Abstract
Simplicity has made C. elegans pharyngeal development a particularly well-studied subject. Nevertheless, here we add the previously uncharacterized homeobox gene F20D12.6/ceh-19 to the set of transcription factor genes involved. GFP reporter assays revealed that ceh-19 is expressed in three pairs of neurons, the pharyngeal pace-maker neurons MC, the amphid neurons ADF and the phasmid neurons PHA. ceh-19(tm452) mutants are viable and fertile, but grow slightly slower, produce less progeny over a prolonged period, and live longer than the wild type. These phenotypes are likely due to the moderately reduced pharyngeal pumping speed arising from the impairment of MC activity. MC neurons are still born in the ceh-19 mutants but display various morphological defects. ceh-19 expression in MC is completely lost in progeny from animals subject to RNAi for pha-4, which encodes an organ-specifying forkhead transcription factor. CEH-19 is required for the activation in MCs of the excitatory FMRFamide-like neuropeptide-encoding gene flp-2. A regulatory pathway from pha-4 through ceh-19 to flp-2 is thereby defined. The resilience of MC identity in the absence of CEH-19 may reflect the buffering qualities of transcription factor regulatory networks. genesis 51:163–178, 2013. © 2013 Wiley Periodicals, Inc.
Keywords: Caenorhabditis elegans, transcription factors, homeodomain, nerve cells, pharynx
INTRODUCTION
The C. elegans pharynx is an anatomically self-contained structure dedicated to ingestion and transportation of bacteria into the intestine (Albertson and Thomson, 1976; Avery and Horvitz, 1989). The entire structure is surrounded by and isolated from the rest of the worm by a basement membrane (Sulston et al., 1983). The feeding behavior is accomplished by two distinct types of muscle contractions, namely pumping and peristalsis. Pumping ingests and concentrates bacteria in the anterior lumen and is followed by peristaltic contractions, which bring ingested bacteria through the isthmus (Avery and Shtonda, 2003). Feeding is regulated and modulated by the 20 pharyngeal neurons, almost completely independently of neuro-muscular activity elsewhere in the animal (Avery and Horvitz, 1989). Extensive laser ablation experiments have shown that the nervous system is not essential for pumping, but is important for normal feeding, growth rate, and fertility (Avery and Horvitz, 1989). Pharyngeal muscles pump even without the stimulation from neurons, but the pace of pumping is controlled by the motor neurons MC and M3, which initiate and terminate the muscle action potentials respectively (Raizen et al., 1995). The MC neuron is an excitatory cholinergic neuron. Its firing triggers a pharyngeal muscle action potential via the release of acetylcholine, which acts on a muscle nicotinic receptor. It is said to be necessary and probably sufficient for rapid pharyngeal pumping (Mukhopadhyay et al., 2007; Raizen et al., 1995). A third motor neuron, M4, is responsible for peristalsis of the isthmus; its inactivation has little effect on pumping but eliminates peristalsis causing animals to arrest with a “stuffed pharynx” (Avery and Horvitz, 1989).
Homeobox genes have been shown to be required for pharynx morphogenesis and cell differentiation. Notably, several NK-2 class homeodomain factors, including PHA-2, CEH-2, CEH-22, and CEH-24, are each required, independently of or in combination with other transcription factors, for target gene expression in and specification of one or a few of the 8 sets of pharyngeal muscles (pm1-pm8) (Harfe and Fire, 1998; Mango, 2009; Morck et al., 2004; Okkema and Fire, 1994; Okkema et al., 1997). NK-2 class homeodomain factors also function in the pharyngeal nervous system. For example, ceh-2 is expressed in the NSM and M3 neurons, and in a ceh-2 deletion mutant M3 was generated but its activity was substantially reduced, indicating ceh-2 acts in a late differentiation step for M3 development (Aspock et al., 2003). Similarly, CEH-28 is required for M4 function; ceh-28 inactivation results in irregularly spaced and sized M4 synapses in the isthmus, and frequent and prolonged peristalses (Ray et al., 2008).
Although MC has long been known to be the pacemaker neuron, regulatory factors critical for its specification and action, other than the FoxA transcription factor PHA-4 which is required broadly for pharyngeal cell fate specification, had not been identified. Here we report that a previously uncharacterized homeobox gene, F20D12.6/ceh-19 (Fig. 1), is specifically expressed in MCs from late embryogenesis through to adulthood. Well-fed, healthy animals bearing the ceh-19(tm452) or ceh-19(tm461) deletion mutations, that both remove most of the homeobox, displayed a moderate reduction of pharyngeal pumping speed. In ceh-19(tm452) mutants, the MC cells are generated, but with obvious axonal morphological defects, suggesting CEH-19 is required for proper specification of the MC neuronal type. Reporter fusions for ceh-19 also directed expression in two pairs of sensory neurons, the amphid and phasmid neurons ADF and PHA. We found that the ceh-19 expression level in ADF was up-regulated in wild type dauer animals, indicating a potential link of this homeoprotein to the C. elegans dauer and/or aging pathways.
FIG. 1.
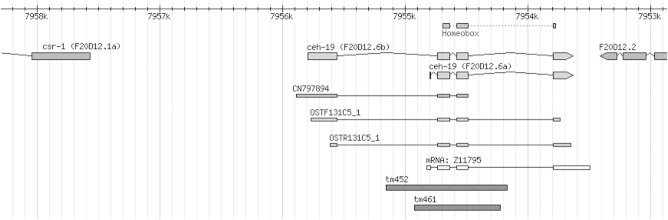
The ceh-19 gene model. In WormBase, ceh-19 is annotated to encode two transcripts, ceh-19a (F20D12.6a) and ceh-19b (F20D12.6b), based on the cDNA clones Z11795 and CN797894, and ORFeome sequence tags (OSTs). The extent of the homeobox and both deletion alleles, tm452 and tm461, is indicated. ceh-19 is located between the genes csr-1 (F20D12.1) and F20D12.2, both transcribed in the opposite direction to ceh-19. The scale is in kb.
RESULTS
F20D12.6/ceh-19 Expression Pattern
In (WormBase) (http://www.wormbase.org), ceh-19 is annotated to encode two transcripts, ceh-19a and ceh-19b. The two transcripts would produce proteins of 122 and 199 amino acids, respectively, with exons 2, 3, and 4, encoding the carboxyl terminal 119 amino acids including the homeodomain, in common, but each with unique first exons (Fig. 1). The C. elegans Promoterome project (Reece-Hoyes et al., 2007) had generated a ceh-19bprom::gfp fusion containing the 1.5 kb region upstream of the ceh-19 b start codon. In strains UL2701, carrying this reporter gene fusion as an extrachromosomal array, and UL2702 and UL2703, containing chromosomally integrated arrays of the same fusion, this assayed promoter region had been described as driving GFP expression in four neurons in the head and two neurons in the tail region, from 3-fold embryos until the adult stage.
Closer inspection of UL2701, UL2702, and UL2703 revealed that the cell bodies of two of the GFP expressing neurons in the head were located in the anterior bulb of the pharynx. There are only 20 neurons within the pharynx and from the axonal morphology these neurons were identified unambiguously as MCL/R (Fig. 2a–c).
FIG. 2.
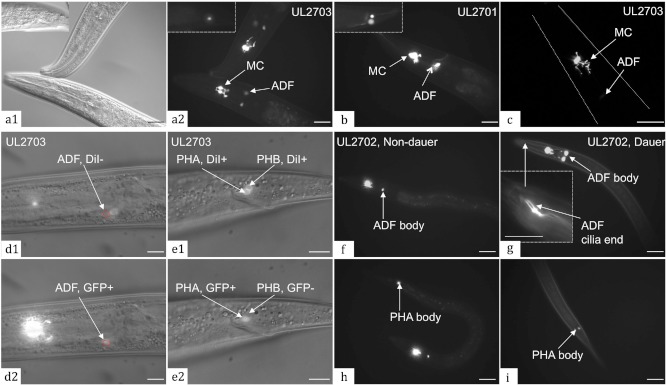
Expression of the ceh-19bprom::gfp fusion. GFP distributions were observed in UL2701 (b), UL2702 (f, g, h, i) and UL2703 (a, c, d, e). Three pairs of neurons, two pairs in the head, MC and ADF, (a, b) and one pair in the tail region, PHA, (a, b insets) expressed GFP. DIC optics (a1) reveals the position of fluorescence (a2) within the same animal. The path of axon projections was determined using confocal Z-sections, as in this projection (c). The amphid neurons ADF (circled in red), expressing the GFP (d2), do not take up DiI (d1). In the tail, DiI stained both phasmids PHA and PHB (e1), whereas only PHA expressed GFP (e2). The red fluorescence (d1, e1) and green fluorescence (d2, e2) are superimposed upon the DIC image. The ADF signal increased in the dauer (g) compared to the L3 (f), both images being captured at the same magnification and with a 1 second exposure. The dual ciliated end of ADF is clearly visible in dauers at high magnification (g inset). The GFP signal in PHA is similar in L3s (h) and dauers (i). Scale bars represent 25 µm in a1/2, b, f, g, h, i, 10 µm in d1/2, e1/2, g inset, and 16 µm in c.
