Abstract
Nonstop decay is the mechanism of identifying and disposing aberrant transcripts that lack in-frame stop codons. It is hypothesized that these transcripts are identified during translation when the ribosome arrives at the 3′ end of the mRNA and stalls. Presumably the ribosome stalling recruits additional cofactors, Ski7 and the exosome complex. The exosome degrades the transcript using either one of is ribonucleolytic activities and the ribosome and the peptide are both released. Additional precautionary measures by the nonstop decay pathway may include translational repression of the nonstop transcript after translation, and proteolysis of the released peptide by the proteasome. This surveillance mechanism protects the cells from potentially harmful truncated proteins, but it may also be involved in mediating critical cellular functions of transcripts that are prone to stop codon read-through. Important advances have been made in the past decade as we learn that nonstop decay may have implications in human disease.
Keywords: nonstop decay, exosome, ski complex, review, surveillance, mRNA degradation, Ski7
Introduction
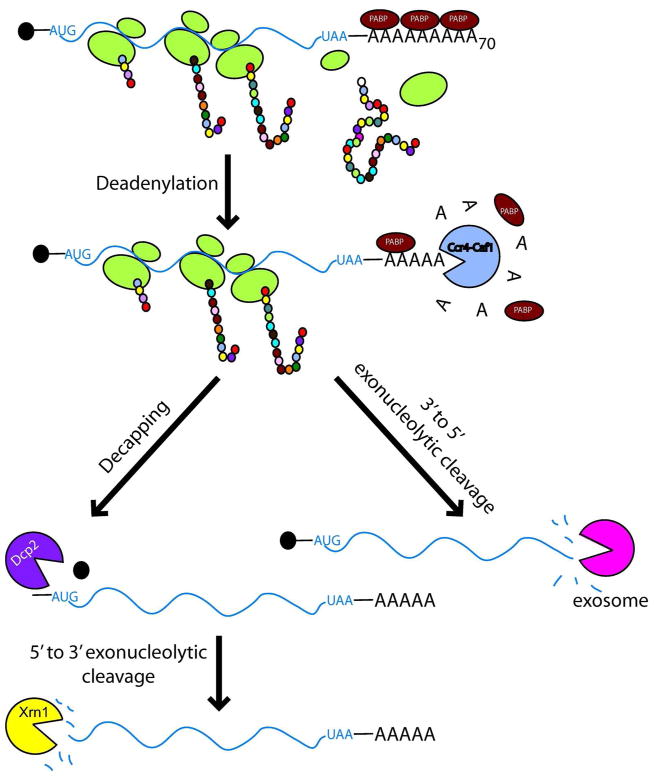
In all three kingdoms of life, gene expression is a precisely regulated process. As a way to control the gene expression in the cell, eukaryotes have evolved two pathways to rid the cell of normal mRNAs. Normal mRNA decay has been characterized in yeast and it is conserved throughout eukaryotes1. The rate limiting step in both pathways includes the deadenylation, or poly(A) shortening, of the target mRNA2, 3. Once the mRNA is deadenylated, it can be degraded in either 5′ to 3′ or 3′ to 5′ directions. Deadenylated mRNAs quickly become prone to decapping by the Dcp2 enzyme and are subsequently degraded by the 5′ to 3′ exonuclease Xrn12, 4–11. The 3′ to 5′ degradation pathway involves the 3′ exonucleolytic digestion of the deadenylated mRNA by the exosome12. The exosome is a complex of ten essential proteins that performs RNA maturation and degradation functions in the nucleus and cytoplasm of eukaryotic cells.
In addition to the exosome, the 3′ to 5′ decay pathway requires a complex of three proteins, Ski2, Ski3, and Ski8, as well as the cofactor Ski78, 12–14. Yeast cells remain viable when either the decapping or exosome pathway is disrupted, however simultaneous inactivation of these mRNA degradation pathways is lethal 12, 15. The observations that mutants of the 3′ to 5′ degradation pathway show little increase in the half-lives of most mRNAs indicated that this pathway may be secondary to the 5′ to 3′ pathway 12. However, for two mRNAs whose decay kinetics have been carefully characterized, the decapping pathway is only 2–5 fold faster than the exosome pathway and genome-wide analysis of the mutants in the 5′ to 3′ decay pathway indicated that 20% of mRNAs depend on this pathway for degradation, arguing that the 3′ to 5′ decay pathway may be the main degradation pathway for some mRNAs 16,17.
In addition to controlling the amount of normal mRNAs, the cell needs to maintain fidelity in the production of such mRNAs. Since mistakes can arise from transcription to translation and anywhere in between, the cell has evolved ways to rapidly identify, target, and eliminate these errors. These aberrancies can occur at the gene, mRNA or protein levels. Through mRNA surveillance mechanisms, the cell distinguishes between transcripts suitable for translation and those that are unsuitable. Examples of aberrant mRNAs include those with premature stop codons, rare codons, or those that lack a poly(A) tail 18–22. Each of these different types of aberrant mRNAs triggers a different surveillance response. Transcripts with premature stop codons activate nonsense-mediated decay 23 and mRNAs that contain rare codons or secondary structures that promote long translations pauses trigger no-go decay 24. Transcripts that lack a poly(A) tail are degraded through an exosome-mediated decay pathway 22. Moreover, transcripts that promote translation beyond the normal stop codon are degraded through ribosome-extension mediated decay 25, 26. As more of these pathways become unveiled, we expect that we will discover more aberrant substrates, which may be degraded through a variety of decay pathways. Finally, this review focuses on the degradation pathway that targets transcripts that lack in-frame stop codons, the nonstop decay pathway. Interestingly, some of these mRNA surveillance pathways are related and use similar trans-acting factors. We refer the interested reviewer to thorough and recent reviews on this relatedness27, 28.
NONSTOP mRNAs ARE COMMON PRODUCTS OF MISTAKES IN GENE EXPRESSION
Nonstop transcripts can arise in instances when transcription aborts, when polyadenylation occurs prematurely or through point mutations that disrupt the stop codon. It is also possible that certain transcripts are prone to nonstop decay in order to maintain low levels of gene expression under certain/normal physiological conditions.
In silico analysis provided evidence that premature polyadenylation occurs in about 1% of random yeast and human cDNA clones 20, 29. Furthermore, species like yeast and plants have poorly conserved poly(A) signals, indicating that polyadenylation may be occurring at alternative sites more often than thought 29 Through premature polyadenylation, nonstop mRNAs may be generated perhaps 5 to 10% of the time 20, 30. Cryptic poly(A) sites have been found in about 1% of S. cerevisiae cDNAs upstream of the termination codon 20.
