ABSTRACT
Uniparental inheritance of mitochondrial DNA is pervasive in nonisogamic higher eukaryotes during sexual reproduction, and postzygotic and/or prezygotic factors are shown to be important in ensuring such an inheritance pattern. Although the fungus Cryptococcus neoformans undergoes sexual production with isogamic partners of opposite mating types a and α, most progeny derived from such mating events inherit the mitochondrial DNA (mtDNA) from the a parent. The homeodomain protein complex Sxi1α/Sxi2a, formed in the zygote after a-α cell fusion, was previously shown to play a role in this uniparental mtDNA inheritance. Here, we defined the timing of the establishment of the mtDNA inheritance pattern during the mating process and demonstrated a critical role in determining the mtDNA inheritance pattern by a prezygotic factor, Mat2. Mat2 is the key transcription factor that governs the pheromone sensing and response pathway, and it is critical for the early mating events that lead to cell fusion and zygote formation. We show that Mat2 governs mtDNA inheritance independently of the postzygotic factors Sxi1α/Sxi2a, and the cooperation between these prezygotic and postzygotic factors helps to achieve stricter uniparental mitochondrial inheritance in this eukaryotic microbe.
IMPORTANCE
Mitochondrial DNA is inherited uniparentally from the maternal parent in the majority of eukaryotes. Studies done on higher eukaryotes such as mammals have shown that the transmission of parental mitochondrial DNA is controlled at both the prefertilization and postfertilization stages to achieve strict uniparental inheritance. However, the molecular mechanisms underlying such uniparental mitochondrial inheritance have been investigated in detail mostly in anisogamic multicellular eukaryotes. Here, we show that in a simple isogamic microbe, Cryptococcus neoformans, the mitochondrial inheritance is controlled at the prezygotic level as well as the postzygotic level by regulators that are critical for sexual development. Furthermore, the cooperation between these two levels of control ensures stricter uniparental mitochondrial inheritance, echoing what has been observed in higher eukaryotes. Thus, the investigation of uniparental mitochondrial inheritance in this eukaryotic microbe could help advance our understanding of the convergent evolution of this widespread phenomenon in the eukaryotic domain.
Introduction
Organelle genomes are different from nuclear genomes in their ability to replicate multiple times per cell cycle, to segregate during both mitosis and meiosis, and to be inherited uniparentally (1, 2). Uniparental inheritance of organelle genomes is pervasive in sexual eukaryotes and is observed across a wide variety of organisms, including fungi, protists, plants, and animals (1–5). Uniparental inheritance of organelle genomes is shown to help prevent the spread of deleterious mutations present in organelle genomes as well as harmful parasites present in the cytoplasm (2). The discordance of mitochondrial and nuclear genomes could cause senescence in fungi (6) and male sterility in plants (7). Thus, uniparental mitochondrial inheritance has been a subject of intense research. However, despite the widely accepted view regarding the importance of uniparental mitochondrial inheritance, the timing and the underlying mechanisms of such an inheritance pattern during sexual reproduction are highly debated.
In the fungus Cryptococcus neoformans, bisexual reproduction commences when yeast cells of opposite mating types (a and α) are in close proximity under appropriate conditions. Mating in this fungus involves the formation of conjugation tubes by α cells in response to the pheromone produced by nearby a cells (8) (Fig. 1). Cytological studies showed that there is unidirectional nuclear migration from the α cell to the a cell during conjugation (8). The cell fusion product contains the nuclear genomes of both parents and is equivalent to the zygote of plants and animals (5,9). The two parental nuclei congress but do not fuse after the α-a cell fusion event. A dikaryotic mating hypha emerges from the zygote by the side of the original a parental cell, followed by septation between the zygote and the growing dikaryotic hyphae (8). The dikaryon can be maintained in the hyphal form indefinitely until the formation of basidia, where nuclear fusion and meiosis occur. Recombinant spores are generated subsequently from the basidia, forming four spore chains (Fig. 1). If the environmental temperature is high, nuclear fusion takes place in the dikaryon to form a stable diploid, which can differentiate to form monokaryotic diploid hyphae and give rise to haploid spores after meiosis in basidia when the temperature decreases (10).
FIG 1 .
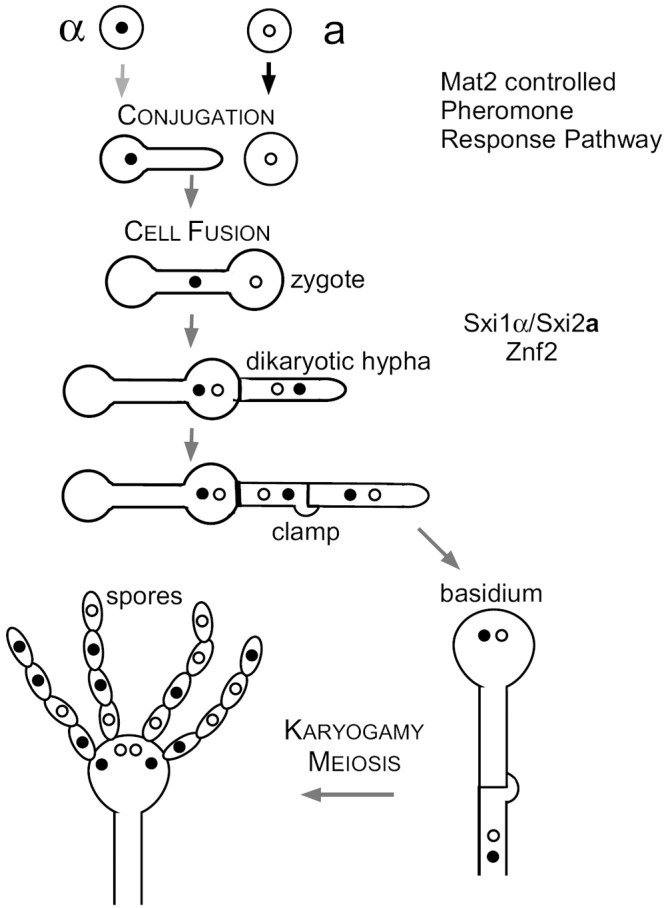
Bisexual mating of Cryptococcus neoformans. Under mating-inducing conditions, a and α cells undergo cell fusion. The early events during mating are controlled by the pheromone sensing and response pathway. Following cell fusion, the original zygote sends out the dikaryotic hypha, which grows filamentously as heterokaryotic dikaryons. Nuclear fusion and meiosis occur in the basidium formed at the tip of the aerial hypha. Four chains of spores are then generated from the basidium. Transcription factor Mat2 is essential for cell fusion (26), the mating-type-specific homeodomain complex Sxi1α/Sxi2a is specifically required for dikaryotic hypha formation and the completion of sexual development (15, 16), and the transcription factor Znf2 is essential for hyphal formation under all conditions (26, 27).
It has been well established that the progeny resulting from α-a sexual reproduction inherit mitochondrial DNA (mtDNA) predominantly from the a parent (11, 12). The uniparental mtDNA inheritance pattern appears to be established early during sexual reproduction, as intermediate cell types examined predominantly inherit the a parental mtDNA (11, 12). However, the timing of such an event has not been defined. The protein complex Sxi1α/Sxi2a formed after the cell fusion event was shown to control the uniparental mtDNA inheritance postzygotically (9, 13). This protein complex is specifically required for the production of dikaryotic hyphae from the original zygote and the completion of the subsequent sexual development (15, 16). Neither Sxi1α in α cells nor Sxi2a in a cells has any apparent role in the early mating events before the cell fusion. How the Sxi1α/Sxi2a complex that is formed in the zygote helps to control the uniparental mtDNA inheritance is currently unknown.