The other two GFP expressing cells in the head had cell bodies outside of the pharynx, just anterior to the posterior pharyngeal bulb, with dendrites extending to the head tip and therefore were identified as amphids. Specific identity was narrowed down by their dual ciliated sensory endings (Fig. 2g inset), a morphological feature of only ADFL/R and ADLL/R. Furthermore as the GFP expressing cells did not take up externally delivered DiI (Fig. 2d1), which stains ASI, ADL, ASK, AWB, ASH, and ASJ, the expressing cells were identified as ADFL/R. In addition, the GFP expression increased in this pair of amphids in the dauer stage (Fig. 2g) and ADFs control entry into the dauer stage (Bargmann and Horvitz, 1991b), suggesting a connection between ceh-19 regulation and dauer formation. The GFP signal was much higher in MCs than in ADFs during normal developmental stages (Fig. 2a, c, d2, f) whereas the strength of signal in the two neuron types was approximately the same in the dauer stage due to the considerable increase of signal within ADFs (Fig. 2g). In UL2701, which contains the transgene as an extrachromosomal array, the difference in intensity in non-dauer stages was not as apparent (Fig. 2b).
The two cells in the tail showing GFP expression have cell bodies located just behind the rectum and short processes in both anterior and posterior directions (Fig. 2b inset, h, i), a morphology suggestive of the phasmids neurons, PHAL/R or PHBL/R. DiI filling, which stains both of these phasmid neuron types revealing their relative positions, confirmed that the pair of neurons expressing GFP in the tail is the more anterior pair, PHAL/R (Fig. 2, E1/2). The extrachromosomal array in UL2701 directed stronger GFP expression in PHA than the integrated array in UL2702 and UL2703. In all three strains there was no apparent difference of signal in PHA between non-dauer and dauer animals (Fig. 2h, i). Although the tail neural anatomy is substantially modified in the male, no male specific expression was observed for the ceh-19bprom::gfp fusion in any of the three transgenic strains.
We further generated two gfp fusions for ceh-19b in a large genomic background by recombineering the fosmid WRM0620bD04. This fosmid contains 18,586 bp upstream and 15,366 bp downstream of ceh-19 and would be expected to contain all ceh-19 regulatory elements. Strains UL3010, UL3011, and UL3012 independently transformed with the fosmid fUL#HF005.1, with the ceh-19b protein-coding region in WRM0620bD04 replaced precisely by gfp, showed consistent GFP expression in ADFL/R, MCL/R, and PHAL/R (Fig. 3a1/2), as obtained with the promoter reporter fusion. GFP was distributed throughout the cell body and along the neuronal axon as expected for a transcriptional fusion, but the signal was lower than with the plasmid-based fusion. The recombineered C-terminal translational fusion, with gfp inserted immediately before the ceh-19 termination codon (fUL#HF004.1) to tag both ceh-19a and ceh-19b, drove GFP expression in apparently the same pairs of neurons, i.e. ADFL/R, MCL/R, and PHAL/R, in 2 independent strains (UL3014, UL3308) (Fig. 3b1/2). GFP was localized to the cell nucleus only, as expected for a transcription factor fusion protein, and at very low level, making cell identification less certain. For both recombineered reporter fusions, expression in ADFL/R was very low through the normal life cycle but increased greatly in dauer animals (Fig. 3a3, b3), as for the plasmid-based fusions, allowing the cell processes to be seen more clearly (Fig. 3a3). This increase was not observed in animals that had simply been starved (data not shown). DiI filling on these strains was consistent with ADF and PHA identification.
FIG. 3.
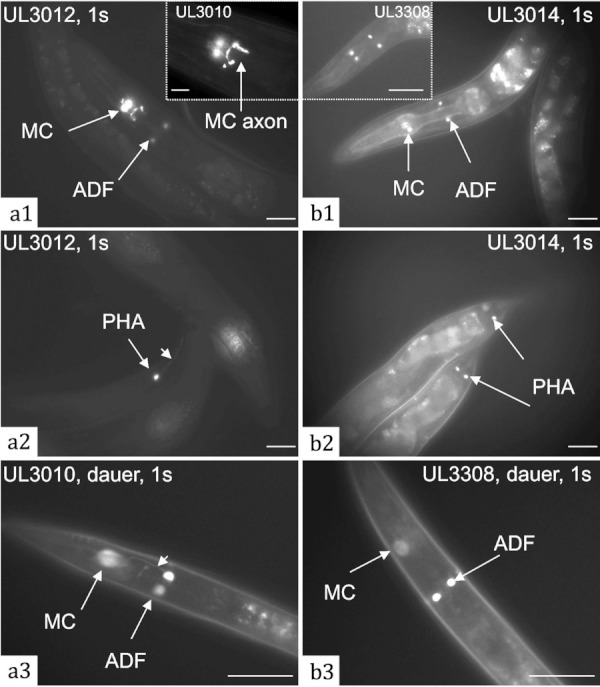
GFP expression of fosmid-based reporter fusions for ceh-19b. The recombineered fosmid fUL#HF005.1, with the ceh-19b protein coding region replaced by gfp, drove GFP expression in MCL/R, ADFL/R and PHAL/R in UL3010 and UL3012 (a1–3). Expression was detectable in axons of MC and PHA (a1 inset, a2 arrow head). The recombineered fosmid fUL#HF004.1, with gfp inserted immediately before the stop codon of ceh-19b, also drove GFP expression in MCL/R, ADFL/R and PHAL/R, in UL3014 and UL3308 (b1–3), but to a low level and with nuclear-localization. Expression in ADFs increased in dauer animals such that the axon of ADF was visible in UL3010 (a3, arrow head) and ADF nuclei became much brighter in UL3308 (b3). Bars represent 25 µm in a1–3, b1–3 and inset, and 10 µm in a1 inset.
A reporter fusion was constructed to assay the expression pattern of the annotated ceh-19a transcript, supported by the single EST Z11795 and SAGE tags for exon 1 (Naito et al., 1992). The gfp reporter was fused to the 2770 bp region upstream of the ceh-19a start codon, including the start of transcript b and the entire intergenic region, by Gateway recombinational cloning, creating pUL#HF053. However, in two strains transformed with this plasmid no GFP expression was observed, throughout the normal life cycle or in the dauer, suggesting the ceh-19a transcript may not be functional or may only be expressed in circumstances not assayed. The absence of additional expressing cells for the C-terminal translational fusion is also consistent with ceh-19a not adding to the expression arising from ceh-19b.
Molecular Phylogeny of CEH-19 and Its Homologues
Within the C. elegans genome, ceh-30, ceh-31, tab-1, and ceh-1 have the highest similarity to ceh-19b by BLAST. Each of the five paralogues has one orthologue (best reciprocal BLAST match) in Caenorhabditis briggsae and Caenorhabditis remanei, suggesting conserved functional importance. CG13424 and CG10604 (bsh, brain specific homeobox) are the Drosophila melanogaster genes most similar to ceh-19b. CG13424 has not been functionally characterized. bsh is expressed in approximately 30 cells in each brain hemisphere of the Drosophila embryo but a bsh deletion showed no dramatic changes in embryonic brain morphology and therefore its function in the Drosophila brain is unknown (Jones and McGinnis, 1993). Two BarH-like homeobox genes, BARHL1 and BARHL2 are the closest homologues of ceh-19b in the human genome.
WormBase has CEH-19 placed in the BarH-like TF family, presumably based on its closest human homologues. However, BarH family members normally possess a highly conserved tyrosine (Y) residue at position 49 of the third helix of the homeodomain, as in the homeodomains of Caenorhabditis CEH-30 and CEH-31 and human BARHL1 and BARHL2 but not CEH-19 (Fig. 4) (Peden et al., 2007; Reig et al., 2007). In addition, typical BarH type homeoproteins have one or more FIL domains, enriched in phenylalanine (F), isoleucine (I), and leucine (L) residues, within the amino terminal region (Germán Reig et al., 2007) and a conserved 22-amino-acid BARC motif, immediately downstream of the homeodomain (Schwartz and Horvitz, 2007), which are also absent from CEH-19. Therefore CEH-19 does not appear to be a C. elegans orthologue of vertebrate BarH-like proteins. With no function yet determined for the potential CEH-19 orthologues in Drosophila either, no clues as to CEH-19 function can be inferred from other species.
FIG. 4.
