Abstract
Regulation of gene transcription is vitally important for the maintenance of normal cellular homeostasis. Failure to correctly regulate gene expression, or to deal with problems that arise during the transcription process, can lead to cellular catastrophe and disease. One of the ways cells cope with the challenges of transcription is by making extensive use of the proteolytic and nonproteolytic activities of the ubiquitin-proteasome system (UPS). Here, we review recent evidence showing deep mechanistic connections between the transcription and ubiquitin-proteasome systems. Our goal is to leave the reader with a sense that just about every step in transcription—from transcription initiation through to export of mRNA from the nucleus—is influenced by the UPS and that all major arms of the system—from the first step in ubiquitin (Ub) conjugation through to the proteasome—are recruited into transcriptional processes to provide regulation, directionality, and deconstructive power.
Keywords: chaperone, chromatin, coactivators, gene expression, proteolysis
INTRODUCTION
Eukaryotic gene transcription is a phenomenally complex process. For a gene to be transcribed, a large cohort of proteins need to descend upon DNA in a highly orchestrated manner, dealing not only with the physical constraints of transcribing a template that is encased in chromatin, but also ensuring that genes are turned on only at the right place, at the right time, and in response to the right signals. Cells employ a myriad of processes to ensure that these challenges are met appropriately; one of the most recently discovered of these processes is by engaging the proteolytic and nonproteolytic capabilities of the ubiquitin-proteasome system (UPS).
The aim of this review is discuss how ubiquitin (Ub), Ub-dependent transactions, and the proteasome impact eukaryotic transcription by RNA polymerase II (Pol II). This is an area of intense investigation, and one in which many surprising and counterintuitive discoveries have been made. Our review centers on the UPS—other reviews have dealt with roles of Ub-like modifications like SUMO (1)—and focuses on advances that have been made in this arena in the past 10 years (for reviews of earlier work see References 2–4). We highlight four major arms of intersection between the transcription and ubiquitin-proteasome systems, discuss the mechanisms that are at work, and present working models of how the UPS intervenes at key stages in transcriptional processes.
CHALLENGES OF EUKARYOTIC GENE TRANSCRIPTION
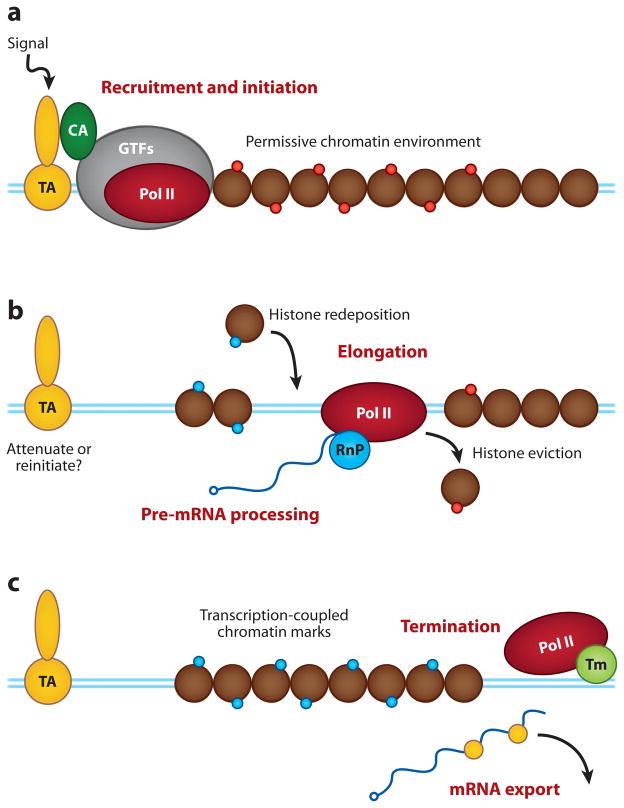
At its simplest level, mRNA transcription occurs when Pol II engages DNA upstream of the open reading frame of a gene and synthesizes a complementary strand of RNA that serves as a template for protein synthesis. But numerous elaborations on this scheme both present obstacles that must be overcome and provide critical regulatory opportunities (Figure 1). Unlike bacterial RNA polymerases, Pol II cannot initiate promoter-specific transcription but relies on interactions with a host of general transcription factors (GTFs) that engage polymerase at core promoter elements and facilitate both initiation and elongation of transcription (5). In vivo Pol II and the GTFs are recruited to chromatin by the action of transcriptional activators (TAs), proteins that possess a DNA-binding domain that tethers them to promoter DNA, and a transcriptional activation domain (TAD) that either directly or via coactivators brings GFTs and Pol II to sites of transcription. Transcriptional activators and coactivators (and conversely repressors and corepressors) are a major point of regulation of gene expression, and these proteins are heavily regulated by processes that control their abundance, localization, and activity.
Figure 1.
Transcriptional regulation. (a) Activation of transcription. Within a permissive chromatin environment, a transcriptional activator (TA) binds to promoter sequences, often in response to environmental signals. The activator typically requires a coactivator (CA), and together they recruit general transcription factors (GTFs) and RNA polymerase II (Pol II), leading to the initiation of transcription. (b) As Pol II enters the elongation phase, histones are evicted ahead of the transcribing Pol II and redeposited in its wake. At this stage, components of the RNA processing (RnP) machinery join Pol II, carrying out premessenger RNA maturation. This is also a stage when transcription can reinitiate, or the signal to initiate is terminated. (c) Transcription-coupled chromatin marks persist on the recently transcribed gene, and transcription terminates by association of Pol II with the termination machinery (Tm). mRNA is then exported from the nucleus. Blue lines represent the template DNA duplex.
The compaction of DNA into chromatin also has a major impact on transcription (6). Nucleosomes present a barrier to the binding of transcriptional regulators and are incompatible with passage of Pol II (7), meaning that histones must either be locally remodeled for transcription to occur or evicted from DNA ahead of the elongating Pol II complex (8). Importantly, because nucleosomes also act to conceal cryptic promoter elements present throughout the genome (9, 10), histones must be redeposited appropriately in the wake of Pol II to prevent transcription of nonauthentic RNA from occurring. The ability of nucleosomes to control template accessibility affords tremendous regulatory potential, and numerous chromatin modifying and remodeling factors (6, 11) act to not only control transcriptional processes but also to signal to the cell the transcriptional state of a given piece of chromatin.
Finally, it is worth emphasizing that each transcript is synthesized by a single Pol II complex and that events such as premessenger RNA processing and mRNA export are tightly coupled to transcription. The net effect of this arrangement is that Pol II has to continually change its entourage of interacting proteins: one set for initiation; another for elongation and RNA processing; and yet another for termination, 3′ processing, and mRNA export. Many of these events are coordinated by the so-called CTD code (12), which corresponds to distinct patterns of phosphorylation within the C-terminal domain of the largest subunit of Pol II and acts as a molecular beacon to broadcast specific points in the transcription process. The ability of transcriptional proteins to “sense” the activity of Pol II and other factors involved in transcription is a key mechanism through which order is brought to each of the stages in gene expression.
FLEXIBILITY AND UTILITY OF THE UBIQUITIN-PROTEASOME SYSTEM
The most recognizable role of the UPS is in protein turnover. Ub-mediated proteolysis is a highly specific process in which covalent linkage of substrates to the small protein molecule Ub signals their destruction by the 26S proteasome. Regulated protein destruction by the UPS is instrumental in processes ranging from cell cycle regulation through to antigen presentation, but it is important to remember that proteolysis is only one of many possible outcomes for a ubiquitylated protein. First and foremost, ubiquitylation is a posttranslational modification that provides a complex and nuanced way to alter the localization, activity, or stability of a linked protein substrate. Similarly, the complexities of the proteasome endow it with capabilities that extend well beyond simple protein breakdown. As we describe below, these noncanonical functions of the UPS feature prominently in the control of gene activity, and transcription makes full use of all of the capabilities of the UPS to insure appropriate transcriptional outcomes.
Protein Ubiquitylation
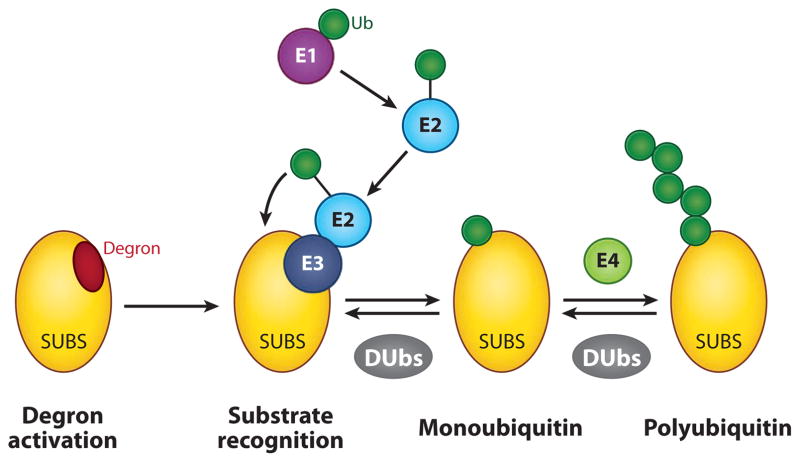
The conjugation of Ub is a multistep process (Figure 2) that begins when Ub forms a high-energy thioester linkage with a Ub-activating enzyme, also known as an E1. The “activated” Ub is then transferred to the active-site cysteine of a Ub-conjugating enzyme (E2), which functions in concert with a Ub-protein ligase (E3) to conjugate Ub to an amino group on the substrate, typically at a lysine residue. Specificity in this process is governed by the interaction between an E3 and its target site on the substrate, known as a degron. There are many hundreds of E3s in the mammalian genome, and degrons themselves are often activated by posttranslational modifications, providing a high level of selectivity and control to the process of ubiquitylation. Although numerous proteins are monoubiquitylated, most substrates undergo additional rounds of ubiquitylation on Ub (perhaps catalyzed by an additional class of enzymes known as E4s), leaving them in a heavily modified and polyubiquitylated state.
Figure 2.
Ubiquitylation. A destruction element in the substrate (a degron) is activated, often by phosphorylation in response to a specific signal. The degron is then bound by a ubiquitin (Ub)-protein ligase (E3), which acts in conjunction with a Ub-activating enzyme (E1) and Ub-conjugating enzyme (E2) to transfer Ub (green circle) to the substrate (SUBS), typically at a lysine residue. Repeated rounds of this process, perhaps catalyzed by a Ub chain elongation factor (E4), give rise to a polyubiquitylated substrate. Ubiquitylation can be opposed by the action of deubiquitylating enzymes (DUbs), which can remove all Ub or trim the length of the Ub chain.
A few characteristics of protein ubiquitylation are worth highlighting. First, Ub carries eight potential sites of ubiquitylation, meaning that Ub chains acquire unique characteristics not just via their length, but also by their topology (13). It is generally thought that proteins targeted for proteasomal destruction must carry at least a tetra-Ub chain in which Ub groups are linked via lysine 48 of Ub [K48-linkage (14)]; whereas other linkages, such as lysine 63, adopt distinct conformations that are recognized by Ub-binding receptor proteins for nonproteolytic outcomes (15). This diversity in chain recognition generates a range of signals far more diverse than might be expected from simple iteration of the Ub moiety itself. Second, ubiquitylation is a dynamic process that can be reversed by the action of deubiquitylating enzymes (DUbs) that either trim Ub chains or remove them entirely (16). The dynamic nature of ubiquitylation means that Ub conjugation is not a terminal process and that DUbs can act to modify the fate of a ubiquitylated substrate. Finally, ubiquitylation is unusual in that it is a very large posttranslational modification, and it is likely that the bulkiness of Ub can itself serve to alter the inherent characteristics of a ubiquitylated substrate protein.
The Proteasome
The proteasome is a self-compartmentalized protease (17) that carries its proteolytic activities deep within its interior. This arrangement means that for a protein to be destroyed by the proteasome, it not only typically needs to be ubiquitylated, but also presented in an appropriate manner to gain access to the central proteolytic chamber. Delivery of substrates to the proteasome is often facilitated by Ub-dependent chaperones and shuttling factors (18, 19), but once a substrate arrives on its doorstep, much of the actions of the proteasome are devoted to feeding substrates to the inner protease sites.