Given that cryptic polyadenylation occurs, one should expect that organisms use this to regulate genes under certain physiological conditions. Indeed, the premature polyadenylation of four yeast genes has been shown to be regulated by nutrient source 31. Transcripts for CBP1 AEP2/ATP13 are known to be involved in respiration, so it is no surprise that these would be regulated with different nutrient sources. However RNA14 and SIR1 have no apparent involvement in respiration and, interestingly, respond to different types of carbon sources. It is still unclear mechanistically how gene expression is regulated through premature/alternative polyadenylation, and furthermore how nonstop decay may be coupled.
Premature polyadenylation is not restricted to yeast, but has also been described in Arabidopsis using a transcriptome sequencing 32. Most transcriptome sequencing studies focus on functional mRNAs and therefore filter out mRNAs that result from polyadenylation within the coding region. However, the study from Hunt and coworkers indicates that such premature polyadenylation sites reflect widespread premature polyadenylation. In Arabidopsis about 10% of all the mapped poly(A) sites mapped within coding regions of more than 4000 genes. We anticipate that what is true for yeast and Arabidopsis is also true for other eukaryotes, but most of the available transcriptome data have not been analyzed in way that would discover these.
In addition to premature polyadenylation of endogenous genes, this phenomenon has also been shown to be a problem in expressing exogenous genes. For example, transgenic crops expressing a toxin from Bacillus thuringiensis are widespread in agriculture. These crops all contain codon optimized genes because the original gene failed to be expressed. Premature polyadenylation, and subsequent rapid degradation of the mRNA severely limits the expression of the Bt toxin 33, 34.
THE MECHANISM OF NONSTOP DEGRADATION IN YEAST
Nonstop mRNAs are degraded in the cytoplasm
An initial hint that nonstop mRNAs are rapidly degraded came in 1989 when Herrick and Jacobson described two chimeric mRNAs between the yeast ACT1 and HIS3 genes 35. One of these mRNAs contained the HIS3 stop codon and was stable (half-life of 79 minutes), while the other lacked an in-frame stop codon due to a 4-nucleotide deletion and was unstable (half-life of 9 minutes). Independently, this was also shown for yeast PGK1 mRNA, and this reporter was used for a more mechanistic analysis of nonstop mRNA decay 20, 21. The PGK1-nonstop reporter was shown to degrade with a half-life of less than 2 minutes in wild-type yeast cells, while it is stabilized to a half-life of 15 minutes in a yeast strain with a point mutation in the exosome. The exosome, a complex of ten essential proteins, contains only one catalytic subunit, Rrp44. Recently, it was shown that the exosome has two catalytic sites in Rrp44, an exonucleolytic and an endonucleolytic domain 36–38. Surprisingly, either the endo- or the exonuclease activity can promote the degradation of the nonstop transcript 39.
In addition to requiring the exosome, nonstop mRNA decay requires the Ski7 protein and the Ski complex. The Ski complex is composed of the DEVH-box RNA helicase Ski2, the tetratricopeptide repeat protein Ski3 and two copies of the WD repeat protein Ski813, 40–43. The SKI genes were initially identified as part of an antiviral mechanism of the yeast S. cerevisiae and later identified as cofactors required for the cytoplasmic functions of the exosome 7, 12, 14, 44–46. Specifically, Ski2, Ski3, Ski8, and Ski7 physically and functionally interact with the exosome and promote the exosome-mediated degradation of normal and aberrant mRNAs in the cytoplasm 21, 22, 46. It is not completely clear what the specific function of each of these Ski proteins in nonstop decay, however all four are required for nonstop mRNA degradation.
The nonstop decay pathway is different from the general mRNA decay mechanism and the nonsense-mediated degradation (NMD) pathway. Dcp2, Xrn1, and Upf1, which are proteins involved in general cytoplasmic mRNA decay and NMD, are not required for nonstop decay (see fig 1) 20. The mechanism for nonstop decay is also distinct from the exosome-mediated decay of normal mRNAs in three aspects (see figs 1 and 2 for comparison). First, nonstop decay occurs without prior deadenylation of the mRNA 21. Second, nonstop mRNA decay requires the C-terminal domain of Ski7, which is dispensable for exosome-mediated decay of normal mRNAs 21 (see below). Third, as mentioned above, nonstop mRNAs can be efficiently degraded by either the endo- or the exonuclease activities of the exosome, while exosome-mediated decay of normal mRNAs requires only the exonuclease activity 39.
Figure 1.
Pathways for the degradation of normal yeast mRNAs. See text for details.
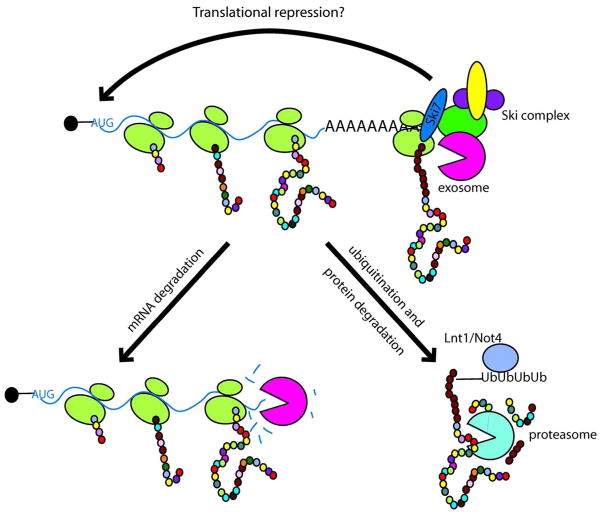
Figure 2.
nonstop mRNAs are rapidly degraded, and may also trigger translational repression and proteolysis. See text for details.
Nonstop mRNA degradation may be initiated when Ski7 recognizes a stalled ribosome
Translation of the nonstop transcripts is required for initiation of the nonstop decay pathway 20. This conclusion is based on the observation that treatment with cycloheximide which prevents translation elongation, stabilizes nonstop transcripts 20. Furthermore, addition of a stop codon just upstream of the poly(A) tail also blocked nonstop decay 20. During in vitro translation, nonstop transcripts form stable complex with ribosomes 47, 48 indicating that unlike RNA or DNA polymerases, the ribosome does not dissociate from its template when it reaches the end. Furthermore in vivo sucrose gradient experiments confirm that nonstop mRNAs remain associated with polysomes 26. These observations suggest a model where nonstop decay requires at least one round of translation, whereby the ribosome travels to the 3′ end of the transcript and stalls.
