistance associated mutations in HIV-infected patients falling efavirenz or nevirapine in the Spanish antiretroviral resistance database. AIDS. 2010;24:469–471. doi: 10.1097/QAD.0b013e328331a4b8. [DOI] [PubMed] [Google Scholar]
- 24.Manosuthi W, Butler DM, Chantratita W, Sukasem C, Richman DD, Smith DM. Patients infected with HIV Type 1 Subtype CRF01_AE and failing first-line nevirapine- and Efavirenz-based regimens demonstrate considerable cross-resistance to etravirine. AIDS Res Hum Retroviruses. 2010;26:609–611. doi: 10.1089/aid.2009.0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richman DD, Havlir D, Corbeil J, Looney D, Ignacio C, Spector SA, et al. Nevirapine resistance mutations of human immunodeficiency virus type 1 selected during therapy. J Virol. 1994;68:1660–1666. doi: 10.1128/jvi.68.3.1660-1666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lapadula G, Calabresi A, Castelnuovo F, Costarelli S, Quiros-Roldan E, Paraninfo G, et al. Prevalence and risk factors for etravirine resistance among patients failing on non-nucleoside reverse transcriptase inhibitors. Antivir Ther. 2008;13:601–605. [PubMed] [Google Scholar]
- 27.Bunupuradah T, Ananworanich J, Chetchotisakd P, Kantipong P, Jirajariyavet S, Sirivichayakul S, et al. Prevalence and predictors of etravirine resistance in Thai HIV-infected adults failing first-line NNRTI-based regimens [Abstract]. XVIII International AIDS Conference; Vienna, Austria. July 18–23, 2010; p. Abstract no. MOPDB104. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