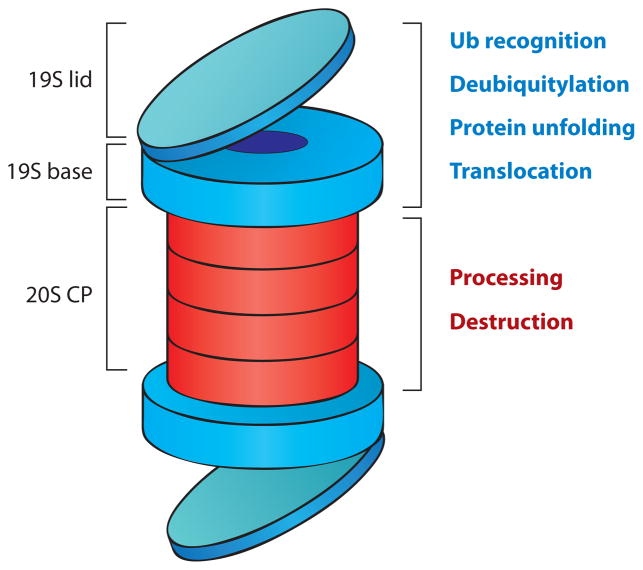
The 26S proteasome (Figure 3) consists of a 20S proteolytic core particle, capped at one or both ends by a 19S regulatory particle, the latter of which can be further subdivided into lid and base subcomplexes (20). The lid of the 19S regulatory particle is composed of ~10 subunits, whereas the base is composed of six AAA-type ATPases and two non-ATPase subunits. Ubiquitylated proteins delivered to the proteasome are recognized by receptors in the 19S complex, deubiquitylated, unfolded by ATPases in the base, and translocated into the 20S complex for destruction. Within the 20S complex, three distinct proteoltyic activities—chymotryptic-, tryptic-, and caspase-like—attack the target, typically cleaving it into peptides of between 4–25 amino acids in length (17), but occasionally, these activities clip the substrate at specific sites, generating functional protein fragments (21). The proteasome thus carries at least six distinct biochemical activities: (a) recognition of ubiquitylated proteins, (b) deubiquitylation, (c) protein unfolding/chaperone, (d) protein translocation, (e) protein destruction, and (f) protein processing, which are typically devoted to proteolysis but could theoretically be diverted to other processes as well. As we discuss below, the potential moonlighting of these functions has received particular attention in the field of transcriptional regulation.
Figure 3.
Architecture of the 26S proteasome. The proteasome is a self-compartmentalized protease that consists of three main subcomplexes: a 20S core particle (CP) that houses three distinct proteolytic activities in its inner core, a 19S lid that recognizes and deubiquitylates substrates, and a 19S base structure that uses the energy of ATP hydrolysis to unfold proteins and pass them into the 20S CP for destruction. 26S proteasomes can be capped at one or both ends by a 19S complex.
REGULATING TRANSCRIPTIONAL ACTIVATORS AND COACTIVATORS
Given their importance in setting gene expression levels, it is not surprising that TAs and coactivators are a major point of intervention of the UPS in transcriptional control. The most obvious way in which the UPS controls these proteins is by fine-tuning their steady-state levels or by destroying them when their function is no longer appropriate (2), but examples also exist in which the UPS controls the localization of activators (22) or processes them into a functional state via limited proteolysis (23). These actions are in line with traditional views of how the UPS regulates protein activity, acting either upstream or downstream of the point at which its substrates function. In transcription, however, the UPS has inserted itself in a prominent way directly into the heart of gene regulatory mechanisms, controlling activators and coactivators on chromatin and during the process of transcriptional regulation. The ability of the UPS to modulate activators at their workplace endows the system with tremendous potential to micromanage the proteins that control gene activity.
Nonproteolytic Functions of Ubiquitin in Controlling Transcriptional Activators
Many TAs and coactivators can be regulated by nonproteolytic ubiquitylation. A consensus has yet to emerge on the effects of ubiquitylation on transcription factor activity, but on the basis of published examples, we can define two general ways in which Ub acts, independent of proteolysis, to influence activator and coactivator function.
The first group is the least mechanistically developed and encompasses examples where monoubiquitylation stimulates TA activity through unknown means (Figure 4a). A collection of natural (24–27) and synthetic (28, 29) transcription factors are regulated by this mechanism, and although exactly how is unclear, the phenomenon does appear to be both biologically relevant and a point of regulation. In the case of FOXO4, for example, a transcription factor important for the cellular response to stress, oxidative stress induces both monoubiquitylation and deubiquitylation of the activator. Monoubiquitylation of FOXO4 causes it to enter the nucleus and stimulates its transcriptional activity, whereas deubiquitylation by the DUb Usp7 reverses this process. By having both ubiquitylation and deubiquitylation under the control of oxidative stress, cells inherently limit FOXO4 activity, ensuring that ongoing rounds of FOXO4-driven transcription are responsive to the continual presence of the activating signal. Another interesting example centers on SRC-3, a coactivator for hormone-liganded transcription factors (Figure 4b). SRC-3 is activated by monoubiquitylation (30) but destroyed by the proteasome once the Ub chain on SRC-3 is extended beyond a certain threshold. The time it takes for SRC-3 to transition from a monoubiquitylated to a polyubiquitylated species thus acts as a “molecular clock,” defining a specific period of time during which SRC-3 can function before it is destroyed. In this way, ubiquitylation acts as a self-limiting mechanism that prevents renegade transcription factors from uncontrollably activating transcription. In the section titled Proteolytic Control of Activators and Coactivators, we discuss an even more intimate coupling of activator turnover and function that likely achieves the same objective.
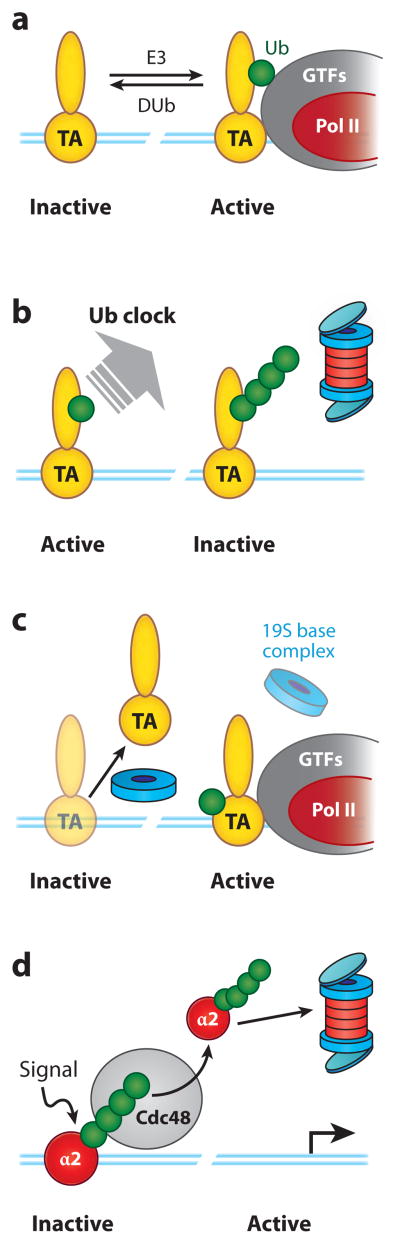
Figure 4.
Roles for ubiquitylation in controlling transcriptional regulators on chromatin.
(a) Activation by monoubiquitylation. In this view, monoubiquitylation of DNA-bound activators stimulates their inherent activation properties, a process that is promoted by Ub ligases (E3s) and antagonized by deubiquitylating enzymes (DUbs). (b) The Ub clock model. In this view, the period of activity of a transcriptional regulator is governed by the length of time it takes to transition from the monoubiquitylated and active form to a polyubiquitylated form that is rapidly destroyed by the proteasome. (c) Control of promoter stripping by activator ubiquitylation. In this model, activators are aggressively removed from chromatin by resident ATPases in the 19S base complex (blue ring). The presence of Ub on the activator blocks this activity, allowing the TA to stably associate with its cognate DNA element. (d) Extraction of ubiquitylated transcription factors from chromatin by Ub-selective chaperones. Here, polyubiquitylation of the α2 repressor causes it to be extracted from promoter DNA by the ATPase activities of Cdc48 (p97), a process that allows immediate cessation of function before α2 is subsequently destroyed by the proteasome. Note that these models are not mutually exclusive, and it is possible that multiple mechanisms contribute to transcription factor control at a specific promoter. Abbreviations: GTFs, general transcription factors; Pol II, RNA polymerase II; TA, transcriptional activator. Blue lines represent the template DNA duplex.
The second group includes a growing number of examples in which ubiquitylation affects how a transcription factor binds to its target sites in the genome, both positively and negatively. At one end of the spectrum, monoubiquitylation of the yeast activator Gal4 appears to prevent it from being stripped off chromatin by ATPases resident in the 19S base complex (Figure 4c) (31), suggesting that ubiquitylation could act to lock an activator onto chromatin in an active configuration. At the other extreme, ubiquitylation can also play a proactive role in dissociating transcriptional regulators from their target sites (Figure 4d) (32, 33). The best-understood example in this group is the yeast transcriptional repressor α2, which controls the transition between different mating types. Although Ub-mediated proteolysis of α2 is important for mating-type switching (34), the critical point of regulation in this instance occurs at the level of α2 binding to chromatin and precedes proteolysis. When the signal to switch mating types is received, α2 is ubiquitylated on chromatin, and this ubiquitylation recruits the Ub-selective chaperone Cdc48 (p97), a AAA-type ATPase that mobilizes ubiquitylated proteins for destruction by the proteasome (19). Once Cdc48 encounters ubiquitylated α2, it uses the energy of ATP hydrolysis to extract α2 from chromatin, immediately terminating its function and subsequently directing it to the proteasome for permanent inhibition of α2 activity. The α2 study is particularly important because it demonstrates that steps in the linear sequence of Ub-mediated proteolysis may be both temporally and spatially separated to effect immediate versus long-term outcomes and that just because α2 is destroyed by the UPS does not mean that proteolysis per se is the rate-limiting step.
Two final points are worth noting here. First, it is entirely possible that some of the first class of activators, where the mechanism of activation by ubiquitylation is unknown, are regulated by Ub at the level of promoter occupancy. Second, the principal that ubiquity-lation can control transcription factor activity separate from proteolysis may not be restricted to monoubiquitylation, nor to TAs. Atypical poly-Ub chain topologies, which do not lead to proteasomal proteolysis, have been found to stimulate transcription factor activity under some circumstances (35), and the activity of the repressive histone deacetylase complex mSin3 is stimulated by formation of lysine 63-linked Ub chains on its Sds3 subunit (36). Thus, the rules that are distilled from transcriptional regulators may be broadly applicable to other components of the transcriptional apparatus, and similar mechanisms may be at work.
Proteolytic Control of Activators and Coactivators
The concept that transcriptional regulators can be made to accumulate or disappear by manipulating their Ub-mediated proteolysis is well established (2, 3). There is, however, another more intriguing side to this story: Ub-mediated proteolysis can promote the activity of the transcriptional regulators it destroys. This counterintuitive role of the UPS in controlling transcription has only been appreciated in recent years, but this role appears to apply to a growing number of transcriptional regulators and provides insight into how other processes may be controlled by an “activation by destruction” mechanism (37).
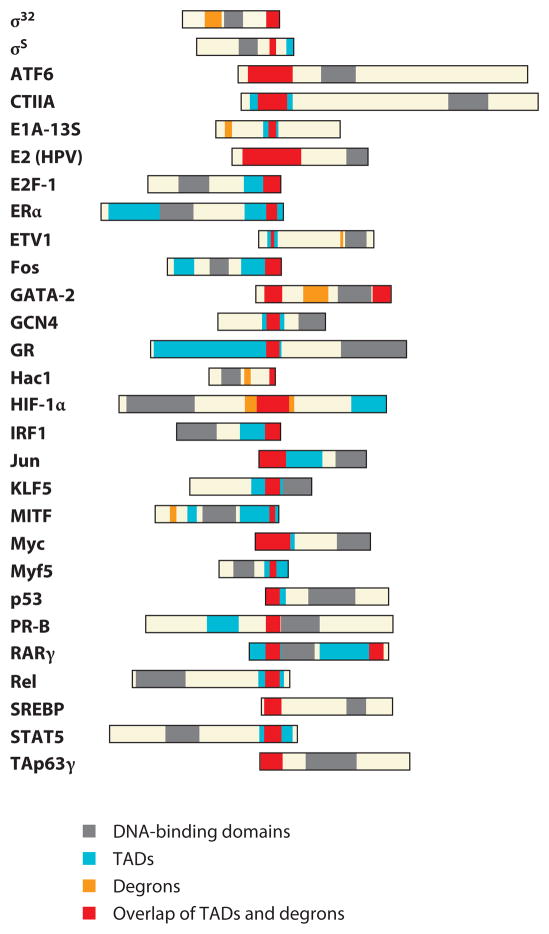
The first clue to the existence of this phenomenon came from the discovery that TADs often overlap with degrons (38). This overlap is widespread—there are now nearly 30 transcription factors that have overlapping TADs and degrons (Figure 5)—and is intimate, as manipulations that affect protein turnover typically have corresponding effects on TAD activity [e.g., (28, 37, 39, 40)]. The coupling of these two functions within TAs [and coactivators (41)] raises an intriguing question: Why are two apparently opposing functions—activation and destruction—linked in these proteins?