Genetic evidence and sequence similarity suggests that Ski7 recognizes ribosomes stalled on nonstop mRNA through its C-terminal domain, and promotes the exosome-mediated degradation of the transcript 21. The C-terminus of Ski7 is a member of a family of ribosome-associated GTPases, which also include the eukaryotic elongation factor 1α (eEF1α), the eukaryotic release factor 3 (eRF3), and the no-go decay protein Hbs149 All three of these factors recognize the ribosome, depending on what codon is in the ribosomal A-site. During translation elongation the A-site contains a sense codon, which is recognized by an aminoacyl-tRNA/eEF1a complex. During translation termination the A-site contains a stop codon, which is recognized by the eRF1/eRF3 complex. Finally, when the ribosome stalls during translation elongation, it is recognized by the Dom34/Hbs1 complex 19, 50–53. (Please see 24 for more details on no-go decay)
Based on the similarity between Ski7 and these translation factors, it was proposed that the C-terminal domain of Ski7 recognizes a stalled ribosome on a nonstop mRNA 21. As predicted, C-terminal truncations of Ski7 affect nonstop decay without disrupting the general cytoplasmic function of the exosome 21. Alternatively, mutations in SKI7 that disrupt Ski7 binding to the exosome and the Ski complex (N-terminal truncations) affect both normal and nonstop mRNA degradation 21. Thus, genetic evidence suggests that the C-terminal part of Ski7 may be responsible for recognizing ribosomes stalled on nonstop mRNAs.
Although Ski7 shares characteristics with the translation factors eEF1a, eRF3 and the no-go decay factor Hbs1, two differences are striking. First, eEF1a, eRF3, and Hbs1 each act as a heterodimer, with an interacting partner responsible for recognizing the state of the ribosome (tRNA, eRF1 and Dom34, respectively). No such interacting partner has been identified for Ski7, and thus it is not clear why Ski7 is specific for ribosome stalled on nonstop mRNAs. Second, GTPase activity is critical for the specificity of eEF1a, eRF3, and Hbs1, and requires a conserved active site His residue 54–56. This residue is not conserved in Ski7, and thus Ski7 may accomplish this function without the use of energy provided by GTP. Biochemical characterization of Ski7 and its interaction with the ribosome will be needed to further understand the recognition of nonstop mRNAs during translation.
Additional mechanisms may down-regulate nonstop gene expression
As described above, one consequence of detecting the lack of a stop codon in an mRNA is the rapid degradation of the mRNA This aspect is fairly well understood, but may not be the only manner in which the cell limits the consequences of these aberrant mRNAs. For example, translation of the nonstop transcript may be repressed, possibly due to the translation of the poly(A) tract 26, 57 and several studies have argued that the protein translated from nonstop mRNA is also very rapidly degraded (see below). One caveat of this statement is that it is difficult to decipher whether these consequences are due to the absence of a stop codon in the mRNA or unrelated to the absence of the stop codon. For example, nonstop reporter mRNAs are typically generated by mutating the normal sop codon in the mRNA, causing the 3′ UTR and poly(A) to become part of the open reading frame. The protein encoded by such transcripts has a C-terminal extension with a poly-Lysine tail. The encoded protein is generally unstable, which in several cases has been attributed to the absence of a stop codon, but contribution of the C-terminal extension or poly(Lys) tail have not been ruled out in all cases. For example, it has been shown that a poly(Lys) tail can contribute to rapid protein decay, but a poly(Arg) tail, and several other Komopolymers were also effective 57, 58.
The abundance of the protein translated from nonstop mRNAs is much lower than can be explained simply from the lower mRNA levels 57. Protein degradation significantly contributes to the low levels of reporter expression. It has been shown that the proteasome inhibitor MG132 increases expression of the protein encoded by the nonstop mRNA 58. Wilson and coworkers showed that mutations of a proteasome subunit (pre9Δ), two proteasome assembly factors (umplΔ and irc25Δ, a.k.a. poc3Δ, pba3Δ and dmp2Δ) and a RING ubiquitin E3 ligase (IntIΔ, a.k.a. rkr1Δ) cause the stabilization of proteins translated from two different nonstop reporter mRNAs 59. The function of the Lnt1 was confirmed by a subsequent study that also showed that Lnt1 specifically promotes the ubiquitination and proteasome-dependent degradation of the nonstop product 60. Additional results indicate that Lnt1 mediates cotranslational recognition of the poly(Lys) tail on the nascent peptide. Notably, this recognition occurred both when poly(Lys) was encoded by a poly(A) tail, or by 12 AAA or AAG codons followed by a stop codon 58. Thus, recognition of the nascent peptide triggers Lnt1-mediated protein decay, independent of the lack of a stop codon. In addition, the effect of ski7Δ is additive with the pre9Δ or IntIΔ effect, indicating that the exosome-mediated decay of nonstop mRNAs and the proteasome-mediated decay of the encoded peptide are independent of each other 59, 60. Interestingly, the poly(Lys) tail would be inside the protein exit tunnel of the ribosome, and thus unavailable for direct co-translational recognition. A reasonable model appears that some peptide sequences mediate ribosome stalling, and the stalled ribosome is recognized by Lnt1, which mediates ubiquitination and protein degradation. This phenomenon might also depend on protein context, since a different set of experiments using different reporters suggest that (Lys)12-triggered ribosome stalling led to degradation of the nascent peptide mediated by E3 enzyme Not458. Ribosome stalling Triggered by features in the RNA is known to activate no-go decay, so it will be interesting to see whether this generates additional substrates for Lnt1 or Not4.
NONSTOP DECAY IN HUMAN CELLS
Nonstop decay factors are conserved throughout eukaryotes and important or human health
The exosome and its cytoplasmic cofactors are conserved in all eukaryotes 61–64. Most of the ten exosome subunits and four cytoplasmic cofactors each have one corresponding human ortholog, suggesting that cytoplasmic exosome functions are also conserved between yeast and humans. In addition the capacity of Ski2, Ski3, and Ski8 to form a Ski complex is also conserved in humans 65. The molecular function of human Ski2, 3, and 8 has not been studied in detail, but the function of the drosophila homologs is similar to the yeast genes. Specifically, both the yeast and fly proteins have shown to be responsible for the degradation of 5′ mRNA fragments generated by various endonucleases 22, 66–69. Thus the function of the Ski complex and presumably the exosome in cytoplasmic mRNA decay was already established in the common ancestor of fungi and animals.
The physiological function of the human Ski complex has been revealed by the observation that loss of function mutations in the human SKI3 (TTC37) or SKI2 gene causes a rare congenital disorder (Tricho-Hepato-Enteric Syndrome) 70–72 The major defect in Tricho-Hepato-Enteric syndrome occurs in the small intestine, and therefore the initial presentation of the syndrome is diarrhea starting shortly after birth, which can be treated by long-term parenteral nutrition. These findings, suggest that some function of the human ski complex is especially important in the small intestine, although other symptoms can not be explained defects in the small intestine. Whether Tricho-Hepato-Enteric syndrome is caused by a defect in nonstop decay, or a more general cytoplasmic exosome disruption, remains to be determined.