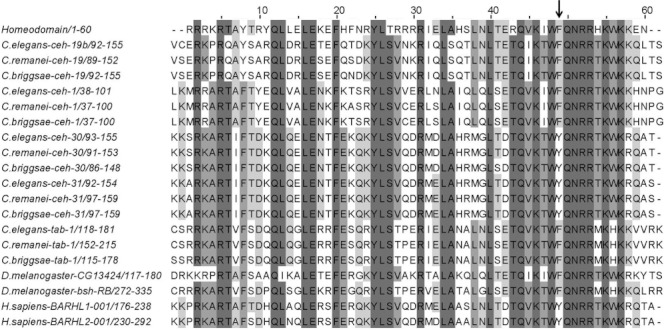
Protein sequence alignment of the homeodomains of C. elegans CEH-19 and its homologues from C. briggsae, C. remanei, D. melanogaster, and H. sapiens, in comparison with the more general consensus homeodomain. The arrow points to position 49 of the homeodomain, which is a characteristic tyrosine (Y) residue for the typical BarH homeoproteins rather than the more common phenylalanine (F). Alignment was performed using ClustalW software and edited with Jalview 2.5.1.
Instead of the FIL domain, the N-termini of CEH-19 across the Caenorhabditid species are enriched for the acidic residues aspartate (D) and glutamate (E) typical of a transcription activation domain (Willmore, 2004). Acidic activation domains from many transcription factors have been shown to interact with the general transcription factors TFIIB, TFIID, and TFIIH of the basal transcription machinery (Xiao et al., 1994). Lack of this N-terminal region in CEH-19a makes it less likely that this short isoform, if expressed, could function as a transcription activator, but it may still make sequence specific DNA interactions and form homodimeric complexes, or heterodimeric complexes with other factors, using its homeodomain and carboxy-terminal region.
ceh-19 (tm452) Has Reduced Pharyngeal Pumping Speed
To investigate the function of ceh-19, two C. elegans ceh-19 mutant alleles were obtained and examined for a phenotype altered from the wild type. The ceh-19(tm452) and ceh-19(tm461) alleles, generated by TMP/UV mutagenesis, have 993 bp and 701 bp deletions, respectively. Both alleles lack the two middle exons of ceh-19b and the first three exons of ceh-19a (Fig. 1). This was verified by PCR and sequencing. The mutations delete most of the ceh-19 homeobox and potential splicing together of the remaining two exons of transcript ceh-19b would cause a shift to an incorrect translational reading frame for the last exon. The truncated peptide encoded by such a hypothetical transcript would retain the potential acidic transcriptional activation domain but none of the homeodomain. Therefore, tm452 and tm461 are likely to be null alleles. The tm452 allele was backcrossed to the N2 wild type seven times to generate UL3128, to remove other mutations that might be present in the original strain.
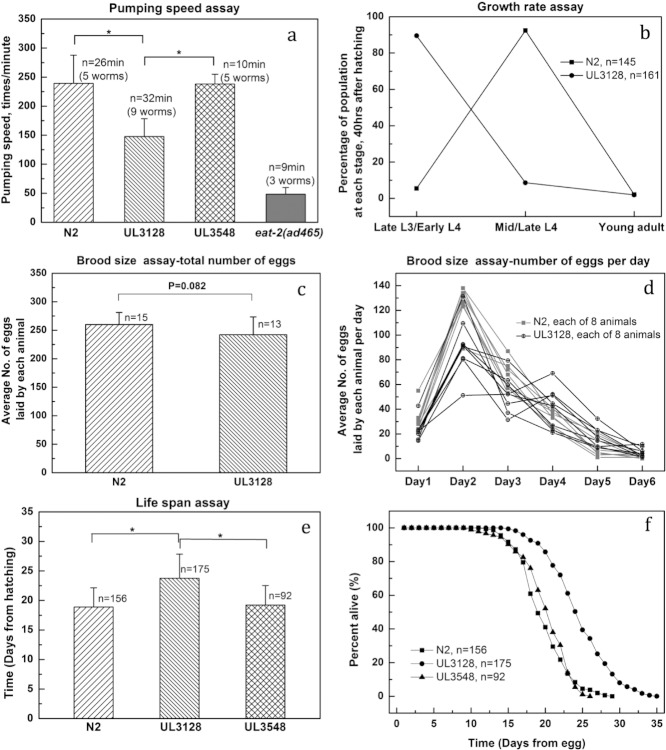
The original strains bearing the tm452 and tm461 alleles and UL3128 are viable and fertile and have no immediately obvious morphological, physiological, or locomotion defects. However, the specific expression of ceh-19::gfp in the pharyngeal motor neuron MC, which is necessary for rapid pharyngeal pumping (Avery and Horvitz, 1989), suggested the ceh-19 mutants might be defective in pharynx function. Any such defect would need to be to a degree that is not strong enough to cause other more obvious defects, such as a starved body appearance. Closer examination of pumping behavior indicated that pharyngeal pumping rate of UL3128 was indeed reduced by more than 30% compared to N2 animals (Fig. 5a). In N2 individuals the pharynx pumped 239 ± 48.5 times per minute (mean ± s.d.) compared to 148 ± 30 times per minute for UL3128. The unbackcrossed tm461 mutant animals have an almost identical pumping speed to UL3128 (data not shown), consistent with the two ceh-19 alleles having essentially the same consequences. On an agar plate in the presence of abundant bacteria, pharynxes of N2 pump continuously at a near steady frequency. In contrast pharynxes of UL3128 pump in alternate cycles of 7-10 pumps at high and low frequencies, the slower-pumping phase responsible for reducing the average speed. This defect is not as severe as when both MC neurons were killed with a laser, which reduced the speed to 45 ± 6 pumps/minute (Raizen et al., 1995), indicating that MC function is not completely inactivated in the ceh-19 mutants. The ceh-19 mutant defect is not even as severe as that for eat-2(ad465) (Fig. 5a) which affects synapses between the pharyngeal muscles pm4 and pm5, post-synaptic to MC (Mckay et al., 2004). As for N2, pharyngeal pumping is absent in UL3128 animals on plates with no bacterial food, and when pumping is restored by supplementation with 10 mM serotonin UL3128 still pumps at a slower rate than N2. This suggests that the compromised pumping rate of UL3128 is due to a defect in the pharynx, and presumably MC itself, rather than in the ability to sense food. UL3128 was transformed with the fosmid fUL#HF004.1 bearing the full-length ceh-19::gfp translational fusion to rescue the ceh-19 defect. The rescued strain, UL3548, had recovered pumping speed (241 ± 19.2 pumps/minute), very close to that of wild type worms, indicating that the pharynx defect in UL3128 is indeed due to the absence of ceh-19 (Fig. 5a).
FIG. 5.
Effect of ceh-19 deletion on life-history traits. (a) Mean pumping speeds ±s.d. are presented for N2, UL3128 (the backcrossed ceh-19 mutant), UL3548 (the ceh-19 rescued strain) and eat-2 (ad465) mutants. (b) The growth rates of N2 and UL3128 were compared under standard culturing conditions by recording the number of individuals that had become late L3/early L4s, mid/late L4s or young adults, 40 hrs after hatching. (c) UL3128 hermaphrodites appeared to produce slightly fewer progeny than N2 but the difference was not statistically significant (P=0.082) according to a one-way ANOVA. (d) Daily egg-laying of 8 UL3128 hermaphrodites was scored (black) and compared with that of 8 N2 hermaphrodites (grey) over the six-day reproductive period. (e) The average mean life span (with standard deviation) of N2, UL3128 and UL3548 individuals is plotted in days. (f) The percentage of individuals surviving on each day was determined for N2, UL3128 and UL3548. “*” indicates a statistically significant difference by a one-way ANOVA test, P<0.0001.
Consequences of ceh-19(tm452) for Other Life-History Traits
Reduced food intake can influence other life-history parameters such as the development rate, and UL3128 animals showed slightly slower growth than N2, presumably as a result of the slower pharyngeal pumping. Growth rate was assessed in two ways. First, when two synchronized L1s were placed on each of six seeded agar plates, and incubated for 70 h at 20°C, 10 of 12 N2 worms had developed into mature adults, and there were scores of laid eggs with a few hatched L1 larvae on each plate. In contrast, when UL3128 L1s were used, each plate had only around ten eggs and no L1s had appeared. By the tenth day of incubation, the N2 populations had exhausted the bacteria but the UL3128 populations required another 20–24 h to use up the food source. In a second assessment, after 40 hr growth at 20°C, 3 of 145 synchronized N2 L1s had grown into young adults, 8 were late L3s/early L4s, and the remainder were mid/late L4s. For 161 UL3128 L1s, after 40 hr there were also 3 young adults, but the vast majority were late L3/early L4s and only 14 were mid/late L4s, in clear contrast with the N2s (Fig. 5b).