Figure 5.
Transcriptional activation domains (TADs) often overlap with degrons. The domain structure of 28 transcription factors is shown. Although the overlap of TADs and degrons is not always perfect, it is worth remembering that different studies have typically defined each type of element and that different sets of mutations (which define the boundaries) have been used in these studies.
Since this overlap was first noted, significant progress has been made in both expanding the range of transcription factors that fall into this category and exposing its significance. Although deep mechanistic insight is still lacking, a few important conclusions can be made. Destruction of activators is likely to be a direct consequence of their ability to activate transcription (39, 42). Activator turnover requires DNA binding and ongoing transcriptional activity (43), and this turnover is often signaled by kinases that are integral parts of the transcriptional apparatus (44–46). Ub ligases are recruited to sites of activator function (2, 44, 46–50). And the ongoing activity of these proteins is dependent on their ubiquitylation and on the proteolytic capacity of the proteasome (2, 3, 37, 39, 41, 44, 51–58). Taken together with the widespread overlap of activation and destruction elements, these observations indicate that Ub-mediated destruction of activators is inherently linked to the way in which they stimulate transcription.
A number of models have been proposed to explain the connection between activator destruction and function (2, 3, 59); the majority of these models invoke a “suicide” theme in which destruction of the activator is an obligate part of the activation mechanism that both drives the process forward and is guaranteed to terminate the activation signal. This notion is closely aligned with the Ub clock hypothesis, and it is possible that a similar mechanism underlies both processes, the only difference being the number of rounds of transcription an activator can stimulate before it is terminated. There are, however, a number of issues that need to be noted in any critical evaluation of a suicide-type model. First, the mechanism must apply only to a certain mode of transcriptional activation, as there are TADs that stimulate robust levels of transcription without triggering proteolysis (38). Second, the mechanism must apply only in certain instances, as there are clear examples of individual transcription factors that can be both activated and inhibited by Ub-mediated proteolysis (44). Third, there must be a way for the UPS to attack activators only after they have functioned; otherwise, proteolysis could precede activation and would obligatorily inhibit TA activity. Finally, transcription-coupled destruction of an activator needs to serve a functional purpose such that, if it is blocked, repeated rounds of transcriptional activation cannot occur.
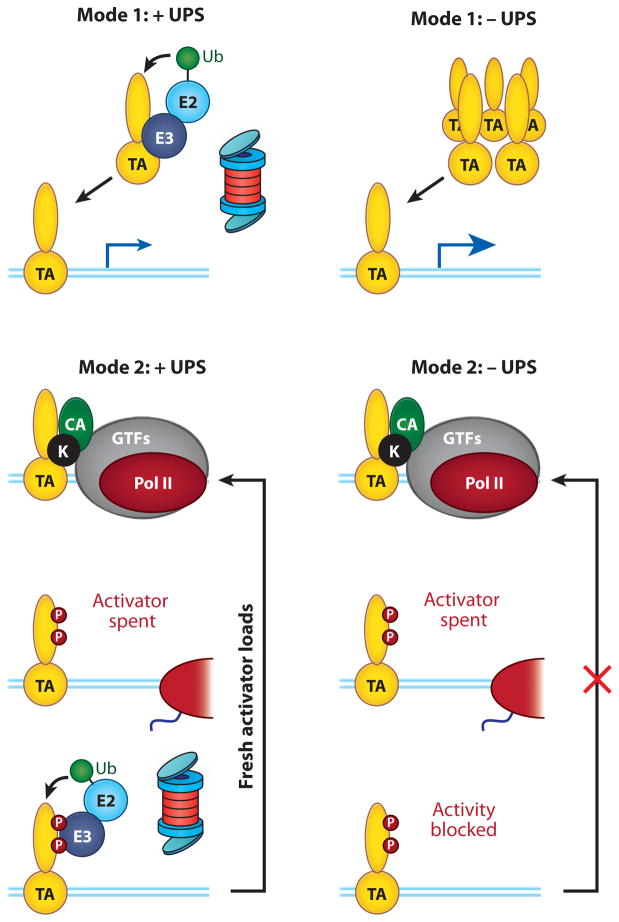
Taking these issues into account, we suggest a revised model (Figure 6) in which TAs are regulated by two distinct modes of Ub-mediated proteolysis. Mode 1 occurs off chromatin, and in this case, the UPS limits the activity of a transcription factor by restricting its abundance. If proteolysis does not occur, the TA accumulates, and its function is enhanced. Mode 2, however, occurs on chromatin and during the process of gene induction. We posit that kinases associated with the transcriptional machinery (44–46) serve to mark an activator as “spent,” trapping it in an inactive state that cannot stimulate further rounds of transcription. At the same time, these phosphorylation events bring in components of the UPS that destroy the activator in situ, clearing the deck for promoter association with a fresh activator molecule. In this mode, proteolysis serves a positive role in transcription by allowing pristine activators to access the promoter and stimulate additional rounds of transcription. This model, which is a refinement of one originally proposed by Lipford et al. (3, 37), predicts that the UPS is only required when the activator is locked in its phosphorylated state. Thus, if the mode of activation does not lead to activator phosphorylation, or if the sites of phosphorylation on the TA are blocked (37), the UPS becomes dispensable for activator function.
Figure 6.
Two modes of activator regulation by ubiquitin-mediated proteolysis. The figure shows how a transcription factor can be regulated in disparate ways by proteolysis and the consequences of disrupting the ubiquitin-proteasome system (UPS) on those modes of regulation. Mode 1 occurs off chromatin, and the UPS is used to limit the concentration of available activators. When the UPS is disrupted in this case, the transcription activator (TA) accumulates, and transcription is induced. Mode 2 occurs on promoter DNA and during the course of transcriptional activation. We posit that kinases (K) associated with the general transcriptional machinery phosphorylate activators at some point during transcriptional activation. This phosphorylation (P, red circles) has two functions. Its marks the activator as spent and incapable of stimulating further rounds of transcription. Concurrently, it also recruits a Ub ligase that ubiquitylates the transcription activator (TA), allowing it to be destroyed and a fresh activator to reach the promoter. In this mode, if the UPS is perturbed, the inactive TA remains on chromatin, and subsequent rounds of transcription are blocked. Abbreviations: CA, coactivator; E2, ubiquitin-conjugating enzyme; E3, ubiquitin-protein ligase. Blue lines represent the template DNA duplex.
Linking the destruction of TAs directly to their activity installs a set of mandatory checkpoints to the function of any particular transcription factor molecule and provides a powerful way for the cell to tightly control transcription factor activity. Our model gives mechanistic insight into how this may occur, but it also raises some thought-provoking questions. Does this type of regulation apply to activators only, or are coactivators and repressors also subject to this type of control? What are the steps in transcription that depend on activator phosphorylation and proteolysis, and are other transcription proteins, besides coactivators, also destroyed in the process? And—more broadly—does this type of inherently self-limiting mechanism apply to other cellular events in which the UPS has a regulatory foothold?
REGULATION OF HISTONES BY THE UBIQUITIN-PROTEASOME SYSTEM
One of the most high-profile intersections between the transcription and ubiquitin-proteasome systems centers on ubiquitylation of histones and the impact this has on chromatin structure and function. It is now clear that all histones can be ubiquitylated (60), that this modification is abundant [almost 10% of H2B, for example, is present in the ubiquitylated form (61)], and that histone ubiquitylation acts nonproteolytically to control gene activity. The best-studied examples of histone ubiquitylation are H2A and H2B. H2A ubiquitylation is typically associated with chromatin compaction and transcriptional repression, whereas H2B ubiquitylation is associated with gene activation (60). Numerous excellent reviews have been written on the subject of histone modifications, so rather than cover this subject comprehensively here, we focus on just one example, H2B, that illustrates some of the ways in which the presence of Ub on a core histone can influence gene activities (Figure 7).
Figure 7.
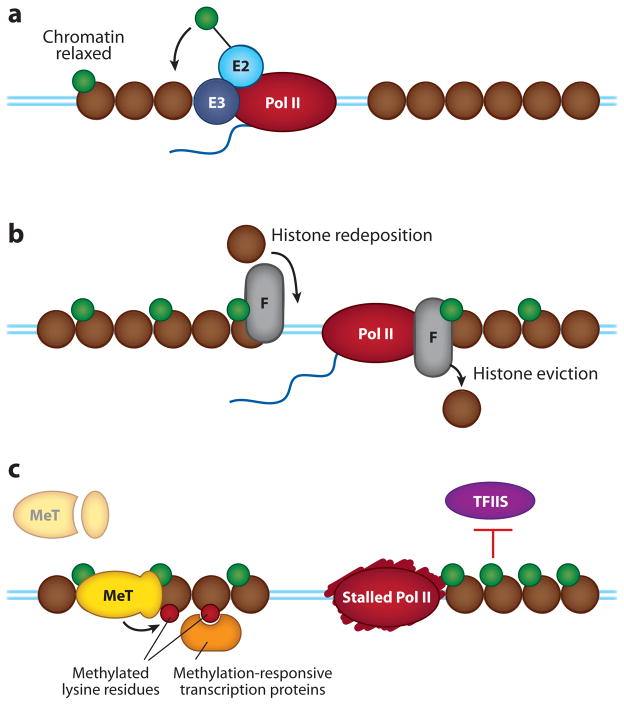
Functions of H2B ubiquitylation. (a) During transcriptional elongation, the ubiquitylation machinery for H2B (Rad6/Bre1) is recruited to transcriptional complexes, where it ubiquitylates H2B, most prominently on lysine 120. This modification does not impact nucleosome structure, but it does relax higher-order chromatin configurations. (b) As transcription proceeds, ubiquitylation of H2B selectively recruits the FACT elongation complex (F), which not only displaces H2A/H2B dimers ahead of RNA polymerase II (Pol II), but also insures that histones are redeposited afterward. (c) H2B ubiquitylation recruits and activates Dot1- and Set1-containing histone methyltransferase complexes (MeT), which promote the di- and trimethylation of histone H3 at lysine residues 4 and 79. These methylation events, in turn, recruit other factors to chromatin and repel binding of transcriptional silencing complexes (not shown). If levels of ubiquitylated H2B accumulate, elongation factors such as TFIIS are excluded from chromatin, preventing the rescue of stalled polymerase molecules. Abbreviations: E2, ubiquitin-conjugating enzyme; E3, ubiquitin-protein ligase. Blue lines represent the DNA duplex. Brown circles represent nucleosomes. Green circles represent ubiquitylated histone H2B.
Although H2B can be ubiquitylated at multiple sites (62), the major modification of H2B is monoubiquitylation at lysine 120, a process carried out by the Rad6 Ub-conjugating enzyme (63) and the Bre1 Ub-protein ligase (64). This modification is tightly coupled to active chromatin (65), is likely signaled by combinatorial processes that are linked to transcriptional elongation (66–68), and depends on transcription-coupled alterations to chromatin structure (66, 69). Thus, ubiquitylation of H2B marks the template as having recently been transcribed and sets forth a process that impacts transcriptional events. But what does H2B ubiquitylation do?
Inherently, modification of H2B by Ub does not disrupt nucleosome architecture (70), but does impair chromatin fiber compaction, and can work with other histone modifications to relax higher-order chromatin structure (71). This phenomenon is consistent with the bulky nature of the Ub moiety, which presumably prevents tight compaction of chromatinized DNA strands, and has obvious ramifications for influencing the ability of Pol II to access its template. In vivo, however, H2B ubiquitylation is unlikely to act solely through this mechanism. The effects of H2B ubiquitylation cannot be recapitulated by replacing Ub with another bulky Ub-like modification, such as SUMO (72) or HUb1 (71), suggesting that components in the transcriptional machinery recognize the Ub modification on H2B to elicit a biological response.