The two exceptions of a one-to-one correspondence between human and yeast cytoplasmic exosome subunits and cofactors are the two proteins most central in the yeast nonstop decay, namely the Ski7 nonstop mRNA recognition protein, and the Rrp44 RNase responsible for degrading nonstop mRNAs. The human genome includes three homologs of Rrp44, named hDis3, Dis3L1, and Dis3L273, 74. The functions of hDis3 and hDis3L1 have been studied to some extent, and are more specialized than the single yeast homolog. While hDis3 functions in the nuclear exosome, hDis3L1 localizes to the cytoplasm 73, 74. The reverse is true for Ski7, which is a more specialized protein in a subset of the saccharomycete yeasts, but whose function is carried out by a more general protein in other eukaryotes 75, 76.
Whole genome duplication (WGD) in Saccharomyces cerevisiae led to a specialized nonstop decay factor-Ski7
Approximately 100 million years ago a genome duplication occurred in an ancestor of Saccharomyces cerevisiae 77, 78 Initially the whole genome of S. cerevisiae was duplicated, but subsequently most duplicated genes were lost, and a minority retained. Genomic and molecular analyses that compared ancestors of S. cerevisiae pre-duplication with S. cerevisiae have shed light to the gene specialization that occurred after this WGD 77, 79–81. Prior to genome duplication, the genome of ancestors of the Saccharomyces complex contained an ancestral gene, which we will refer to as SKI7/HBS1. WGD gave rise to separate HBS1 and SKI7 genes, with HBS1 functioning in no-go decay and SKI7 in nonstop decay. Lachancea kluyveri (previously named Saccharomyces kluveryi) diverged from S. cerevisiae just before genome duplication and thus has only a single SKI7/HBS1 gene. The L. kluyveri SKI7/HBS1 gene can complement both a ski7Δ and an hbs1Δ, strongly suggesting that this gene functions in both no-go and nonstop decay. Furthermore, other eukaryotes including humans, Drosophila, Arabidopsis, and Trypanosoma each have a single SKI7/HBS1 gene like L. kluyveri, which presumably functions in both nonstop and no-go decay. It remains to be elucidated whether and how this gene specialization of SKI7 and HBS1 conferred an increased fitness for the Saccharomyces species.
Nonstop mRNAs are rapidly degraded in mammalian cells without prior deadenylation
Nonstop reporter mRNA are also preferentially degraded in human cells, but whether all aspects of the nonstop decay mechanism are conserved, remains to be elucidated. An initial analysis of the stability of a nonstop mRNA reporter in HeLa cells showed that its cytoplasmic steady state levels were reduced about 3-fold 20. A second paper used a nonstop α-globin-nonstop reporter mRNA as a control in a study of a different mRNA decay pathway 25. α-globin-nonstop mRNA was very rapidly degraded in human HeLa cells, as well as mouse MEL and C127 cells (half-life of 2h versus 11.5h). Similarly to yeast nonstop mRNA decay, the rapid degradation of the nonstop transcript was not preceded by deadenylation and dominant negative versions of the deadenylase enzyme had no effect on the stability of α-globin-nonstop mRNA 25. This study however did not analyze whether rapid decay of these nonstop reporter mRNAs requires ongoing translation. A different study using a nonstop version of the human RPS19 mRNA, showed that the rapid degradation of the nonstop transcript is blocked by cycloheximide 82. Thus, translation-dependent, but deadenylation-independent rapid decay of nonstop mRNA seems to be conserved in yeast and mammals. Whether the mammalian decay also required the exosome and its cofactors has not been tested, but it was shown to not require the nonsense-mediated decay factor Upf125.
Rapid degradation of nonstop mRNAs contributes to several human diseases
With the understanding that the steady state level of RNAs contributes greatly to gene expression, researchers are paying more attention to RNA stability. Gene therapy that attempts to disrupt the normal stability of mRNAs through miRNAs continues to be a hot topic in the cancer research field. Likewise, mutated genes that produce mRNAs with altered stability have been shown to contribute to human diseases.
As explained above, mutations in the human ski complex cause Tricho-Hepato-Enteric Syndrome. A second connection between nonstop decay and disease results from a growing number of reports that link nonstop mutations in specific genes to disease. By far the most prevalent nonstop mutation, perhaps affecting 100’s of millions of people, was reported for the DEFB126 gene 83. The encoded β-defensin protein is important for human sperm function 83–86, but 20% of males are homozygous for a nonstop mutation in DEFB12683. A population study of Chinese couples with and without this polymorphism showed that the nonstop mutation in the DEFB126 gene causes lower fertility which manifests as a longer time to achieve pregnancy 83. More rare instances of nonstop mutations are listed in table 182, 83, 87–95. In most of these cases the nonstop mutation correlates with reduced mRNA levels. Importantly, the nonstop mutation leads to reduced levels of a C-terminally extended protein and in these cases the lack of protein is the main cause for the symptoms. In some cases the mutant protein is functional, so that increased stability of the nonstop proteins and increased production of the protein could provide therapeutic benefit.
Table 1.
Human diseases caused by nonstop mutations.
| Gene | Disease/symptoms | Reduction in mRNA level | reference |
|---|---|---|---|
| DEFB126 | Reduced fertility | Y | [81] |
| Dysferlin | Muscular dystrophy | [92] | |
| RPS19 | Diamond Blackfan anemia | Y | [80,90,91] |
| Factor X | Haemophilia | Y | [89] |
| GPR54 | idiopathic hypogonadotrophic hypogonadism, infertility | Y | [88] |
| TYMP | Mitochondrial neurogastrointestinal encephalomyophathy | N | [93] |
| APRT | 2,8-dihydroxyadenine urolithiasis, kidney stones, kidney failure | Y | [87] |
| FOXE3 | Anterior segment dysgenesis, Peters anomaly | Y | [86] |
| HSD3B2 | Adrenal hyperplasia | N | [85] |
Two different genome-wide analyses have been performed on either mutations that cause disease or on SNPs 96, 97. Each of them identified over 100 mutations where a stop codon is changed to a sense codon. In a subset of these, no other in frame stop codon is found in the 3′UTR, and thus these mutations generate a nonstop mRNA, which may cause disease 96. In contrast, mutating stop codon that was followed by an additional next in frame stop codon within 50nt appears less likely to cause deleterious/clinically relevant phenotype 97. More careful characterization of these cases will be required to fully understand the impact of nonstop mRNA decay on human genetic diseases.
CONCLUSIONS AND FUTURE DIRECTIONS
Great advances have been made in the past ten years, unveiling and deciphering the nonstop degradation pathway. We have only begun to scratch the surface in understanding the mechanisms of nonstop decay. Other surveillance pathways such as no-go and the prokaryotic trans-translation pathway have detailed structural and functional mechanisms elucidated, the lack of structural and biochemical data on Ski7, the SKI complex and their interactions with the stalled ribosome and the exosome limit our understanding of nonstop decay at the molecular level.
