The detection of biomolecules has been a major research focus in science and technology for many decades given its myriad applications in molecular diagnostics,[1,2] environmental monitoring,[3,4] drug discovery,[5] and biological warfare agents detection,[6] among others. A large majority of modern detection assays rely on a labeling strategy where the target molecule is either tagged with a label (e.g., a fluorescent dye[7,8] or a radioisotope[9]) or coupled to an enzyme[10] to generate a detectable signal. Unfortunately, this labeling process often increases the time and expense required to perform the assay and can alter the functionality of the target biomolecules.[5] As such, a detection strategy that can directly target biomolecules without labeling would greatly facilitate the development of reliable, rapid, cost-effective, and easy-to-use biomolecular detection assays. To this end, a wide range of technologies have been evaluated for the direct detection of biomolecules, using surface plasmon resonance (SPR) spectroscopy,[11] microcantilever,[12] and quartz crystal microbalance (QCM);[13] however, the biomolecular detection process in all of these techniques must occur on a stationary solid substrate. This requirement leads to several shortcomings, including cumbersome surface-immobilization of capture molecules, slow binding kinetics due to diffusion limitations, and a distortion of intrinsic biomolecular interactions (the surface-bound nature of the capture molecules can result in undesirable orientation for binding, reduce the degrees of freedom available for capturing the target, and induce non-specific binding of targets),[14,15] all of which can deteriorate detection performance (reliability, reproducibility, sensitivity, and specificity).
In contrast to the aforementioned stationary surface-incorporated techniques, solution detection using nanomaterials (e.g., molecular beacons,[14] carbon nanotubes,[16,17] and inorganic nanoparticles[18,19]) as capture platforms can offer a more reliable, rapid, specific, and convenient assay by eliminating the slow diffusion kinetics and increasing the degrees of freedom available for capturing the targets. Recently, graphene oxide, a two-dimensional oxygenated graphene sheet, has emerged as a promising biosensing platform for the detection of DNAs and proteins.[20-22] This nanosheet features a variety of chemically reactive functionalities, with epoxy and hydroxyl groups decorating the basal plane and carbonyl and carboxylic acid groups lining the sheet edge;[23,24] all of which can be orthogonally functionalized for biomolecular detection.[25,26] In addition to these surface moieties, graphitic regions containing sp2-hybridized carbons remain on the basal plane,[27] having survived the harsh oxidative conditions of the synthesis. Such regions allow the graphene oxide sheet to retain an overall high degree of planarity (as demonstrated in thin-film deposition and fabrication experiments),[28,29] a correspondingly high surface-to-volume ratio,[30] and strong quenching of fluorescence from adsorbed organic dyes.[31-33] Similar fluorescence quenching has been observed for other carbonaceous nanoparticles like graphene and carbon nanotubes,[34-36] allowing these particles to be used as platforms for “turn-on” sensing of DNAs,[17] proteins,[37,38] and other biomolecules.
Of the few reports that utilize graphene oxide for biomolecular detection via turn-on fluorescence detection,[20-22,39-41] no study to date has considered the effects that the extent of oxidation on graphene oxide (as quantified by its carbon-to-oxygen (C/O) ratio) would affect its performance as a biosensor. In fact, it is quite surprising that none of the aforementioned reports have even documented the C/O ratio of the graphene oxide materials that were used, which can vary greatly depending on the extent of oxidation.[27,30,33,42] As the C/O ratio is a direct indicator of the abundance of the graphitic domains on graphene oxide (i.e., the higher the C/O ratio, the more abundant the graphitic domains), which in turn are primarily responsible for fluorescence quenching of adsorbed dyes near their surface via energy transfer,[34,35] one would expect that this parameter will greatly affect the sensitivity and scope of the detection assay. While a few research groups have recently begun to compare the electrochemical properties[43-45] and fluorescence quenching efficiencies[46,47] of reduced graphene oxide and graphene oxide and attributed the observed property changes to the drastic differences in the abundance of the graphitic domains between these two very different materials, the effect of fine-tuning the C/O ratio of graphene oxide has not been explored. We hypothesize that such an investigation into the property changes that accompany the extent of oxidation in graphene oxide can be quite beneficial in biological applications where solutions of reduced graphene oxide cannot be used directly due to their tendency to aggregate in electrolytes.[33]
Herein, we report that small variations in the level of oxidation of graphene oxide not only strongly affect its fluorescence quenching ability but also significantly influence its binding interactions to single-stranded oligodeoxyribonucleotide (ssODN), leading to a broad range of DNA detection sensitivity and media dependence. Among the graphene oxide samples that were examined, the least-oxidized sample (C/O = 1.9) exhibited nearly four times higher ssODN loading and ~30% enhanced fluorescence quenching efficiency in comparison to the most-oxidized one (C/O ratio = 1.1), attributable to a stronger interaction between the less-oxidized graphene oxide and the capture DNA strands. This high loading, together with a more-facile capture-and-release kinetics in serum, allows the least-oxidized graphene oxide to have ~400% higher sensitivity than the more-oxidized materials and enhance its sensitivity in serum by over 30 folds above that in Tris-HCl buffer at low target concentrations (10-50 nm), making this platform highly suitable for use in applications with biological fluids. Interestingly, the moderately oxidized graphene oxide (C/O = 1.6) platform exhibits the best DNA detection sensitivity in Tris-HCl buffer as compared to the least- and the most-oxidized graphene oxides.