Ubiquitylation of H2B is closely connected to the function of the FACT nucleosome-remodeling complex, which is important for displacing H2A/H2B dimers ahead of Pol II and for reassembling them after polymerase has passed (73). H2B ubiquitylation stimulates the activity of FACT (74) and appears to negatively act against other nucleosome-restoring complexes, making FACT the preferred nucleosome rebuilder at transcribed genes where H2B ubiquitylation occurs (75). The ability of ubiquitylated H2B to select for FACT implies that a specific molecular recognition event, signaled by Ub on H2B, acts to stimulate FACT activity at specific sites in the genome. In a similar vein, recognition of Ub by histone methyltransferase complexes (76–78) is likely involved in another essential function of H2B ubiquitylation—signaling processive methylation of histone H3 (79). In this instance, the presence of H2B ubiquitylation at active sites of transcription allows localized assembly and activation of methyltransferase complexes (80–83), which then go on to modify their respective sites in H3. The linking in trans of these two sets of histone modifications may seem like a lot of effort, but it provides the cell with a way to amplify and extend the actions of H2B ubiquitylation by tying it into a host of processes that respond to H3 methylation (84–87).
Finally, like many ways in which the UPS controls transcription, H2B ubiquitylation is a dynamic process that acts both positively and negatively, and the timing and extent of this modification determines the biological outcome. Removal of Ub from H2B, which is mediated by a DUb that is an integral part of the SAGA coactivator complex, is important for optimal levels of transcription (88). If ubiquitylated H2B accumulates, Pol II cannot recruit kinases important for transcriptional elongation (89), and stalled Pol II complexes cannot be reactivated by factors such as TFIIS (90). It seems likely, therefore, that H2B ubiquitylation is not a static event but that H2B is cycling between its modified and unmodified states, giving the cell a set of constant updates on transcriptional processes and providing critical opportunities for regulatory intervention.
CONNECTIONS BETWEEN THE TRANSCRIPTION AND UBIQUITIN-PROTEASOME SYSTEMS
Given the growth of this field in recent years, we cannot hope to cover all of the various examples that have surfaced of how Ub-dependent processes impact transcription. We can, however, make two important points that illustrate the extent to which the UPS regulates gene activity and the physical connections between the two processes.
The first point is that activators, coactivators, and histones are not the sole venue of intervention of the UPS in transcriptional processes. Termination of transcription depends on proteasome function (91). Core components of the transcriptional machinery are regulated by Ub-dependent processes (36, 47). And events that occur commensurate with transcription, such as premRNA splicing and mRNA export, are also impacted by Ub and the proteasome (92, 93). Indeed, given that Ub-dependent processes are also important for translation (94), it seems that the UPS has inserted itself into just about every stage in the expression of the genetic information.
The second point is that the extensive functional links between the transcription and ubiquitin-proteasome systems are now supported by a host of physical interactions between components in the two pathways (Table 1). Molecules that directly connect the transcription and ubiquitin-proteasome systems influence processes ranging from gene repression through to chromatin modifications and to elongation of transcription. The understanding that has come from studying these molecules gives important insight into how the UPS is such an efficient regulator of transcriptional processes.
Table 1.
Proteins that physically link the transcription and ubiquitin-proteasome systems
| Component | Linkage between the transcription and ubiquitin-proteasome systems | Reference |
|---|---|---|
| Asr1 | Ub ligase that binds directly to the C-terminal domain of RNA polymerase II in response to S5 phosphorylation | 98 |
| Atf1 | Sequence-specific DNA-binding transcription factor that activates the anaphase-promoting complex (APC) Ub ligase complex | 100 |
| BAF250 | Component of the SWI/SNF-A chromatin-remodeling complex that associates with elongin C to form an E3 that ubiquitylates histone H2B | 103 |
| CBP/p300 | Transcriptional coactivator and coactivator of the APC Ub ligase complex | 101 |
| CCR4–Not | Transcriptional repressor complex containing an E3 | 143 |
| CSN | COP9 signalosome; prevalent Ub ligase complex and transcriptional regulator | 144 |
| Elongin B,C | Ub ligase complex that associates with RNA polymerase II (Pol II); isolated as a factor that stimulates transcriptional elongation | 145 |
| GCN5 | Histone acetyltransferase and Ub ligase cofactor | 146 |
| Med8 | Component of the Pol II-associated mediator complex; forms a Ub ligase complex with elongins B,C | 145 |
| Met4 | Transcriptional activator that acts as a substrate-specificity factor for the SCFMet30 Ub ligase | 104 |
| Rpn4 | Substrate, ligand, and transcriptional regulator of the proteasome | 147 |
| Ssl1 | Ub ligase and core component of the TFIIH general transcription factor | 96 |
| TAFI | Part of the basal transcription factor TFIID, carries on both E1 and E2 activities; ubiquitylates histones and activators | 95, 148 |
| TBL1/TBLR1 | Ubiquitin ligases that are part of the N-CoR corepressor complex | 52 |
| Tfb3 | RING-finger component of TFIIH, stimulates Ub-like modification on the Cullin-type Ub ligase | 97 |
| TIF1 γ | Ub ligase that is activated by binding to modified histone tails; destroys Smad4 on chromatin | 102 |
| Tom1 | Ub ligase associated with the SAGA chromatin-remodeling complex; ubiquitylates the mRNA export factor Yra1 | 149, 150 |
| Ubp8, SGf11, Sus1, Sgf73 | Deubiquitylating enzyme module of the SAGA complex; deubiquitylates histone H2B | 88 |
| Uch37 | Deubiquitylating enzyme that is an integral part of the Ino80 chromatin remodeler; activated by the proteasome | 151 |
Notably, many proteins that connect the transcription and ubiquitin-proteasome systems are integral components of the transcriptional apparatus. TAFI, for example, is a core component of the TFIID complex and is unusual in that it has combined E1 and E2 activities (95), allowing it to single-handedly activate Ub and conjugate it to a substrate. The basal factor TFIIH has two potential ways it can influence ubiquitylation, acting as a direct Ub ligase (96) and by modulating the activity of other ligases that are regulated by the Ub-like modification Nedd8 (97). And as mentioned above, the SAGA chromatin-remodeling complex has a built in DUb (88) that deubiquitylates H2B, and potentially other substrates. The setting of Ub-conjugating and deconjugating enzymes within components of the transcriptional apparatus gives the UPS extraordinary opportunities to influence gene expression mechanisms.
One of the more interesting features to emerge from the study of proteins that connect these systems is that many of them have evolved the ability to sense the activity or environment of molecules involved in gene regulation. In this regard, three general mechanisms have emerged to explain how the UPS zeros in on its active transcriptional targets. First, the UPS can read activity-dependent modifications or configurations of transcription proteins (Figure 8a). This concept is at the heart of our model of how Ub-mediated proteolysis controls TAs (Figure 6), but examples also exist in which an E3 can directly interpret the CTD code on Pol II (98) or in which the CTD is read together with other structural changes in polymerase that only occur when the molecule is active (99). Second, components of the UPS can be activated specifically within a transcriptional context (Figure 8b) (100, 101). One of the best examples of this is the Ub ligase TIF1γ, which is activated by modified histone tails (102), allowing it to select only its chromatin-bound pool of substrate (Smad4) for ubiquitylation. Finally, Ub ligase selectivity can be directly reprogrammed by transcription proteins to target new substrates (Figure 8c) (103, 104). In one surprising example, the transcription factor Met4 inserts itself within a Ub ligase complex (104), changing E3 selectivity and allowing Met4 to direct the proteolysis of its transcriptional coregulators. By these three mechanisms, the activity of the UPS can be co-opted and tuned with laser-like precision toward transcriptional processes. The close interplay between molecules important for transcription and Ub-dependent transactions, and the ability of transcription proteins to inherently alter the function of proteins such as Ub ligases, makes the UPS an effective tool for controlling transcriptional processes.
Figure 8.
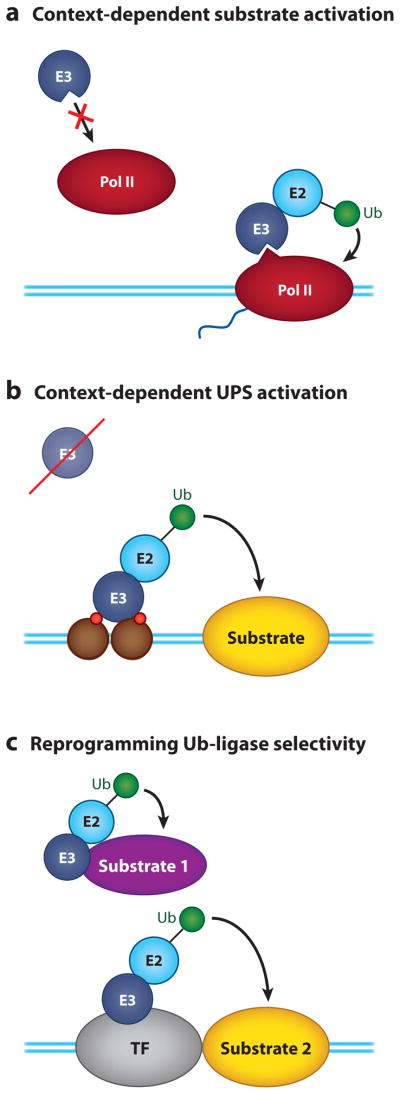
Ways in which the ubiquitin-proteasome system (UPS) can sense the activity of the transcriptional machinery. (a) By recognizing the specific context of the substrate. This example shows how the ubiquitylation machinery detects RNA polymerase II (Pol II) only when it is engaged in transcription, via a combination of phosphorylation events and structural changes in polymerase that are induced by transcription. (b) By context-specific activation of the ubiquitylation machinery. In this example, the ubiquitin-protein ligase, E3, is not active off chromatin, but it is activated by histone tails, insuring that it ubiquitylates its target only within a transcription setting. (c) By reprogramming Ub ligase selectivity. Here, a transcription factor (TF) serves as a substrate adapter for the ubiquitylation machinery, reprogramming E3 specificity from substrate 1 to substrate 2 and allowing selective modification of the second substrate in a transcriptional context. Abbreviation: E2, ubiquitin-conjugating enzyme. Blue lines represent the DNA duplex. Green circles represent ubiquitin. Brown circles represent nucleosomes. Red circles represent covalent histone modifications.
THE PROTEASOME IN TRANSCRIPTION
Implicit in much of our discussion is the concept that the proteasome is available to process ubiquitylated substrates within the immediate vicinity of chromatin, either directly or via chaperones, such as Cdc48. This indeed appears to be the case. Proteasome subunits are clearly recruited to sites of transcription (105–112), and genome-wide studies suggest that one or more components of the proteasome interact with the majority of highly active genes in yeast (109), raising the intriguing possibility that few Pol II–transcribed genes, and perhaps those transcribed by RNA polymerase I (113), are expressed without proteasomes being present. Studies have implicated the proteasome in just about every step in gene transcription (Table 2), from control of activators and how they associate with chromatin through to regulated exchange of coactivators (52, 114), elongation (115, 116), termination (91), covalent histone modifications (114, 117–120), and repressing cryptic transcription (111, 121). However, the nature of the proteasome that is involved in gene regulation and the exact contributions of 19S versus 20S activities are unclear.
Table 2.
Transcriptionally relevant activities of the proteasome
| Component of the proteasome | Activity | Reference |
|---|---|---|
| 19S | Regulates transcriptional activator loading onto chromatin | 152 |
| Strips activators off chromatin | 31, 153 | |
| Promotes activator/coactivator association | 106, 114 | |
| Promotes transcriptional elongation | 115, 116 | |
| Regulates histone modifications | 107, 114, 117–120 | |
| 26S | Promotes dynamic association of activators with chromatin | 154–157 |
| Activity-coupled destruction of transcription factors | 37, 44, 47, 54, 55, 57 | |
| Promotes coactivator exchange | 52, 158 | |
| Terminates transcription appropriately | 91 | |
| Represses cryptic transcription | 111, 121 | |
| Resolves permanently stalled RNA polymerase II complexes at sites of DNA damage | 99, 129 |
One school of thought posits that the transcriptionally relevant form of the proteasome is an independent 19S base complex (122), which uses the energy of ATP hydrolysis to regulate dynamic exchanges of key proteins at sites of transcription. Support for this notion comes from genetic evidence linking 19S base components—but not those in 20S—to activities of the yeast Gal4 activator (123–126), from the finding that TADs and GTFs can selectively interact with 19S proteins in vitro (117, 127, 128), and from some notable discrepancies in the distribution of 19S and 20S components on chromatin, as measured by chromatin immunoprecipitation (105, 106, 108, 109, 111). Importantly, biochemical studies have clearly indicated that 19S complexes are sufficient for promoting events such as transcriptional elongation (116), and stimulating coactivator binding (114), lending further support to the notion that ATPases in the 19S base are all that is needed for transcriptionally relevant processes.
