Abstract
In angiosperms, fertilization and subsequent zygotic development occur in embryo sacs deeply embedded in the ovaries; therefore, these processes are poorly elucidated. In this study, microarray-based transcriptome analyses were conducted on rice sperm cells, egg cells, and zygotes isolated from flowers to identify candidate genes involved in gametic and/or early zygotic development. Cell type-specific transcriptomes were obtained, and up- or down-regulated genes in zygotes after fertilization were identified, in addition to genes enriched in male and female gametes. A total of 325 putatively up-regulated and 94 putatively down-regulated genes in zygotes were obtained. Interestingly, several genes encoding homeobox proteins or transcription factors were identified as highly up-regulated genes after fertilization, and the gene ontology for up-regulated genes was highly enriched in functions related to chromatin/DNA organization and assembly. Because a gene encoding methyltransferase 1 was identified as a highly up-regulated gene in zygotes after fertilization, the effect of an inhibitor of this enzyme on zygote development was monitored. The inhibitor appeared partially to affect polarity or division asymmetry in rice zygotes, but it did not block normal embryo generation.
Key words: Egg cell, fertilization, microarray, Oryza sativa, sperm cell, zygote.
Introduction
In angiosperms, the sporophytic generation is initiated by double fertilization, resulting in the formation of seeds (reviewed in Raghavan, 2003). In double fertilization, one sperm cell from the pollen grain fuses with the egg cell, and the resultant zygote develops into an embryo that transmits genetic material from the parents to the next generation. The central cell fuses with the second sperm cell to form a triploid primary endosperm cell, which develops into the endosperm that nourishes the developing embryo/seedling (Nawaschin, 1898; Guignard, 1899; Russell, 1992).
To date, three molecular factors, GENERATIVE CELL SPECIFIC 1/HAPLESS 2 (GCS1/HAP2), EGG CELL 1 (EC1), and ANKYRIN REPEAT PROTEIN 6 (ANK6), are known to play critical roles in male–female gamete recognition and/or fusion in angiosperms (Mori et al., 2006; von Besser et al., 2006; Yu et al., 2010; Sprunck et al., 2012). GCS1/HAP2 was identified as a key male membrane protein with a single transmembrane domain and a histidine-rich domain in the extracellular region. Sperm cells with a gcs mutation remained attached to the egg cell without cell fusion (Mori et al., 2006). Although the mechanism by which GCS1/HAP2 acts is not known, both the N- and C-terminal domains appear to be essential for its function (Mori et al., 2010; Wong et al., 2010). Recently, Sprunck et al. (2012) indicated that small cysteine-rich EC1 proteins accumulated in storage vesicles in Arabidopsis egg cells are secreted via exocytosis upon sperm cell attachment to the egg cell, and that the secreted EC1 proteins function in redistribution of GCS1/HAP2 proteins to the sperm cell surface, resulting in successful gamete fusion. ANK6 is a mitochondrial ankyrin-repeat protein with high expression in both male and female gametophytes and appears to play a role in male–female recognition by regulating mitochondrial gene expression (Yu et al., 2010). In addition to these three proteins, other players need to be identified in order to understand the mechanisms of gamete recognition and fusion.
When a sperm and egg cell fuse, plasmogamy is followed by the sperm nucleus moving toward the egg nucleus. Subsequently, karyogamy, the fusion of nuclei, occurs to form a zygotic nucleus. Ultrastructural and cytological observations of the zygote produced by in vitro fertilization (IVF) using maize gametes revealed that karyogamy in maize occurs within 1h of gametic fusion (Faure et al., 1993). In angiosperms, it has been supposed that the zygotic genome appears to be activated during embryogenesis through ‘maternal to zygotic transition’, as in animals (Schier, 2007; Tadros and Lipshitz, 2009; Autran et al., 2011; Pillot et al., 2010). However, recent studies on early development in maize, tobacco, and Arabidopsis have indicated that the zygotic genome switches on within hours of fertilization (Meyer and Scholten, 2007; Zhao et al., 2011; Nodine and Bartel, 2012), and the zygotic genome is generally considered to be activated almost immediately after fertilization in angiosperms (Hale and Jacobsen, 2012; Nodine and Bartel, 2012).
In addition to de novo transcription/translation, a notable feature of zygote development is remodelling of cell polarity. For example, the vacuole in Arabidopsis zygotes becomes fragmented after fertilization, and the zygote elongates 2- to 3-fold before a large vacuole is re-assembled (Faure et al., 2002). In rice zygotes, the vacuoles and nucleus, which are localized at the apical pore (chalazal side) and basal region (micropyle side) in egg cells, respectively, are re-positioned to opposite poles in the cell (Sato et al., 2010). Although various polarity remodelling processes occur in zygotes of different species, each creates a cytologically polarized cell, with its nucleus positioned at the apical pore and its vacuoles at the basal region (reviewed in Jeong et al., 2011). The polarized zygote divides asymmetrically into a two-celled proembryo consisting of an apical and a basal cell, which develop into the embryo proper and the suspensor/hypophysis, respectively (Pritchard, 1964; Schulz and Jensen, 1968; Tykarska, 1976; Schel et al., 1984; reviewed in Lindsey and Topping, 1993). These facts strongly suggest that cellular polarity in the zygote is tightly linked to the establishment of the initial apical–basal axis in plants. However, the molecular machinery generating and maintaining cellular polarity in zygotes remains poorly understood (Abrash and Bergmann, 2010; Jeong et al., 2011).
In contrast to animals and lower plants, which have free-living gametes, angiosperm fertilization and subsequent events, such as embryogenesis and endosperm development, occur in the embryo sac, which is deeply embedded in ovular tissue. Difficulties in directly researching the biology of the embedded female gametophyte, zygote, and early embryo have impeded investigations into the molecular mechanisms of fertilization and embryogenesis. Therefore, such studies have been conducted predominantly through analyses of Arabidopsis mutants. Alternatively, direct analyses using isolated gametes or zygotes are possible, because procedures for isolating viable gametes have been reported for a wide range of plant species, including maize, wheat, tobacco, rape, rice, barley, Plumbago zeylanica, and Alstroemeria (Dupuis et al., 1987; Kranz et al., 1991; Holm et al., 1994; Kovács et al., 1994; Cao and Russell, 1997; Katoh et al., 1997; Tian and Russell, 1997; Hoshino et al., 2006; Uchiumi et al., 2006). Moreover, IVF using isolated gametes can be used to observe and analyse fertilization and post-fertilization processes directly (reviewed in Wang et al., 2006; Okamoto, 2011).
Using isolated gametes and/or embryos, previous studies have successfully identified genes specifically expressed in male gametes, female gametes, or early embryos (Kasahara et al., 2005; Márton et al., 2005; Sprunck et al., 2005; Ning et al., 2006; Yang et al., 2006; Steffen et al., 2007; Borges et al., 2008; Amien et al., 2010; Ohnishi et al. 2011; Wang et al., 2010; Wuest et al., 2010). However, as far as is known, microarray-based transcriptome analyses of sperm cells, egg cells, and zygotes using the same experimental platform have not been conducted, probably because of difficulties in preparing zygote samples. A procedure to isolate rice gametes and an IVF system to produce zygotes that could develop into fertile plants were established previously (Uchiumi et al., 2006, 2007b ). In this study, microarray analyses of rice gametes and zygotes were performed to examine their gene expression profiles and to monitor changes in gene expression from pre-fertilization to post-fertilization phases.
Materials and methods
Plant materials, isolation of gametes and zygotes, and electrofusion of gametes
Oryza sativa L. cv. Nipponbare was grown in environmental chambers (K30-7248, Koito Industries, Yokohama, Japan) at 26 °C in a 13/11h light/dark cycle. Rice egg and sperm cells were isolated according to Uchiumi et al. (2006), except for using mannitol solution adjusted to 370 mOsmol/kg H2O instead of 0.3M mannitol. Electrofusion of egg and sperm cells and culture of the subsequent zygote were conducted as described by Uchiumi et al. (2007b).
To isolate zygotes from pollinated rice ovaries, ovaries were harvested from flowers 2–3h after flowering, and zygotes were obtained by the above-mentioned procedure for egg cell isolation, with some modifications. Briefly, the pollinated ovaries were transferred into plastic dishes (φ3.5cm) containing 3ml of mannitol adjusted to 450 mOsmol/kg H2O and cut transversely at their midpoints with razor blades (Supplementary Fig. S1 available at JXB online). The zygote was obtained from a dissected ovary by gently pushing the basal portion of the cut ovary with a glass needle under an inverted microscope. The isolated zygotes were either cultured or immediately used for RNA extraction.