Based on these previous observations, three nonexclusive hypotheses can be envisioned to explain the uniparental mtDNA inheritance from the a parent in Cryptococcus: (i) blockage of the α mitochondria from entering the zygote (prezygotic control), (ii) incomplete cytoplasmic mixing and preferential mating hyphal formation from the a parental side (postzygotic control due to position effect), or (iii) selective degradation of α mitochondria and/or selective preservation of the a mitochondria in the zygote due to prezygotic marking (cooperation between the prezygotic and the postzygotic control).
In this study, we employed genetic approaches to examine the timing and genetic factors that control the mtDNA inheritance in Cryptococcus. We specifically focused on the key components that function during sexual development as uniparental mtDNA inheritance is observed only during a-α matings (9). Our findings argue against the hypothesis that postzygotic control due to position effect is important for the determination of the uniparental mtDNA inheritance pattern. The data suggest that the mtDNA inheritance pattern is determined in the original mature zygote and α mitochondria can efficiently enter the zygote but they are subsequently degraded during zygote maturation. Our observations support the role of the prezygotic factor Mat2 in the control of uniparental mtDNA inheritance. We propose that the a mtDNA inheritance in Cryptococcus is determined through selective preservation of a mitochondria and selective elimination of α mitochondria in the zygote. This is achieved by differential marking by Mat2 before the cell fusion event and the degradation of unpreserved α mitochondria in the zygote assisted by Sxi1α/Sxi2a. Thus, uniparental mtDNA inheritance in Cryptococcus is controlled both at the postzygotic level and at the prezygotic level.
RESULTS
Uniparental mitochondrial DNA inheritance is determined in the original zygote before the formation of mating hyphae.
Previous studies on mtDNA inheritance in Cryptococcus demonstrated that intervariety matings (hybrid matings between strains of two different serotypes) or intravariety matings (matings between strains of the same serotype) both lead to uniparental mtDNA inheritance from the a parent (9, 11, 12, 17). Here, we also chose to use intervarietal matings between serotype A and serotype D strains to investigate the factors involved in determining mtDNA inheritance in this fungus for the following reasons. The intervarietal mating shows the same hallmarks as does the intravariety mating (see Fig. S1 in the supplemental material); the intervarietal mating also displays uniparental a mtDNA inheritance. The meiosis-associated problems due to genome divergence occur later at basidia during the late stage of sexual development, which is long after the mtDNA inheritance pattern is established (9, 11–13, 17). Hybrid matings occur commonly in nature (18–22), and studying mtDNA inheritance in hybrid matings would help our understanding of the genetic makeup of cryptococcal natural populations. Importantly, AD hybrid mating allows us to track the origin of the progeny’s mtDNA based on the serotype-specific size polymorphism of the gene residing in the mitochondria that encodes cytochrome b subunit 1 (COB1) (17, 23). The serotype A congenic pair KN99a and KN99α are designated Aa and Aα, and the serotype D congenic pair JEC20a and JEC21α are designated Da and Dα in this study. The Aa/Aα strains do not self-filament, and they mate poorly with each other (Fig. 2). The Dα strain self-filaments sporadically after long incubation, and the Da strain does not self-filament. However, the Da and Dα strains mate with each other robustly. Furthermore, they also mate with the Aα or the Aa strain robustly (Fig. 2). These strains also differ in other phenotypes, such as thermotolerance and melanization (29) (Fig. 2).
FIG 2 .
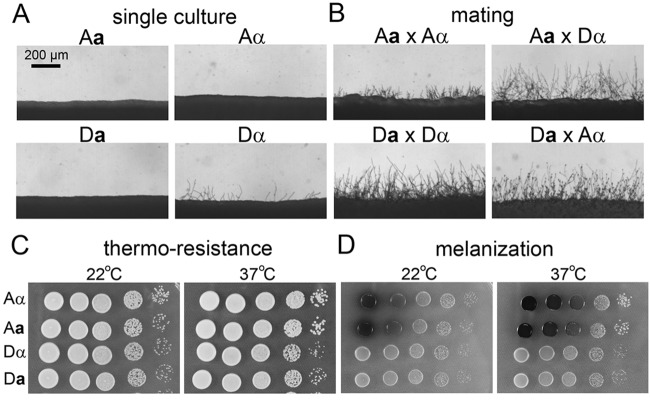
Phenotypic differences of the strains used. (A) Strains were incubated alone on V8 agar medium for 48 h. A and D represent variety serotype A and serotype D, respectively. a and α indicate the mating types of the strains. (B) Coculture of cells of opposite mating types on a V8 agar plate for 48 h to visualize filamentation, which reflects successful mating. (C and D) Strains with equal numbers of cells with serial dilutions were inoculated on YPD medium (C) or l-DOPA medium (D) and incubated at the indicated temperature for 48 h to analyze their thermotolerance and melanin production, respectively.
Previous studies have shown that the mtDNA inheritance pattern is established at an early but unidentified stage of sexual development. Sexual spores, hyphae, cells budded from the hyphae, or diploids derived from dikaryons all inherit mtDNA predominantly from the a parent (9, 12, 13). Here, we used previously established methods and tested the mtDNA of cells derived from the hybrid crosses. As shown in Table 1 and also in Fig. S2 in the supplemental material, cells derived from the reciprocal hybrid matings showed mtDNA inheritance predominantly from the a parent (Table 1, crosses 1 and 4).
TABLE 1 .
Mitochondrial inheritance from wild-type crosses and crosses involving znf2∆ strainsa
| Cross no. | Cross | % mtDNA from parent (no. positive/total no.) |
|
|---|---|---|---|
| α | a | ||
| 1 | A(α) × D(a) | 4 (2/50) | 82 (41/50) |
| 2 | A(α) × D(a) znf2Δ | 0 (0/50) | 100 (50/50) |
| 3 | A(α) znf2Δ × D(a) znf2Δ | 2 (1/48) | 87.5 (42/48) |
| 4 | D(α) × A(a) | 4 (2/49) | 92 (45/49) |
| 5 | D(α) znf2Δ × A(a) | 2 (1/49) | 98 (48/49) |
| 6 | D(α) znf2Δ × A(a) znf2Δ | 13 (6/45) | 87 (39/45) |
A minor portion of the progeny in certain crosses were found to inherit mtDNA from both the a and the α parent. It is also possible that recombination could also occur in these rare cases (43). Such a phenomenon was also observed previously (9, 13, 17). The number of progeny examined from each cross is indicated in parentheses. Inheritance of mitochondrial DNA from a and α parents in unilateral crosses or bilateral crosses involving znf2∆ mutant strains is not statistically different from that in the respective wild-type crosses.
It has been proposed that the postzygotic control of uniparental mtDNA inheritance could be caused by the incomplete cytoplasmic mixing and subsequent filamentation from the a parental side of the original zygote (12). This hypothesis is reasonable given that in Saccharomyces cerevisiae, mtDNA inheritance in daughter cells depends on the budding position on the zygote. If the first bud arises from either end of the zygote, then those cells contain mtDNA from only one parent, depending on which parental side the bud arises from. If the first bud arises from the middle of the zygote, then it contains mtDNA from both the parents (24, 25). If such a position effect is critical in Cryptococcus mtDNA inheritance, one would predict that progeny would be more likely to inherit mtDNA from the parental strain with an enhanced ability to filament. Alterations of the ability to undergo filamentation in the parental strains would then affect the mtDNA inheritance pattern.