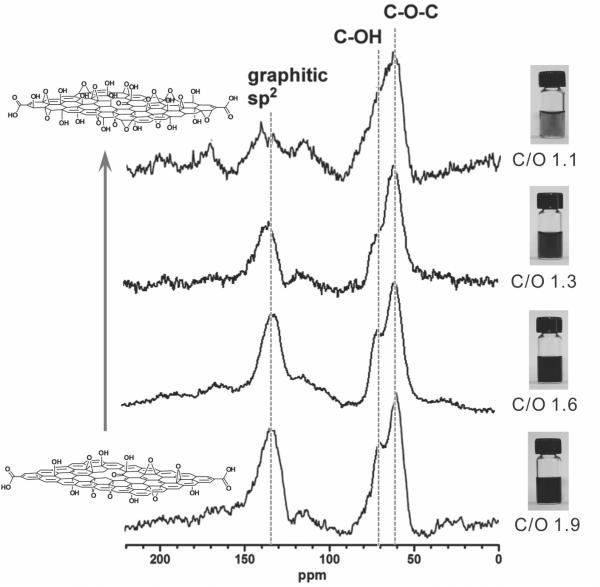
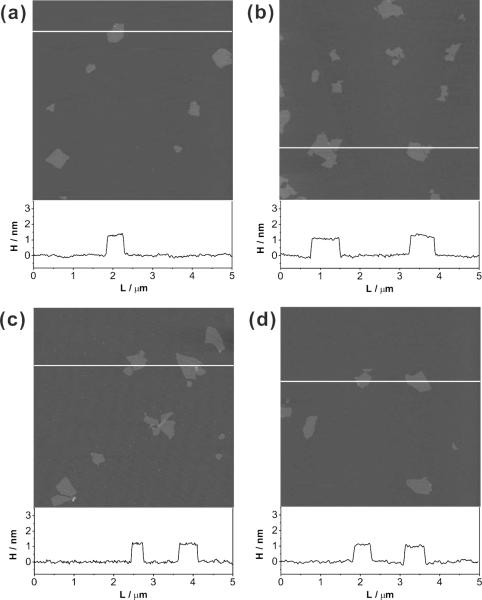
Graphene oxide samples were prepared from graphite using a combination of Hummers oxidation[48] and exfoliation. Samples with different C/O ratios (1.1, 1.3, 1.6, and 1.9) were obtained either by varying the synthetic conditions,[30,49] or by exposing graphene oxide to an additional cycle of oxidation.[33,50] Analysis of the solid-state 13C NMR spectra (Figure 1 and Figure S1 in Supporting Information (SI)) of these materials shows a near-linear relationship between their C/O ratios and the ratios of the peak areas for the graphitic sp2-hybridized carbons to those for the oxygenated carbons (Figure S2 in the SI). It is clear that graphene oxide nanosheets with higher C/O ratios have more sp2-hybridized carbons that contribute to the graphitic domains of graphene oxide than those with lower C/O ratios. In spite of the different chemical compositions between our samples, their topographic morphology remains relatively unchanged as shown by atomic force microscopy (AFM), which reveals that the nanosheets in all samples are similar in size and thickness (Figure 2).
Figure 1.
Left column: Solid-state 13C NMR spectra of graphene oxide samples having the C/O ratios of 1.1, 1.3, 1.6, and 1.9, illustrating that more oxidized graphene oxide has less graphitic sp2 signal. Right column: Digital images of vials containing the corresponding solutions of graphene oxide nanosheets. At the same concentration (0.5 mg mL-1), the solution becomes progressively lighter as the C/O ratios of the graphene oxide decreases, a consequence of the reduced abundance of sp2 graphitic domains. A color reproduction of this figure is available in the SI as Figure S1.