RNA isolation from egg cells, zygotes, sperm cells, and pollen grains
Isolated egg cells, sperm cells, and zygotes were washed three times by transferring the cells into fresh droplets of mannitol solution on coverslips. The washed cells were submerged in 5 μl of the extraction buffer supplied in a PicoPure RNA Isolation Kit (Life Technologies, Carlsbad, CA, USA) and stored at –80 °C until use. Pollen grains were released from mature anthers, transferred into 5 μl of extraction buffer, and stored at –80 °C until use. For RNA extraction, cells stored in the extraction buffer were pooled, and total RNAs were isolated using the PicoPure RNA Isolation Kit according to the manufacturer’s instructions. The quality of the total RNAs used for microarray analyses was checked with an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
Microarray and data analysis
Total RNAs prepared from 31–111 egg cells (31, 34, and 111 egg cells; three biological replicates), 30 and 33 zygotes (two biological replicates), ~3000 sperm cells (two biological replicates), and ~100 pollen grains (three biological replicates) were labelled using a Quick Amp Labeling Kit One-Color (Agilent) in the presence of cyanine-3 (Cy3)-CTP for 6h according to the manufacturer’s protocol. The Cy3-labelled cRNA was purified with an RNeasy Mini Kit (Qiagen, Venlo, The Netherlands), and the quantity was examined with a NanoDrop ND-1000 UV-VIS spectrophotometer (Thermo Scientific, Waltham, MA, USA). A total of 1000–1200ng of Cy3-labelled cRNA was fragmented, and hybridized on a rice 4×44K oligoDNA microarray slide (G2519F#15241; Agilent) at 65 °C for 17h. After washing according to the manufacturer’s protocol, the hybridized slides were scanned on a DNA microarray scanner (G2505BA; Agilent). Background correction of the Cy3 raw signals was performed using Feature Extraction software 10.5 (Agilent). Subsequent data processing was performed using GeneSpring GX 11.0 software (Agilent). Raw signal intensities of all probes were subjected to 75th percentile normalization and were log2 transformed. Next, probes were filtered by signal intensity values from the 20th to the 100th percentile and further filtered by Present/Absent flags and by standard deviation (SD <0.5). Two different cell types were compared with an unpaired Student’s t-test. To correct the false discovery rate (FDR), the P-value was adjusted for multiple testing by the Benjamini–Hochburg procedure. The cut-off-corrected P-value was 0.05.
A hierarchical clustering tree was constructed based on genes expressed in egg cells, zygotes, sperm cells, and pollen grains by the centred Pearson correlation algorithm with complete linkage. Gene ontology terms enriched in the 286 genes with 2-fold higher expression levels in egg cells than in zygotes and in the 325 genes whose expression levels in zygotes were 3-fold higher than in egg cells were obtained from the AgriGO database (http://bioinfo.cau.edu.cn/agriGO/index.php). A user-driven tool that displays large genomics data sets, MapMan (Thimm et al., 2004; Usadel et al., 2005), was used to overview the metabolism and regulation in zygotes, compared with egg cells.
RT–PCR and quantitative PCR
cDNAs were synthesized from total RNAs of 10 egg cells, 10 zygotes, and 100 sperm cells using the High Capacity RNA-to-cDNA™ Kit (Life Technologies) according to the manufacturer’s instructions. For reverse transcription–PCR (RT–PCR), 0.2 μl of first-strand cDNA was used as template in a 20 μl PCR with 0.3 μM primers using KOD-FX DNA polymerase (Toyobo, Osaka, Japan) as follows: 35 or 40 cycles of 94 °C for 1min, 55 °C for 30 s, and 72 °C for 1min. Expression of the ubiquitin gene (Os02g0161900) was monitored as an internal control. For quantitative PCR analysis, 0.5 μl of first-strand cDNA was used with LightCycler 480 SYBR Green I Master (Roche Applied Science, Penzberg, Germany) according to the manufacturer’s protocol. Each PCR cycle was conducted as follows: 94 °C for 10 s, 55 °C for 10 s, and 72 °C for 10 s, and relative quantification was calculated with ubiquitin as a referece by the ΔΔCt method. Primer sequences used for PCR analyses are listed in Supplementary Table S1 at JXB online.
Vector construction and preparation of transformants
The genomic sequence of the promoter region positioned 2558bp upstream of the translation start site of a ubiquitin gene (Os02g0161900) was PCR amplified using the primers 5’-AAGCTTTGAATCAGCATAGGCTGCCG-3’ and 5’- TCTAGAC TGCAAGAAATAATCACCAAACAG -3’. The amplified PCR product was subcloned into pGEM-T Easy Vector (Promega, Fitchburg, WI, USA) and its sequence verified. The plasmid harbouring the ubiquitin promoter was cut with HindIII and XbaI, and the excised DNA fragment was subcloned into the HindIII–XbaI site of pIG121-Hm, a binary plasmid vector harbouring a β-glucuronidase (GUS) coding sequence. The resulting vector was termed the Ubi promoter::GUS vector.
cDNA for Arabidopsis histone H2B protein (At5g22880.1) was amplified by PCR using gene-specific primers (5’-CACCAT GGCGAAGGCAGATAAGAAACCAG-3’ and 5’-AGAACTCG TAAACTTCGTAACCG-3’) and cloned into the entry vector pENTR/D-TOPO (Life Technologies). To generate the DNA construct for H2B–green fluorescent protein (GFP) fusion protein, the cloned cDNA was transferred from the entry vector to the destination vector pGWB405 (Nakagawa et al., 2007) by an LR reaction using Gateway LR Clonase enzyme mix (Invitrogen) according to the manufacturer’s instructions. This constructed vector was used as template for PCR amplification of the DNA region encoding H2B–GFP fusion protein using the specific primers 5’-GCTAGCATGGCGAAGGCAGATAAGAA-3’ and 5’- GAGCTCTTACTTGTACAGCTCGTCCA-3’. The amplified PCR product was subcloned into pGEM-T Easy Vector, and the plasmid harbouring the H2B–GFP sequence was cut with NheI and SacI. Then, the excised DNA fragment was subcloned into the XbaI–SacI site of the Ubi promoter::GUS vector, replacing the GUS sequence with that of H2B–GFP. The vector was used as the Ubi promoter::H2B-GFP vector to transform rice plants.
Agrobacterium tumefaciens LBA4404 was transformed with the Ubi promoter::H2B-GFP vector, and transformed rice plants were prepared by co-cultivation of scutellum tissue with A. tumefaciens according to Toki et al. (2006).
Microscopic observation
Cells and embryos were observed using a BX-71 inverted microscope (Olympus, Tokyo, Japan). Fluorescent cells/embryos expressing H2B–GFP were visualized with a BX-71 inverted fluorescence microscope with 460–490nm excitation and 510–550nm emission wavelengths (U-MWIBA2 mirror unit; Olympus). Digital images of egg cells, zygotes, and their resulting embryos were obtained through a cooled charge-coupled device camera (Penguin 600CL; Pixcera, Los Gatos, CA, USA) and InStudio software (Pixcera).
Effects of RG108 on DNA methylation status of some transposon-related elements in cultured rice cells and zygotic development
Based on the methods of Yin et al. (2008), DNA methylation of transposon-related elements on chromosome 4 was assayed by McrBC digestion of genomic DNA and subsequent PCR amplification. A rice suspension cell culture was initiated from scutellum-derived callus according to Kyozuka et al. (1987), and the suspension cells were cultured with or without 100 μM RG108 for 4 d. Genomic DNA (250ng) isolated from cultured cells was digested with 20U of McrBC for 16h at 37 °C. Water was substituted for enzyme as a negative control. PCR was performed with gene-specific primers (Supplementary Table S1 at JXB online) and the products were visualized by gel electrophoresis.
Zygotes were produced by electrofusion of sperm cells with egg cells expressing H2B–GFP and cultured with or without 100 μM RG108.
Results and Discussion
Isolation of gametes and zygotes
Rice egg cells and zygotes were isolated from transversely cut unpollinated and pollinated ovaries, respectively (Supplementary Fig. S1 at JXB online). The isolation efficiency of zygotes was much lower than that of egg cells; on average, 8–15 egg cells and 1–3 zygotes were obtained from 30 unpollinated and pollinated flowers, respectively. This difference can be explained thus: because egg cells have an incomplete cell wall (Russell, 1992), they were easily released from cut ovaries. However, upon fertilization, zygotes immediately begin forming a complete cell wall (Kranz et al., 1995), which tightly attaches the zygote to other cells in the embryo sac and/or to the embryo sac membrane. Such attachment could explain why zygotes were so much more difficult to isolate.