By contrast, an equally compelling case can be made for the role of 20S proteins in critical transcriptional events. 20S components bind active genes (91, 109, 111, 112); are required for activator and coactivator function (described above), as well as for transcriptional termination (91) and repression of cryptic transcription (111, 121); and are recruited to chromatin in response to DNA damage (129), where they facilitate removal of Pol II complexes that have terminally stalled at DNA lesions. It appears, therefore, that the proteolytic activity of the proteasome is required for normal transcriptional processes and also to cope with problems that arise during expression of the genetic information.
How can we reconcile these apparently disparate observations? It needs to be stated that evidence for the existence of free 19S base complexes is lacking and is confounded by the inherent instability of proteasome complexes during extraction (130). Most in vivo evidence supporting a 20S-free role of proteasome components in transcription is based on chromatin immunoprecipitation studies, which are susceptible to issues of epitope accessibility, and on studies using pharmacological inhibitors of the chymotryptic site of the proteasome, which can leave other proteoltyic activities intact (57). It should also be stressed that there is no conceptual need to physically segregate the proteasome for its nonproteolytic activities to act; protein unfolding occurs on the surface of the 19S complex (131), and 26S proteasomes can disrupt protein complexes independent of proteolysis (132). Depending on their characteristics, some proteins are inexorably shuttled into the proteolytic chamber, whereas others escape degradation (133). Thus, canonical 26S proteasomes certainly have the capability to act both proteolytically and nonproteolytically during the transcription process.
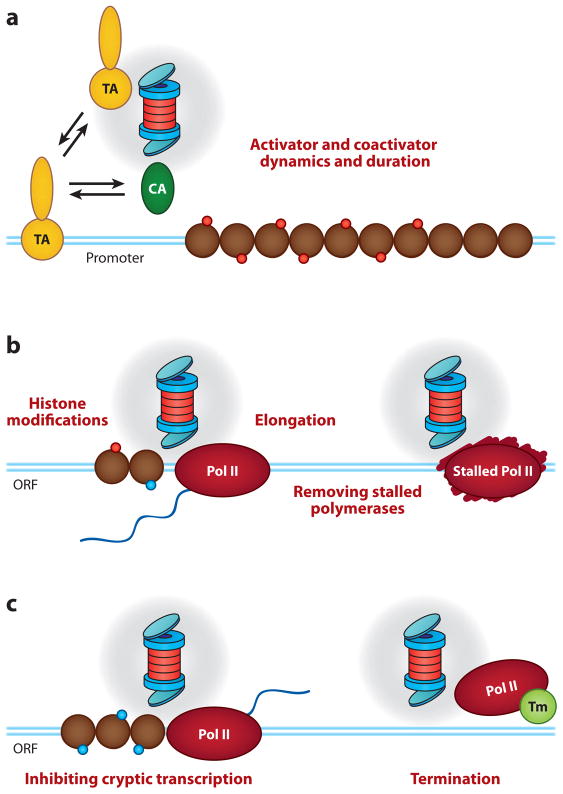
We propose that transcriptionally active genes recruit the entire proteasome and use it as a kind of “Swiss army knife” that carries an integrated set of biological functions (Figure 9). During the early stages of transcription, ATPases in 19S complex regulate activator occupancy (134) and coactivator recruitment (52, 114), and the entire proteasome is poised to destroy the activator once its suicide mechanism is activated. After Pol II has initiated transcription, proteasomes enter the transcribed portion of the gene and again put their proteolytic and nonproteolytic activities to work, remodeling the chromatin template to facilitate transcriptional elongation (115, 116) and linked histone modifications (107, 114, 117–120), as well as serving to remove failed transcriptional complexes or those that have initiated at inappropriate sites in the genome (109, 111, 135). And as transcription nears completion, the proteasome uses its activities to drive changes in the composition of Pol II complexes necessary for appropriate termination of transcription (91). Thus, just as one might carry a Swiss army knife—both for routine tasks and to cope with emergencies—transcription complexes carry proteasomes and sample their various activities depending on the stage or challenge to transcription.
Figure 9.
The Swiss army knife model of proteasome function in transcription. This model posits that the entire 26S proteasome is recruited into transcriptional processes, and its various activities are used depending on the molecular requirements. (a) The proteasome uses a combination of proteolytic and nonproteolytic functions to control activator binding, residency time, and coactivator (CA) exchange on promoter DNAs. (b) We posit that proteasomes enter the open reading frame (ORF) of transcribed genes, where their ATPase activities stimulate elongation, while their proteolytic functions serve to remove terminally stalled RNA polymerase II (Pol II) complexes that may arise. (c) Proteolytic activity of the proteasome is also important for attenuating cryptic, nonauthentic, and transcription complexes as well as for processes required for appropriate termination of the transcription event. Abbreviation: Tm, termination machinery. Blue lines represent the DNA duplex. Brown circles represent nucleosomes. Red circles represent covalent histone modifications present before transcription. Cyan circles represent covalent histone modifications present after transcription. The spool is the 26S proteasome (as in Figure 3).
Notably, our model does not exclude the possibility that individual functions of the proteasome may be transiently inhibited or that proteasomes may be fragmented to achieve gene-specific transcriptional outcomes. For example, the human immunodeficiency virus 1 (HIV-1) promoter recruits both 19S and 20S components when in the process of synthesizing short, nonproductive transcripts but, in response to the activator Tat, recruits a proteasome disassembly protein, PAAF1, that displaces 20S proteins, allowing the 19S functions to predominate in synthesis of long, productive mRNAs (136). These results lend support to the notion that proteasomes are recruited en masse into transcriptional processes, but also demonstrate that proteasome subcomplexes can be tailored to meet specific transcription requirements on select promoter DNAs.
How proteasomes are recruited into transcriptional complexes is largely unresolved. This could occur by direct interaction with TADs (105, 117, 137–140), by histone modifications specific to active chromatin (107), or by specific adapter proteins (112, 141). Alternatively, if the canonical 26S proteasome is the form that is involved in transcription, there is no reason to believe that proteasomes could not simply come in response to the presence of specific ubiquitylated substrates—a notion supported by the finding that the Ub ligase activity of the CCR4-Not complex is required for specific recruitment of proteasome proteins to the active PMA1 gene in yeast (142).
The Swiss army knife model makes a number of frank predictions about how proteasomes associate with chromatin, and these predictions need to be challenged experimentally, the most important of which is the tracking of proteasome subunit association across genomes. This research will likely require combined efforts that use multiple approaches to systematically monitor proteasome interaction with chromatin as well as functional approaches that compare the effects of disrupting both 19S and 20S functions on patterns of authentic and nonauthentic transcription. Only once unambiguous assignment of how these subunits interact with active genes is made—or once a transcriptionally relevant activity of the 19S base complex that cannot function within a 26S setting has been found—can these issues be resolved with certainty.
CONCLUSIONS
We hope to make it clear in this review that the UPS is intimately involved, physically and functionally, in gene regulatory mechanisms. Both proteolytic and nonproteolytic activities of the system impact transcriptional processes by targeting multiple steps in gene activity, from controlling activators through to export of a finished transcript from the nucleus, and possibly beyond. We emphasize that, because of the nature of the UPS and the linkage between many of its activities, it is likely that the examples we have cited here represent points on a spectrum of possibilities rather than absolute ways in which the UPS is always involved in gene regulation. It is easy to imagine, for example, how lines between proteolytic versus non-proteolytic regulation of a specific transcription protein could be blurred by events that change the type of Ub linkage or that transiently inactivate one or more functions in downstream UPS components, such as the proteasome. Indeed, it may very well be that transcription is a perfect venue in which to expose the various intricacies of the UPS in protein regulation. On a related note, it will be very interesting to learn whether processes other than transcription can be regulated in so many interesting ways by Ub, Ub-dependent processes, and the proteasome.
SUMMARY POINTS.
The ubiquitin-proteasome system (UPS) interacts with chromatin and regulates multiple steps in gene transcription, from controlling activators through to mRNA export.
Proteolytic and nonproteolytic actions of the UPS are important for transcriptional regulation. This includes proteolytic and nonproteolytic roles for Ub as well as the proteasome.
Transcriptional regulators are a major point of intervention of the UPS in transcriptional processes. Nonproteolytic ubiquitylation can modulate activator binding and activity, whereas proteolytic ubiquitylation can both inhibit and promote activator function.
The UPS can paradoxically be required for the function of transcription proteins it destroys. This likely reflects the action of the UPS in promoting activator turnover on chromatin and provides the cell with a way to maintain tight control over activators by inexorably linking their destruction to their activity.
Histone H2B ubiquitylation is a dynamic process that controls higher-order chromatin compaction; that influences transcription by recruiting effectors, promoting nucleosome disassembly and reassembly; and that signals additional histone modifications.
The extraordinary ability of the UPS to impact transcription stems from close physical connections between the two processes and from the ability of the UPS to detect the activity of proteins that are functioning in a transcription context.
The proteasome associates with chromatin and uses chaperone functions in the ATPase base, as well as its proteolytic capabilities, to modulate activator binding, coactivator recruitment, transcriptional elongation, and transcriptional termination. The proteasome also plays an important role in clearing RNA polymerase complexes that have stalled or initiated at incorrect sites in the genome.
The proteasome is an integrated multifunctional machine that has the potential to bring at least six distinct biochemical activities to bear in transcriptional processes.
FUTURE ISSUES.
The mechanism through which monoubiquitylation regulates the binding and activity of TAs and coactivators needs to be exposed.
Suicide models for activator function need to be challenged biochemically to determine the steps in transcriptional activation that require the UPS, the role of phosphorylation in this process, and whether regulatory proteins are the sole molecules being destroyed during transcriptional induction.
The role of ubiquitylation on histones H1, H3, and H4 needs to be understood both it terms of its impact on chromatin organization and its influence on transcriptional processes.
Receptors for ubiquitylated proteins in the transcriptional apparatus need to be identified. Given the large number of transcription proteins regulated by Ub, it seems likely that Ub-specific receptors exist, but few examples are known.
The influence of Ub chain length and topology on the function of transcription proteins needs to be understood. Understanding how differences in Ub chains are established and recognized could help resolve instances where it is uncertain whether proteolytic or nonproteolytic ubiquitylation is at work.
Clear resolution of the relative contribution of 19S versus 20S activities of the proteasome in each step of transcription is needed. The unambiguous assignment of the association of various components of the proteasome on chromatin, and their mode of recruitment to transcription sites, also needs to be resolved.
Biochemical analysis of transcriptionally relevant functions of the proteasome is required to reveal the steps in transcription that are directly impacted and to understand the molecular actions of chaperone proteins in the 19S base.
Other processes heavily controlled by the UPS need to be carefully examined to see if similar mechanisms, such as activation by destruction, recognition on the basis of activity, and functionally independent 19S chaperone action, are at work.
Acknowledgments
We thank members of the Tansey laboratory, past and present, for useful discussions. Research on connections between the transcription and ubiquitin-proteasome systems in the laboratory is supported by a grant from the National Institutes of Health (GM067728).
Glossary
- Ubiquitin-proteasome system (UPS)
includes all proteins required for ubiquitylation, deubiquitylation, recognition, and processing (proteoltyic or otherwise) of ubiquitylated proteins
- Ub
ubiquitin
- RNA polymerase II (Pol II)
a 12-subunit enzyme responsible for mRNA synthesis
- General transcription factors (GTFs)
a group of ancillary proteins necessary for promoter-driven transcription by Pol II
- TA
transcriptional activator
- Transcriptional activation domain (TAD)
a transferable element present in transcriptional activators that is responsible for recruiting Pol II and GTFs to chromatin
- C-terminal domain (CTD) of RNA polymerase II
a repeating motif in Pol II that is phosphorylated depending on the transcription stage
- Ubiquitylation
covalent linkage of Ub onto an accessible amino group of an acceptor protein
- Ubiquitin-activating enzyme (E1)
the enzyme that carries out the first stage in protein ubiquitylation
- Ubiquitin-conjugating enzyme (E2)
accepts Ub from E1 and carries out the chemistry of protein ubiquitylation, usually with an E3
- Ubiquitin-conjugating enzyme (E3)
recognizes substrates and works with an E2 to promote substrate ubiquitylation
- Deubiquitylating enzyme (DUb)
a class of enzymes that remove Ub modification from substrates
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
Fuqiang Geng, Email: fuqiang.geng@vanderbilt.edu.