We decided to examine the impact of alterations in the ability to undergo self-filamentation on the mtDNA inheritance pattern through genetic mutations of the ZNF2 gene. Znf2 is the master regulator of filamentation, and it does not control cell fusion events during mating (26, 27). Deletion of ZNF2 completely abolishes the cryptococcal ability to filament (26, 27) (Fig. 3). To examine the position effect of mating hyphal formation, we analyzed mtDNA inheritance in crosses involving znf2Δ mutants in the a or the α mating partner. If the position effect controls the mtDNA inheritance, one would predict a decrease in the inheritance from the a parent when ZNF2 is deleted in the a parental strain. However, blocking the ability to filament in the Da parental strain did not diminish the predominance of a mtDNA inheritance in the unilateral cross A(α) × D(a) znf2Δ (Table 1, crosses 2 and 1; see also Fig. S2 in the supplemental material). Similarly, we did not observe any significant change in the mtDNA inheritance pattern in the unilateral cross D(α) znf2Δ × A(a), where the ZNF2 gene was deleted in the α parent compared to the control cross (Table 1, crosses 5 and 4; see also Fig. S2). Thus, it appears that the position of hyphal formation does not affect the uniparental mtDNA inheritance pattern.
FIG 3 .
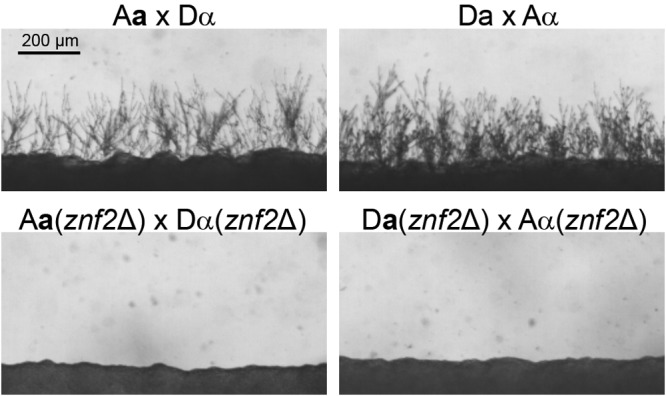
Deletion of ZNF2 abolishes filamentation. Robust filamentation is produced in the control matings after 48 h. Production of filamentation is completely eliminated in the bilateral matings with the znf2∆ mutants.
However, because the ZNF2 gene is deleted in only one parent, the ZNF2 gene from the other wild-type parent could compensate for the loss once the two parental cells are fused. It has been shown previously that the deletion of ZNF2 abolishes mating hypha formation only in bilateral crosses (znf2Δ × znf2Δ), not in unilateral crosses (znf2Δ × wild type) (26). Thus, we decided to examine mtDNA inheritance in crosses where the ZNF2 gene is deleted in both parental strains. If the position effect of mating hypha formation is responsible for the uniparental mtDNA inheritance from the a parent, abolishing the formation of mating hyphae from the zygote should result in a biparental mtDNA inheritance pattern. Interestingly, bilateral crosses with Znf2 disrupted in both parents yielded uniparental mtDNA inheritance from the a parent (Table 1, crosses 3 and 6; see also Fig. S2 in the supplemental material), a pattern similar to that for the wild-type crosses (Table 1, crosses 1 and 4). Again, the results suggest that the formation of mating hyphae is not important for the establishment of mtDNA inheritance pattern. Consequently, the position of mating hyphal formation does not determine the uniparental mtDNA inheritance in Cryptococcus.
Taken together, these results refute the original hypothesis and indicate that mtDNA inheritance is not dependent on the position of the mating hypha formation from the original zygotes. These results also suggest that the pattern of mtDNA inheritance is established in the original zygotes before the formation of mating hyphae. This new hypothesis offers a plausible explanation for the previous observations that various intermediate cell types obtained from bisexual matings predominantly inherited mtDNA from the a parent (9, 11, 12).
Prezygotic control of uniparental mitochondrial DNA inheritance.
The results presented above indicate that a uniparental mtDNA inheritance pattern is established in the original zygote before the generation of dikaryotic hyphae. The formation of dikaryotic hyphae requires the homeodomain protein complex Sxi1α/Sxi2a (15, 16), which is demonstrated to affect the uniparental mtDNA inheritance in Cryptococcus (9, 13). Because this intact protein complex can form only after the cell fusion event, deletion of either the SXI1α gene from the α parent or the SXI2a gene from the a parent (unilateral matings) has the same effect as does the deletion of both genes (bilateral matings). A biparental mtDNA inheritance pattern was observed in these unilateral or bilateral crosses, and surprisingly with modestly more progeny inheriting the α parental mtDNA (9, 13). We further confirmed these previous observations in our study (see below for details). These observations suggest that the original zygote contains ample mitochondria that originate from the α parent.
The observation that zygotes derived from crosses involving sxi1α∆ or sxi2a∆ mutants are more likely to contain mtDNA from the α parent than the a parent makes it unlikely that uniparental mitochondrial inheritance in Cryptococcus is due to blockage of α mitochondria from entering the zygote. We envision that rapid and selective degradation of α mitochondria in the zygote could achieve the uniparental a mtDNA inheritance and be consistent with the presence of both a and α mitochondria in the newly formed zygote. The retention of a mtDNA in the zygote can be achieved by prezygotically marking either the a mitochondria for preservation or the α mitochondria for annihilation, followed by rapid degradation of the α mitochondria in the zygote after Sxi1α and Sxi2a form the functional complex.
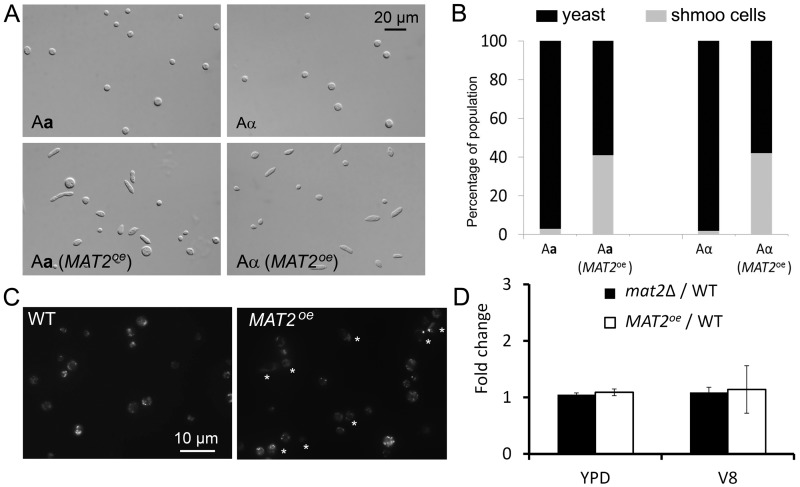
Such a model predicts that efficient selective elimination of the α mtDNA from the zygote relies on selective marking of parental mitochondria (or parental mtDNA) before the cell fusion event. Thus, some prezygotic factors must be involved in the control of mtDNA inheritance in Cryptococcus. We decided to investigate the role of Mat2 in mtDNA inheritance, given its essential role in early stages of mating. Mat2 is the key transcription factor in the pheromone sensing and response cascade (26, 28). It directs the conjugation process, and it must be present in both mating partners for the cell fusion event during α-a mating (26, 27). The essential role of Mat2 in cell fusion presents a challenge, as no zygote can be generated even in unilateral matings where Mat2 is disrupted in only one parent. Hence, we decided to use MAT2 overexpression (MAT2oe) strains where the MAT2 gene is placed under the control of the constitutively active promoter from the GPD1 gene (27). We generated MAT2oe strains in both Aα and Aa backgrounds. As shown in Fig. 4A and B, overexpression of MAT2 increases the competency of cells to mate as indicated by increased production of shmoo cells in both Aα and Aa backgrounds. This is consistent with the established roles for Mat2 (26, 27): stimulating pheromone production and driving the formation of mating-competent shmoo cells.
FIG 4 .