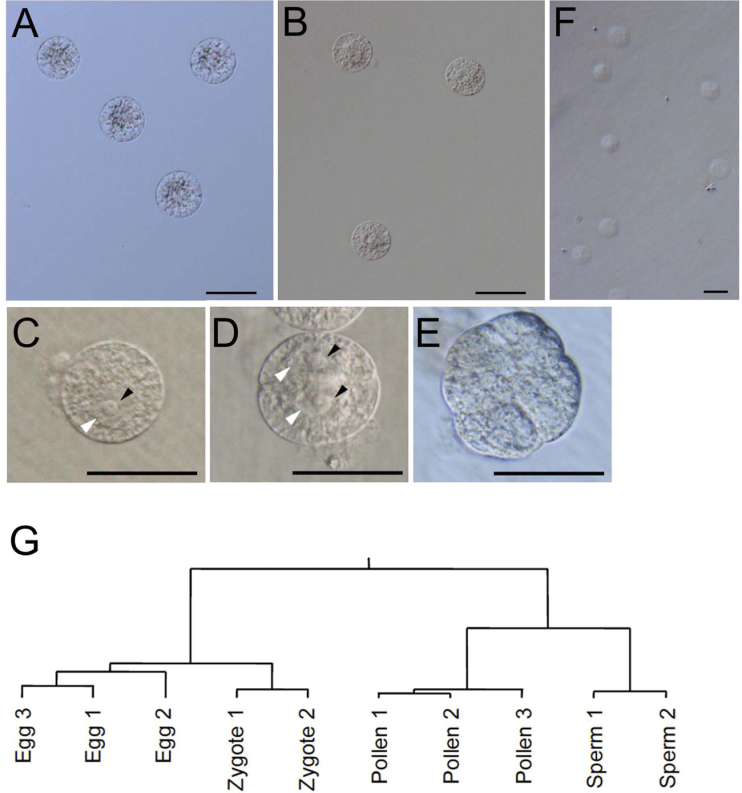
The isolated egg cells showed typical cytological characters, with the cytoplasm, mitochondria, and possible starch granules densely distributed around the nucleus, and with vacuoles at the peripheries of the cells (Fig. 1A; Uchiumi et al., 2006). In the zygotes isolated from pollinated ovaries, cytoplasm and granule structures were observed throughout the cells and vacuoles were undeveloped (Fig. 1B, C). When isolated zygotes were further cultured, they divided into globular-like embryos via asymmetric two-celled proembryos (Fig. 1C–E), as is known to occur in IVF-produced and in planta zygotes (Uchiumi et al., 2007b ; Sato et al., 2010). This finding suggests that the gene expression profiles in the isolated zygotes probably reflect those of zygotes embedded in embryo sacs.
Fig. 1.
Isolated rice cells, cultured zygotes, and hierarchical clustering of expressed genes in the cell types. (A) Egg cells. (B) Zygotes. Zygotes were further cultured for 0 (C), 14 (D), and 37h (E). Black and white arrowheads indicate nucleoli and nuclei, respectively. (F) Sperm cells. (G) Hierarchical clustering tree of egg cells, zygotes, sperm cells, and pollen grains. Bars=50 μm in A–E; 10 μm in F.
Rice pollen grains are tricellular, with one vegetative cell and two sperm cells (Raghavan, 1988). Sperm cells released by bursting pollen grains in mannitol solution were collected with almost homogeneity (Fig. 1F).
Microarrays using RNA from isolated gametes and zygotes
In total RNA fractions prepared from egg cells, zygotes, or sperm cells, 28S and 18S rRNAs were clearly visible (Supplementary Fig. S2 at JXB online). The RNAs were labelled and hybridized to a rice 44K oligo microarray. Correlation plots for the microarray data sets supported the correlation between samples of the same cell type (Supplementary Fig. S3). The correlation cofficient between two sperm cell sets (0.68) was lower than that between other data sets. It may be a reason that sperm cells degenerate after isolation more easily than egg cells and zygotes. Hierarchical sample clustering also grouped the biological replicates together (Fig. 1G), indicating that cell type-specific transcriptomes were obtained. Interestingly, the egg cells and zygotes formed a tighter cluster and showed higher correlation than did the sperm cells and pollen grains, consistent with previous reports that male gametes and gametophytes show very different gene expression profiles from somatic tissue (Borges et al., 2008) and female gametic cells, including eggs, synergids, and central cells (Wuest et al., 2010).
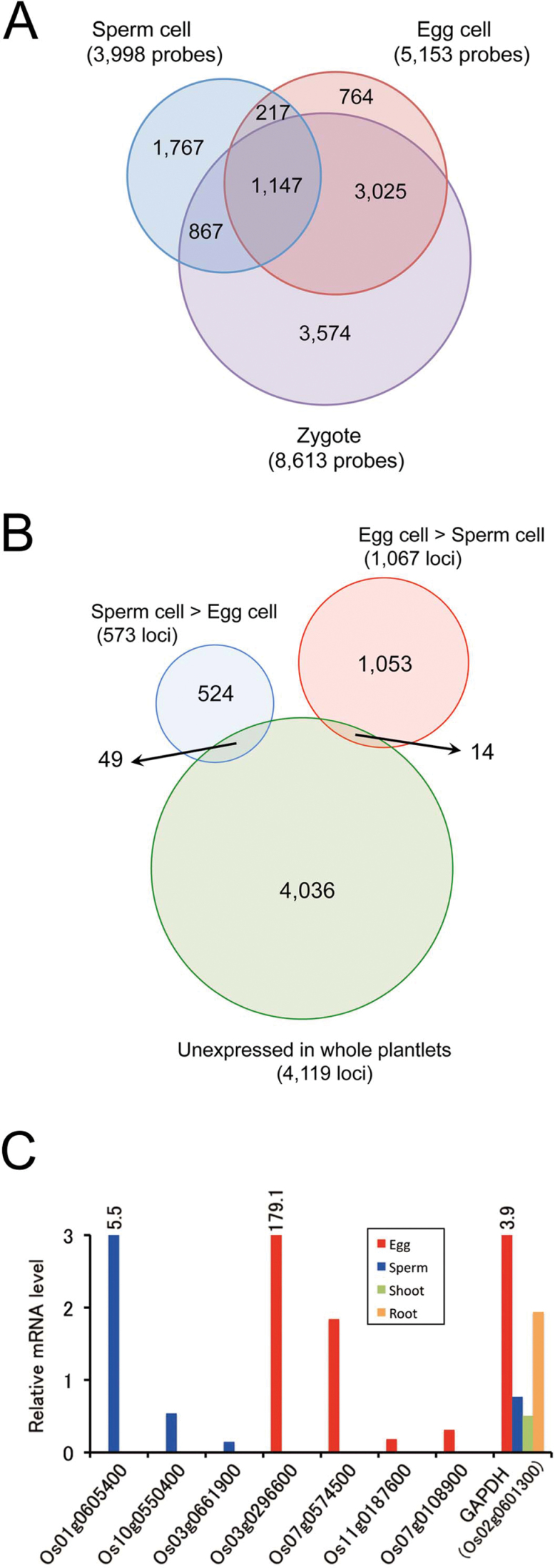
After processing, signals from 3998, 5153, and 8613 probes were judged to be positive in sperm cells, egg cells, and zygotes, respectively (Supplementary Tables S2–S4 at JXB online). Notably, 81% of positive probes in egg cells were also positive in zygotes (Fig. 2A). In contrast, only 26.5% of positive probes in egg cells were positive in sperm cells. These results were consistent with those of the clustering analysis (Fig. 1G) and correlation plots (Supplementary Fig. S3).
Fig. 2.
Gene expression in rice gametes and zygotes. (A) Venn diagram of gene expression overlap in sperm cells, egg cells, and zygotes. Numbers indicate genes with significant t-tests (P <0.05) in each cell type. (B) Venn diagram of genes with substantially higher (>10-fold) expression levels in sperm versus egg cells and genes whose expression was clearly suppressed in whole plantlets. Data for whole plantlets were from Ohnishi et al. (2011). (C) Transcript levels of seven genes enriched in egg or sperm cells and of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) in somatic tissues and gametes were monitored by quantitative PCR and normalized to ubiquitin mRNA. The numbers at the bars indicate values of relative mRNA levels. See Supplementary Table S1 at JXB online for primer sequences.
More than 5000 genes have been reported to be genes expressed in Arabidopsis sperm cells (Borges et al., 2008). Deveshwar et al. (2011) identified 4152 rice genes orthologous to those in Arabidopsis. Oligonucleotide probes for 3965 of these orthologous genes are printed on the Agilent rice 44K oligo microarray chip. In rice sperm cells, 807 of these 3965 genes were expressed (Supplementary Table S2 at JXB online).
To find genes enriched in sperm or egg cells, gene expression profiles were first compared between male and female gametes. A total of 573 genes had expression levels in sperm cells that were 10-fold higher than in egg cells (Supplementary Table S5 at JXB online), and 1067 genes had 10-fold higher expression levels in egg cells than in sperm cells (Supplementary Table S6). Next, to identify gamete-specific genes, these differentially expressed genes were compared with 4119 genes whose expression was clearly suppressed in somatic tissues in the whole-plantlet rice transcriptome of Ohnishi et al. (2011). This analysis found that 49 and 14 genes were enriched in sperm and egg cells, respectively (Fig. 2B). Thirty of the 49 sperm-specific genes had relatively high expression levels in pollen grains (>5000 raw signal value) and were omitted; the remaining 19 genes are listed in Table 1. Table 2 includes the 14 potentially egg-specific genes. Using quantitative PCR, expression levels of seven genes enriched in gametes and a control gene encoding glyceraldehyde 3-phosphate dehydrogenase in somatic tissues and gametes were monitored. All seven genes showed gamete-specific expression (Fig. 2C), supporting the possibility that the genes listed in Tables 1 and 2 have gamete-specific expression profiles.
Table 1.