Learning how to suppress the nonstop decay pathway will have important implications in treating the aforementioned diseases. Recently, genetic therapies that suppress NMD, are being analyzed in clinical trials to prevent/treat Cystic fibrosis and Duchenne’s muscular dystrophy among other genetic diseases 98. Likewise, nonstop decay could be targeted to prevent mRNA and/or protein degradation. There is still a long and exciting road ahead to fully understand and learn how to control the mechanism of nonstop mRNA and peptide degradation.
Sidebar. Trans-translation versus nonstop decay.
Similarly to the eukaryotic nonstop decay, bacteria have a surveillance mechanism to relieve the cell of stalled ribosomes. Trans-translation is the process whereby mRNAs that lack a termination codon are released from the ribosome and targeted for degradation. The recycling of ribosomes is a necessary feature for the cells to continue efficient translation of other transcripts. The details of the trans-translation have been reviewed by Barends et al. 99. Briefly, trans-translation uses a tmRNA molecule, which functions as both a tRNA and a mRNA. The tmRNA is required for recognition of the stalled ribosome with the peptidyl-tRNA bound to the P-site. The “tRNA” side of the tmRNA binds the A-site of the ribosome and donates its charged alanine to the still bound polypeptide, which triggers the release of the tRNA from the P-site. While tmRNA is now bound to the polypeptide, the ribosome continues to translate the “mRNA” side of the tmRNA, which encodes a protein degradation tag and a stop codon. Translation termination occurs normally with the stop codon from the tmRNA and the tagged polypeptide is targeted for degradation by specific proteases.
Recognition and degradation of bacterial and eukaryotic nonstop mRNAs requires homologous factors
Both trans-translation and nonstop decay are triggered by stalled ribosomes caused by the lack of stop codons. tmRNA is delivered to the ribosome by EF-tu, the same GTPase that delivers tRNAs during translation elongation. EF-tu is a distant relative of Ski7. Whether this involvement of a GTPase is a conserved feature of bacterial trans-translation and eukaryotic nonstop mRNA decay or whether bacteria and eukaryotes have independently recruited a GTPase to these processes is not clear. Additionally, there is no evidence of Ski7 associating with a homologous eukaryotic tmRNA molecule. Furthermore, there is no evidence of a transpeptidation reaction occurring in eukaryotic nonstop decay like it does in trans-translation. After transpeptidation the tmRNA is positioned in a way that allows the release of the aberrant mRNA and proper placement of the “mRNA” side of the tmRNA in decoding center of the ribosome. The released mRNA is then targeted for degradation by RNase R, a homolog of Rrp44, the catalytic subunit of the exosome 100. SimNar to EF-tu and Ski7 involvement RNase R and Rrp44 involvement may reflect conservation from an ancient ancestor, or a case where bacteria and eukaryotes have recruited similar proteins for similar functions.
Release and degradation of nonstop peptide in trans-translation and eukaryotic nonstop decay
One clear difference between the trans-translation and nonstop decay mechanism is that fact that in the bacterial system addition of a proteolysis tag is intimately linked recognition of the stalled ribosome. In contrast to bacterial mRNAs eukaryotic mRNAs carry a long poly(A) tail. This distinction may explain why the eukaryotic mechanism does not require a tmRNA, since this C-terminal poly(lysine) tail may provide the tag for protein.
Finally, it is unclear how the mRNA, ribosomal subunits and the peptide are released in the eukaryotic nonstop decay pathway. In the trans-translation translation termination occurs normally with the help of the stop codon encoded by the tmRNA molecule.
Acknowledgments
The authors are grateful for grants from NIH (R01GM099790) NSF (MCB1020739) and the Welch foundation (AU-1773) that made work in their lab possible.
References
- 1.Meyer S, Temme C, Wahle E. Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit Rev Biochem Mol Biol. 2004;39:197–216. doi: 10.1080/10409230490513991. [DOI] [PubMed] [Google Scholar]
- 2.Muhlrad D, Decker CJ, Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′->3′ digestion of the transcript. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- 3.Shyu AB, Belasco JG, Greenberg ME. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991;5:221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- 4.Dunckley T, Parker R. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 1999;18:5411–5422. doi: 10.1093/emboj/18.19.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muhlrad D, Parker R. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev. 1992;6:2100–2111. doi: 10.1101/gad.6.11.2100. [DOI] [PubMed] [Google Scholar]
- 6.Decker CJ, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 7.Hsu CL, Stevens A. Yeast cells lacking 5′->3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol Cell Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muhlrad D, Decker CJ, Parker R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol Cell Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beelman CA, Stevens A Caponigro G, LaGrandeur TE, Hatfield L, Fortner DM, Parker R. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature. 1996;382:642–646. doi: 10.1038/382642a0. [DOI] [PubMed] [Google Scholar]
- 10.van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Seraphin B. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002;21:6915–6924. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steiger M, Carr-Schmid A, Schwartz DC, Kiledjian M, Parker R. Analysis of recombinant yeast decapping enzyme. RNA. 2003;9:231–238. doi: 10.1261/rna.2151403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobs Anderson JS, Parker RP. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown JT, Bai X, Johnson AW. The yeast antiviral proteins Ski2p, Ski3p, and Ski8p exist as a complex in vivo. RNA. 2000;6:449–457. doi: 10.1017/s1355838200991787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Hoof A, Staples RR, Baker RE, Parker R. Function of the ski4p (Csl4p) and Ski7p proteins in 3′-to-5′ degradation of mRNA. Mol Cell Biol. 2000;20:8230–8243. doi: 10.1128/mcb.20.21.8230-8243.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson AW, Kolodner RD. Synthetic lethality of sep1 (xrn1) ski2 and sep1 (xrn1) ski3 mutants of Saccharomyces cerevisiae is independent of killer virus and suggests a general role for these genes in translation control. Mol Cell Biol. 1995;15:2719–2727. doi: 10.1128/mcb.15.5.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao D, Parker R. Computational modeling of eukaryotic mRNA turnover. RNA. 2001;7:1192–1212. doi: 10.1017/s1355838201010330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He F, Li X, Spatrick P, Casillo R, Dong S, Jacobson A. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol Cell. 2003;12:1439–1452. doi: 10.1016/s1097-2765(03)00446-5. [DOI] [PubMed] [Google Scholar]