Overexpression of MAT2 drives shmoo cell formation but does not alter mitochondrial morphology or the mtDNA copy number. (A) The images in the top panel show the round yeast cells of the wild-type (WT) Aa and Aα strains after incubation in V8 medium for 48 h. The images in the bottom panel show the cell morphology (round yeasts and pear-shaped or elongated shmoo cells) of Aa and Aα strains when MAT2 is overexpressed. (B) Quantification of the distribution of yeast cell morphology and shmoo cell morphology in different populations as shown in panel A. (C) Mitotracker CMXRox-labeled wild-type cells and the MAT2oe cells. Stars indicate shmoo cells. (D) The comparison of the COB1/TEF1 ratios between the wild type, the MAT2oe strain, and the mat2Δ mutant cultured in YPD and V8 media. All strains shown here were in the Aα background. Similar results were obtained for strains in the Aa background (see Fig. S3 in the supplemental material).
Surprisingly, when MAT2 was overexpressed in the α parent, a modest dominance of α mtDNA inheritance (60%) was observed [Table 2, cross 2, A(α) MAT2oe × D(a); also see Fig. S2B in the supplemental material]. Thus, it appears that the overexpression of MAT2 in the α parent was able to change the progeny inheritance pattern from predominantly a mtDNA to biparental with modest dominance by the α mtDNA. When MAT2 was overexpressed in the a parent, mtDNA inheritance remained uniparental from the a parent (Table 2, crosses 5 and 6; see also Fig. S2B). To determine whether the dominance of the mtDNA originating from the parental MAT2oe strain is caused by an increased number of copies of mtDNA in the MAT2oe strains, we compared the ratio of mtDNA (the COB1 gene) and nuclear DNA (the TEF1 gene) between the MAT2oe strains and the wild-type strains. No difference in the COB1/TEF1 ratio was observed between the MAT2oe strains and the wild-type strains when cells were cultured in either yeast extract-peptone-dextrose (YPD) medium or V8 juice medium (Fig. 4D; see also Fig. S3). Similarly, the deletion of the MAT2 gene did not alter the ratio of COB1/TEF1 either (Fig. 4D). Thus, mutations of Mat2, either disruption or overexpression, do not appear to alter the copy number of mtDNA per cell. Furthermore, we found no apparent alterations in mitochondrial morphology in the MAT2oe strain compared to the wild type based on microscopic examination (Fig. 4C). Thus, Mat2 promotes the mtDNA inheritance through a means other than increasing the copy number of mtDNA in the cell.
TABLE 2.
Effect of mutations of SXI1α, SXI2a, and MAT2 on mitochondrial inheritancea
| Cross no. | Cross | % mtDNA from parent (no. positive/total no.) |
|
|---|---|---|---|
| α | a | ||
| 1 | A(α) × D(a) | 4 (2/50) | 82 (41/50) |
| 2 | A(α) MAT2oe × D(a) | 60 (52/87) | 35 (31/87) |
| 3 | A(α) × D(a) sxi2aΔ | 67 (20/30) | 20 (6/30) |
| 4 | A(α) MAT2oe × D(a) sxi2aΔ | 84 (36/44) | 12 (5/44) |
| 5 | D(α) × A(a) | 4 (2/49) | 92 (45/49) |
| 6 | D(α) × A(a) MAT2oe | 2 (1/46) | 98 (45/46) |
| 7 | D(α) sxi1αΔ × A(a) | 75 (36/48) | 25 (12/48) |
| 8 | D(α) sxi1αΔ × A(a) MAT2oe | 23.8 (10/42) | 73.8 (31/42) |
| 9 | D(α) × A(a) sxi2aΔ | 69.4 (34/49) | 24.5 (12/49) |
| 10 | D(α) × A(a) sxi2aΔ MAT2oe | 6 (3/49) | 83.6 (41/49) |
Effect of MAT2, SXI1α, and SXI2a on mitochondrial inheritance. The control crosses 1 and 5 are the same as crosses 1 and 4 in Table 1. They are included here for easy comparison and are shown with shading.
Prezygotic control and postzygotic control cooperate to determine mitochondrial DNA inheritance.
Next we decided to investigate whether the prezygotic control of mtDNA inheritance regulated by Mat2 is dependent on the postzygotic control regulated by Sxi1α/Sxi2a. As observed in previous studies (9, 13), we found that the deletion of either the SXI1α gene in the α parent or the SXI2a gene in the a parent results in biparental mtDNA inheritance in the unilateral matings (Table 2, crosses 3, 7, and 9; see also Fig. S2B in the supplemental material). A modest domination by the α mtDNA inheritance was observed in all of these crosses (67%, 75%, and 69%, respectively). Modest dominance of α mtDNA among the progeny was also observed previously when either the SXI1α gene or the SXI2a gene was disrupted in the parental strains. We found that this altered mtDNA inheritance pattern is independent of the serotype or the mating type where the mutation occurs. This is consistent with our hypothesis that the newly formed zygote contains both a and α mtDNA, with α mtDNA being modestly more abundant.
To examine whether mtDNA inheritance controlled by Mat2 is dependent on the Sxi1α/Sxi2a complex, we analyzed the mtDNA inheritance pattern of crosses involving one parent overexpressing MAT2 and the other parent disrupted with the homeodomain complex (Table 2, crosses 4 and 8). When MAT2 is overexpressed in the a parent, crossing with the α sxi1αΔ strain produced a modest dominance of a mtDNA inheritance among the progeny (74%), compared to the mere 25% a mtDNA inheritance in the cross between Aa and D(α) sxi1αΔ strains (Table 2, crosses 7 and 8). Thus, it appears that the overexpression of MAT2 in the a parent partially restores the dominance of the a mtDNA inheritance pattern, although the restoration is not to the level comparable to that of the wild-type cross where 92% of the progeny inherited the a mtDNA (Table 2, cross 5). In contrast, the overexpression of MAT2 in the α parent worsened the alteration caused by the deletion of SXI2a and increased the dominance of α mtDNA inheritance (84%) in the cross A(α) MAT2oe × D(a) sxi2aΔ compared to 67% observed in the cross A(α) × D(a) sxi2aΔ (Table 2, crosses 3 and 4). Thus, it appears that the parental origin of the MAT2 overexpression determines the type of mitochondria to be preserved independently of Sxi1α/Sxi2a: active Mat2 in the a parent leads to the preservation of the a mtDNA in the progeny and active Mat2 in the α parent results in the preservation of the α mtDNA in the progeny. However, a stricter mtDNA inheritance is achieved when the Sxi1α/Sxi2a complex works together with Mat2.
To further confirm the cooperation between Mat2 and Sxi1α/Sxi2a in the control of mtDNA inheritance, we constructed a MAT2oe strain in the sxi2a∆ mutant background. Consistent with our hypothesis, the mtDNA inheritance pattern in the cross D(α) × A(a) sxi2aΔ MAT2oe is similar to the cross D(α) sxi1αΔ × A(a) MAT2oe (Table 2, crosses 8 and 10; see also Fig. S2B in the supplemental material), where a modestly predominant proportion of the progeny (84% and 74%, respectively) inherited mtDNA from the a parent where MAT2 is overexpressed. In contrast, in the control crosses D(α) × A(a) sxi2aΔ and D(α) sxi1αΔ × A(a), only 24.5% and 25% of the progeny inherited mtDNA from the a parent, respectively. This again supports our hypothesis that the parental origin of the MAT2 overexpression determines the type of mitochondria to be preserved in the progeny, and a tighter uniparental mtDNA inheritance pattern can be achieved with the cooperation between Mat2 and Sxi1α/Sxi2a.