Genes enriched in rice sperm cells. Values are the average of binary log values of two biological replicates.
| Locus | RAPDB description | Sperm cell | Egg cell |
|---|---|---|---|
| Os01g0605400 | Quinonprotein alcohol dehydrogenase-like domain-containing protein | 6.21 | –1.22 |
| Os06g0715300 | Similar to CEL5 | 5.72 | –1.74 |
| Os02g0664400 | Hypothetical conserved gene | 5.04 | 0.18 |
| Os01g0180900 | Similar to Oxidoreductase | 5.08 | –2.23 |
| Os07g0634100 | Hypothetical conserved gene | 4.60 | –1.56 |
| Os10g0550400 | Hypothetical conserved gene | 4.13 | 0.51 |
| Os03g0809200 | Similar to Transcription factor EmBP-1 | 4.22 | –2.14 |
| Os01g0876100 | Similar to Chloride channel | 3.61 | –0.52 |
| Os01g0853600 | Conserved hypothetical protein | 3.47 | –1.93 |
| Os03g0661900 | Peptidase, trypsin-like serine and cysteine domain-containing protein | 3.18 | –1.27 |
| Os11g0601600 | Protein of unknown function DUF248, methyltransferase putative family protein | 3.14 | –1.89 |
| Os02g0177400 | Conserved hypothetical protein | 3.03 | –0.94 |
| Os05g0393800 | Protein of unknown function DUF221 domain-containing protein | 2.45 | –1.81 |
| Os08g0266700 | Rad21/Rec8 like protein, C-terminal domain-containing protein | 2.28 | –1.19 |
| Os09g0483200 | Similar to UBQ13 (ubiquitin 13) | 2.12 | –2.17 |
| Os09g0244200 | Conserved hypothetical protein | 1.96 | –2.19 |
| Os11g0620800 | Hypothetical conserved gene | 1.46 | –1.98 |
| Os02g0628100 | Hypothetical gene | 1.22 | –2.22 |
| Os08g0474400 | Hypothetical conserved gene | 1.19 | –2.28 |
Mean t-test P-values < 0.05.
Table 2.
Genes enriched in rice egg cells. Values are average binary log values of two biological replicates.
| Locus | RAPDB description | Sperm cell | Egg cell |
|---|---|---|---|
| Os03g0296600 | Similar to ECA1 protein | –1.53 | 8.82 |
| Os05g0491400 | Similar to LRR protein | 0.14 | 6.25 |
| Os07g0574500 | Ubiquitin domain-containing protein | –1.75 | 5.58 |
| Os04g0289600 | Allergen V5/Tpx-1 related family protein | –1.65 | 4.65 |
| Os01g0299700 | 3’–5’ exonuclease domain-containing protein | –1.80 | 4.45 |
| Os06g0602400 | Similar to DEAD-box protein 3, X-chromosomal | –1.63 | 4.26 |
| Os11g0187600 | Similar to Heat shock protein 70 | –1.86 | 3.65 |
| Os07g0108900 | Similar to MADS-box transcription factor 15 | –1.84 | 3.02 |
| Os07g0136300 | Conserved hypothetical protein | –1.69 | 2.80 |
| Os03g0679800 | Similar to TPR Domain-containing protein | –1.79 | 2.95 |
| Os10g0560200 | Protein of unknown function Cys-rich family protein | –1.02 | 2.93 |
| Os01g0350500 | Conserved hypothetical protein | –1.76 | 2.66 |
| Os11g0579900 | Armadillo-like helical domain-containing protein | –1.44 | 2.38 |
| Os05g0153200 | Region of unknown function XH domain-containing protein | –1.85 | 2.28 |
Mean t-test P-values < 0.05.
Five of the 19 genes enriched in sperm cells (Os01g0180900, Os10g0550400, Os03g0661900, Os09g0483200, and Os11g0620800) were reported to be sperm cell specific by Russell et al. (2012). Nine of the 19 enriched genes were annotated as hypothetical proteins or genes, consistent with a previous report indicating enrichment of genes encoding protein with unknown function in sperm-specific genes (Russell et al., 2012). In animals, serine proteases in the trypsin family can be expressed in sperm and involved in fertilization, although their molecular mechanisms during the fertilization process remain unknown (Sawada et al., 1984, 1996; Baba et al., 1994; Adham et al., 1997). Interestingly, Os03g0661900 encodes a trypsin-like serine protease (Table 1). Trypsin-like protease may be expressed in male gametes of both plants and animals and perhaps have similar roles in gamete attachment, recognition, or fusion, although the fertilization systems are largely divergent in the kingdoms.
Three genes in Table 2 (Os03g0296600, Os01g0299700, and Os11g0187600) were reported as enriched in egg cells by Ohnishi et al. (2011). Os03g0296600 encodes an EARLY CULTURE ABUNDANCE (ECA) family protein; ECA1 expression in egg cells was first revealed by expressed sequence tag analysis of wheat egg cells (Sprunck et al., 2005). Os11g0187600 encodes heat shock protein 70 (HSP70). In addition to HSP70, HSP90 was identified as a major protein component of rice egg cells by proteomic analysis (Uchiumi et al., 2007a). HSP90 proteins are evolutionarily conserved molecular chaperones that promote the folding of client proteins with various co-chaperones (reviewed in Sangster and Queitsch, 2005). Notably, among its pleiotropic functions, HSP90 plays a role in buffering the expression of genetic variation when divergent ecotypes are crossed, and profoundly affects developmental plasticity in response to environmental cues (Queitsch et al., 2002). Furthermore, Calvert et al. (2003) revealed that mouse eggs contain molecular chaperones, including HSP90, HSP70, and protein disulphide isomerase (PDI), as major protein components. Because HSP70 and HSP90 both appear to be major protein components of rice egg cells as well, an abundance of these proteins may be a common characteristic of mammalian and plant eggs. These HSPs in egg cells may function following fertilization by a sperm cell, because conversion of an egg cell into a zygote represents major genetic and environmental changes.
MADS-box proteins bind to specific DNA sites as homo- and/or heterodimers to regulate their own transcription and that of target genes (West et al., 1998), and act early in organ development (Riechmann and Meyerowitz, 1997; Theißen et al., 2000). Os07g0108900, encoding MADS-box transcription factor 15 (OsMADS15), was enriched in eggs in this study. In maize, ZmMADS3, which is orthologous to OsMADS15, is strongly expressed in maize egg cells, although its function in egg cells is still unclear (Heuer et al., 2001). MADS-box proteins that accumulate in female gametes may have roles in egg cell differentiation during gametophytogenesis and/or zygotic development after fertilization. A gene enriched in eggs, Os04g0289600, encodes allergen V5/Tpx-1-related protein, which belongs to the CAP superfamily, a family of cysteine-rich secretory proteins (Gibbs et al., 2008). The gene is known to be expressed in synergids as well as egg cells at a high level (Ohnishi et al., 2011), suggesting that the protein functions in the egg apparatus and may be related to the reproductive process.
Down-regulated genes in zygotes
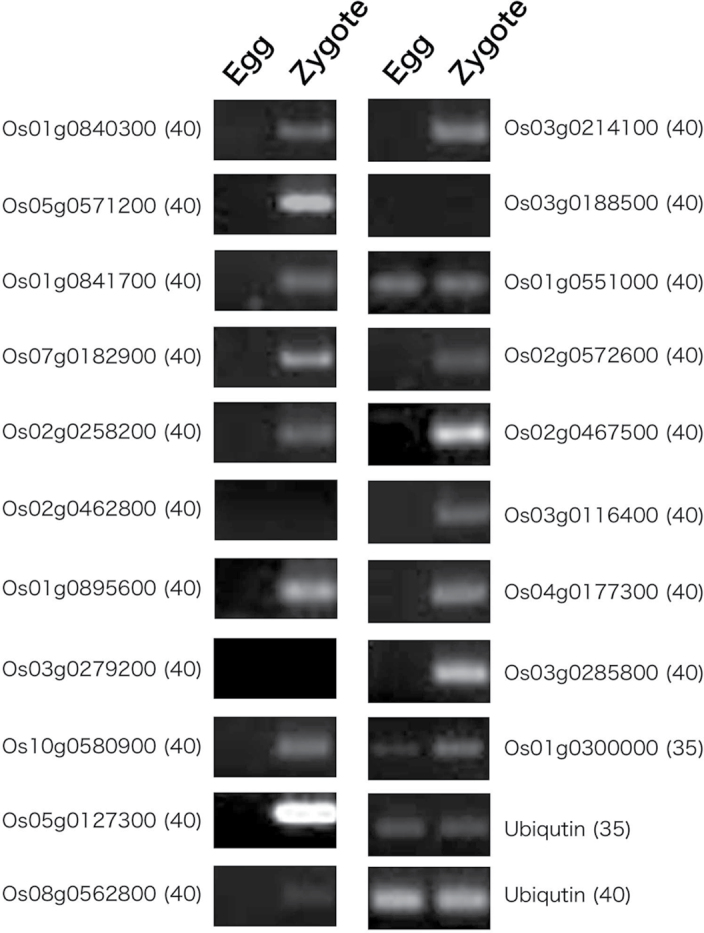
Egg cells are developmentally quiescent, a state that is broken after fertilization and subsequent egg activation. Because the expression of genes involved in maintaining egg cell quiescence should be suppressed in zygotes, the gene expression profiles of egg cells and zygotes were compared to detect genes that were down-regulated after fertilization. A total of 94 genes that had 3-fold lower expression levels in zygotes than in egg cells were identified (Supplementary Table S7 at JXB online). No gene ontologies could be found for these genes, so 286 genes with 2-fold higher expression levels were examined. Most ontologies were related to metabolic or biosynthetic processes (Supplementary Table S8). A comprehensive overview of metabolism in zygotes, compared with egg cells, indicated that several metabolic pathways, including terpene, flavonoid, and amino acid synthetic pathways, appear to be down-regulated after fertilization (Supplementary Fig. S4A). Expression profiles of the 10 most strongly suppressed genes in zygotes (Table 3) were confirmed using semi-quantitative PCR (Fig. 3). Seven genes showed clear down-regulation in zygotes; however, the expression of three genes was not detected in egg cells, suggesting that they may have low expression levels in egg cells. Alternatively, this may be due to using a small amount of RNAs from gametes or zygotes for microarray and RT–PCR analyses.