Sabine Wenzel, Email: sabine.a.wenzel@vanderbilt.edu.
William P. Tansey, Email: william.p.tansey@vanderbilt.edu.
LITERATURE CITED
- 1.Ouyang J, Valin A, Gill G. Regulation of transcription factor activity by SUMO modification. Methods Mol Biol. 2009;497:141–52. doi: 10.1007/978-1-59745-566-4_9. [DOI] [PubMed] [Google Scholar]
- 2.Muratani M, Tansey WP. How the ubiquitin-proteasome system controls transcription. Nat Rev Mol Cell Biol. 2003;4:192–201. doi: 10.1038/nrm1049. [DOI] [PubMed] [Google Scholar]
- 3.Lipford JR, Deshaies RJ. Diverse roles for ubiquitin-dependent proteolysis in transcriptional activation. Nat Cell Biol. 2003;5:845–50. doi: 10.1038/ncb1003-845. [DOI] [PubMed] [Google Scholar]
- 4.Dennis AP, O’Malley BW. Rush hour at the promoter: how the ubiquitin-proteasome pathway polices the traffic flow of nuclear receptor-dependent transcription. J Steroid Biochem Mol Biol. 2005;93:139–51. doi: 10.1016/j.jsbmb.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 5.Baumann M, Pontiller J, Ernst W. Structure and basal transcription complex of RNA polymerase II core promoters in the mammalian genome: an overview. Mol Biotechnol. 2010;45:241–47. doi: 10.1007/s12033-010-9265-6. [DOI] [PubMed] [Google Scholar]
- 6.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–19. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 7.Hartzog GA. Transcription elongation by RNA polymerase II. Curr Opin Genet Dev. 2003;13:119–26. doi: 10.1016/s0959-437x(03)00024-8. [DOI] [PubMed] [Google Scholar]
- 8.Park YJ, Luger K. Histone chaperones in nucleosome eviction and histone exchange. Curr Opin Struct Biol. 2008;18:282–89. doi: 10.1016/j.sbi.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–92. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 10.Kaplan CD, Laprade L, Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301:1096–99. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- 11.Suganuma T, Workman JL. Signals and combinatorial functions of histone modifications. Annu Rev Biochem. 2011;80:473–99. doi: 10.1146/annurev-biochem-061809-175347. [DOI] [PubMed] [Google Scholar]
- 12.Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280–88. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 13.Behrends C, Harper JW. Constructing and decoding unconventional ubiquitin chains. Nat Struct Mol Biol. 2011;18:520–28. doi: 10.1038/nsmb.2066. [DOI] [PubMed] [Google Scholar]
- 14.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals ‘Protein Modifications: Beyond the Usual Suspects’ review series. EMBO Rep. 2008;9:536–42. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–63. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 17.Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–68. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 18.Madura K. Rad23 and Rpn10: Perennial wallflowers join the melee. Trends Biochem Sci. 2004;29:637–40. doi: 10.1016/j.tibs.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 19.Stolz A, Hilt W, Buchberger A, Wolf DH. Cdc48: a power machine in protein degradation. Trends Biochem Sci. 2011;36:515–23. doi: 10.1016/j.tibs.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 20.Sauer RT, Baker TA. AAA+ proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem. 2011;80:587–612. doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- 21.Liu CW, Corboy MJ, DeMartino GN, Thomas PJ. Endoproteolytic activity of the proteasome. Science. 2003;299:408–11. doi: 10.1126/science.1079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoppe T, Matuschewski K, Rape M, Schlenker S, Ulrich HD, Jentsch S. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell. 2000;102:577–86. doi: 10.1016/s0092-8674(00)00080-5. [DOI] [PubMed] [Google Scholar]
- 23.Palombella VJ, Rando OJ, Goldberg AL, Maniatis T. The ubiquitin-proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell. 1994;78:773–85. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 24.Burgdorf S, Leister P, Scheidtmann KH. TSG101 interacts with apoptosis-antagonizing transcription factor and enhances androgen receptor-mediated transcription by promoting its monoubiquitination. J Biol Chem. 2004;279:17524–34. doi: 10.1074/jbc.M313703200. [DOI] [PubMed] [Google Scholar]
- 25.Brés V, Kiernan RE, Linares LK, Chable-Bessia C, Plechakova O, et al. A non-proteolytic role for ubiquitin in Tat-mediated transactivation of the HIV-1 promoter. Nat Cell Biol. 2003;5:754–61. doi: 10.1038/ncb1023. [DOI] [PubMed] [Google Scholar]
- 26.van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, et al. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol. 2006;8:1064–73. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- 27.Moren A, Hellman U, Inada Y, Imamura T, Heldin CH, Moustakas A. Differential ubiquitination defines the functional status of the tumor suppressor Smad4. J Biol Chem. 2003;278:33571–82. doi: 10.1074/jbc.M300159200. [DOI] [PubMed] [Google Scholar]
- 28.Salghetti SE, Caudy AA, Chenoweth JG, Tansey WP. Regulation of transcriptional activation domain function by ubiquitin. Science. 2001;293:1651–53. doi: 10.1126/science.1062079. [DOI] [PubMed] [Google Scholar]
- 29.Kurosu T, Peterlin BM. VP16 and ubiquitin; binding of P-TEFb via its activation domain and ubiquitin facilitates elongation of transcription of target genes. Curr Biol. 2004;14:1112–16. doi: 10.1016/j.cub.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 30.Wu RC, Feng Q, Lonard DM, O’Malley BW. SRC-3 coactivator functional lifetime is regulated by a phospho-dependent ubiquitin time clock. Cell. 2007;129:1125–40. doi: 10.1016/j.cell.2007.04.039. [DOI] [PubMed] [Google Scholar]
- 31.Archer CT, Delahodde A, Gonzalez F, Johnston SA, Kodadek T. Activation domain-dependent monoubiquitylation of Gal4 protein is essential for promoter binding in vivo. J Biol Chem. 2008;283:12614–23. doi: 10.1074/jbc.M801050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilcox AJ, Laney JD. A ubiquitin-selective AAA-ATPase mediates transcriptional switching by remodelling a repressor-promoter DNA complex. Nat Cell Biol. 2009;11:1481–86. doi: 10.1038/ncb1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang B, Suzuki H, Kato M. Roles of monoubiquitinated Smad4 in the formation of Smad transcriptional complexes. Biochem Biophys Res Commun. 2008;376:288–92. doi: 10.1016/j.bbrc.2008.08.143. [DOI] [PubMed] [Google Scholar]
- 34.Laney JD, Hochstrasser M. Ubiquitin-dependent degradation of the yeast Mat(alpha)2 repressor enables a switch in developmental state. Genes Dev. 2003;17:2259–70. doi: 10.1101/gad.1115703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adhikary S, Marinoni F, Hock A, Hulleman E, Popov N, et al. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell. 2005;123:409–21. doi: 10.1016/j.cell.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 36.Ramakrishna S, Suresh B, Lee EJ, Lee HJ, Ahn WS, Baek KH. Lys-63-specific deubiquitination of SDS3 by USP17 regulates HDAC activity. J Biol Chem. 2011;286:10505–14. doi: 10.1074/jbc.M110.162321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lipford JR, Smith GT, Chi Y, Deshaies RJ. A putative stimulatory role for activator turnover in gene expression. Nature. 2005;438:113–16. doi: 10.1038/nature04098. [DOI] [PubMed] [Google Scholar]
- 38.Salghetti SE, Muratani M, Wijnen H, Futcher B, Tansey WP. Functional overlap of sequences that activate transcription and signal ubiquitin-mediated proteolysis. Proc Natl Acad Sci USA. 2000;97:3118–23. doi: 10.1073/pnas.050007597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, Muratani M, Tansey WP, Ptashne M. Proteolytic instability and the action of nonclassical transcriptional activators. Curr Biol. 2010;20:868–71. doi: 10.1016/j.cub.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhat KP, Greer SF. Proteolytic and non-proteolytic roles of ubiquitin and the ubiquitin proteasome system in transcriptional regulation. Biochim Biophys Acta. 2011;1809:150–55. doi: 10.1016/j.bbagrm.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 41.Amazit L, Roseau A, Khan JA, Chauchereau A, Tyagi RK, et al. Ligand-dependent degradation of SRC-1 is pivotal for progesterone receptor transcriptional activity. Mol Endocrinol. 2011;25:394–408. doi: 10.1210/me.2010-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leung A, Geng F, Daulny A, Collins G, Guzzardo P, Tansey WP. Transcriptional control and the ubiquitin-proteasome system. Ernst Schering Found Symp Proc. 2008;2008:75–97. [PubMed] [Google Scholar]
- 43.Sundqvist A, Ericsson J. Transcription-dependent degradation controls the stability of the SREBP family of transcription factors. Proc Natl Acad Sci USA. 2003;100:13833–38. doi: 10.1073/pnas.2335135100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muratani M, Kung C, Shokat KM, Tansey WP. The F box protein Dsg1/Mdm30 is a transcriptional coactivator that stimulates Gal4 turnover and cotranscriptional mRNA processing. Cell. 2005;120:887–99. doi: 10.1016/j.cell.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 45.Chi Y, Huddleston MJ, Zhang X, Young RA, Annan RS, et al. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 2001;15:1078–92. doi: 10.1101/gad.867501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chymkowitch P, Le May N, Charneau P, Compe E, Egly JM. The phosphorylation of the androgen receptor by TFIIH directs the ubiquitin/proteasome process. EMBO J. 2011;30:468–79. doi: 10.1038/emboj.2010.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andress EJ, Holic R, Edelmann MJ, Kessler BM, Yu VP. Dia2 controls transcription by mediating assembly of the RSC complex. PLoS ONE. 2011;6:e21172. doi: 10.1371/journal.pone.0021172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gammoh N, Gardiol D, Massimi P, Banks L. The Mdm2 ubiquitin ligase enhances transcriptional activity of human papillomavirus E2. J Virol. 2009;83:1538–43. doi: 10.1128/JVI.01551-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Punga T, Bengoechea-Alonso MT, Ericsson J. Phosphorylation and ubiquitination of the transcription factor sterol regulatory element-binding protein-1 in response to DNA binding. J Biol Chem. 2006;281:25278–86. doi: 10.1074/jbc.M604983200. [DOI] [PubMed] [Google Scholar]
- 50.von der Lehr N, Johansson S, Wu S, Bahram F, Castell A, et al. The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol Cell. 2003;11:1189–200. doi: 10.1016/s1097-2765(03)00193-x. [DOI] [PubMed] [Google Scholar]
- 51.Greer SF, Zika E, Conti B, Zhu XS, Ting JP. Enhancement of CIITA transcriptional function by ubiquitin. Nat Immunol. 2003;4:1074–82. doi: 10.1038/ni985. [DOI] [PubMed] [Google Scholar]
- 52.Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116:511–26. doi: 10.1016/s0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- 53.Higazi A, Abed M, Chen J, Li Q. Promoter context determines the role of proteasome in ligand-dependent occupancy of retinoic acid responsive elements. Epigenetics. 2011;6:202–11. doi: 10.4161/epi.6.2.13658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaluz S, Kaluzova M, Stanbridge EJ. Proteasomal inhibition attenuates transcriptional activity of hypoxia-inducible factor 1 (HIF-1) via specific effect on the HIF-1alpha C-terminal activation domain. Mol Cell Biol. 2006;26:5895–907. doi: 10.1128/MCB.00552-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spoel SH, Mou Z, Tada Y, Spivey NW, Genschik P, Dong X. Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell. 2009;137:860–72. doi: 10.1016/j.cell.2009.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chae E, Tan QK, Hill TA, Irish VF. An Arabidopsis F-box protein acts as a transcriptional co-factor to regulate floral development. Development. 2008;135:1235–45. doi: 10.1242/dev.015842. [DOI] [PubMed] [Google Scholar]
- 57.Collins GA, Gomez TA, Deshaies RJ, Tansey WP. Combined chemical and genetic approach to inhibit proteolysis by the proteasome. Yeast. 2010;27:965–74. doi: 10.1002/yea.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yin H, Jiang Y, Li H, Li J, Gui Y, Zheng XL. Proteasomal degradation of myocardin is required for its transcriptional activity in vascular smooth muscle cells. J Cell Physiol. 2011;226:1897–906. doi: 10.1002/jcp.22519. [DOI] [PubMed] [Google Scholar]