DISCUSSION
Previous studies have demonstrated the predominantly uniparental a mtDNA inheritance pattern in α-a crosses, and such an inheritance pattern is suggested to be established during the early stages of sexual development (11, 12). In this study, we investigated the roles of the postzygotic factors Znf2 and Sxi1α/Sxi2a and the prezygotic factor Mat2 in mtDNA inheritance in this fungus. Our findings indicate that mtDNA inheritance is established in the original zygote, before the formation of dikaryotic hyphae or subsequent developmental stages that lead to the production of meiotic spores (Fig. 1). In addition, we have shown the existence of a prezygotic control by Mat2. Although Mat2 is known to be highly upregulated during mating in both partners (26, 27), we posit that the timing and the level of Mat2 activation in the parental strains are critical in determining the origin of mitochondria to be inherited. It was shown previously that wild-type a cells express pheromone even in the absence of the α partner while α cells produce pheromone as a response to the pheromone produced by a cells (8). Therefore, under natural a-α matings, a cells likely initiate the mating process and their mitochondria are preserved to be inherited in the progeny. When the MAT2 gene is constitutively expressed using the GPD1 promoter, cells with MAT2oe act, in effect, as the parent initiating the mating process and their mitochondria are preferentially inherited in the progeny.
To confirm that Mat2 is differentially regulated in a and α cells when they are cultured alone and when they are cocultured together during mating, we measured the expression level of the MAT2 gene and the pheromone genes controlled by Mat2 (MFa and MFα) at multiple time points during the process. The pheromone genes were included as they are highly sensitive to the change of the MAT2 expression level (27) and thus can robustly reflect the activity of Mat2 even when the change in the MAT2 expression level itself is subtle. As shown in Fig. S4A in the supplemental material, the expression level of MFa increased more than 70-fold by 4 h in the culture of a cells alone. In contrast, the expression level of MFα increased less than 3-fold by 4 h in the culture of α cells alone and the level gradually increased to about 18-fold by 10 h. The patterns of MFa and MFα expression levels are largely correlated with the MAT2 expression level in these cultures (see Fig. S4B). During a-α mating, the expression levels of both MFa and MFα were drastically higher due to the positive feedback regulation of the pheromone sensing and response cascade in both α and a cells. Nonetheless, the expression level of MFa again increased earlier and stronger than that of MFα during the mating process (see Fig. S4C). Thus, a cells initiate the mating process through activation of the pheromone pathway controlled by Mat2.
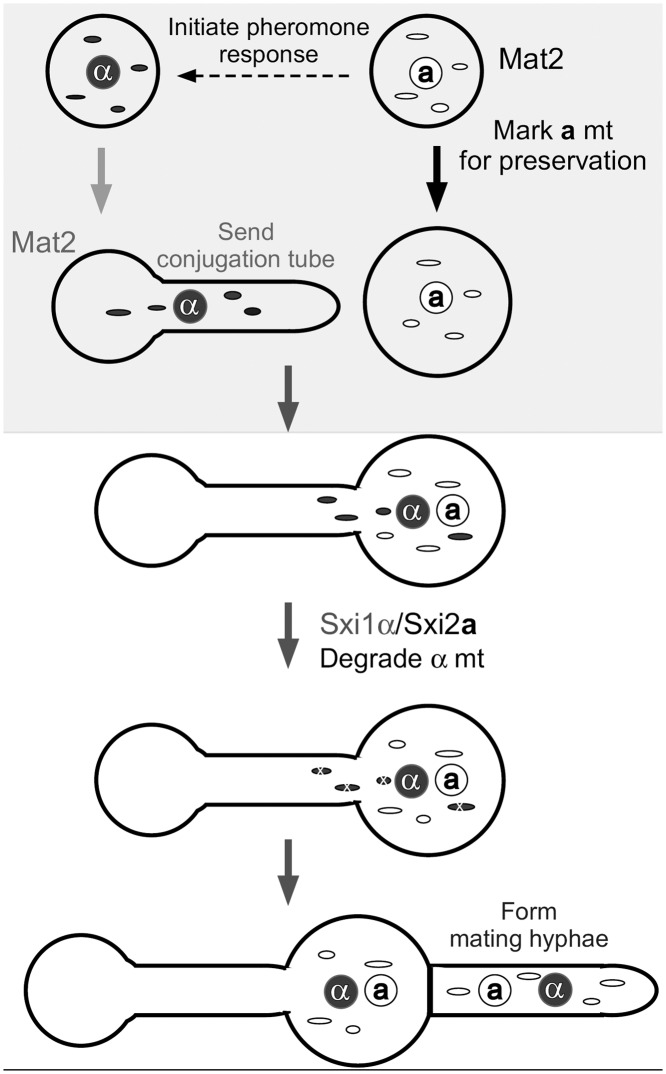
Based on these observations, we propose the following model to explain mtDNA inheritance in Cryptococcus (Fig. 5). During natural α-a matings, the activation of Mat2 in a cells leads to the marking of a mitochondria for preservation. In response to the pheromone produced by a cells, α cells also activate their Mat2 and send conjugation tubes toward a cells. After the α cell fuses with the a cell, the nuclei and mitochondria from both parental cells coexist in this newly formed zygote, which allows the formation of the Sxi1α/Sxi2a complex. This protein complex then mediates downstream factors to help eliminate α mitochondria before the zygote matures and sends out the dikaryotic hyphae. Thus, the cooperation between Mat2 and the Sxi1α/Sxi2a complex ensures stricter uniparental mtDNA inheritance from the a parent. Based on this model, one would predict that variations in the timing and the relative level of Mat2 activation between the mating pair, due to genetic differences or environmental factors, might cause variations in the tightness of the uniparental mtDNA inheritance. For instance, high temperatures inhibit the expression of MAT2 in both α and a cells (27) and may diminish the differences in the initial Mat2 activity between the mating pair, which could lead to the leakage in the uniparental mtDNA inheritance observed previously (14). In contrast to the α-a bisexual mating, the unisexual α-α mating yields a biparental mtDNA inheritance pattern (9). This could result from similar regulations on Mat2 in the two α partners. Further investigation with a large number of natural isolates with different Mat2 expression levels or with strains where Mat2 expression level and timing can be precisely controlled is necessary to validate this model.
FIG 5 .
Model of mtDNA inheritance in Cryptococcus. Under mating-inducing conditions, the a cell activates Mat2 and produces high levels of the MFa pheromone (8). In response to MFa, the α cell activates Mat2 and produces a conjugation tube. The earlier induction of Mat2 at a higher level in the a cell marks its mitochondria for preservation. Immediately after cell fusion, the Sxi1α/Sxi2a complex forms, and it helps degrade the unprotected α mitochondria in the newly formed zygote. Thus, the a mitochondria dominate in the mature zygote and cells subsequently generated from that zygote (e.g., hyphae and spores).
The cooperation between the prezygotic and postzygotic control to ensure a stricter uniparental mtDNA inheritance has been studied in other systems, although there are cases where either the prezygotic control or the postzygotic control prevails. In mammals, sperm mitochondria were tagged with ubiquitin during spermatogenesis, and they rapidly disappeared during zygote development (30). In the fish Oryzias latipes, the abundance of sperm mtDNA decreases 5-fold during spermatogenesis and the rapid elimination of remaining paternal mtDNA takes place within 2 h after fertilization (31). A 10-fold reduction in paternal mtDNA occurs during mouse spermatogenesis (32). The size difference between sperms and oocytes in these systems results in unequal contributions of paternal and maternal mtDNA in the zygote that facilitate a highly stringent uniparental mtDNA inheritance pattern (1, 30, 33). In the fruit fly Drosophila melanogaster, paternal mtDNA is completely eliminated during sperm development (34), and thus, the prezygotic control is sufficient to guarantee a uniparental mtDNA inheritance pattern.