Table 3.
Ten genes whose expression levels were most putatively down-regulated in zygotes after fertlization.
| Locus | RAPDB description | Fold change (egg/zygote) |
|---|---|---|
| Os01g0360200 | Similar to Respiratory burst oxidase homologue | 15.44 |
| Os01g0936200 | Lipase, class 3 family protein | 11.75 |
| Os03g0760200 | Cytochrome P450 family protein | 11.32 |
| Os03g0154000 | Aromatic-ring hydroxylase family protein | 10.08 |
| Os08g0453200 | Dormancyauxin associated family protein | 8.30 |
| Os02g0530100 | Heavy metal transport/detoxification protein domain containing | 6.82 |
| Os05g0231700 | Similar to Tonoplast membrane integral protein ZmTIP4-2 | 6.67 |
| Os04g0503500 | Leucine-rich repeat, cysteine-containing subtype-containing protein | 6.52 |
| Os03g0100800 | Plasma membrane H+-ATPase | 6.50 |
| Os08g0299400 | Similar to MGDG synthase type A | 6.40 |
Mean t-test P-values < 0.05.
Fig. 3.
Expression patterns of 10 genes whose expression levels were putatively suppressed after fertilization in rice zygotes. Semi-quantitative RT–PCR was performed on total RNAs isolated from egg cells and zygotes using primers specific for the putatively down-regulated genes listed in Table 3. Ubiquitin mRNA was used as an internal control. Numbers in parentheses indicate the number of PCR cycles. See Supplementary Table S1 at JXB online for primer sequences.
Up-regulated genes in zygotes
Upon fertilization, the developmentally quiescent egg cell converts to an active zygote, and expression of genes involved in zygotic development should be induced. Comprehensive overviews of metabolism and regulation in zygotes, compared with egg cells, suggested that synthetic pathways for the cell wall, auxin, and ethylene appeared to be activated in zygotes after fertilization (Supplementary Fig. S4A, B at JXB online). In addition, several receptor kinase-related genes appeared to be up-regulated in zygotes, suggesting that signal transduction pathways are also activated via fertilization. A search for genes that were up-regulated in zygotes identified 325 genes whose expression levels in zygotes were 3-fold higher than in egg cells (Supplementary Table S9). Enriched ontologies were obtained for these genes (Supplementary Table S10). Interestingly, genes related to chromatin/DNA organization and assembly were well represented among the up-regulated genes. In addition, several genes encoding homeobox protein or transcription factors were strongly induced in zygotes (Table 4). Rice plants grown under the conditions used in this study undergo the first zygotic cleavage 14–19h after pollination (Sato et al., 2010). In this study, zygotes were isolated 2–3h post-pollination, implying that zygotes were in the early developmental stage. The developmental stage, the enriched gene ontologies, and the putative functions of up-regulated genes in zygotes together indicate that the zygotic genome is activated early in rice development, as reported for other plant zygotes (Meyer and Scholten, 2007; Zhao et al., 2011; Nodine and Bartel, 2012).
Table 4.
Twenty genes whose expression levels were most putatively up-regulated in zygotes after fertlization.
| Locus | RAPDB description | Fold change (zygote/egg) |
|---|---|---|
| Os01g0840300 | Similar to WUSCHEL-related homeobox 5 | 44.9 |
| Os05g0571200 | Similar to WRKY transcription factor 19 | 41.1 |
| Os01g0841700 | Similar to Isoform ERG1b of Elicitor-responsive protein 1 | 39.6 |
| Os07g0182900 | Similar to Cytosine-5 DNA methyltransferase MET1 | 35.7 |
| Os02g0258200 | High mobility group, HMG1/HMG2 domain-containing protein | 35.2 |
| Os02g0462800 | WRKY transcription factor 42 | 29.9 |
| Os01g0895600 | Similar to Calreticulin 3 | 29.1 |
| Os03g0279200 | Similar to Histone H2A | 27.5 |
| Os10g0580900 | Conserved hypothetical protein | 24.8 |
| Os05g0127300 | Serine/threonine protein kinase domain-containing protein | 23.0 |
| Os08g0562800 | Similar to Transparent testa 12 protein | 19.4 |
| Os03g0214100 | Replication protein A1 | 18.7 |
| Os03g0188500 | Glutelin family protein | 18.6 |
| Os01g0551000 | Conserved hypothetical protein | 16.3 |
| Os02g0572600 | Protein kinase PKN/PRK1, effector domain-containing protein | 16.0 |
| Os02g0467500 | Hypothetical conserved gene | 15.6 |
| Os03g0116400 | Similar to Membrane protein | 15.2 |
| Os04g0177300 | DEAD-like helicase, N-terminal domain-containing protein | 14.6 |
| Os03g0285800 | MAP kinase | 14.6 |
| Os01g0300000 | 3’–5’ exonuclease domain-containing protein | 14.4 |
Mean t-test P-values < 0.05.
Semi-quantitative PCR was conducted for the up-regulated genes listed in Table 4. Although 16 were clearly induced in zygotes (Fig. 4), Os01g0551000 expression was detected at equal levels in egg cells and zygotes, and no expression was detected for three genes. Although why such a difference occurs between semi-quantitative PCR and microarray analyses cannot be explained, it may due to using a small amount of gamete/zygote RNA for these analyses.
Fig. 4.
Expression patterns of 20 genes whose expression levels were putatively induced after fertilization in rice zygotes. Semi-quantitative RT–PCR was performed on total RNAs isolated from egg cells and zygotes using primers specific for the putatively up-regulated genes listed in Table 4. Ubiquitin mRNA was used as an internal control. The numbers in parentheses indicate the number of PCR cycles. See Supplementary Table S1 at JXB online for primer sequences.
Fertilization-induced expression of Os01g0840300, encoding a Wuschel-related homeobox (WOX) protein, was confirmed by PCR (Fig. 4). WOX proteins are key regulators in determining cell fate in plants (Haecker et al., 2004; Park et al., 2005; Mayer et al., 1998; Zhao et al., 2009), and 15 WOX genes, including WUSCHEL, have been identified in Arabidopsis. Interestingly, Os01g0840300 has been reported to be a rice orthologue of Arabidopsis WOX2 (Deveaux et al., 2008). WOX2 transcripts accumulate in Arabidopsis zygotes and are restricted to the apical cell of two-celled proembryos (Haecker et al., 2004). In addition, WOX2 has been proposed to be the predominant regulator of apical patterning (Jeong et al., 2011). In this study, Os01g0840300, a putative rice orthologue of AtWOX2, was identified as a fertilization-induced gene, which may have a role in determining cell fate during early embryogenesis in rice.
Effects of inhibitors of DNA methyltransferase on zygotic development and early embryogenesis
The gene Os07g0182900, putatively encoding DNA methyltransferase 1 (MET1), which functions in maintaining CG DNA methylation (Kankel et al., 2003), was identified among the highly up-regulated genes in early zygotes (Table 4, Fig. 4). In Arabidopsis, expression of MET1 is suppressed in egg and central cells at the end of female gametophyte development via the RETINOBLASTOMA-RELATED 1 (RBR1) pathway (Jullien et al., 2008, 2012). After fertilization, MET1 expression resumes in young embryos to maintain DNA methylation (Jullien and Berger, 2010; Jullien et al., 2012). In addition, Xiao et al. (2006) showed that Arabidopsis embryos with loss-of-function mutants in both MET1 and CHROMOMETHYLASE 3 (CMT3) develop incorrectly and that genes specifying cell identity are misexpressed in abnormal met1 embryos. These reports suggest that induction of MET1 expression in zygotes would be important for zygotic development and early embryogenesis in rice as well. Therefore, to observe the function of MET1 during zygotic development immediately after gamete fusion, zygotes produced by electrofusion of a rice sperm cell with an egg cell were cultured with or without RG108, a specific inhibitor of MET1 (Brueckner et al., 2005; Stresemann et al., 2006).
Whether RG108 affected GC-methylation status was first checked using cultured rice cells. Two of seven tested loci appeared to show decreased methylation levels when RG108 was applied to the culture medium, although differences in methylation status with and without RG108 could not be detected for the remaining five loci (Supplementary Fig. S5 at JXB online). These data suggest that RG108 partially or weakly affected the CG-methylation status of some loci in cultured rice cells.