Figure 2.
AFM topographic images of graphene oxide nanosheets with C/O ratios of (a) 1.1, (b) 1.3, (c) 1.6, and (d) 1.9 demonstrating that the sheets in all the samples are similar in sample sizes and thicknesses. The height profiles for the white lines in each AFM image are included below the panels. A color reproduction of this figure is available in the SI as Figure S8.
It has generally been accepted that the nucleobases of ssODNs can bind strongly to the basal plane of graphene oxide sheets, presumably via multiple π-π stacking interactions[20-22] that are similar to those observed in DNA-carbon nanotube “complexes”.[51] This mode of binding forces the ssODNs to remain “lying down” in close contact with the sheet, which can effectively quench the fluorescence of any ssODN-tethered chromophores.[20-22] As such, we hypothesize that the degree of binding interaction between graphene oxide nanosheets and fluorescently labeled ssODNs, as well as the extent of fluorescent quenching, can be tuned by varying the C/O ratios of the graphene oxide sample: varying the C/O ratios of graphene oxide would change the amount of residual graphitic domains on the basal planes of the graphene oxide sheets and modulate their interactions with ssODNs. In addition, reducing the extent of conjugation on the graphene oxide sheets would also lead to less-efficient quenching of the tethered fluorescent labels.
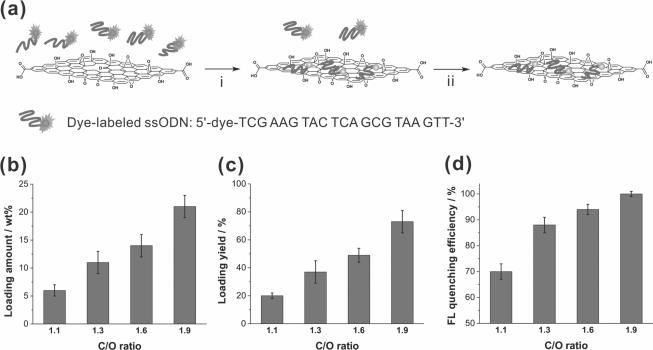
To test the aforementioned hypothesis, we incubated four samples of graphene oxide nanosheets (C/O ratios = 1.1, 1.3, 1.6, and 1.9) with ssODNs that were pre-labeled with one of three different organic dyes (Cy5, Cy3, and Alexa Fluor546). After 2 h, these twelve different mixtures were subjected to centrifugation-filtering to remove unbound ssODNs (Figure 3a). Subjecting the remaining ssODN-graphene oxide complexes to three cycles of [resuspension in water + centrifugation-filtering] leave no observable free ssODNs in the filtrate solution, as monitored by UV-vis spectroscopy, suggesting that the graphene oxide-loaded ssODNs were strongly bound to all samples. The complexes were then redispersed in water, and the amounts of dye-tagged ssODNs loaded on the graphene oxide nanosheets were quantified using UV-vis spectroscopy.
Figure 3.
(a) Scheme illustrating the isolation of ssODN-graphene oxide complexes. Step i: physically combining of the dye-tagged ssODNs (dye = Cy5, Cy3, and Alexa Fluor546) with the graphene oxide nanosheet. Step ii: removal of unbound ssODNs and isolation of the ssODN-graphene oxide complexes through centrifugation and filtration. (b and c) Loading amount (b) and loading yield (c) of dye-tagged ssODNs when exposed to graphene oxide samples (50 μg) with different C/O ratios (1.1, 1.3, 1.6, and 1.9) for 2 h. (d) The corresponding fluorescence quenching efficiency of these four samples of graphene oxide nanosheets (50 μg) against dye-tagged ssODNs that has been physisorbed on the nanosheet surfaces. Each experiment was repeated three times with three different dye-tagged ssODNs. A color reproduction of this figure is available in the SI as Figure S9.
Along with the absorption spectra of the graphene oxide complexes (Figure S3 in the SI), plots of ssODN loading amount (Figure 3b) and ssODN loading yield (Figure 3c) as a function of C/O ratio reveal a consistently increased loading of ssODNs onto the graphene oxide nanosheets as their C/O ratio increases. For the sample with the highest C/O ratio (1.9), ~20 wt% of dye-tagged ssODNs was loaded onto graphene oxide nanosheets, corresponding to a loading yield of ~70%. In stark contrast, the sheets having the lowest C/O ratio (1.1) exhibit a loading amount of only ~6 wt%, equivalent to a loading yield of ~20%. Together with the solid-state NMR data for the four graphene oxide samples (see discussion above), this data clearly suggests a strong correlation between the C/O ratio of graphene oxide and its strength of interaction to ssODNs: the presence of higher concentrations of graphitic regions enables graphene oxide with higher C/O ratios to bind more strongly to ssODNs.