Our study reinforces the concept that there are intricate connections between factors controlling sexual development and organellar inheritance. The high-mobility group (HMG) domain transcription factors are critical regulators controlling sexual development in fungi (Mat2 in Cryptococcus and sexP/sexM in Phycomyces) (35), and they are critical factors controlling mtDNA inheritance during sexual reproduction (this study and personal communication with Alexander Idnurm). Sex is proposed to enhance species fitness in changing and stressful environments. Uniparental mitochondrial inheritance is proposed to enhance the coadaptation of mitochondrial and nuclear genomes and therefore to improve species fitness more rapidly (36). Thus, intricate connections between the regulation of sexual development and mtDNA inheritance might have coevolved during the development of different eukaryotes. The highly virulent Cryptococcus gattii strains responsible for the cryptococcosis outbreak in British Columbia and the northwestern United States might have increased their fitness (virulence) in the temperate climate due to changes in their mitochondrial and nuclear genomes (37, 38). Given the robust genetic and molecular tools available to Cryptococcus and the vast ecological and epidemiological knowledge regarding its natural distribution and genotypes, Cryptococcus could serve as an excellent model to study the evolution of uniparental mitochondrial inheritance, the ecological and medical importance of this phenomenon, and the underlying molecular mechanisms.
MATERIALS AND METHODS
Strains and growth conditions.
Strains used in this study are listed in Table S1 along with their genotypes. Strains were maintained on YPD medium (2% Bacto peptone, 1% yeast extract, 2% dextrose, 2% Bacto agar) at 30°C before mating. Mating was carried out on V8 juice medium (5% V8 juice, 0.5 g/liter KH2PO4, 4% Bacto agar) at 22°C in the dark as described previously (26).
Generation of MAT2 overexpression strains.
The MAT2oe in the mating-type a strain was obtained by crossing LW80 (MATα, PGPD1-MAT2-NEO) (27) and JF99 (MATa, ura5). Mating was carried out for 14 days on V8 juice agar medium (pH 5). The mating type of the spores generated was confirmed by crossing the derived colonies with reference strain JEC20a or JEC21α. Colonies that mated with JEC21α but not with JEC20a and were resistant to G418 were selected. Auxotrophic strains were confirmed by the absence of growth on SC-URA medium. The MAT2 overexpression in the sxi2a∆ strain was obtained by introducing the construct PGPD1-MAT2-NEO (27) into the sxi2a∆ mutant as described previously (39). Stable transformants were selected, and the ability of these strains to produce filamentation on their own and the enhanced mating filaments were confirmed on V8 juice medium as described previously (27).
Generation of ura5 auxotrophic strains.
To generate ura5 auxotrophic strains, freshly grown strains on YPD were collected and washed twice with water. Strains were then plated on 5-fluoroorotic acid (5-FOA) plates (6.7 g/liter or 0.67% yeast nitrogen base without amino acids, 50 mg/liter uracil, 2% Bacto agar, 2% dextrose, 1 g/liter 5-fluoroorotic acid, SC-URA medium). Strains that grew on 5-FOA plates but failed to grow on SC-URA plates were selected as ura5 auxotrophic strains as described previously (40). Only strains showing a stable auxotrophic phenotype after several passages were used in this study.
Mating assays.
Parental serotype D and serotype A strains of different mating types were grown on YPD agar at 30°C. Mating was performed on V8 juice medium (pH 7) by mixing equal numbers of cells of opposite mating types. The coculture was incubated at 22°C in the dark for 2 to 3 days. Mating mixtures were then scraped off the V8 agar plates, washed with water, and plated on selective medium. To select products of the cell fusion events between two auxotrophic strains, cells were plated on minimal YNB medium (6.7 g/liter or 0.67% yeast nitrogen base with no amino acids, 2% Bacto agar, 2% dextrose) and grown for an additional 3 days at 37°C. To select products of the cell fusion events between two dominant marked strains, cells were plated on YPD medium supplemented with G418 and NAT (Nourseothricin or clonNAT) and incubated for an additional 3 days at 37°C. To select products of the cell fusion events between a ura5 auxotrophic strain and a dominant marked strain (NAT or NEO/G418), cells were plated on proline plus NAT or proline plus G418 agar medium and incubated for an additional 3 days at 37°C as described previously (16).
Determining the mitochondrial genotype.
The natural size polymorphism present in the cytochrome b subunit 1 gene (COB1) between serotype D and serotype A strains was used to determine the mtDNA genotype of the fusion product as described previously (17). In serotype A, COB1 contains 1 intron, whereas in the case of serotype D, it carries 2 introns. Thus, PCR amplification of the COB1 gene using the forward primer 5′-CCACAACCTATTAACATTAGCTACGC-3′ and the reverse primer 5′-CGTCTCCATCTACAAAGCCAGCAAAC-3′ yielded products of 610 bp in serotype A and 1,585 bp in serotype D (17). Total DNA extracted from the fusion products and their parental strains was treated with RNase A to get rid of RNA contamination. The treated DNA samples then served as the templates for PCRs. PCR amplicons were separated on agarose gels and stained with ethidium bromide for visualization under UV light.
Statistical tests.
Statistical tests were performed using GraphPad Prism, version 5.04. Fisher’s exact test was used to analyze if the inheritance pattern of mitochondrial DNA from the a and the α parent is statistically different between various crosses and the control crosses. A P value lower than 0.05 is considered statistically significant.
Phenotypic assays.
Phenotypic assays were performed as previously described (29). Briefly, yeast cells of tested strains were grown on YPD medium overnight and washed three times with water. The optical density of the cultures was measured at 600 nm, and all cultures were adjusted to the same cell density. Cells were then serially diluted by 10-fold. To examine melanin production, cells were spotted on melanin-inducing medium containing l-dihydroxyphenylalanine (l-DOPA) (100 mg/liter) and incubated at 22°C and 37°C in the dark for 2 to 6 days. Melanization was visualized as the colonies developed a brown color. To analyze growth at different temperatures, cells were spotted on YPD medium and incubated at the indicated temperatures for 2 days.
Quantification of the ratio between mitochondrial DNA and nuclear DNA.
Strains KN99α, XL1598 (Aα mat2Δ), LW80 (Aα MAT2oe), KN99a, and RG40 (Aa MAT2oe) were collected after incubation in YPD or V8 medium for 24 h. The cell number of each strain was calculated based on hemocytometer counting, and total DNA was extracted from the same number of cells for each strain using the cetyltrimethylammonium bromide (CTAB) method as previously described (41). DNA samples were treated with RNase A, and then the RNA-free DNA samples served as the templates for real-time PCRs using Kapa SYBR Fast PCR mix. The quantification of relative levels of the mitochondrial genome-borne COB1 (primers 5′ GCAAACACCACCTTCAATT 3′ and 5′ AGAAGGCAAATCGTGTTAGA 3′) and the nuclear genome-borne TEF1 (Lin lab 329/330; 5′-CGTCACCACTGAAGTCAAGT-3′ and 5′-AGAAGCAGCCTCCATAGG-3′) was used to compare mutant strains with the wild-type strains cultured under the same conditions.
Microscopic examination of mitochondrial morphology.
Cells were grown in V8 (pH 7) or YPD medium overnight, washed, and then suspended in phosphate-buffered saline (PBS). Mitotracker CMXRox stain was added to the cell suspension to a 100 nM final concentration as described previously (42). The cells were incubated with Mitotracker for 1 h at 30°C before they were washed three times with PBS. Washed cells were then visualized using a Zeiss Axioplan2 microscope.