- 59.Thomas D, Tyers M. Transcriptional regulation: kamikaze activators. Curr Biol. 2000;10:R341–43. doi: 10.1016/s0960-9822(00)00462-0. [DOI] [PubMed] [Google Scholar]
- 60.Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29:653–63. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 61.Osley MA. Regulation of histone H2A and H2B ubiquitylation. Brief Funct Genomics Proteomics. 2006;5:179–89. doi: 10.1093/bfgp/ell022. [DOI] [PubMed] [Google Scholar]
- 62.Geng F, Tansey WP. Polyubiquitylation of histone H2B. Mol Biol Cell. 2008;19:3616–24. doi: 10.1091/mbc.E08-01-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–4. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- 64.Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, Madhani HD. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol Cell. 2003;11:261–66. doi: 10.1016/s1097-2765(02)00826-2. [DOI] [PubMed] [Google Scholar]
- 65.Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat Cell Biol. 2008;10:483–88. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- 66.Kim J, Roeder RG. Direct Bre1-Paf1 complex interactions and RING finger-independent Bre1-Rad6 interactions mediate histone H2B ubiquitylation in yeast. J Biol Chem. 2009;284:20582–92. doi: 10.1074/jbc.M109.017442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, et al. RAD6-mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–71. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang F, Yu X. WAC, a functional partner of RNF20/40, regulates histone H2B ubiquitination and gene transcription. Mol Cell. 2011;41:384–97. doi: 10.1016/j.molcel.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zheng S, Wyrick JJ, Reese JC. Novel trans-tail regulation of H2B ubiquitylation and H3K4 methylation by the N terminus of histone H2A. Mol Cell Biol. 2010;30:3635–45. doi: 10.1128/MCB.00324-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davies N, Lindsey GG. Histone H2B (and H2A) ubiquitination allows normal histone octamer and core particle reconstitution. Biochim Biophys Acta. 1994;1218:187–93. doi: 10.1016/0167-4781(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 71.Fierz B, Chatterjee C, McGinty RK, Bar-Dagan M, Raleigh DP, Muir TW. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat Chem Biol. 2011;7:113–19. doi: 10.1038/nchembio.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chandrasekharan MB, Huang F, Sun ZW. Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc Natl Acad Sci USA. 2009;106:16686–91. doi: 10.1073/pnas.0907862106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Formosa T. The role of FACT in making and breaking nucleosomes. Biochim Biophys Acta. 2012;1819:247–55. doi: 10.1016/j.bbagrm.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pavri R, Zhu B, Li G, Trojer P, Mandal S, et al. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–17. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 75.Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell. 2008;31:57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 76.Chatterjee C, McGinty RK, Fierz B, Muir TW. Disulfide-directed histone ubiquitylation reveals plasticity in hDot1L activation. Nat Chem Biol. 2010;6:267–69. doi: 10.1038/nchembio.315. [DOI] [PubMed] [Google Scholar]
- 77.Wu L, Zee BM, Wang Y, Garcia BA, Dou Y. The RING finger protein MSL2 in the MOF complex is an E3 ubiquitin ligase for H2B K34 and is involved in crosstalk with H3 K4 and K79 methylation. Mol Cell. 2011;43:132–44. doi: 10.1016/j.molcel.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Oh S, Jeong K, Kim H, Kwon CS, Lee D. A lysine-rich region in Dot1p is crucial for direct interaction with H2B ubiquitylation and high level methylation of H3K79. Biochem Biophys Res Commun. 2010;399:512–17. doi: 10.1016/j.bbrc.2010.07.100. [DOI] [PubMed] [Google Scholar]
- 79.Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–8. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 80.Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, et al. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell. 2007;131:1084–96. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- 81.Vitaliano-Prunier A, Menant A, Hobeika M, Geli V, Gwizdek C, Dargemont C. Ubiquitylation of the COMPASS component Swd2 links H2B ubiquitylation to H3K4 trimethylation. Nat Cell Biol. 2008;10:1365–71. doi: 10.1038/ncb1796. [DOI] [PubMed] [Google Scholar]
- 82.McGinty RK, Kohn M, Chatterjee C, Chiang KP, Pratt MR, Muir TW. Structure-activity analysis of semisynthetic nucleosomes: mechanistic insights into the stimulation of Dot1L by ubiquitylated histone H2B. ACS Chem Biol. 2009;4:958–68. doi: 10.1021/cb9002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature. 2008;453:812–16. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pinskaya M, Gourvennec S, Morillon A. H3 lysine 4 di- and trimethylation deposited by cryptic transcription attenuates promoter activation. EMBO J. 2009;28:1697–707. doi: 10.1038/emboj.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chandrasekharan MB, Huang F, Sun ZW. Histone H2B ubiquitination and beyond: Regulation of nucleosome stability, chromatin dynamics and the trans-histone H3 methylation. Epigenetics. 2010;5:460–68. doi: 10.4161/epi.5.6.12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Leung A, Cajigas I, Jia P, Ezhkova E, Brickner JH, et al. Histone H2B ubiquitylation and H3 lysine 4 methylation prevent ectopic silencing of euchromatic loci important for the cellular response to heat. Mol Biol Cell. 2011;22:2741–53. doi: 10.1091/mbc.E11-05-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Terzi N, Churchman LS, Vasiljeva L, Weissman J, Buratowski S. H3K4 trimethylation by Set1 promotes efficient termination by the Nrd1-Nab3-Sen1 pathway. Mol Cell Biol. 2011;31:3569–83. doi: 10.1128/MCB.05590-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ingvarsdottir K, Krogan NJ, Emre NC, Wyce A, Thompson NJ, et al. H2B ubiquitin protease Ubp8 and Sgf11 constitute a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol Cell Biol. 2005;25:1162–72. doi: 10.1128/MCB.25.3.1162-1172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wyce A, Xiao T, Whelan KA, Kosman C, Walter W, et al. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Mol Cell. 2007;27:275–88. doi: 10.1016/j.molcel.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 90.Shema E, Kim J, Roeder RG, Oren M. RNF20 inhibits TFIIS-facilitated transcriptional elongation to suppress pro-oncogenic gene expression. Mol Cell. 2011;42:477–88. doi: 10.1016/j.molcel.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gillette TG, Gonzalez F, Delahodde A, Johnston SA, Kodadek T. Physical and functional association of RNA polymerase II and the proteasome. Proc Natl Acad Sci USA. 2004;101:5904–9. doi: 10.1073/pnas.0305411101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Song EJ, Werner SL, Neubauer J, Stegmeier F, Aspden J, et al. The Prp19 complex and the Usp4Sart3 deubiquitinating enzyme control reversible ubiquitination at the spliceosome. Genes Dev. 2010;24:1434–47. doi: 10.1101/gad.1925010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gwizdek C, Iglesias N, Rodriguez MS, Ossareh-Nazari B, Hobeika M, et al. Ubiquitin-associated domain of Mex67 synchronizes recruitment of the mRNA export machinery with transcription. Proc Natl Acad Sci USA. 2006;103:16376–81. doi: 10.1073/pnas.0607941103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shcherbik N, Pestov DG. Ubiquitin and ubiquitin-like proteins in the nucleolus: multitasking tools for a ribosome factory. Genes Cancer. 2010;1:681–89. doi: 10.1177/1947601910381382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pham AD, Sauer F. Ubiquitin-activating/conjugating activity of TAFII250, a mediator of activation of gene expression in Drosophila. Science. 2000;289:2357–60. doi: 10.1126/science.289.5488.2357. [DOI] [PubMed] [Google Scholar]
- 96.Takagi Y, Masuda CA, Chang WH, Komori H, Wang D, et al. Ubiquitin ligase activity of TFIIH and the transcriptional response to DNA damage. Mol Cell. 2005;18:237–43. doi: 10.1016/j.molcel.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 97.Rabut G, Le Dez G, Verma R, Makhnevych T, Knebel A, et al. The TFIIH subunit Tfb3 regulates cullin neddylation. Mol Cell. 2011;43:488–95. doi: 10.1016/j.molcel.2011.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Daulny A, Geng F, Muratani M, Geisinger JM, Salghetti SE, Tansey WP. Modulation of RNA polymerase II subunit composition by ubiquitylation. Proc Natl Acad Sci USA. 2008;105:19649–54. doi: 10.1073/pnas.0809372105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Somesh BP, Sigurdsson S, Saeki H, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Communication between distant sites in RNA polymerase II through ubiquitylation factors and the polymerase CTD. Cell. 2007;129:57–68. doi: 10.1016/j.cell.2007.01.046. [DOI] [PubMed] [Google Scholar]
- 100.Ors A, Grimaldi M, Kimata Y, Wilkinson CR, Jones N, Yamano H. The transcription factor Atf1 binds and activates the APC/C ubiquitin ligase in fission yeast. J Biol Chem. 2009;284:23989–94. doi: 10.1074/jbc.M109.018309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Turnell AS, Stewart GS, Grand RJ, Rookes SM, Martin A, et al. The APC/C and CBP/p300 cooperate to regulate transcription and cell-cycle progression. Nature. 2005;438:690–95. doi: 10.1038/nature04151. [DOI] [PubMed] [Google Scholar]
- 102.Agricola E, Randall RA, Gaarenstroom T, Dupont S, Hill CS. Recruitment of TIF1gamma to chromatin via its PHD finger-bromodomain activates its ubiquitin ligase and transcriptional repressor activities. Mol Cell. 2011;43:85–96. doi: 10.1016/j.molcel.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 103.Li XS, Trojer P, Matsumura T, Treisman JE, Tanese N. Mammalian SWI/SNF-A subunit BAF250/ARID1 is an E3 ubiquitin ligase that targets histone H2B. Mol Cell Biol. 2010;30:1673–88. doi: 10.1128/MCB.00540-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ouni I, Flick K, Kaiser P. A transcriptional activator is part of an SCF ubiquitin ligase to control degradation of its cofactors. Mol Cell. 2010;40:954–64. doi: 10.1016/j.molcel.2010.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gonzalez F, Delahodde A, Kodadek T, Johnston SA. Recruitment of a 19S proteasome subcomplex to an activated promoter. Science. 2002;296:548–50. doi: 10.1126/science.1069490. [DOI] [PubMed] [Google Scholar]
- 106.Malik S, Shukla A, Sen P, Bhaumik SR. The 19S proteasome subcomplex establishes a specific protein interaction network at the promoter for stimulated transcriptional initiation in vivo. J Biol Chem. 2009;284:35714–24. doi: 10.1074/jbc.M109.035709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ezhkova E, Tansey WP. Proteasomal ATPases link ubiquitylation of histone H2B to methylation of histone H3. Mol Cell. 2004;13:435–42. doi: 10.1016/s1097-2765(04)00026-7. [DOI] [PubMed] [Google Scholar]
- 108.Sikder D, Johnston SA, Kodadek T. Widespread, but non-identical, association of proteasomal 19 and 20 S proteins with yeast chromatin. J Biol Chem. 2006;281:27346–55. doi: 10.1074/jbc.M604706200. [DOI] [PubMed] [Google Scholar]
- 109.Auld KL, Brown CR, Casolari JM, Komili S, Silver PA. Genomic association of the proteasome demonstrates overlapping gene regulatory activity with transcription factor substrates. Mol Cell. 2006;21:861–71. doi: 10.1016/j.molcel.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 110.Tran K, Mahr JA, Spector DH. Proteasome subunits relocalize during human cytomegalovirus infection, and proteasome activity is necessary for efficient viral gene transcription. J Virol. 2010;84:3079–93. doi: 10.1128/JVI.02236-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Szutorisz H, Georgiou A, Tora L, Dillon N. The proteasome restricts permissive transcription at tissue-specific gene loci in embryonic stem cells. Cell. 2006;127:1375–88. doi: 10.1016/j.cell.2006.10.045. [DOI] [PubMed] [Google Scholar]