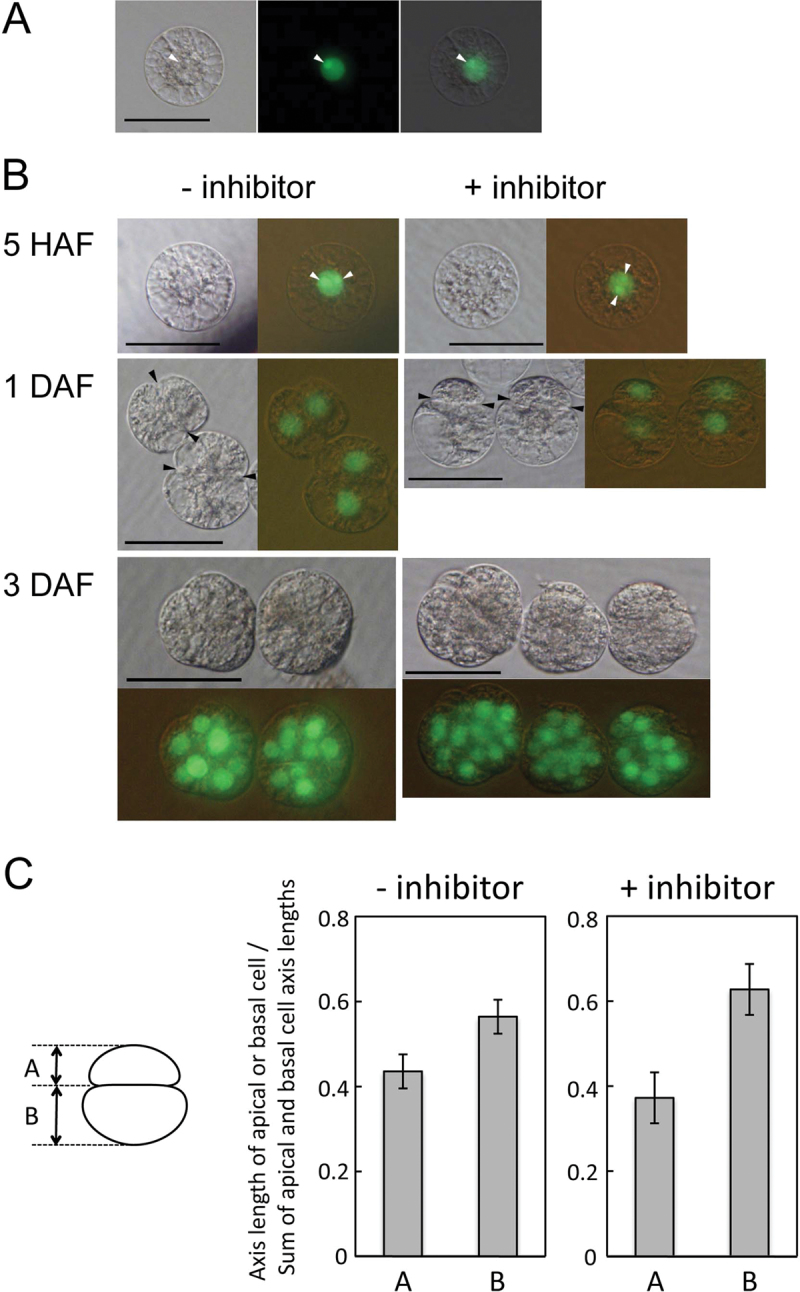
Next, rice zygotes were produced by IVF in which H2B–GFP fusion protein was heterologously expressed, and the nuclei were visualized (Fig. 5A). Expression of the putative MET1 gene (Os07g0182900) in IVF-produced zygotes was confirmed by PCR (Supplementary Fig. S6 at JXB online). The zygotes expressing H2B–GFP were cultured with or without 100 μM RG108, and zygotic development was compared. Two nucleoli were observed in zygotes cultured with and without RG108 at 5h after fusion (Fig. 5B). Approximately 1 d after fusion, the zygotes cultured with the inhibitor divided unequally into two-celled proembryos with small apical cells and large basal cells, similar to zygotes cultured without inhibitor. Notably, among 14 two-celled proembryos cultured with inhibitor, several proembryos clearly showed extreme asymmetry (Fig. 5B), and the apical and basal cells appeared to be more asymmetric in volume in inhibitor-treated two-celled proembryos than in untreated proembryos. Therefore, the axis lengths of the apical and basal cells in two-celled proembryos were next measured. As for two-celled proembryos cultured without inhibitor, the ratio of axis length between the apical and the basal cells was determined as 0.44:0.56 (n=6) when the combined axis length of both cells was considered as 1 (Fig. 5C). In the case of proembryos cultured with inhibitor, the ratio of axis length was calculated to be 0.37:0.63 (n=14), indicating the effect of the inhibitor on asymmetry of the two-celled proembryo. The two-celled proembryos cultured with inhibitor, however, grew into normal globular embryos of 10–20 cells during subsequent culture for 1–2 d (Fig. 5B). These results suggested that establishment of polarity or asymmetric cell division was partly affected by the MET inhibitor, but cells can recover from the unusual asymmetry during early embryonic development. These findings may be consistent with the phenotype of the homologous met1 mutant in Arabidopsis. In the mutant, ~13% of two-celled embryos had abnormal division patterns but eventually grew into mature embryos (Xiao et al., 2006). This phenotypic recovery during Arabidopsis and rice embryogenesis can be explained by redundancy of DNA methylation, including CNG methylation by CHROMOMETHYLASE 3 (Xiao et al., 2006; Jullien et al., 2012), de novo CHH methylation by DOMAINS REARRANGED METHYLTRANSFERASES (Jullien et al., 2012), and the RNA-directed DNA methylation (RdDM) pathway (Mathieu et al., 2007). Alternatively, it is also suggested that MET1 is not a key regulator for zygote development, although it is highly up-regulated in zygotes after fertilization.
Fig. 5.
Effects of RG108 on rice zygote development. (A) Egg cell isolated from transgenic rice expressing histone H2B–GFP under the control of the ubiquitin promoter. The nucleus was fluorescently labelled, and a strong fluorescent signal was detected in nucleoli (arrowheads). Bright-field and fluorescent images are presented in the left-hand and middle panels, respectively. The right-hand panel merges bright-field and fluorescent images. (B) Egg cells labelled as in A were used for in vitro fusion with sperm cells. The resulting zygotes were cultured with or without RG108 inhibitor. Bright-field images (left) were merged with fluorescent images (right). White arrowheads indicate nucleoli. Black arrowheads indicate possible traces of the first division plane. (C) An illustration showing how the axis lengths in an apical cell, A, and a basal cell, B, of a two-celled proembryo were measured is presented on the left. On the right, the ratio of axis length between an apical cell, A, and a basal cell, B, in a two-celled proembryo cultured with or without RG108 is presented. The data are the mean ±SD of six and 14 two-celled proembryos cultured without and with inhibitor, respectively. HAF, hours after gamete fusion. DAF, days after gamete fusion. Bars=50 μm.
Conclusion
In this study, up- or down-regulated genes in rice zygotes after fertilization and genes enriched in gametes were identified. Addressing the functions of these genes during gametic and/or early zygotic developmental processes will improve our knowledge of these processes. Analyses using mutant plants for several of these genes are currently underway in the authors’ laboratories.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Isolation of rice egg cells from ovaries.
Figure S2. Assessment of RNA extracted from rice gametes and zygotes.
Figure S3. Microarray correlation plot within and among rice cell types.
Figure S4. Change of transcript levels in zygotes, compared with egg cells. Overview display of genes assigned to metabolism and regulation.
Figure S5. Effects of RG108 on DNA methylation status on some transposon-related elements in cultured rice cells.
Figure S6. Expression of a putative MET1 gene in a zygote produced by IVF.
Table S1. DNA primers used for RT–PCR and quantitative PCR of rice reproductive cells or cultured cells.
Table S2. Genes expressed in rice sperm cells.
Table S3. Genes expressed in rice egg cells.
Table S4. Genes expressed in rice zygotes.
Table S5. Rice genes whose expression levels in sperm cells were at least 10-fold higher than in egg cells.
Table S6. Rice genes whose expression levels in egg cells were at least 10-fold higher than in sperm cells.
Table S7. Rice genes putatively down-regulated in zygotes after fertilization.
Table S8. Enriched gene ontologies among rice genes that are putatively down-regulated in zygotes.
Table S9. Rice genes putatively up-regulated in zygotes after fertilization.
Table S10. Enriched gene ontologies among rice genes that are putatively up-regulated in zygotes.
Acknowledgements
We thank Dr Y. Nagamura and Ms. R.Motoyama (National Institute of Agrobiological Sciences, Tsukuba, Japan) for assistance with the microarray experiment, Ms T. Mochizuki (Tokyo Metropolitan University) for isolating rice egg cells, Dr T. Kinoshita (NAIST, Japan) for advice on methylation analyses, and RIKEN Bio Resource Center (Tsukuba, Japan) for providing cultured rice cells (Oc line). This work was supported, in part, by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan (no. 21112007 to TO) and from the Japan Society for the Promotion of Science (no. 20570206 to TO).