SUPPLEMENTAL MATERIAL
Supplemental methods. Download
AD hybrid matings exhibit the same hallmarks as do intravarietal matings. (A) The hybrid mating between Aa and Dα strains was examined at multiple time points during sexual development. At early stage, yeast cells become shmoo cells (first column) and then send conjugation tubes (second column). The length of the conjugation tubes depends on the distance between the mating partner cells. The mating partners fuse to produce the original zygote cells (third, fourth, and fifth columns). The original zygote then sends out a heterokaryotic dikaryon with fused clamp cells (sixth column). Some dikaryotic hyphae develop into aerial hyphae (seventh column) and generate swollen basidia (eighth column) where spore chains form. Asterisks, clamp cells; black arrow, basidium. (B) The dikaryotic hyphae with fused clamp cells contain two parental nuclei as visualized through the 4′,6-diamidino-2-phenylindole (DAPI) stain. Asterisks, clamp cells. Download
Graphic representation of mitochondrial inheritance in the crosses listed in Tables 1 and 2. (A) Inheritance of mitochondrial DNA from a and α parents in unilateral crosses or bilateral crosses involving znf2∆ mutant strains is not statistically different from that in the respective wild-type crosses. (B) Graphic representation for the effect of MAT2oe on the mitochondrial inheritance. *, the inheritance of mtDNA from a and α parents is statistically significant across the groups (P value, <0.05). Download
The mtDNA/nuclear DNA in the MAT2oe strain is the same as that in the wild-type strain in the Aa background. The comparison of the COB1/TEF1 ratio between the wild-type strain and the MAT2oe strain in the Aa background cultured in YPD or V8 juice medium. Download
MFa is induced earlier and to a higher level than is MFα. (A) The pheromone expression level of the a and α cells at various time points when cultured alone on V8 agar medium. (B) The MAT2 expression level of the a and α strains at various time points when cultured alone on V8 agar medium. (C) The expression level of the pheromone MFa and MFα at various time points during a-α coculture (mating) on V8 medium. Download
Strains used in this study.
ACKNOWLEDGMENTS
We gratefully acknowledge financial support from the Norman Hackerman Advanced Research Program (grant 01957 to X.L.) and the National Institute of Allergy and Infectious Diseases (grant R01AI097599 to X.L.).
We thank Linqi Wang and Srijana Upadhyay for their helpful suggestions on this project and Alexander Idnurm and Dylan Foyle for critical reading.
Footnotes
Citation Gyawali R, Lin X. 2013. Prezygotic and postzygotic control of uniparental mitochondrial DNA inheritance in Cryptococcus neoformans. mBio 4(2):e00112-13. doi:10.1128/mBio.00112-13.
REFERENCES
- 1. Birky CW. 2001. The inheritance of genes in mitochondria and chloroplasts: laws, mechanisms, and models. Annu. Rev. Genet. 35:125–148 [DOI] [PubMed] [Google Scholar]
- 2. Birky CW. 1995. Uniparental inheritance of mitochondrial and chloroplast genes: mechanisms and evolution. Proc. Natl. Acad. Sci. USA 92:11331–11338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barr CM, Neiman M, Taylor DR. 2005. Inheritance and recombination of mitochondrial genomes in plants, fungi and animals. New Phytol. 168:39–50 [DOI] [PubMed] [Google Scholar]
- 4. Basse CW. 2010. Mitochondrial inheritance in fungi. Curr. Opin. Microbiol. 13:712–719 [DOI] [PubMed] [Google Scholar]
- 5. Xu J. 2005. The inheritance of organelle genes and genomes: patterns and mechanisms. Genome 48:951–958 [DOI] [PubMed] [Google Scholar]
- 6. Bonawitz ND, Shadel GS. 2007. Rethinking the mitochondrial theory of aging: the role of mitochondrial gene expression in lifespan determination. Cell Cycle 6:1574–1578 [DOI] [PubMed] [Google Scholar]
- 7. Chase CD. 2007. Cytoplasmic male sterility: a window to the world of plant mitochondrial–nuclear interactions. Trends Genet. 23:81–90 [DOI] [PubMed] [Google Scholar]
- 8. McClelland CM, Chang YC, Varma A, Kwon-Chung KJ. 2004. Uniqueness of the mating system in Cryptococcus neoformans. Trends Microbiol. 12:208–212 [DOI] [PubMed] [Google Scholar]
- 9. Yan Z, Hull CM, Sun S, Heitman J, Xu J. 2007. The mating type-specific homeodomain genes SXI1 alpha and SXI2a coordinately control uniparental mitochondrial inheritance in Cryptococcus neoformans. Curr. Genet. 51:187–195 [DOI] [PubMed] [Google Scholar]
- 10. Sia RA, Lengeler KB, Heitman J. 2000. Diploid strains of the pathogenic basidiomycete Cryptococcus neoformans are thermally dimorphic. Fungal Genet. Biol. 29:153–163 [DOI] [PubMed] [Google Scholar]
- 11. Xu J, Ali RY, Gregory DA, Amick D, Lambert SE, Yoell HJ, Vilgalys RJ, Mitchell TG. 2000. Uniparental mitochondrial transmission in sexual crosses in Cryptococcus neoformans. Curr. Microbiol. 40:269–273 [DOI] [PubMed] [Google Scholar]
- 12. Yan Z, Xu J. 2003. Mitochondria are inherited from the MATa parent in crosses of the basidiomycete fungus Cryptococcus neoformans. Genetics 163:1315–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yan Z, Hull CM, Heitman J, Sun S, Xu J. 2004. SXI1alpha controls uniparental mitochondrial inheritance in Cryptococcus neoformans. Curr. Biol. 14:R743–R744 http://dx.doi.org/10.1016/j.cub.2004.09.008 [DOI] [PubMed] [Google Scholar]
- 14. Yan Z, Sun S, Shahid M, Xu J. 2007. Environment factors can influence mitochondrial inheritance in the fungus Cryptococcus neoformans. Fungal Genet. Biol. 44:315–322 [DOI] [PubMed] [Google Scholar]
- 15. Hull CM, Boily MJ, Heitman J. 2005. Sex-specific homeodomain proteins Sxi1alpha and Sxi2a coordinately regulate sexual development in Cryptococcus neoformans. Eukaryot. Cell 4:526–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hull CM, Davidson RC, Heitman J. 2002. Cell identity and sexual development in Cryptococcus neoformans are controlled by the mating-type-specific homeodomain protein Sxi1 alpha. Genes Dev. 16:3046–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Toffaletti DL, Nielsen K, Dietrich F, Heitman J, Perfect JR. 2004. Cryptococcus neoformans mitochondrial genomes from serotype A and D strains do not influence virulence. Curr. Genet. 46:193–204 [DOI] [PubMed] [Google Scholar]
- 18. Bovers M, Hagen F, Kuramae EE, Diaz MR, Spanjaard L, Dromer F, Hoogveld HL, Boekhout T. 2006. Unique hybrids between the fungal pathogens Cryptococcus neoformans and Cryptococcus gattii. FEMS Yeast Res. 6:599–607 [DOI] [PubMed] [Google Scholar]
- 19. Bovers M, Hagen F, Kuramae EE, Hoogveld HL, Dromer F, St-Germain G, Boekhout T. 2008. AIDS patient death caused by novel Cryptococcus neoformans × C. gattii hybrid. Emerg. Infect. Dis. 14:1105–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cogliati M, Esposto MC, Tortorano AM, Viviani MA. 2006. Cryptococcus neoformans population includes hybrid strains homozygous at mating-type locus. FEMS Yeast Res. 6:608–613 [DOI] [PubMed] [Google Scholar]