- 18.Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- 19.Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frischmeyer PA, van Hoof A, O’Donnell K, Guerrerio AL, Parker R, Dietz HC. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002;295:2258–2261. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- 21.van Hoof A, Frischmeyer PA, Dietz HC, Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- 22.Meaux S, van Hoof A. Yeast transcripts cleaved by an internal ribozyme provide new insight into the role of the cap and poly(A) tail in translation and mRNA decay. RNA. 2006;12:1323–1337. doi: 10.1261/rna.46306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicholson P, Muhlemann O. Cutting the nonsense: the degradation of PTC-containing mRNAs. Biochem Soc Trans. 2010;38:1615–1620. doi: 10.1042/BST0381615. [DOI] [PubMed] [Google Scholar]
- 24.Harigaya Y, Parker R. No-go decay: a quality control mechanism for RNA in translation. Wiley Interdiscip Rev RNA. 2010;1:132–141. doi: 10.1002/wrna.17. [DOI] [PubMed] [Google Scholar]
- 25.Kong J, Liebhaber SA. A cell type-restricted mRNA surveillance pathway triggered by ribosome extension into the 3′ untranslated region. Nat Struct Mol. 2007;14:670–676. doi: 10.1038/nsmb1256. 5/0. [DOI] [PubMed] [Google Scholar]
- 26.Inada T, Aiba H. Translation of aberrant mRNAs lacking a termination codon or with a shortened 3′-UTR is repressed after initiation in yeast. EMBO J. 2005;24:1584–1595. doi: 10.1038/sj.emboj.7600636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Hoof A, Wagner EJ. A brief survey of mRNA surveillance. Trends in biochemical sciences. 2011;36:585–592. doi: 10.1016/j.tibs.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shoemaker CJ, Green R. Translation drives mRNA quality control. Nat Struct Mol Biol. doi: 10.1038/nsmb.2301. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graber JH, Cantor CR, Mohr SC, Smith TF. In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc Natl Acad Sci U S A. 1999;96:14055–14060. doi: 10.1073/pnas.96.24.14055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson MA, Meaux S, van Hoof A. Diverse aberrancies target yeast mRNAs to cytoplasmic mRNA surveillance pathways. Biochim Biophys Acta. 2008;1779:550–557. doi: 10.1016/j.bbagrm.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sparks KA, Dieckmann CL. Regulation of poly(A) site choice of several yeast mRNAs. Nucleic Acids Res. 1998;26:4676–4687. doi: 10.1093/nar/26.20.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu X, Liu M, Downie B, Liang C, Ji G, Li QQ, Hunt AG. Genome-wide landscape of polyadenylation in Arabidopsis provides evidence for extensive alternative polyadenylation. Proc Natl Acad Sci U S A. 2011;108:12533–12538. doi: 10.1073/pnas.1019732108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diehn SH, Chiu WL, De Rocher EJ, Green PJ. Premature polyadenylation at multiple sites within a Bacillus thuringiensis toxin gene-coding region. Plant Physiol. 1998;117:1433–1443. doi: 10.1104/pp.117.4.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Rocher EJ, Vargo-Gogola TC, Diehn SH, Green PJ. Direct evidence for rapid degradation of Bacillus thuringiensis toxin mRNA as a cause of poor expression in plants. Plant Physiol. 1998;117:1445–1461. doi: 10.1104/pp.117.4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herrick D. Structural determinants of mRNA turnover in yeast: a thesis. Molecular Genetics and Microbiology. 1989 Vol. Ph.D. [Google Scholar]
- 36.Lebreton A, Tomecki R, Dziembowski A, Seraphin B. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature. 2008;456:993–996. doi: 10.1038/nature07480. [DOI] [PubMed] [Google Scholar]
- 37.Schaeffer D, Tsanova B, Barbas A, Reis FP, Dastidar EG, Sanchez-Rotunno M, Arraiano CM, van Hoof A. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat Struct Mol Biol. 2009;16:56–62. doi: 10.1038/nsmb.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneider C, Leung E, Brown J, Tollervey D. The N-terminal PIN domain of the exosome subunit Rrp44 harbors endonuclease activity and tethers Rrp44 to the yeast core exosome. Nucleic Acids Res. 2009;37:1127–1140. doi: 10.1093/nar/gkn1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schaeffer D, van Hoof A. Different nuclease requirements for exosome-mediated degradation of normal and nonstop mRNAs. Proc Natl Acad Sci U S A. 2011;108:2366–2371. doi: 10.1073/pnas.1013180108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rhee SK, Icho T, Wickner RB. Structure and nuclear localization signal of the SKI3 antiviral protein of Saccharomyces cerevisiae. Yeast. 1989;5:149–158. doi: 10.1002/yea.320050304. [DOI] [PubMed] [Google Scholar]
- 41.Widner WR, Wickner RB. Evidence that the SKI antiviral system of Saccharomyces cerevisiae acts by blocking expression of viral mRNA. Mol Cell Biol. 1993;13:4331–4341. doi: 10.1128/mcb.13.7.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng Z, Liu Y, Wang C, Parker R, Song H. Crystal structure of Ski8p, a WD-repeat protein with dual roles in mRNA metabolism and meiotic recombination. Protein Sci. 2004;13:2673–2684. doi: 10.1110/ps.04856504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Synowsky SA, Heck AJ. The yeast Ski complex is a hetero-tetramer. Protein Sci. 2008;17:119–125. doi: 10.1110/ps.073155908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toh EA, Guerry P, Wickner RB. Chromosomal superkiller mutants of Saccharomyces cerevisiae. J Bacteriol. 1978;136:1002–1007. doi: 10.1128/jb.136.3.1002-1007.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ridley SP, Sommer SS, Wickner RB. Superkiller mutations in Saccharomyces cerevisiae suppress exclusion of M2 double-stranded RNA by L-A-HN and confer cold sensitivity in the presence of M and L-A-HN. Mol Cell Biol. 1984;4:761–770. doi: 10.1128/mcb.4.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Araki Y, Takahashi S, Kobayashi T, Kajiho H, Hoshino S, Katada T. Ski7p G protein interacts with the exosome and the Ski complex for 3′-to-5′ mRNA decay in yeast. EMBO J. 2001;20:4684–4693. doi: 10.1093/emboj/20.17.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akimitsu N, Tanaka J, Pelletier J. Translation of nonSTOP mRNA is repressed post-initiation in mammalian cells. EMBO J. 2007;26:2327–2338. doi: 10.1038/sj.emboj.7601679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perara E, Rothman RE, Lingappa VR. Uncoupling translocation from translation: implications for transport of proteins across membranes. Science. 1986;232:348–352. doi: 10.1126/science.3961485. [DOI] [PubMed] [Google Scholar]