The period of fecundity is prolonged in UL3128 animals. Although the average number of embryos produced by each UL3128 hermaphrodite (brood size=242 ± 31, n=13) appeared slightly less than that of N2 (brood size=260 ± 21, n=15) (Fig. 5c) this difference was not found to be statistically significant. However, the period of fecundity of UL3128 seemed prolonged (Fig. 5d). N2 hermaphrodites laid most of their eggs (83.2 ± 4.1%, n=8) within the first 3 days of the reproductive period with a peak on the second day (Fig. 5d). UL3128 animals laid a smaller proportion of eggs during the first 3 days (72.1 ± 9.4%, n=8) and laid more eggs on subsequent days, especially on Day 4. In addition, UL3128 showed considerably more variation in individual egg-laying curves than N2. When the number of eggs retained in the uterus was checked at 90 h after hatching (on the 2nd day of the reproductive period), N2 adults retained on average 13 ± 4.6 (n=10, mean ± s.d.) eggs in the uterus and UL3128 had 8.7 ± 4.2 (n=10, mean ± s.d.). At 96 h after hatching, N2 had 9.5 ± 1.5 eggs within the uterus of each hermaphrodite (n=14, mean ± s.d.) and UL3128 had 8.2 ± 2.3 (n=13, mean ± s.d.). This provided further evidence of the slight difference of brood size and egg-laying pattern between N2 and UL3128.
UL3128 has an extended life span. Reduced food intake lengthens life span in C. elegans as observed for many eat mutants with their defects in pharyngeal function (Lakowski and Hekimi, 1998). UL3128 is also essentially a weak eat mutant and the life span assay for this strain was performed twice, independently (Fig. 5e,f). N2 worms on average lived 18.9 ± 3.3 days (n=156, mean ± s.d.), very close to the 19.5–21.6 days reported previously by Lakowski and Hekimi (1998) who also fed the worms on live OP50 bacteria. For UL3128 the lifespan was 23.8 ± 4.1 days (n=175, mean ± s.d.), a lifespan extension similar to that of eat-1(e2343) (23.9 ± 0.9 days, mean ± SEM) and eat-6(ad997) (23.6 ± 0.7 days, mean ± SEM) mutants, when assayed with the same protocol (Lakowski and Hekimi, 1998). The pharyngeal pumping speed of eat-1(e2343) mutants is not published, but the pharyngeal pumping rate of eat-6 (ad997) mutants (∼150 pumps/minute) (Doi and Iwasaki, 2008) was very close to that of UL3128. These observations are consistent with the good correlation between life span extension and the severity of the eating defect, as has been observed previously (Lakowski and Hekimi, 1998).
Rescue of ceh-19 in UL3548 shortened lifespan compared to that of the ceh-19 mutant, towards that of the wild type (Fig. 5e,f). The rescued animals had a lifespan of 19.2 ± 3.3 days (n=92, mean ± s.d.). Statistically, the UL3548 lifespan was significantly shorter than for UL3128 and was not significantly different from that for N2. Transformation with CEH-19 did appear to rescue the lifespan extension phenotype of the ceh-19 mutant, as for the decreased pharyngeal pumping speed phenotype.
UL3128 animals enter into and exit from the dauer stage normally. Expression of ceh-19 in ADF and PHA neurons might suggest a role connected with the dauer stage. Although UL3128 animals develop to adults at a slower rate, dauer formation was not observed in normal growth conditions; the ceh-19 deletion does not confer a dauer formation constitutive (Daf-c) phenotype. Efficiency of dauer recovery for N2 and UL3128 was also compared and no difference was observed. All dauer animals (n=∼200) resumed development within 24 hrs after food became available again. Possible further roles of ceh-19 in chemosensation, suggested from expression in these sensory neurons, were not investigated.
Finally, defecation did not appear affected in the ceh-19 mutant. The period of the defecation cycle of UL3128 remained at 40 ± 1 s, similar to, but apparently even more regular than, N2 at 42 ± 3.8 s (mean ± s.d., n=5 worms each, 10 min of observation for each worm). This is consistent with previous observations that bacteria intake has only a minor effect on the period of the defecation cycle (Thomas, 1990).
ceh-19 Is a Novel Eat Gene
Previous screens for pharyngeal pumping defects had yielded the Eat mutants (Avery, 1993). Almost all Eat mutations characterized so far result in severe impairment to the pharynx and most of them also affect tissues outside the pharynx (Avery, 1993; Shibata et al., 2000). The molecular nature of some of these genetically identified Eat genes, including eat-1, eat-8, eat-9, eat-10, eat-13, eat-14, eat-15, and eat-17, remain to be determined. Among these, eat-1 and eat-10 have been mapped onto linkage group IV where ceh-19 is found. eat-10(ad606) is on the left arm of LGIV (IV: −26.74 ± 0.306cM), far from ceh-19 (IV:3.51 ± 0.001cM). However, eat-1 was mapped onto LGIV at 4.8 ± 6.395cM, and both its recessive alleles, ad427 and e2343, have slow and irregular pharyngeal pumping, like tm452. The very large confidence interval of the eat-1 location presumably reflects problems with the genetic mapping but does cover the locus of ceh-19. We found, however, that all three eat-1 mutants have even slower pumping speed than the ceh-19 deletion mutants: DA531[eat-1(ad427)IV] at 89.1 ± 21.7, CB4394 [eat-1(e2343) unc-31(e928)IV] at 87 ± 17.9, and DA449[eat-1(e2343) dpy-20(e1282)IV] at 106 ± 17.4 times per minute, respectively (mean ± s.d, n=10). And direct tests revealed ceh-19 and eat-1 are not allelic; PCR amplification of ceh-19 from genomic DNA of eat-1 mutants yielded products of wild type sizes and tm452 complements genetically all three eat-1 mutant alleles. Therefore, the molecular nature of eat-1 remains to be determined and ceh-19 is a newly identified Eat gene, a homeobox gene that is necessary for C. elegans pharynx development and proper pumping.
UL3128 Animals Have Morphological Defects in MC
To assess whether the specification and development of MC, ADF and PHA were affected by the ceh-19 deletion and also whether CEH-19 is required for its own expression in these neurons, their morphology was examined in the ceh-19(tm452) deletion background using ceh-19bprom::gfp fusions. The chromosomally integrated transgenic array leIs2703[ceh-19bprom::gfp, unc-119(+)] was crossed from strain UL2703 into the ceh-19(tm452) mutant background of UL3128 to give strain UL3413. The extrachromosomal array leEx3011 and leEx3012 both carrying the recombineered fosmid fUL#HF005.1 with the ceh-19b protein coding region replaced with gfp, in the strains UL3011 and UL3012 respectively, were also crossed into the ceh-19(tm452) mutant background of UL3128 to give strains UL3013, and UL3019 and UL3022.
In each strain with the ceh-19(tm452) deletion background, GFP expression in all three pairs of neurons remained at a similar level to that in the wild type background, suggesting MC, ADF, and PHA are still born normally in the absence of CEH-19, and ceh-19 expression is not self-regulated. The ceh-19bprom::gfp expression was also examined in dauer animals in the ceh-19 mutant background as the change in strength of expression in ADF in this stage might have depended on functional CEH-19, but again no difference from wild type levels was observed (data not shown). Although apparently lacking a role in the fundamental generation of these nerve cells, CEH-19 could still be functional in their specific terminal differentiation. Indeed, in 90% (n>100) of mutant animals, there were axonal defects in the MC neurons to varying degrees. MCs in wild type animals have symmetrical, well-organized cell bodies and consistent axonal projections as revealed under epifluorescence microscopy (Fig. 6a, b) and in confocal microscopy sections (Fig. 6c, d). Although usually both MC cell bodies were still present in the ceh-19 mutant background, they were often different sizes and shapes (Fig. 6h-l). Moreover, severe abnormalities were observed for MC axons, by both epifluorescence and confocal microscopy (Fig. 6e-m), which presumably would result in abnormal inter-neuronal and neuro-muscular connections. Occasionally, in the mutant background, GFP was only apparent in one MC (Fig. 6m). These defects are consistent with the moderate pharyngeal pumping defect observed for the ceh-19 mutant animals.
FIG. 6.
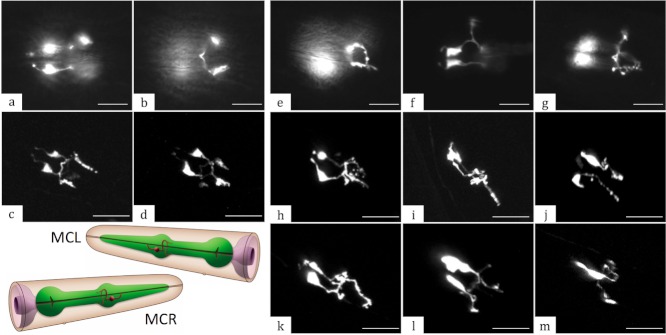
The ceh-19 deletion causes axonal defects in MC neurons. In the wild type, MCs have symmetrically located cell bodies and well-organized, consistent axonal projections, as observed by both epifluorescence (a, b) and confocal (c, d) microscopy. In the ceh-19(tm452) mutant background, the ceh-19b::gfp transcriptional reporter fusions were still expressed in the MCs, but revealed various subtle to moderate axonal defects upon epifluorescence (e–g) and confocal microscopy (h–m). All images were captured in the region of anterior pharyngeal bulb with anterior towards the upper left. Strains presented are: UL3012 (a–d), UL3022 (e–g), UL3013 (h–i), UL3019 (j) and UL3022 (k–m). A panel depicting the morphology of MCL and MCR (red) with respect to the pharynx (green) in the wild type is included for comparison (from WormAtlas). Scale bars represent 10 µm in all panels.