- 21. Lin X, Litvintseva AP, Nielsen K, Patel S, Floyd A, Mitchell TG, Heitman J. 2007. αADα hybrids of Cryptococcus neoformans: evidence of same-sex mating in nature and hybrid fitness. PLoS Genet. 3:1975–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lin X, Patel S, Litvintseva AP, Floyd A, Mitchell TG, Heitman J. 2009. Diploids in the Cryptococcus neoformans serotype A population homozygous for the α mating type originate via unisexual mating. PLoS Pathog. 5:e1000283 http://dx.doi.org/10.1371/journal.ppat.1000283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Litter J, Keszthelyi A, Hamari Z, Pfeiffer I, Kucsera J. 2005. Differences in mitochondrial genome organization of Cryptococcus neoformans strains. Antonie Van Leeuwenhoek 88:249–255 [DOI] [PubMed] [Google Scholar]
- 24. Strausberg RL, Perlman PS. 1978. The effect of zygotic bud position on the transmission of mitochondrial genes in Saccharomyces cerevisiae. Mol. Gen. Genet. 163:131–144 [DOI] [PubMed] [Google Scholar]
- 25. Berger KH, Yaffe MP. 2000. Mitochondrial DNA inheritance in Saccharomyces cerevisiae. Trends Microbiol. 8:508–513 [DOI] [PubMed] [Google Scholar]
- 26. Lin X, Jackson JC, Feretzaki M, Xue C, Heitman J. 2010. Transcription factors Mat2 and Znf2 operate cellular circuits orchestrating opposite- and same-sex mating in Cryptococcus neoformans. PLoS Genet. 6:e1000953 http://dx.doi.org/10.1371/journal.pgen.1000953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang L, Zhai B, Lin X. 2012. The link between morphotype transition and virulence in Cryptococcus neoformans. PLoS Pathog. 8:e1002765 http://dx.doi.org/10.1371/journal.ppat.1002765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kruzel EK, Giles SS, Hull CM. 2012. Analysis of Cryptococcus neoformans sexual development reveals rewiring of the pheromone-response network by a change in transcription factor identity. Genetics 191:435–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lin X, Nielsen K, Patel S, Heitman J. 2008. Impact of mating type, serotype, and ploidy on the virulence of Cryptococcus neoformans. Infect. Immun. 76:2923–2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sutovsky P, Moreno RD, Ramalho-Santos J, Dominko T, Simerly C, Schatten G. 2000. Ubiquitinated sperm mitochondria, selective proteolysis, and the regulation of mitochondrial inheritance in mammalian embryos. Biol. Reprod. 63:582–590 [DOI] [PubMed] [Google Scholar]
- 31. Nishimura Y, Yoshinari T, Naruse K, Yamada T, Sumi K, Mitani H, Higashiyama T, Kuroiwa T. 2006. Active digestion of sperm mitochondrial DNA in single living sperm revealed by optical tweezers. Proc. Natl. Acad. Sci. U. S. A. 103:1382–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hecht NB, Liem H, Kleene KC, Distel RJ, Ho SM. 1984. Maternal inheritance of the mouse mitochondrial genome is not mediated by a loss or gross alteration of the paternal mitochondrial DNA or by methylation of the oocyte mitochondrial DNA. Dev. Biol. 102:452–461 [DOI] [PubMed] [Google Scholar]
- 33. Smith LC, Alcivar AA. 1993. Cytoplasmic inheritance and its effects on development and performance. J. Reprod. Fertil. Suppl. 48:31–43 [PubMed] [Google Scholar]
- 34. DeLuca SZ, O’Farrell PH. 2012. Barriers to male transmission of mitochondrial DNA in sperm development. Dev. Cell 22:660–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Idnurm A, Walton FJ, Floyd A, Heitman J. 2008. Identification of the sex genes in an early diverged fungus. Nature 451:193–196 [DOI] [PubMed] [Google Scholar]
- 36. Hadjivasiliou Z, Pomiankowski A, Seymour RM, Lane N. 2012. Selection for mitonuclear co-adaptation could favour the evolution of two sexes. Proc. Biol. Sci. 279:1865–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ma H, Hagen F, Stekel DJ, Johnston SA, Sionov E, Falk R, Polacheck I, Boekhout T, May RC. 2009. The fatal fungal outbreak on Vancouver Island is characterized by enhanced intracellular parasitism driven by mitochondrial regulation. Proc. Natl. Acad. Sci. U. S. A. 106:12980–12985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ma H, May RC. 2010. Mitochondria and the regulation of hypervirulence in the fatal fungal outbreak on Vancouver Island. Virulence 1:197–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Varma A, Kwon-Chung KJ. 1994. Formation of a minichromosome in Cryptococcus neoformans as a result of electroporative transformation. Curr. Genet. 26:54–61 [DOI] [PubMed] [Google Scholar]
- 40. Kwon-Chung KJ, Varma A, Edman JC, Bennett JE. 1992. Selection of ura5 and ura3 mutants from the two varieties of Cryptococcus neoformans on 5-fluoroorotic acid medium. J. Med. Vet. Med. Mycol. 30:61–69 [PubMed] [Google Scholar]
- 41. Pitkin JW, Panaccione DG, Walton JD. 1996. A putative cyclic peptide efflux pump encoded by the TOXA gene of the plant-pathogenic fungus Cochliobolus carbonum. Microbiology 142:1557–1565 [DOI] [PubMed] [Google Scholar]
- 42. Ingavale SS, Chang YC, Lee H, McClelland CM, Leong ML, Kwon-Chung KJ. 2008. Importance of mitochondria in survival of Cryptococcus neoformans under low oxygen conditions and tolerance to cobalt chloride. PLoS Pathog. 4:e1000155 http://dx.doi.org/10.1371/journal.ppat.1000155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bovers M, Hagen F, Kuramae EE, Boekhout T. 2009. Promiscuous mitochondria in Cryptococcus gattii. FEMS Yeast Res. 9:489–503 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods. Download
AD hybrid matings exhibit the same hallmarks as do intravarietal matings. (A) The hybrid mating between Aa and Dα strains was examined at multiple time points during sexual development. At early stage, yeast cells become shmoo cells (first column) and then send conjugation tubes (second column). The length of the conjugation tubes depends on the distance between the mating partner cells. The mating partners fuse to produce the original zygote cells (third, fourth, and fifth columns). The original zygote then sends out a heterokaryotic dikaryon with fused clamp cells (sixth column). Some dikaryotic hyphae develop into aerial hyphae (seventh column) and generate swollen basidia (eighth column) where spore chains form. Asterisks, clamp cells; black arrow, basidium. (B) The dikaryotic hyphae with fused clamp cells contain two parental nuclei as visualized through the 4′,6-diamidino-2-phenylindole (DAPI) stain. Asterisks, clamp cells. Download
Graphic representation of mitochondrial inheritance in the crosses listed in Tables 1 and 2. (A) Inheritance of mitochondrial DNA from a and α parents in unilateral crosses or bilateral crosses involving znf2∆ mutant strains is not statistically different from that in the respective wild-type crosses. (B) Graphic representation for the effect of MAT2oe on the mitochondrial inheritance. *, the inheritance of mtDNA from a and α parents is statistically significant across the groups (P value, <0.05). Download
The mtDNA/nuclear DNA in the MAT2oe strain is the same as that in the wild-type strain in the Aa background. The comparison of the COB1/TEF1 ratio between the wild-type strain and the MAT2oe strain in the Aa background cultured in YPD or V8 juice medium. Download
MFa is induced earlier and to a higher level than is MFα. (A) The pheromone expression level of the a and α cells at various time points when cultured alone on V8 agar medium. (B) The MAT2 expression level of the a and α strains at various time points when cultured alone on V8 agar medium. (C) The expression level of the pheromone MFa and MFα at various time points during a-α coculture (mating) on V8 medium. Download
Strains used in this study.