References
- Abrash EB, Bergmann DC. 2010. Asymmetric cell divisions: a view from plant development. Developmental Cell 16, 783–796. [DOI] [PubMed] [Google Scholar]
- Adham IM, Nayernia K, Engel W. 1997. Spermatozoa lacking acrosin protein show delayed fertilization. Molecular Reproduction and Development 46, 370–376. [DOI] [PubMed] [Google Scholar]
- Amien S, Kliwer I, Márton ML, Debener T, Geiger D, Becker D, Dresselhaus T. 2010. Defensin-like ZmES4 mediates pollen tube burst in maize via opening of the potassium channel KZM1. PLoS Biology 8, e1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autran D, Baroux C, Raissig MT, et al. 2011. Maternal epigenetic pathways control parental contributions to Arabidopsis early embryogenesis. Cell 145, 707–719. [DOI] [PubMed] [Google Scholar]
- Baba T, Azuma S, Kashiwabara S, Toyoda Y. 1994. Sperm from mice carrying a targeted mutation of the acrosin gene can penetrate the oocyte zona pellucida and effect fertilization. Journal of Biological Chemistry 269, 31845–31849. [PubMed] [Google Scholar]
- Borges F, Gomes G, Gardner R, Moreno N, McCormick S, Feijó JA, Becker JD. 2008. Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiology 148, 1168–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brueckner B, Boy RG, Siedlecki P, Musch T, Kliem HC, Zielenkiewicz P, Suhai S, Wiessler M, Lyko F. 2005. Epigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferases. Cancer Research 65, 6305–6311. [DOI] [PubMed] [Google Scholar]
- Calvert ME, Digilio LC, Herr JC, Coonrod SA. 2003. Oolemmal proteomics—identification of highly abundant heat shock proteins and molecular chaperons in the mature mouse egg and their localization on the plasma membrane. Reproductive Biology and Endocrinology 14, 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Russell SD. 1997. Mechanical isolation and ultrastructural characterization of viable egg cells in Plumbago zeylanica . Sexual Plant Reproduction 10, 368–373. [Google Scholar]
- Deveaux Y, Toffano-Nioche C, Claisse G, Thareau V, Morin H, Laufs P, Moreau H, Kreis M, Lecharny A. 2008. Genes of the most conserved WOX clade in plants affect root and flower development in Arabidopsis. BMC Evolutionary Biology 8, 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveshwar P, Bovill WD, Sharma R, Able JA, Kapoor S. 2011. Analysis of anther transcriptomes to identify genes contributing to meiosis and male gametophyte development in rice. BMC Plant Biology 11, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis I, Roeckel P, Matthys-Rochon E, Dumas C. 1987. Procedure to isolate viable sperm cells from corn (Zea mays L.) pollen grains. Plant Physiology 85, 876–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure JE, Mogensen HL, Dumas C, Lörz H, Kranz E. 1993. Karyogamy after electrofusion of single egg and sperm cell protoplasts from maize: cytological evidence and time course. The Plant Cell 5, 747–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure JE, Rotman N, Fortune P, Dumas C. 2002. Fertilization in Arabidopsis thaliana wild type: developmental stages and time course. The Plant Journal 30, 481–488. [DOI] [PubMed] [Google Scholar]
- Gibbs GM, Roelants K, O’Bryan MK. 2008. The CAP superfamily: cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins. Roles in reproduction, cancer, and immune defense. Endocrine Reviews 29, 865–897. [DOI] [PubMed] [Google Scholar]
- Guignard ML. 1899. Sur les antherozoides et la double copulation sexuelle chez les vegetaux angiosperms. Revue Générale de Botanique 11, 129–135. [DOI] [PubMed] [Google Scholar]
- Haecker A, Gross-Hardt R, Geiges B, Sarkar A, Breuninger H, Herrmann M, Laux T. 2004. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131, 657–668. [DOI] [PubMed] [Google Scholar]
- Hale CJ, Jacobsen SE. 2012. Equal-parenting policy. Nature 482, 42–44. [DOI] [PubMed] [Google Scholar]
- Heuer S, Hansen S, Bantin J, Brettschneider R, Kranz E, Lörz H, Dresselhaus T. 2001. The maize MADS box gene ZmMADS3 affects node number and spikelet development and is co-expressed with ZmMADS1 during flower development, in egg cells, and early embryogenesis. Plant Physiology 127, 33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm PB, Knudsen S, Mouritzen P, Negri D, Olsen FL, Roué C. 1994. Regeneration of fertile barley plants from mechanically isolated protoplasts of the fertilized egg cell. The Plant Cell 6, 531–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino Y, Murata N, Shinoda K. 2006. Isolation of individual egg cells and zygotes in Alstroemeria followed by manual selection with a microcapillary-connected micropump. Annals of Botany 97, 1139–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S, Bayer M, Lukowitz W. 2011. Taking the very first steps: from polarity to axial domains in the early Arabidopsis embryo. Journal of Experimental Botany 62, 1687–1697. [DOI] [PubMed] [Google Scholar]
- Jullien PE, Berger F. 2010. DNA methylation reprogramming during plant sexual reproduction? Trends in Genetics 26, 394–399. [DOI] [PubMed] [Google Scholar]
- Jullien PE, Mosquna A, Ingouff M, Sakata T, Ohad N, Berger F. 2008. Retinoblastoma and its binding partner MSI1 control imprinting in Arabidopsis. PLoS Biology 6, e194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien PE, Susaki D, Yelagandula R, Higashiyama T, Berger F. 2012. DNA methylation dynamics during sexual reproduction in Arabidopsis thaliana. Current Biology 22, 1825–1830. [DOI] [PubMed] [Google Scholar]
- Kankel MW, Ramsey DE, Stokes TL, Flowers SK, Haag JR, Jeddeloh JA, Riddle NC, Verbsky ML, Richards EJ. 2003. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics 163, 1109–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara RD, Portereiko MF, Sandaklie-Nikolova L, Rabiger DS, Drews GN. 2005. MYB98 is required for pollen tube guidance and synergid cell differentiation in Arabidopsis. The Plant Cell 17, 2981–2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh N, Lörz H, Kranz E. 1997. Isolation of viable egg cells of rape (Brassica napus L.). Zygote 5, 31–33. [DOI] [PubMed] [Google Scholar]
- Kovács M, Barnabás B, Kranz E. 1994. The isolation of viable egg cells of wheat (Triticum aestivum L.). Sexual Plant Reproduction 7, 311–312. [Google Scholar]
- Kranz E, Bautor J, Lörz H. 1991. In vitro fertilization of single, isolated gametes of maize mediated by electrofusion. Sexual Plant Reproduction 4, 12–16. [Google Scholar]
- Kranz E, von Wiegen P, Lörz H. 1995. Early cytological events after induction of cell division in egg cells and zygote development following in vitro fertilization with angiosperm gametes. The Plant Journal 8, 9–23. [Google Scholar]
- Kyozuka J, Hayashi Y, Shimamoto K. 1987. High frequency plant regeneration from rice protoplasts by novel nurse culture methods. Molecular Genetics and Genomics 206, 408–413. [Google Scholar]
- Lindsey K, Topping JE. 1993. Embryogenesis: a question of pattern. Journal of Experimental Botany 259, 359–374. [Google Scholar]
- Márton ML, Cordts S, Broadhvest J, Dresselhaus T. 2005. Micropylar pollen tube guidance by egg apparatus 1 of maize. Science 307, 573–576. [DOI] [PubMed] [Google Scholar]
- Mathieu O, Reinders J, Čaikovski M, Smathajitt C, Paszkowski J. 2007. Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell 130, 851–862. [DOI] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jurgens G, Laux T. 1998. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815. [DOI] [PubMed] [Google Scholar]
- Meyer S, Scholten S. 2007. Equivalent parental contribution to early plant zygotic development. Current Biology 17, 1686–1691. [DOI] [PubMed] [Google Scholar]
- Mori T, Hirai M, Kuroiwa T, Miyagishima S. 2010. The functional domain of GCS1-based gamete fusion resides in the amino terminus in plant and parasite species. PloS One 5, e15957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Kuroiwa H, Higashiyama T, Kuroiwa T. 2006. GENERATIVE CELL SPECIFIC 1 is essential for angiosperm fertilization. Nature Cell Biology 8, 64–71. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, et al. 2007. Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Bioscience, Biotechnology, and Biochemistry 71, 2095–2100. [DOI] [PubMed] [Google Scholar]
- Nawaschin S. 1898. Revision der Befruchtungsvorgange bei Lilium martagon und Fritillaria tenella . Bulletin Scientifique Académie Impériale des Sciences de Saint Pétersbourg 9, 377–382. [Google Scholar]
- Ning J, Peng X-B, Qu L-H, Xin H-P, Yan T-T, Sun M. 2006. Differential gene expression in egg cells and zygotes suggests that the transcriptome is restructed before the first zygotic division in tobacco. FEBS Letters 580, 1747–1752. [DOI] [PubMed] [Google Scholar]