In combination with the ssODN loading amount data (Figure 3b), the fluorescence intensity of the dye-tagged ssODN-graphene oxide complexes allows us to quantify the fluorescence quenching efficiency (FQeff) of graphene oxide for physisorbed dye-tagged ssODNs using the equation FQeff = (AUV – AFL)/AUV, where AUV and AFL are the amounts of dye-tagged ssODNs loaded onto the graphene oxide sheet, as calculated respectively from the UV-vis and the fluorescence data. As the C/O ratio of graphene oxide increases, an increased fluorescence quenching efficiency is observed (Figure 3d). Graphene oxide with the highest C/O ratio (1.9) exhibits a fluorescence quenching efficiency of > 99% for all dyes, in contrast to the poorer quenching efficiency (~70%) by graphene oxide with the lowest C/O ratio (1.1). Similar to the aforementioned trends in loading amount and yield of dye-tagged ssODNs, the increased quenching efficiency at high C/O ratios can be directly linked to the extent of oxidation of the nanosheets, which directly affects the area of residual graphitic domains on the sheet surface.
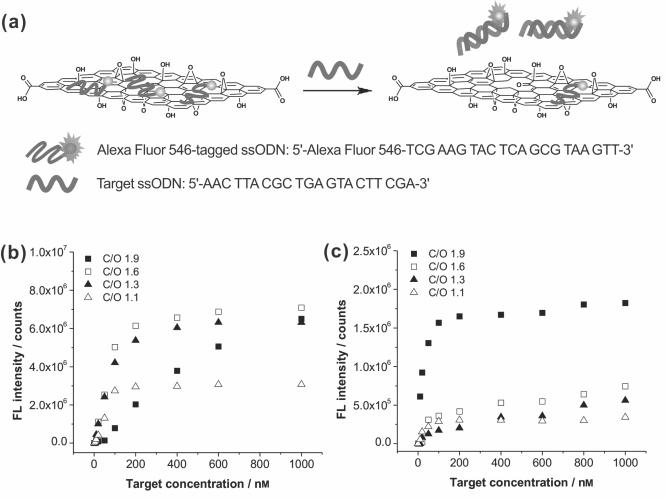
Our results thus far suggest that the level of oxidation in graphene oxide can be readily tuned to influence its fluorescence quenching ability and binding interaction with ssODNs, and thus different DNA sensing performances. To this end, complexes of Alexa Fluor 546-tagged capture ssODNs and graphene oxide nanosheets were incubated with the complementary target DNAs at different concentrations in Tris-HCl buffer solution and fetal bovine serum (FBS) for 1 hour, respectively. In the presence of target DNAs, the dye-labeled capture ssODNs that were originally bound to graphene oxide nanosheets will hybridize to the target DNAs in solution and be released from the sheets, leading to restoration of the previously quenched dye fluorescence (Figure 4a).[20-22] This then provides a ready means for detecting the target DNAs via fluorescence spectroscopy. We note that if the attractive interaction between the dye-labeled capture ssODNs and the graphene oxide nanosheets is stronger than the hybridization interaction between the capture ssODNs and the target DNAs, as that occurs in reduced graphene oxide, most of the capture ssODNs would stay on the graphene oxide surface even in the presence of the target DNAs. This is indeed what happened when a lightly reduced graphene oxide nanosheet (C/O = ~4) is used in our experiment.
Figure 4.
(a) Schematic illustration of the hybridization between target ssODNs and the Alexa Fluor 546-tagged capture ssODNs that have been pre-complexed with a graphene oxide sheet, leading to restoration of the quenched fluorescence from the dyes after 1 h of incubation. (b and c) Plots of corrected fluorescence intensity at emission maximum wavelength of Alexa Fluor 546-tagged ssODNs coupled with the graphene oxide nanosheets (5 μg/mL) in 20 mm Tris-HCl buffer solution (b) and FBS (c) as a function of the concentration of the target ssODNs. The graphene oxide samples have C/O ratios of 1.1 (△), 1.3 (▲), 1.6 (□), and 1.9 (■). For each plot, measured fluorescence intensity data were corrected by subtracting against the baseline solution fluorescence prior to the addition of target DNA. In Tris-HCl buffer, the recovered fluorescence signal for the least-oxidized sample still did not reach saturation after 1 h, suggesting that there are still more ssODNs being bound to the surface. A color reproduction of this figure is available in the SI as Figure S10.