- 112.Morris MC, Kaiser P, Rudyak S, Baskerville C, Watson MH, Reed SI. Cks1-dependent proteasome recruitment and activation of CDC20 transcription in budding yeast. Nature. 2003;424:1009–13. doi: 10.1038/nature01720. [DOI] [PubMed] [Google Scholar]
- 113.Fatyol K, Grummt I. Proteasomal ATPases are associated with rDNA: the ubiquitin proteasome system plays a direct role in RNA polymerase I transcription. Biochim Biophys Acta. 2008;1779:850–59. doi: 10.1016/j.bbagrm.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 114.Lee D, Ezhkova E, Li B, Pattenden SG, Tansey WP, Workman JL. The proteasome regulatory particle alters the SAGA coactivator to enhance its interactions with transcriptional activators. Cell. 2005;123:423–36. doi: 10.1016/j.cell.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 115.Ferdous A, Kodadek T, Johnston SA. A nonproteolytic function of the 19S regulatory subunit of the 26S proteasome is required for efficient activated transcription by human RNA polymerase II. Biochemistry. 2002;41:12798–805. doi: 10.1021/bi020425t. [DOI] [PubMed] [Google Scholar]
- 116.Ferdous A, Gonzalez F, Sun L, Kodadek T, Johnston SA. The 19S regulatory particle of the proteasome is required for efficient transcription elongation by RNA polymerase II. Mol Cell. 2001;7:981–91. doi: 10.1016/s1097-2765(01)00250-7. [DOI] [PubMed] [Google Scholar]
- 117.Truax AD, Koues OI, Mentel MK, Greer SF. The 19S ATPase S6a (S6′/TBP1) regulates the transcription initiation of class II transactivator. J Mol Biol. 2010;395:254–69. doi: 10.1016/j.jmb.2009.10.035. [DOI] [PubMed] [Google Scholar]
- 118.Koues OI, Mehta NT, Truax AD, Dudley RK, Brooks JK, Greer SF. Roles for common MLL/COMPASS subunits and the 19S proteasome in regulating CIITA pIV and MHC class II gene expression and promoter methylation. Epigenetics Chromatin. 2010;3:5. doi: 10.1186/1756-8935-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Koues OI, Dudley RK, Truax AD, Gerhardt D, Bhat KP, et al. Regulation of acetylation at the major histocompatibility complex class II proximal promoter by the 19S proteasomal ATPase Sug1. Mol Cell Biol. 2008;28:5837–50. doi: 10.1128/MCB.00535-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Koues OI, Dudley RK, Mehta NT, Greer SF. The 19S proteasome positively regulates histone methylation at cytokine inducible genes. Biochim Biophys Acta. 2009;1789:691–701. doi: 10.1016/j.bbagrm.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 121.Cheung V, Chua G, Batada NN, Landry CR, Michnick SW, et al. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 2008;6:e277. doi: 10.1371/journal.pbio.0060277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kodadek T. No splicing, no dicing: non-proteolytic roles of the ubiquitin-proteasome system in transcription. J Biol Chem. 2010;285:2221–26. doi: 10.1074/jbc.R109.077883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Russell SJ, Johnston SA. Evidence that proteolysis of Gal4 cannot explain the transcriptional effects of proteasome ATPase mutations. J Biol Chem. 2001;276:9825–31. doi: 10.1074/jbc.M010889200. [DOI] [PubMed] [Google Scholar]
- 124.Russell SJ, Sathyanarayana UG, Johnston SA. Isolation and characterization of SUG2. A novel ATPase family component of the yeast 26 S proteasome. J Biol Chem. 1996;271:32810–17. doi: 10.1074/jbc.271.51.32810. [DOI] [PubMed] [Google Scholar]
- 125.Xu Q, Singer RA, Johnston GC. Sug1 modulates yeast transcription activation by Cdc68. Mol Cell Biol. 1995;15:6025–35. doi: 10.1128/mcb.15.11.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Swaffield JC, Melcher K, Johnston SA. A highly conserved ATPase protein as a mediator between acidic activation domains and the TATA-binding protein. Nature. 1995;374:88–91. doi: 10.1038/374088a0. [DOI] [PubMed] [Google Scholar]
- 127.Sun L, Johnston SA, Kodadek T. Physical association of the APIS complex and general transcription factors. Biochem Biophys Res Commun. 2002;296:991–99. doi: 10.1016/s0006-291x(02)02026-0. [DOI] [PubMed] [Google Scholar]
- 128.Rasti M, Grand RJ, Yousef AF, Shuen M, Mymryk JS, et al. Roles for APIS and the 20S proteasome in adenovirus E1A-dependent transcription. EMBO J. 2006;25:2710–22. doi: 10.1038/sj.emboj.7601169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Verma R, Oania R, Fang R, Smith GT, Deshaies RJ. Cdc48/p97 mediates UV-dependent turnover of RNA Pol II. Mol Cell. 2011;41:82–92. doi: 10.1016/j.molcel.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Verma R, Chen S, Feldman R, Schieltz D, Yates J, et al. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol Biol Cell. 2000;11:3425–39. doi: 10.1091/mbc.11.10.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Navon A, Goldberg AL. Proteins are unfolded on the surface of the ATPase ring before transport into the proteasome. Mol Cell. 2001;8:1339–49. doi: 10.1016/s1097-2765(01)00407-5. [DOI] [PubMed] [Google Scholar]
- 132.Nishiyama A, Tachibana K, Igarashi Y, Yasuda H, Tanahashi N, et al. A nonproteolytic function of the proteasome is required for the dissociation of Cdc2 and cyclin B at the end of M phase. Genes Dev. 2000;14:2344–57. doi: 10.1101/gad.823200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fishbain S, Prakash S, Herrig A, Elsasser S, Matouschek A. Rad23 escapes degradation because it lacks a proteasome initiation region. Nat Commun. 2011;2:192. doi: 10.1038/ncomms1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ferdous A, Sikder D, Gillette T, Nalley K, Kodadek T, Johnston SA. The role of the proteasomal ATPases and activator monoubiquitylation in regulating Gal4 binding to promoters. Genes Dev. 2007;21:112–23. doi: 10.1101/gad.1493207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Daulny A, Tansey WP. Damage control: DNA repair, transcription, and the ubiquitin-proteasome system. DNA Repair. 2009;8:444–48. doi: 10.1016/j.dnarep.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 136.Lassot I, Latreille D, Rousset E, Sourisseau M, Linares LK, et al. The proteasome regulates HIV-1 transcription by both proteolytic and nonproteolytic mechanisms. Mol Cell. 2007;25:369–83. doi: 10.1016/j.molcel.2006.12.020. [DOI] [PubMed] [Google Scholar]
- 137.Chang C, Gonzalez F, Rothermel B, Sun L, Johnston SA, Kodadek T. The Gal4 activation domain binds Sug2 protein, a proteasome component, in vivo and in vitro. J Biol Chem. 2001;276:30956–63. doi: 10.1074/jbc.M102254200. [DOI] [PubMed] [Google Scholar]
- 138.Archer CT, Burdine L, Kodadek T. Identification of Gal4 activation domain-binding proteins in the 26S proteasome by periodate-triggered cross-linking. Mol BioSyst. 2005;1:366–72. doi: 10.1039/b510019d. [DOI] [PubMed] [Google Scholar]
- 139.Satoh T, Ishizuka T, Tomaru T, Yoshino S, Nakajima Y, et al. Tat-binding protein-1 (TBP-1), an ATPase of 19S regulatory particles of the 26S proteasome, enhances androgen receptor function in cooperation with TBP-1-interacting protein/Hop2. Endocrinology. 2009;150:3283–90. doi: 10.1210/en.2008-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Schwarz T, Sohn C, Kaiser B, Jensen ED, Mansky KC. The 19S proteasomal lid subunit POH1 enhances the transcriptional activation by Mitf in osteoclasts. J Cell Biochem. 2010;109:967–74. doi: 10.1002/jcb.22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chaves S, Baskerville C, Yu V, Reed SI. Cks1, Cdk1, and the 19S proteasome collaborate to regulate gene induction-dependent nucleosome eviction in yeast. Mol Cell Biol. 2010;30:5284–94. doi: 10.1128/MCB.00952-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Laribee RN, Shibata Y, Mersman DP, Collins SR, Kemmeren P, et al. CCR4/NOT complex associates with the proteasome and regulates histone methylation. Proc Natl Acad Sci USA. 2007;104:5836–41. doi: 10.1073/pnas.0607996104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yao T, Song L, Jin J, Cai Y, Takahashi H, et al. Distinct modes of regulation of the Uch37 deubiquitinating enzyme in the proteasome and in the Ino80 chromatin-remodeling complex. Mol Cell. 2008;31:909–17. doi: 10.1016/j.molcel.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Saleh A, Collart M, Martens JA, Genereaux J, Allard S, et al. TOM1p, a yeast hect-domain protein which mediates transcriptional regulation through the ADA/SAGA coactivator complexes. J Mol Biol. 1998;282:933–46. doi: 10.1006/jmbi.1998.2036. [DOI] [PubMed] [Google Scholar]
- 145.Iglesias N, Tutucci E, Gwizdek C, Vinciguerra P, Von Dach E, et al. Ubiquitin-mediated mRNP dynamics and surveillance prior to budding yeast mRNA export. Genes Dev. 2010;24:1927–38. doi: 10.1101/gad.583310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Boutet SC, Biressi S, Iori K, Natu V, Rando TA. Taf1 regulates Pax3 protein by monoubiquitination in skeletal muscle progenitors. Mol Cell. 2010;40:749–61. doi: 10.1016/j.molcel.2010.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Xie Y, Varshavsky A. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc Natl Acad Sci USA. 2001;98:3056–61. doi: 10.1073/pnas.071022298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Brower CS, Sato S, Tomomori-Sato C, Kamura T, Pause A, et al. Mammalian mediator subunit mMED8 is an Elongin BC-interacting protein that can assemble with Cul2 and Rbx1 to reconstitute a ubiquitin ligase. Proc Natl Acad Sci USA. 2002;99:10353–58. doi: 10.1073/pnas.162424199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mao X, Gluck N, Li D, Maine GN, Li H, et al. GCN5 is a required cofactor for a ubiquitin ligase that targets NF-kappaB/RelA. Genes Dev. 2009;23:849–61. doi: 10.1101/gad.1748409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chamovitz DA. Revisiting the COP9 signalosome as a transcriptional regulator. EMBO Rep. 2009;10:352–58. doi: 10.1038/embor.2009.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Albert TK, Hanzawa H, Legtenberg YI, de Ruwe MJ, van den Heuvel FA, et al. Identification of a ubiquitin-protein ligase subunit within the CCR4-NOT transcription repressor complex. EMBO J. 2002;21:355–64. doi: 10.1093/emboj/21.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ostendorff HP, Peirano RI, Peters MA, Schluter A, Bossenz M, et al. Ubiquitination-dependent cofactor exchange on LIM homeodomain transcription factors. Nature. 2002;416:99–103. doi: 10.1038/416099a. [DOI] [PubMed] [Google Scholar]
- 153.Zhang H, Sun L, Liang J, Yu W, Zhang Y, et al. The catalytic subunit of the proteasome is engaged in the entire process of estrogen receptor-regulated transcription. EMBO J. 2006;25:4223–33. doi: 10.1038/sj.emboj.7601306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Reid G, Hübner MR, Métivier R, Brand H, Denger S, et al. Cyclic, proteasome-mediated turnover of unliganded and liganded ERalpha on responsive promoters is an integral feature of estrogen signaling. Mol Cell. 2003;11:695–707. doi: 10.1016/s1097-2765(03)00090-x. [DOI] [PubMed] [Google Scholar]
- 155.Walsh HE, Shupnik MA. Proteasome regulation of dynamic transcription factor occupancy on the GnRH-stimulated luteinizing hormone beta-subunit promoter. Mol Endocrinol. 2009;23:237–50. doi: 10.1210/me.2008-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Stavreva DA, Müller WG, Hager GL, Smith CL, McNally JG. Rapid glucocorticoid receptor exchange at a promoter is coupled to transcription and regulated by chaperones and proteasomes. Mol Cell Biol. 2004;24:2682–97. doi: 10.1128/MCB.24.7.2682-2697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kim YC, Wu SY, Lim HS, Chiang CM, Kodadek T. Non-proteolytic regulation of p53-mediated transcription through destabilization of the activator·promoter complex by the proteasomal ATPases. J Biol Chem. 2009;284:34522–30. doi: 10.1074/jbc.M109.017277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Satoh T, Ishizuka T, Yoshino S, Tomaru T, Nakajima Y, et al. Roles of proteasomal 19S regulatory particles in promoter loading of thyroid hormone receptor. Biochem Biophys Res Commun. 2009;386:697–702. doi: 10.1016/j.bbrc.2009.06.099. [DOI] [PubMed] [Google Scholar]