- Nodine MD, Bartel DP. 2012. Maternal and paternal genomes contribute equally to the transcriptome of early plant embryos. Nature 482, 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T, Takanashi H, Mogi M, et al. 2011. Distinct gene expression profiles in egg and synergid cells of rice as revealed by cell type-specific microarrays. Plant Physiology 155, 881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto T. 2011. In vitro fertilization with isolated rice gametes: production of zygotes and zygote and embryo culture. Methods in Molecular Biology 710, 17–27. [DOI] [PubMed] [Google Scholar]
- Park SO, Zheng Z, Oppenheimer DG, Hauser BA. 2005. FEW SEEDS2 gene encodes an Arabidopsis homeodomain protein that regulates ovule development. Development 132, 841–849. [DOI] [PubMed] [Google Scholar]
- Pillot M, Baroux C, Vazquez MA, Autran D, Leblanc O, Vielle-Calzada JP, Grossniklaus U, Grimanelli D. 2010. Embryo and endosperm inherit distinct chromatin and transcriptional states from the female gametes in Arabidopsis. The Plant Cell 22, 307–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard NH. 1964. A cytochemical study of embryo development in Stellaria media . American Journal of Botany 51, 472–479. [Google Scholar]
- Queitsch C, Sangster TA, Lindquist S. 2002. Hsp90 as a capacitor of phenotypic variation. Nature 416, 618–624. [DOI] [PubMed] [Google Scholar]
- Raghavan V. 1988. Anther and pollen development in rice (Oryza sativa). American Journal of Botany 75, 183–186. [Google Scholar]
- Raghavan V. 2003. Some reflections on double fertilization, from its discovery to the present. New Phytologist 159, 565–583. [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Meyerowitz EM. 1997. MADS domain proteins in plant development. Biological Chemistry 378, 1079–1101. [PubMed] [Google Scholar]
- Russell SD. 1992. Double fertilization. International Review of Cytology 40, 357–390. [Google Scholar]
- Russell SD, Gou X, Wong C, Wang X, Yuan T, Wei X, Bhalla PL, Singh MB. 2012. Genomic profiling of rice sperm cell transcripts reveals conserved and distinct elements in the flowering plant male germ lineage. New Phytologist 195, 560–573. [DOI] [PubMed] [Google Scholar]
- Sangster TA, Queitsch C. 2005. The HSP90 chaperone complex, an emerging force in plant development and phenotypic plasticity. Current Opinion in Plant Biology 8, 86–92. [DOI] [PubMed] [Google Scholar]
- Sato A, Toyooka K, Okamoto T. 2010. Asymmetric cell division of rice zygotes located in embryo sac and produced by in vitro fertilization. Sexual Plant Reproduction 23, 211–217. [DOI] [PubMed] [Google Scholar]
- Sawada H, Yokosawa H, Ishii S. 1984. Purification and characterization of two types of trypsin-like enzymes from sperm of the ascidian (Prochordata) Halocynthia roretzi. Evidence for the presence of spermosin, a novel acrosin-like enzyme. Journal of Biological Chemistry 259, 2900–2904. [PubMed] [Google Scholar]
- Sawada H, Iwasaki K, Kihara-Negishi F, Ariga H, Yokosawa H. 1996. Localization, expression, and the role in fertilization of spermosin, an ascidian sperm trypsin-like protease. Biochemical and Biophysical Research Communications 222, 499–504. [DOI] [PubMed] [Google Scholar]
- Schel JHN, Kieft H, van Lammeren AAM. 1984. Interactions between embryo and endosperm during early developmental stage of maize caryopses (Zea mays). Canadian Journal of Botany 62, 2842–2853. [Google Scholar]
- Schier A. 2007. The maternal–zygotic transition: death and birth of RNAs. Science 316, 406–407. [DOI] [PubMed] [Google Scholar]
- Schulz R, Jensen WA. 1968. Capsella embryogenesis: the egg, zygote and young embryo. American Journal of Botany 55, 807–819. [Google Scholar]
- Sprunck S, Baumann U, Edwards K, Langridge P, Dresselhaus T. 2005. The transcript composition of egg cells changes significantly following fertilization in wheat (Triticum aestivum L.). The Plant Journal 41, 660–672. [DOI] [PubMed] [Google Scholar]
- Sprunck S, Rademacher S, Vogler F, Gheyselinck J, Grossniklaus U, Dresselhaus T. 2012. Egg cell-secreted EC1 triggers sperm cell activation during double fertilization. Science 338, 1093–1097. [DOI] [PubMed] [Google Scholar]
- Steffen JG, Kang IH, Macfarlane J, Drews GN. 2007. Identification of genes expressed in the Arabidopsis female gametophyte. The Plant Journal 51, 281–292. [DOI] [PubMed] [Google Scholar]
- Stresemann C, Brueckner B, Musch T, Stopper H, Lyko F. 2006. Functional diversity of DNA methyltransferase inhibitors in human cancer cell lines. Cancer Research 66, 2794–2800. [DOI] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. 2009. The maternal-to-zygotic transition: a play in two acts. Development 136, 3033–3042. [DOI] [PubMed] [Google Scholar]
- Theißen G, Becker A, DiRosa A, Kanno A, Kim JT, Munster T, Winter KU, Saedler H. 2000. A short history of MADS-box genes in plants. Plant Molecular Biology 42, 115–149. [PubMed] [Google Scholar]
- Thimm O, Blaesing OE, Gibon Y, Nagel A, Meyer S, Krueger P, Selbig J, Mueller LA, Rhee SY, Stitt M. 2004. MapMan: a user-driven tool to display genomics datasets onto diagrams of metabolic pathways and other biological processes. The Plant Journal 37, 914–939. [DOI] [PubMed] [Google Scholar]
- Tian HQ, Russell SD. 1997. Micromanipulation of male and female gametes of Nicotiana tabacum: I. Isolation of gametes. Plant Cell Reports 16, 555–560. [DOI] [PubMed] [Google Scholar]
- Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H. 2006. Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. The Plant Journal 47, 969–976. [DOI] [PubMed] [Google Scholar]
- Tykarska T. 1976. Rape embryogenesis: I. The proembryo development. Acta Societatis Botanicorum Poloniae 45, 3–16. [Google Scholar]
- Uchiumi T, Komatsu S, Koshiba T, Okamoto T. 2006. Isolation of gametes and central cells from Oryza sativa L. Sexual Plant Reproduction 19, 37–45. [Google Scholar]
- Uchiumi T, Shinkawa T, Isobe T, Okamoto T. 2007a. Identification of the major protein components of rice egg cells. Journal of Plant Research 120, 575–579. [DOI] [PubMed] [Google Scholar]
- Uchiumi T, Uemura I, Okamoto T. 2007b. Establishment of an in vitro fertilization system in rice (Oryza sativa L.). Planta 226, 581–589. [DOI] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Thimm O, et al. 2005. Extension of the visualisation tool MapMan to allow statistical analysis of arrays, display of corresponding genes and comparison with known responses. Plant Physiology 138, 1195–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Besser K, Frank AC, Johnson MA, Preuss D. 2006. Arabidopsis HAP2 (GCS1) is a sperm-specific gene required for pollen tube guidance and fertilization. Development 133, 4761–4769. [DOI] [PubMed] [Google Scholar]
- Wang D, Zhang CQ, Hearn DJ, Kang IH, Punwani JA, Skaggs MI, Drews GN, Schumaker KS, Yadegari R. 2010. Identification of transcription factor genes expressed in the Arabidopsis female gametophyte. BMC Plant Biology 10, 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YY, Kuang A, Russell SD, Tian HQ. 2006. In vitro fertilization as a tool for investigating sexual reproduction of angiosperm. Sexual Plant Reproduction 19, 103–115. [Google Scholar]
- West AG, Causier BE, Davies B, Sharrocks AD. 1998. DNA binding and dimerization determinants of Antirrhinum majus MADS-box transcription factors. Nucleic Acids Research 26, 5277–5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JL, Leydon AR, Johnson MA. 2010. HAP2(GCS1)-dependent gamete fusion requires a positively charged carboxy-terminal domain. PLoS Genetics 6, e1000882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuest SE, Vijverberg K, Schmidt A, et al. 2010. Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Current Biology 20, 506–512. [DOI] [PubMed] [Google Scholar]
- Xiao W, Custard KD, Brown RC, Lemmon BF, Harada JJ, Goldberg RB, Fischer RL. 2006. DNA methylation is critical for Arabidopsis embryogenesis and seed viability. The Plant Cell 18, 805–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Kaur N, Kiriakopolos S, McCormick S. 2006. EST generation and analyses towards identifying female gametophyte-specific genes in Zea mays L. Planta 224, 1004–1014. [DOI] [PubMed] [Google Scholar]
- Yin BL, Guo L, Zhang DF, Terzaghi W, Wang XF, Liu TT, He H, Cheng ZK, Deng XW. 2008. Integration of cytological features with molecular and epigenetic properties of rice chromosome 4. Molecular Plant 1, 816–829. [DOI] [PubMed] [Google Scholar]
- Yu F, Shi J, Zhou J, et al. 2010. ANK6, a mitochondrial ankyrin repeat protein, is required for male–female gamete recognition in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 107, 22332–22337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Hu Y, Dai M, Huang L, Zhou DX. 2009. The WUSCHEL-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. The Plant Cell 21, 736–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Xin H, Qu L, Ning J, Peng X, Yan T, Ma L, Li S, Sun MX. 2011. Dynamic changes of transcript profiles after fertilization are associated with de novo transcription and maternal elimination in tobacco zygote, and mark the onset of the maternal-to-zygotic transition. The Plant Journal 65, 131–145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.