No axonal defects were observed by epifluorescence microscopy for ADF and PHA neurons in the ceh-19 deletion background (data not shown). These amphid and phasmid neurons might have minor structural and/or synaptic defects beyond detection from observations of the GFP expression but which could be revealed by electron microscopy.
Regulatory Context of ceh-19 in MC
PHA-4 is the pharyngeal master regulator required for development of all cells of the pharynx (Mango, 2009) and the expression of ceh-19 in MC is dependent on this transcription factor. We assayed the dependence of ceh-19 expression on PHA-4 by applying pha-4 RNAi to gravid UL2703, with the chromosomally integrated ceh-19bprom::gfp fusion, and examining GFP in the progeny. As expected, knocking down PHA-4 resulted in embryonic lethality and/or larval arrest for UL2703. Animals that did manage to hatch out appeared to lack a pharynx with no GFP corresponding to MC, although GFP in ADF and PHA remained (Fig. 7a1/2). Presumably no cell with an MC fate is generated with PHA-4 absent.
FIG. 7.
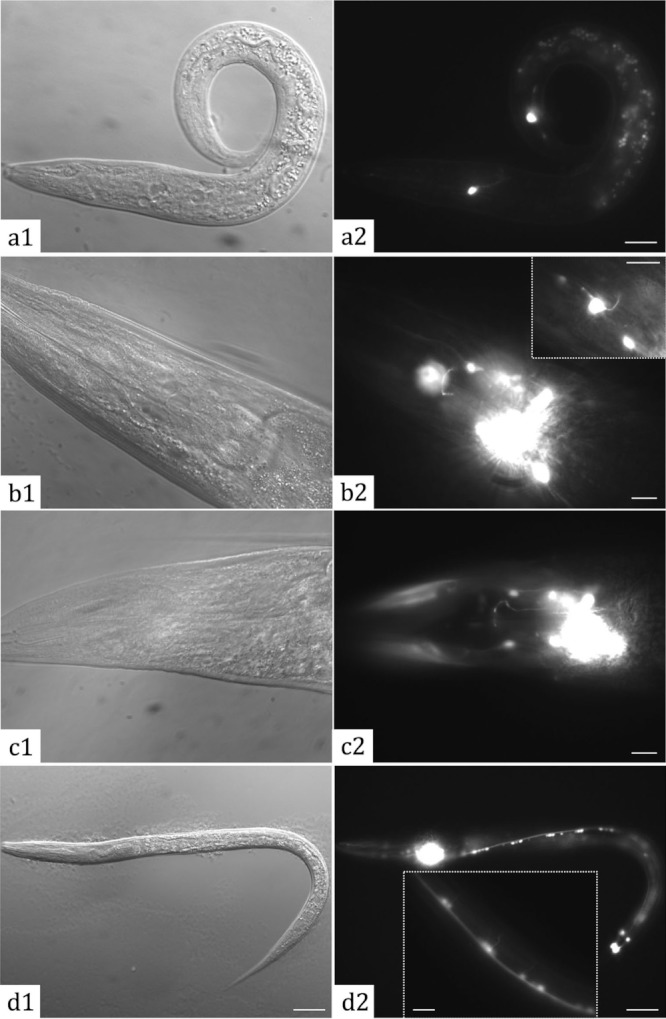
Regulation upstream and downstream of ceh-19. Larva hatched from UL2703 animals transgenic for ceh-19bprom::gfp and subject to RNAi for pha-4 are pharynxless (a1), and although expression in ADF and PHA was maintained, expression in MC was lost (a2). In a wild type background, flp-2prom::gfp is expressed in MC (b2 and b2 inset) and other neurons (b1/2). In the ceh-19(tm452) background, flp-2prom::gfp expression was not detected in MCs but was still present in other neurons at the same level as in wild type (c1/2). flp-2prom::gfp expression was observed in ventral cord motor neurons in the dauer stage only (d1/2) including the axon (d2 inset), here in the wild type. Bars represent 10 µm in a–c, b2 inset and d2 inset, and 25 µm in d. Corresponding images were captured by DIC (a–d1) or epifluorescence (a–d2) microscopy.
CEH-19 is required for flp-2 expression in MC. flp-2 and flp-21 both encode FMRFamide-like peptide neurotransmitters and have been reported to be expressed in MC, in addition to multiple other nerve cells (Kim and Li, 2004). To assay the genetic dependence of these genes on CEH-19, we generated strains UL3795 and UL3793, transgenic for flp-2prom::gfp and flp-21prom::gfp fusions respectively, and indeed detected GFP expression in MC neurons in the wild type background. The reporter fusions were then crossed into UL3128 with the ceh-19 deletion background, resulting in strains UL3881 [ceh-19(tm452); flp-2prom::gfp] and UL3891 [ceh-19(tm452); flp-21prom::gfp], to assess their regulation by CEH-19. While there was no difference in MC expression of flp-21prom::gfp between the wild type and ceh-19 mutant (data not shown), flp-2 expression in MC appeared dependent on CEH-19. In the wild type background, flp-2prom::gfp gave a clear consistent GFP signal in MCs, in UL3795 and 3 other independent transgenic strains, although expression was normally weaker than in other cells (Fig. 7b1/2). In contrast, in the ceh-19 mutant, flp-2prom::gfp expression in MC was not detectable (Fig. 7c1/2), suggesting CEH-19 is required for flp-2 expression in MC. This requirement was specific to MC, the only cells in which ceh-19 and flp-2 expression overlaps, with flp-2 expression in other cells not affected in the more than 50 individuals where expression was carefully and specifically examined in this regard. MC expression of flp-2prom::gfp in dauer animals in both the wild type and ceh-19 mutant backgrounds was also compared. Although ceh-19bprom::gfp was expressed strongly in MC during the dauer stage, MC expression of flp-2prom::gfp in dauers with wild type background was weaker than in non-dauers or even absent. Nevertheless flp-2prom::gfp MC expression was never observed in the ceh-19 mutant dauers, confirming the requirement of CEH-19 for flp-2 expression, even in the dauer. Interestingly, while examining flp-2prom::gfp MC expression, a strong GFP signal was observed in ventral cord motor neurons of dauers in both wild type and ceh-19 mutant backgrounds (Fig. 7d1/2). Such expression for flp-2 was not reported previously and was not seen in non-dauers, indicating a potential involvement of flp-2 in dauer physiology.
Evidence for direct interaction between PHA-4 and ceh-19 and between CEH-19 and flp-2 were sought using the yeast one hybrid (Y1H) approach. The strategy could also have yielded additional targets of CEH-19 and regulators of ceh-19. A previously described C. elegans transcription factor yeast array (Reece-Hoyes et al., 2011) was used in a yeast-one-hybrid screen with the ceh-19b promoter as bait. Out of the 755 transcription factors in the array, three, TBX-8, TBX-9, and MLS-2, appeared to bind the ceh-19b promoter. Although genetically required for ceh-19 expression in MC, PHA-4 did not appear to bind the ceh-19 promoter in these Y1H TF array screens. This was also specifically tested with ceh-19bprom-lacZ bait yeasts directly transformed with the Gal4AD-PHA-4 and Gal4AD-only prey plasmids. Both showed the same level of lacZ expression, yielding no support for specific binding of PHA-4 to the ceh-19b promoter. Twenty genes reported to be expressed in MC, ADF and/or PHA, including flp-2 and two transcription factor genes, lin-11 and lim-4, were selected as potential CEH-19 targets (Supplementary Table 2). Promoters for these genes, either retrieved from the Promoterome or cloned ab initio, were fused with the Y1H lacZ and His3 reporters for screening as baits versus the C. elegans TF yeast array. CEH-19 did not appear to bind to any of these baits, although various other transcription factors did (data not shown).
DISCUSSION
The C. elegans genome encodes ∼100 homeodomain transcription factors, 24 of which lack any other highly conserved motif (Reece-Hoyes et al., 2005). Currently these “homeodomain-only” transcription factors, including ceh-19, are less well characterized although over half are essential, with various developmental and behavioral roles. Reporter gene fusions have been assayed for most and typically revealed expression in a range of cell types, e.g., (Reece-Hoyes et al., 2007). The restricted distribution of reporter expression for ceh-19, however, led to its study here.
The lack of reporter expression with a ceh-19a specific fusion, and lack of additional components for the fusion targeting both transcripts as compared to fusions targeting ceh-19b specifically, suggests ceh-19a is at best poorly expressed under laboratory conditions. The sequence of the donor splicing sites of ceh-19b intron 1 (AGgtaatcat) matches more closely to the well-respected C. elegans consensus sequence (AGgtaagttt) than does that of ceh-19a intron 1 (TTgtatgaaa). Furthermore, the predicted CEH-19a protein would lack the region probably responsible for transcriptional activation in CEH-19b. The significance of ceh-19a, therefore, looks doubtful and the ceh-19b gene model appears to represent the full function of the gene.