- 49.Benard L, Carroll K, Valle RC, Masison DC, Wickner RB. The ski7 antiviral protein is an EF1-alpha homolog that blocks expression of non-Poly(A) mRNA in Saccharomyces cerevisiae. J Virol. 1999;73:2893–2900. doi: 10.1128/jvi.73.4.2893-2900.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen L, Muhlrad D, Hauryliuk V, Cheng Z, Lim MK, Shyp V, Parker R, Song H. Structure of the Dom34-Hbs1 complex and implications for no-go decay. Nat Struct Mol Biol. 2010;17:1233–1240. doi: 10.1038/nsmb.1922. [DOI] [PubMed] [Google Scholar]
- 51.Becker T, Armache JP, Jarasch A, Anger AM, Villa E, Sieber H, Motaal BA, Mielke T, Berninghausen O, Beckmann R. Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome. Nat Struct Mol Biol. 2011;18:715–720. doi: 10.1038/nsmb.2057. [DOI] [PubMed] [Google Scholar]
- 52.Shoemaker CJ, Green R. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc Natl Acad Sci U S A. 2011;108:E1392–1398. doi: 10.1073/pnas.1113956108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pisareva VP, Skabkin MA, Hellen CU, Pestova TV, Pisarev AV. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J. 2011;30:1804–1817. doi: 10.1038/emboj.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carr-Schmid A, Durko N, Cavallius J, Merrick WC, Kinzy TG. Mutations in a GTP-binding motif of eukaryotic elongation factor 1A reduce both translational fidelity and the requirement for nucleotide exchange. J Biol Chem. 1999;274:30297–30302. doi: 10.1074/jbc.274.42.30297. [DOI] [PubMed] [Google Scholar]
- 55.Salas-Marco J, Bedwell DM. GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol Cell Biol. 2004;24:7769–7778. doi: 10.1128/MCB.24.17.7769-7778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shoemaker CJ, Eyler DE, Green R. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science. 2010;330:369–372. doi: 10.1126/science.1192430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ito-Harashima S, Kuroha K, Tatematsu T, Inada T. Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev. 2007;21:519–524. doi: 10.1101/gad.1490207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dimitrova LN, Kuroha K, Tatematsu T, Inada T. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J Biol Chem. 2009;284:10343–10352. doi: 10.1074/jbc.M808840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilson MA, Meaux S, van Hoof A. A genomic screen in yeast reveals novel aspects of nonstop mRNA metabolism. Genetics. 2007;177:773–784. doi: 10.1534/genetics.107.073205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bengtson MH, Joazeiro CA. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature. 2010;467:470–473. doi: 10.1038/nature09371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schilders G, van Dijk E, Raiimakers R, Pruijn GJ. Cell and molecular biology of the exosome: how to make or break an RNA. Int Rev Cytol. 2006;251:159–208. doi: 10.1016/S0074-7696(06)51005-8. [DOI] [PubMed] [Google Scholar]
- 62.Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3′ -> 5′ exonucleases. Genes Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lorentzen E, Basquin J, Tomecki R, Dziembowski A, Conti E. Structure of the active subunit of the yeast exosome core, Rrp44: diverse modes of substrate recruitment in the RNase II nuclease family. Mol Cell. 2008;29:717–728. doi: 10.1016/j.molcel.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 64.Clayton C, Estevez A. The exosomes of trypanosomes and other protists. Adv Exp Med Biol. 2011;702:39–49. doi: 10.1007/978-1-4419-7841-7_4. [DOI] [PubMed] [Google Scholar]
- 65.Zhu B, Mandal SS, Pham AD, Zheng Y, Erdjument-Bromage H, Batra SK, Tempst P, Reinberg D. The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev. 2005;19:1668–1673. doi: 10.1101/gad.1292105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gatfield D, Izaurralde E. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature. 2004;429:575–578. doi: 10.1038/nature02559. [DOI] [PubMed] [Google Scholar]
- 67.Orban TI, Izaurralde E. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA. 2005;11:459–469. doi: 10.1261/rna.7231505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Meaux S, van Hoof A, Baker KE. Nonsense-mediated mRNA decay in yeast does not require PAB1 or a poly(A) tail. Mol Cell. 2008;29:134–140. doi: 10.1016/j.molcel.2007.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meaux S, Lavoie M, Gagnon J, Abou Elela S, van Hoof A. Reporter mRNAs cleaved by Rnt1p are exported and degraded in the cytoplasm. Nucleic Acids Res. 2011;39:9357–9367. doi: 10.1093/nar/gkr627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hartley JL, Zachos NC, Dawood B, Donowitz M, Forman J, Pollitt RJ, Morgan NV, Tee L, Gissen P, Kahr WH, et al. Mutations in TTC37 cause trichohepatoenteric syndrome (phenotypic diarrhea of infancy) Gastroenterology. 2010;138:2388–2398. 2398 e2381–2382. doi: 10.1053/j.gastro.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fabre A, Martinez-Vinson C, Roquelaure B, Missirian C, Andre N, Breton A, Lachaux A, Odul E, Colomb V, Lemale J, et al. Novel mutations in TTC37 associated with tricho-hepato-enteric syndrome. Hum Mutat. 2011;32:277–281. doi: 10.1002/humu.21420. [DOI] [PubMed] [Google Scholar]
- 72.Fabre ACB, Martinez-Vinson C, Roquelaure B, Odul E, Sayar E, Smith H, Colomb V, Andre N, Hugot J, Goulet O, Lacoste C, Sarles J, Royet J, Levy N. Catherine Badens. Mutations in the human Ski complex encoding genes cause Syndromic Diarrhea (Tri-Hepato-Enteric syndrome. The American Journal of Human Genetics. 2012 [Google Scholar]