Although the defect in pharyngeal pumping in the ceh-19 mutant is likely to relate purely to expression in MC, the clear expression of the ceh-19::gfp fusion in ADF and PHA suggests ceh-19 contributes to the sensory roles of these neurons. Defects relating to ADF and PHA sensory function may, however, be subtle due to redundancy with other cells or genes, or may not have been apparent because of ecological specificity. Nevertheless, the striking up-regulation in the ADF of dauers could be significant in this regard. C. elegans dauer entry and exit involve integration and transformation of environmental cues (dauer pheromone, nutrients, and temperature) into endocrine signals by amphid neurons. Amphids ADF, ASI and perhaps ASG act redundantly to prevent dauer formation in favourable conditions, and amphid ASJ is critical to exit from the dauer stage with some minor contributions from ADF or ASI, or ASG (Bargmann and Horvitz, 1991a; Bargmann and Horvitz, 1991b). This redundancy amongst the amphids means the lack of an effect of the ceh-19 deletion on dauer entry or exit is not surprising. The up-regulation suggests increased levels of CEH-19 are important in preparing ADF for a role in the dauer stage, presumably for sensing of conditions for dauer exit or perhaps contributing to the resilience to adverse conditions characteristic of the dauer stage, in this case specifically of ADF. Such roles may only be revealed with ASI, ASG and ASJ inactivated. The increased ceh-19 expression in ADF would itself be a consequence of endocrine signals acting to direct dauer entry. This, along with the flp-2::gfp expression in ventral cord neurons of only the dauer stage, and the expression of this reporter fusion in MC being ceh-19 dependent seems unlikely to be coincidental but the biological significance is not yet apparent. ceh-19 has not been identified previously in global expression analyses seeking genes expressed specifically or with altered levels in the dauer stage (Holt, 2006; Jones et al., 2001; Wang and Kim, 2003). This may be because the problem of low expression, as seen with other transcription factor genes, is further exacerbated by expression being restricted to just a few small cells. Reporter gene fusions provide the localized sensitivity needed to observe such subtle effects.
The minor nature of the ceh-19 mutant's defects, compared with those upon MC ablation, suggests MC function is only partially disrupted. As MC cell processes are abnormal in ceh-19(tm452) mutants, a simple hypothesis would be that receipt or transmission of neural signaling is impaired due to the improperly specified synaptic connections between MC and muscles and/or other neurons. As tm452 and tm461 are likely to be null alleles, ceh-19 may only be required for partial or non-essential aspects of MC specification, with only limited types of terminal differentiation genes being controlled by CEH-19. Alternatively, the robustness of transcription factor regulatory networks may mean that the level of expression of genes expressed for MC terminal differentiation are only moderately perturbed by the complete absence of CEH-19 activity. CEH-19 would contribute to the level of expression of many genes needed for MC fate, but other transcription factors are required for MC differentiation, with roles that remain to be identified, and buffer against ceh-19 loss.
The gene flp-2, the identified target of CEH-19 regulation in MC, encodes two FMRFamide-like neuropeptides (FLPs), FLP-2A and FLP-2B. Different C. elegans pharyngeal neurons express different sets of modulatory FLP neuropeptides (Husson et al., 2007; Kim and Li, 2004). While eleven flp genes encode inhibitors of pharyngeal activity, eight encode excitatory peptides (Papaioannou et al., 2005). FLP-2A, expressed in pharyngeal neurons M4, MC and I5, enhances pharyngeal activity. In contrast, FLP-21, also expressed in MC, suppresses pumping rate (Papaioannou et al., 2005). Hence, in a single neuron, MC, at least two FLPs are expressed with opposing activity, acting in response to different environmental stimuli to fine-tune pumping rate. Presumably, complex transcription factor regulatory networks integrate the subtly distinct nerve cell fates with the different environmental situations encountered so as to achieve the expression of each member of the range of flp genes precisely where, when, and to the levels required. CEH-19 is essential for flp-2 expression in MC. However, interruption of flp-2 by RNAi caused embryonic lethality, slow growth and larval arrest (Simmer et al., 2003), a more severe phenotype than the slow pumping when only inactivated in MC and in accordance with its broad expression and a wider role. Clearly other transcription factors must input into flp-2 such that expression of flp-2 doesn't occur wherever CEH-19 is expressed and can occur in cells beyond where CEH-19 is expressed. As the MC fate is only partially perturbed in ceh-19 mutants, other transcription factors must control other MC terminal differentiation genes, such as flp-21. Furthermore, transcriptional control of flp-2 is not the only role for CEH-19 in MC. The MC morphological defects in ceh-19 mutants indicate transcription of other genes is needed for proper MC differentiation.
Despite the genetic evidence of regulatory dependency, CEH-19 did not bind the flp-2 promoter and PHA-4 did not bind the ceh-19 promoter, in Y1H assays. These regulatory interactions could be indirect in vivo with other intermediate transcription factors, or direct but with co-factors required. A direct interaction of PHA-4 to the ceh-19b promoter might have been expected as PHA-4 directly activates many genes expressed in the different cell types in the pharynx (Gaudet and Mango, 2002). Furthermore, modENCODE ChIP experiments identified PHA-4 binding to the ceh-19 promoter, within the region assayed in the Y1H experiments, but only in L1s and not other stages (Gerstein et al., 2010). Given the lack of any other supporting data, the significance of our finding of TBX-8, TBX-9, and MLS-2 binding to the ceh-19 promoter in Y1H assays, for regulation of ceh-19 expression in vivo is unclear. Although our targeted Y1H screen for potential CEH-19 targets, using 20 candidate gene promoters, was unsuccessful, a larger-scale screen has since revealed Y1H binding of CEH-19 to promoters of cog-1, vha-15, hlh-15 and lbp-8 (Reece-Hoyes et al., 2011). The biological relevance of these interactions for ceh-19 function in vivo is also as yet unclear. Genome-wide screens for direct protein::DNA interactions involving transcription factor combinations would be a huge undertaking even with an assay as easy to apply as the Y1H. Other data, such as from expression pattern determinations or ChIP, may be needed to reduce the number of transcription factors that need to be tested, combinatorially, in such assays before the details of transcription factor regulatory networks can be revealed.
METHODS
C. elegans Strains and Mutant Alleles
All strains were maintained at 20°C on 5 cm NGM agar plates seeded with E. coli OP50 as food source (Sulston and Hodgkin, 1988). C. elegans N2 (Bristol) was used as wild type. Transgenic strains are UL2701 [unc-119(ed3)III; leEx2701(ceh-19bProm::gfp, unc-119(+))], UL2702 [unc-119(ed3)III; leIs2702 (ceh-19bProm::gfp, unc-119(+))], UL2703 [unc-119(ed3)III; leIs2703(ceh-19bProm::gfp, unc-119(+))] (Reece-Hoyes et al., 2007). C. elegans strains with ceh-19(tm452) and ceh-19(tm461) alleles were from the Mitani Laboratory (http://www.shigen.nig.ac.jp). The ceh-19(tm452) allele was backcrossed into N2 seven times to generate the strain UL3128, which was used in most phenotypic assays.
Transgenic strains generated in this work include: UL3010 [leEx3010(fUL#HF005.1, rol-6(su1006))], UL3011 [leEx3011(fUL#HF005.1, rol-6(su1006))], and UL3012 [leEx3012(fUL#HF005.1, rol-6(su1006))], each carrying the recombineered fosmid fusion fUL#HF005.1 in an extrachromosomal array; UL3014 [leEx3014(fUL#HF004.1, rol-6(su1006))] and UL3308 [leEx3308(fUL#HF004.1, rol-6(su1006))], each carrying the recombineered fosmid fusion fUL#HF004.1 in an extrachromosomal array; UL3413 [ceh-19(tm452)IV; leIs2703(ceh-19bProm::gfp, unc-119(+))], created by crossing leIs2703 from UL2703 into UL3128; UL3013 [ceh-19(tm452)IV; leEx3011(fUL#HF005.1, rol-6(su1006))], created by crossing leEx3011 from UL3011 into UL3128; UL3019 [ceh-19(tm452)IV; leEx3012(fUL#HF005.1, rol-6(su1006))] and UL3022 [ceh-19(tm452)IV; leEx3012(fUL#HF005.1, rol-6(su1006))], created by crossing leEx3012 from UL3012 into UL3128; UL3548 [ceh-19(tm452)IV; leEx3014(fUL#HF004.1, rol-6(su1006))], created by crossing leEx3014 from UL3014 into UL3128; UL3795 [leEx3795(flp-2prom::gfp, rol-6(su1006))], UL3793 [leEx3793(flp-21prom::gfp, rol-6(su1006))] and UL3891 [ceh-19(tm452)IV; leEx3891(flp-21prom::gfp, rol-6(su1006))], created by microinjection of the wild type, N2, or the ceh-19 mutant, UL3128, with the plasmid containing the flp-2prom::gfp or flp-21prom::gfp fusions constructed by Gateway recombination; UL3881 [ceh-19(tm452)IV; leEx3795(flp-2prom::gfp, rol-6(su1006))], created by crossing leEx3795 from UL3795 into UL3128.
Reporter Gene Fusions and Expression Pattern Analysis
Recombineering of C. elegans fosmid clones was carried out according to Bamps and Hope (2008). flp-2prom::gfp and flp-21prom::gfp fusions were generated by Gateway recombinational cloning as described by Reece-Hoyes et al. (2007). Primer sequences are provided in Supporting Information Table 1.
A Leica DMR HC microscope fitted with GFP (Chroma 41017), YFP (Chroma 51017), and DAPI/FITC/TexasRed (Chroma 61002) filter sets was used to observe the reporter expression patterns and to identify neuronal identities by DIC. A Hamamatsu ORCA-ER B/W CCD camera was used to capture images with an Improvision Openlab image processing system. A Zeiss LSM 510 confocal system was used to capture z-stacks at 1 µm intervals. Improvision Volocity was used to visualize, organize and export images from Openlab and the confocal system.
Behavioral Assays
Pharyngeal pumping speed was measured by direct inspection at 160× magnification at room temperature. Individuals, 5 per strain, were picked to a fresh NGM plate with OP50 and allowed to settle at room temperature for 10 minutes before counting pharyngeal pumps during 5 periods of 1 minute.
The defecation motor cycle of well-fed, healthy adults was measured manually under 100× magnification at room temperature. Only the posterior body wall muscle contraction (pBoc) and expulsion muscle contraction were followed. The time of each expulsion was recorded over two 10 min windows for each of five individuals per strain.
Brood size was measured by picking L4 animals individually to fresh culture plates, with subsequent serial transfer at 24-hr intervals. The fertilized eggs laid in each 24-hr period were counted until no more eggs were laid. The total number of eggs laid by each hermaphrodite was taken as the brood size.
To compare growth rates, synchronized L1s, hatched overnight on culture plates without bacterial food, were transferred onto a fresh 5 cm NGM plate with an OP50 bacterial lawn grown from 500 μl of an overnight culture and maintained at 20°C. Their development was monitored every day, recording on which day all the bacteria were consumed. At 40 hours after L1s had resumed development upon supply of bacterial food, the numbers of animals in late L3/early L4, late L4, and young adult stage were counted.
To measure life span, about 200 L1s hatched out during a 2-h period were left to grow to the L4 stage on culture plates at 20°C. These L4s were then distributed to fresh seeded plates, 10 per plate. Individuals were transferred to new plates every day during their reproductive period and then examined every day until their death, as determined by lack of movement even in response to physical stimulation. Each day, dead individuals were removed from the plates and the deaths were recorded.
Statistics analyses of the behavioral assays were performed using one-way ANOVA in OriginPro7.5 (Origin Lab Corporation).
RNAi by Feeding
RNAi by feeding was carried out as described by (Kamath and Ahringer, 2003). Standard NGM plates were supplemented with ampicillin (50 µg/ml), tetracycline (10 µg/ml), and IPTG (isopropyl β-D-1-thiogalactopyranoside) (1 mM), and seeded with bacteria, from the RNAi library from Geneservice (Kamath and Ahringer, 2003), verified by restriction enzyme digestion of purified plasmids. A few L3-L4 hermaphrodites of strains transgenic for a gfp reporter fusion, were transferred from an area off of the bacterial lawn of an OP50 seeded NGM plate, first to an unseeded NGM plate for a few minutes, and then to the RNAi plates. The RNAi plates were maintained at 20°C for 3 days before observing the progeny. The negative control was HT115 bacteria containing pL4440, as for clones in the RNAi library, but without an insert between the T7 promoters. An equivalent bacterial strain with a pL4440 insert for unc-22 was used as a positive control.
Yeast One-Hybrid Screens
Yeast one-hybrid (Y1H) screens and assays were performed as described previously (Deplancke et al., 2004; Reece-Hoyes et al., 2011; Vermeirssen et al., 2007; Walhout, 2006). Promoter entry clones were either retrieved from the C. elegans Promoterome library or generated ab initio by Gateway cloning. (Primers are listed in Supplementary Table 1.) Promoter::reporter (HIS3/lacZ) bait fusions were generated by Gateway LR recombination reactions between the promoter entry clones and the pMW2-HIS3 and pMW3-lacZ yeast expression vectors. Promoter::reporter bait fusions were linearized and used in transformation of the YM4271 (MATa) yeast host strain with integration into the genome by homologous recombination. Twelve clones from each integration were assayed for self-activation of HIS3 and lacZ expression and the clone with the lowest level for each bait was used for subsequent screens.
Screens for transcription factors that interact with a promoter (Y1H) bait were performed using the enhanced Y1H (eY1H) approach (Reece-Hoyes et al., 2011). eY1H screens are more efficient than traditional library screens because the transcription factors are presented to the baits as an array of yeast prey strains (Yα1867, MATα), each transformed with a different Gal4AD-TF fusion. 755 C. elegans transcription factors were included in the array, with each TF represented four times and thus retested inherently. A bench top robot (RoToR, Singer Instrument, Somerset, UK) was used to precisely transfer the (up to) 1536 yeast colonies present on each media plate. To prepare for screens, bait yeast cells were propagated as lawn cultures in standard RoToR dishes containing YAPD media. The transcription factor array was maintained on Sc-Trp media in sets of three 1536-colony plates each containing four copies of up to 384 AD-TF prey yeast clones. To set up a mating, each bait lawn and the transcription factor array were sequentially copied to YAPD plates. The yeasts were grown at 30°C for 3 days before being copied to Sc-His-Ura-Trp plates to select for successfully mated diploids. Diploids were grown for two days before being copied to Sc-His-Ura-Trp plus 5 mM 3-AT (3-amino-1,2,4-triazole) and 80 mg/ml X-gal (5-bromo-4-chloro-3-indolyl-β-D-galactoside) plates and incubated at 30°C. Plates were monitored over the next 5–7 days for the expression level of the reporters. Colonies that can grow in the absence of histidine, overcome the inhibitory effects of 3-AT, and turn X-gal into a blue compound are expressing the reporters, indicating a transcription factor-promoter (Y1H) interaction. Positives, with at least two of the four colonies containing a particular transcription factor showing reporter expression, were identified according to their array coordinates. The transcription factor ORFs of positive colonies were PCR amplified using primers corresponding to the vector and sequenced to verify the identity of the transcription factors.
To directly test individual transcription factor - promoter (Y1H) interactions, plasmids encoding Gal4AD-TF prey fusions were transformed into haploid promoter::reporter bait strains and activation of reporters in transformants was assessed with a β-galactosidase assay on overlay filter membranes and from growth on selective plates plus 20, 40, or 60 mM 3-AT (Vermeirssen et al., 2007).
Acknowledgments
The authors thank John Reece-Hoyes and Marian Walhout for facilitating the Y1H experiments and the Mitani Lab for providing us with the ceh-19(tm452) and ceh-19(tm461) strains. Members of the Hope Laboratory and Walhout Laboratory are greatly appreciated for their generous sharing of resources, suggestions and help.