- 73.Tomecki R, Kristiansen MS, Lykke-Andersen S, Chlebowski A, Larsen KM, Szczesny RJ, Drazkowska K, Pastula A, Andersen JS, Stepien PP, et al. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J. 2010;29:2342–2357. doi: 10.1038/emboj.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Staals RH, Bronkhorst AW, Schilders G, Slomovic S, Schuster G, Heck AJ, Raijmakers R, Pruijn GJ. Dis3-like 1: a novel exoribonuclease associated with the human exosome. EMBO J. 2010;29:2358–2367. doi: 10.1038/emboj.2010.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Atkinson GC, Baldauf SL, Hauryliuk V. Evolution of nonstop, no-go and nonsense-mediated mRNA decay and their termination factor-derived components. BMC Evol Biol. 2008;8:290. doi: 10.1186/1471-2148-8-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:el89. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 78.Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 79.van Hoof A. Conserved functions of yeast genes support the duplication, degeneration and complementation model for gene duplication. Genetics. 2005;171:1455–1461. doi: 10.1534/genetics.105.044057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Froyd CA, Rusche LN. The duplicated deacetylases Sir2 and Hst1 subfunctionahzed by acquiring complementary inactivating mutations. Mol Cell Biol. 2011;31:3351–3365. doi: 10.1128/MCB.05175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gordon JL, Byrne KP, Wolfe KH. Additions, losses, and rearrangements on the evolutionary route from a reconstructed ancestor to the modern Saccharomyces cerevisiae genome. PLoS Genet. 2009;5:e1000485. doi: 10.1371/journal.pgen.1000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chatr-Aryamontri A, Angelini M, Garelli E, Tchernia G, Ramenghi U, Dianzani I, Loreni F. Nonsense-mediated and nonstop decay of ribosomal protein S19 mRNA in Diamond-Blackfan anemia. Hum Mutat. 2004;24:526–533. doi: 10.1002/humu.20117. [DOI] [PubMed] [Google Scholar]
- 83.Tollner TL, Venners SA, Hollox EJ, Yudin Al, Liu X, Tang G, Xing H, Kays RJ, Lau T, Overstreet JW, et al. A common mutation in the defensin DEFB126 causes impaired sperm function and subfertility. Sci Transl Med. 2011;3:92ra65. doi: 10.1126/scitranslmed.3002289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tollner TL, Yudin AI, Tarantal AF, Treece CA, Overstreet JW, Cherr GN. Beta-defensin 126 on the surface of macaque sperm mediates attachment of sperm to oviductal epithelia. Biol Reprod. 2008;78:400–412. doi: 10.1095/biolreprod.107.064071. [DOI] [PubMed] [Google Scholar]
- 85.Tollner TL, Yudin AI, Treece CA, Overstreet JW, Cherr GN. Macaque sperm coating protein DEFB126 facilitates sperm penetration of cervical mucus. Hum Reprod. 2008;23:2523–2534. doi: 10.1093/humrep/den276. [DOI] [PubMed] [Google Scholar]
- 86.Yudin AI, Generao SE, Tollner TL, Treece CA, Overstreet JW, Cherr GN. Beta-defensin 126 on the cell surface protects sperm from immunorecognition and binding of anti-sperm antibodies. Biol Reprod. 2005;73:1243–1252. doi: 10.1095/biolreprod.105.042432. [DOI] [PubMed] [Google Scholar]
- 87.Pang S, Wang W, Rich B, David R, Chang YT, Carbunaru G, Myers SE, Howie AF, Smillie KJ, Mason JI. A novel nonstop mutation in the stop codon and a novel missense mutation in the type II 3beta-hydroxysteroid dehydrogenase (3beta-HSD) gene causing, respectively nonclassic and classic 3beta-HSD deficiency congenital adrenal hyperplasia. J Clin Endocrinol Metab. 2002;87:2556–2563. doi: 10.1210/jcem.87.6.8559. [DOI] [PubMed] [Google Scholar]
- 88.Doucette L, Green J, Fernandez B, Johnson GJ, Parfrey P, Young TL. A novel, non-stop mutation in FOXE3 causes an autosomal dominant form of variable anterior segment dysgenesis including Peters anomaly. Eur J Hum Genet. 2011;19:293–299. doi: 10.1038/ejhg.2010.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Taniguchi A, Hakoda M, Yamanaka H, Terai C, Hikiji K, Kawaguchi R, Konishi N, Kashiwazaki S, Kamatani N. A germline mutation abolishing the original stop codon of the human adenine phosphoribosyltransferase (APRT) gene leads to complete loss of the enzyme protein. Hum Genet. 1998;102:197–202. doi: 10.1007/s004390050677. [DOI] [PubMed] [Google Scholar]
- 90.Seminara SB, Messager S, Chatzidaki EE, Thresher RR, Acierno JS, Jr, Shagoury JK, Bo-Abbas Y, Kuohung W, Schwinof KM, Hendrick AG, et al. The GPR54 gene as a regulator of puberty. N Engl J Med. 2003;349:1614–1627. doi: 10.1056/NEJMoa035322. [DOI] [PubMed] [Google Scholar]
- 91.Ameri A, Machiah DK, Tran TT, Channell C, Crenshaw V, Fernstrom K, Khachidze M, Duncan A, Fuchs S, Howard TE. A nonstop mutation in the factor (F)X gene of a severely haemorrhagic patient with complete absence of coagulation FX. Thromb Haemost. 2007;98:1165–1169. doi: 10.1160/th07-02-0125. [DOI] [PubMed] [Google Scholar]
- 92.Willig TN, Draptchinskaia N, Dianzani I, Ball S, Niemeyer C, Ramenghi U, Orfali K, Gustavsson P, Garelli E, Brusco A, et al. Mutations in ribosomal protein S19 gene and diamond blackfan anemia: wide variations in phenotypic expression. Blood. 1999;94:4294–4306. [PubMed] [Google Scholar]
- 93.Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Willig TN, Dianzani I, Ball S, Tchernia G, Klar J, Matsson H, et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet. 1999;21:169–175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- 94.Cacciottolo M, Numitone G, Aurino S, Caserta IR, Fanin M, Politano L, Minetti C, Ricci E, Piluso G, Angelini C, et al. Muscular dystrophy with marked Dysferlin deficiency is consistently caused by primary dysferlin gene mutations. Eur J Hum Genet. 2011;19:974–980. doi: 10.1038/ejhg.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Torres-Torronteras J, Rodriguez-Palmero A, Pinos T, Accarino A, Andreu AL, Pintos-Morell G, Martii R. A novel nonstop mutation in TYMP does not induce nonstop mRNA decay in a MNGIE patient with severe neuropathy. Hum Mutat. 2011;32:E2061–2068. doi: 10.1002/humu.21447. [DOI] [PubMed] [Google Scholar]
- 96.Yamaguchi-Kabata Y, Shimada MK, Hayakawa Y, Minoshima S, Chakraborty R, Goiobori T, Imanishi T. Distribution and effects of nonsense polymorphisms in human genes. PLoS One. 2008;3:e3393. doi: 10.1371/journal.pone.0003393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hamby SE, Thomas NS, Cooper DN, Chuzhanova N. A meta-analysis of single base-pair substitutions in translational termination codons (‘nonstop’ mutations) that cause human inherited disease. Hum Genomics. 2011;5:241–264. doi: 10.1186/1479-7364-5-4-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Welch EM, Barton ER Zhuo J, Tomizawa Y, Friesen WJ, Trifillis P, Paushkin S, Patel M, Trotta CR, Hwang S, et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447:87–91. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- 99.Barends S, Barend K, van Wezel GP. The tmRNA-tagging mechanism and the control of gene expression: a review. Wiley Interdiscip Rev RNA. 2011;2:233–246. doi: 10.1002/wrna.48. [DOI] [PubMed] [Google Scholar]
- 100.Richards J, Mehta P, Karzai AW. RNase R degrades non-stop mRNAs selectively in an SmpB-tmRNA-dependent manner. Mol Microbiol. 2006;62:1700–1712. doi: 10.1111/j.1365-2958.2006.05472.x. [DOI] [PubMed] [Google Scholar]