Supplementary material
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- Albertson DG, Thomson JN. The pharynx of Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1976;275:299–325. doi: 10.1098/rstb.1976.0085. [DOI] [PubMed] [Google Scholar]
- Aspock G, Ruvkun G, Burglin TR. The Caenorhabditis elegans ems class homeobox gene ceh-2 is required for M3 pharynx motoneuron function. Development. 2003;130:3369–3378. doi: 10.1242/dev.00551. [DOI] [PubMed] [Google Scholar]
- Avery L. The genetics of feeding in Caenorhabditis elegans. Genetics. 1993;133:897–917. doi: 10.1093/genetics/133.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L, Horvitz HR. Pharyngeal pumping continues after laser killing of the pharyngeal nervous system of C. elegans. Neuron. 1989;3:473–485. doi: 10.1016/0896-6273(89)90206-7. [DOI] [PubMed] [Google Scholar]
- Avery L, Shtonda BB. Food transport in the C. elegans pharynx. J Exp Biol. 2003;206:2441–2457. doi: 10.1242/jeb.00433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamps S, Hope IA. Large-scale gene expression pattern analysis, in situ, in Caenorhabditis elegans. Briefings in functional genomics and proteomics. 2008;7:175–183. doi: 10.1093/bfgp/eln013. [DOI] [PubMed] [Google Scholar]
- Bargmann CI, Horvitz HR. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron. 1991a;7:729–742. doi: 10.1016/0896-6273(91)90276-6. [DOI] [PubMed] [Google Scholar]
- Bargmann CI, Horvitz HR. Control of larval development by chemosensory neurons in Caenorhabditis elegans. Science. 1991b;251:1243–1246. doi: 10.1126/science.2006412. [DOI] [PubMed] [Google Scholar]
- Deplancke B, Dupuy D, Vidal M, Walhout AJM. A Gateway-compatible yeast one-hybrid system. Genome Res. 2004;14:2093–2101. doi: 10.1101/gr.2445504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi M, Iwasaki K. Na+/K+ ATPase regulates the expression and localization of acetylcholine receptors in a pump activity-independent manner. Mol Cell Neurosci. 2008;38:548–558. doi: 10.1016/j.mcn.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet J, Mango SE. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science. 2002;295:821–825. doi: 10.1126/science.1065175. [DOI] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Nostrand ELV, Cheng C, Arshinoff BI. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, Fire A. Muscle and nerve-specific regulation of a novel NK-2 class homeodomain factor in Caenorhabditis elegans. Development. 1998;125:421–429. doi: 10.1242/dev.125.3.421. [DOI] [PubMed] [Google Scholar]
- Holt SJ. Staying alive in adversity: transcriptome dynamics in the stress-resistant dauer larva. Funct Integr Genomics. 2006;6:285–299. doi: 10.1007/s10142-006-0024-5. [DOI] [PubMed] [Google Scholar]
- Husson SJ, Mertens I, Tom J, Marleen L, Schoofs L. Neuropeptidergic signaling in the nematode Caenorhabditis elegans. Prog Neurobiol. 2007;82:23. doi: 10.1016/j.pneurobio.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Jones B, McGinnis W. A new Drosophila homeobox gene, bsh, is expressed in a subset of brain cells during embryogenesis. Development. 1993;117:793–806. doi: 10.1242/dev.117.2.793. [DOI] [PubMed] [Google Scholar]
- Jones SJ, Riddle DL, Pouzyrev AT, Velculescu VE, Hillier L, Eddy SR, Stricklin SL, Baillie DL, Waterston R, Marra MA. Changes in gene expression associated with developmental arrest and longevity in Caenorhabditis elegans. Genome Res. 2001;11:1346–1352. doi: 10.1101/gr.184401. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Kim K, Li C. Expression and regulation of an FMRFamide-related neuropeptide gene family in Caenorhabditis elegans. J Compar Neurol. 2004;475:540–550. doi: 10.1002/cne.20189. [DOI] [PubMed] [Google Scholar]
- Lakowski B, Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proc Natl Acad Sci USA. 1998;95:13091–13096. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mango SE. The molecular basis of organ formation: insights from the C. elegans foregut. Annu Rev Cell Dev Biol. 2009;25:597–628. doi: 10.1146/annurev.cellbio.24.110707.175411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mckay JP, Raizen DM, Gottschalk A, Schafer WR, Avery L. eat-2 and eat-18 are required for nicotinic neurotransmission in the Caenorhabditis elegans pharynx. Genetics. 2004;166:161–169. doi: 10.1534/genetics.166.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morck C, Rauthan M, Wagberg F, Pilon M. pha-2 encodes the C. elegans ortholog of the homeodomain protein HEX and is required for the formation of the pharyngeal isthmus. Dev Biol. 2004;272:403–418. doi: 10.1016/j.ydbio.2004.05.011. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, Lu Y, Qin H, Lanjuin A, Shaham S, Sengupta P. Distinct IFT mechanisms contribute to the generation of ciliary structural diversity in C. elegans. EMBO J. 2007;26:2966–2980. doi: 10.1038/sj.emboj.7601717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito M, Kohara Y, Kurosawa Y. Identification of a homeobox-containing gene located between lin-45 and unc-24 on chromosome IV in the nematode Caenorhabditis elegans. Nucleic Acids Res. 1992;20:2967–2969. doi: 10.1093/nar/20.12.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkema PG, Fire A. The Caenorhabditis elegans NK-2 class homeoprotein CEH-22 is involved in combinaorial activation of gene expresion in pharyngeal muscle. Development. 1994;120:2175–2186. doi: 10.1242/dev.120.8.2175. [DOI] [PubMed] [Google Scholar]
- Okkema PG, Ha E, Haun C, Chen W, Fire A. The Caenorhabditis elegans NK-2 homeobox gene ceh-22 activates pharyngeal muscle gene expression in combination with pha-1 and is required for normal pharyngeal development. Development. 1997;124:3965–3973. doi: 10.1242/dev.124.20.3965. [DOI] [PubMed] [Google Scholar]
- Papaioannou S, Marsden D, Franks CJ, Walker RJ, Holden-Dye L. Role of a FMRFamide-like family of neuropeptides in the pharyngeal nervous system of Caenorhbditis elegans. J Neurobiol. 2005;65:16. doi: 10.1002/neu.20201. [DOI] [PubMed] [Google Scholar]
- Peden E, Kimberly E, Gengyo-Ando K, Mitani S, Xue D. Control of sex-specific apoptosis in C. elegans by the BarH homeodomain protein CEH-30 and the transcriptional repressor UNC-37/Groucho. Genes Dev. 2007;21:3195–3207. doi: 10.1101/gad.1607807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizen DM, Lee RYN, Avery L. Interacting genes required for pharyngeal excitation by motor neuron MC in Caenorhabditis elegans. Genetics. 1995;141:1365–1382. doi: 10.1093/genetics/141.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray P, Schnabel R, Okkema PG. Behavioral and synaptic defects in C. elegans lacking the NK-2 homeobox gene ceh-28. Dev Neurobiol. 2008;68:421–433. doi: 10.1002/dneu.20599. [DOI] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Deplancke B, Shingles J, Grove CA, Hope IA, Walhout AJM. A compendium of Caenorhabditis elegans regulatory transcription factors: A resource for mapping transcription regulatory networks. Genome Biol. 2005;6:R110. doi: 10.1186/gb-2005-6-13-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Diallo A, Lajoie B, Kent A, Shrestha S, Kadreppa S, Pesyna C, Dekker J, Myers CL, Walhout AJM. Enhanced yeast one-hybrid assays for high-throughput gene-centered regulatory network mapping. Nat Methods. 2011;8:1059–1064. doi: 10.1038/nmeth.1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Shingles J, Dupuy D, Grove CA, Walhout AJM, Vidal M, Hope IA. Insight into transcription factor gene duplication from Caenorhabditis elegans Promoterome-driven expression patterns. BMC Genomics. 2007;8:27. doi: 10.1186/1471-2164-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reig G, Cabrejos ME, Concha ML. Functions of BarH transcription factors during embryonic development. Dev Biol. 2007;302:367–375. doi: 10.1016/j.ydbio.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Schwartz HT, Horvitz HR. The C. elegans protein CEH-30 protects male-specific neurons from apoptosis independently of the Bcl-2 homolog CED-9. Genes Dev. 2007;21:3181–3194. doi: 10.1101/gad.1607007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Fujii T, Dent JA, Fujisawa H, Takagi ST. EAT-20, a novel transmembrane protein with EGF motifs, is required for efficient feeding in Caenorhabditis elegans. Genetics. 2000;154:635–646. doi: 10.1093/genetics/154.2.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F, Moorman C, van der Linder AM, Kuijk E, van der Berghe PVE, Kamath RS, Fraser AG, Ahringer J, Plasterk RH. Genome-Wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 2003;1:e12. doi: 10.1371/journal.pbio.0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J, Hodgkin J. Methods. In: Wood WB, editor. The Nematode Caenorhabditis elegans. New York: Cold Spring Harbor Laboratory, Cold Spring Harbor; 1988. pp. 587–606. [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Thomas JH. Genetic analysis of defecation in Caenorhabditis elegans. Genetics. 1990;124:855–872. doi: 10.1093/genetics/124.4.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeirssen V, Barrasa MI, Hidalgo CA, Babon JAB, Sequerra R, Doucette-Stamm L, Barabási A-L, Walhout AJM. Transcription factor modularity in a gene-centered C. elegans core neuronal protein–DNA interaction network. Genome Res. 2007;17:1061–1071. doi: 10.1101/gr.6148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhout AJM. Unraveling transcription regulatory networks by protein-DNA and protein-protein interaction mapping. Genome Res. 2006;16:1445–1454. doi: 10.1101/gr.5321506. [DOI] [PubMed] [Google Scholar]
- Wang J, Kim SK. Global analysis of dauer gene expression in Caenorhabditis elegans. Development. 2003;130:1621–1634. doi: 10.1242/dev.00363. [DOI] [PubMed] [Google Scholar]
- Willmore WG. Chapter 6: Control of transcription in eukaryotic cells. In: Story KB, editor. Functional metabolism: regulation and adaptation. Hobocken, New Jersey: John Wiley-Liss, Inc; 2004. pp. 153–187. [Google Scholar]
- Altun ZF, Herndon LA, Crocker C, Lints R, Hall DH, editors. WormAtlas. 2002-2012. Available at: http://www.wormatlas.org.
- WormBase. Available at: http://www.wormbase.org.
- Xiao H, Pearson A, Coulombe B, Truant R, Zhang S, Regier JL, Triezenberg SJ, Reinberg D, Flores O, Ingles CJ, Greenblatt J. Binding of basal transcription factor TFIIH to the acidic activation domains of VP16 and p53. Mol Cell Biol. 1994;14:7013–7024. doi: 10.1128/mcb.14.10.7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.