As we suspected at the onset of this work, the DNA detection sensitivity by the graphene oxide complex could be changed significantly by the extent of oxidation of graphene oxide (Figures 4b and 4c), mainly due to the differences in binding interactions between each graphene oxide sample and dye-tagged ssODNs. The graphene oxide nanosheets with the highest C/O ratio (1.9) show superior sensitivity in serum (Figure 4c), where a plethora of extraneous biomolecules such as proteins, antigens, antibodies, and hormones can also interact strongly with graphene oxide and interfere in DNA detection by releasing dye-tagged ssODNs from the graphene oxide surface prematurely. Notably, at low target concentration (10-50 nm), the fluorescence intensity for the experiment in serum is 1000% that in Tris-HCl buffer even though only less than 30% of the original DNA loading is left after 1 h exposure to serum (see Table S5 in the SI). At 10 nm target concentration, the detection sensitivity in serum is enhanced by over 30 fold that in Tris-HCl buffer when calculated on a per-capture-DNA-strand basis (see Figure S6 in the SI).
We attribute the aforementioned phenomenon to a delicate balance between the loading capacity of the graphene oxide sheets and their capture-and-release kinetics (of the target ssODNs): while the total amount of graphene oxide-bound dye-tagged ssODNs greatly decreased in serum, reducing the total overall fluorescence capacity of the platform, the strength of the attractive interaction of ssODNs to the graphene oxide surface also decreased due to competing interactions by the aforementioned extraneous biomolecules. The result is an overall more-facile capture-and-release process that can generate higher fluorescent signals upon addition of the target DNA, if there is still a significant amount of dye-tagged ssODNs bound to the graphene oxide surface. Indeed, control experiments show that dye-tagged ssODNs could be released from the sheets of all ssODN-loaded graphene oxide samples upon addition to serum, even in the absence of their complementary target DNAs (Figure S7 in the SI). In spite of this loading loss, the graphene oxide with the highest C/O ratio (1.9) can still hold more of the dye-tagged ssODNs for DNA detection in serum due to its stronger binding interaction with ssODNs than the more-oxidized graphene oxides (C/O ratio = 1.1, 1.3, and 1.6) (Figure 4, see also Table S5 in the SI). In combination with the more-facile capture-and-release process described above, the net result is an enhanced fluorescent signal for the least-oxidized graphene oxide in serum comparing to that in Tris-HCl buffer (cf. Figures 4c and 4b).
Consistent with the aforementioned explanation, the graphene oxide with the lowest C/O ratio (1.1) tends to exhibit poor sensitivity in both Tris-HCl buffer and serum due to a combination of weak binding interaction with ssODNs and low loading of ssODNs, despite outstanding dispersibility in electrolyte solution and even in serum.[33] With intermediate ssODN loadings and binding interaction, the “moderately oxidized” graphene oxide nanosheets (C/O ratio = 1.3 and 1.6) have the best combination of features for DNA sensing and can exhibit better sensitivity over a broad range of target DNA concentrations in Tris-HCl buffer solution than the nanosheets with the highest and the lowest C/O ratios (1.9 and 1.1, Figure 4b). We note that all ssODN-graphene oxide complexes, where negatively charged ssODNs adsorb on the surface of the graphene oxide nanosheets and aid in their aqueous solubilities, were well-dispersed in Tris-HCl buffer solution without any visible aggregates, as confirmed by dynamic light scattering (DLS) measurement (see Tables S3 and S4 in the SI). In the absence of ssODNs, however, the graphene oxide nanosheets with the highest C/O ratio (1.9) aggregate quickly upon being added into the Tris-HCl buffer solution.
In summary, we have demonstrated the tunable capabilities of graphene oxide for fluorescence quenching against organic dyes and binding interaction with dye-tagged ssODNs. Graphene oxide samples with different C/O ratios (1.1, 1.3, 1.6, and 1.9) can be readily prepared and complexed with ssODNs. Graphene oxide samples with high C/O ratios can bind more strongly to ssODNs and quench the fluorescence of organic chromophores more effectively than those with low C/O ratios. The least-oxidized graphene oxide (C/O = 1.9) examined in this study can be loaded with nearly four times more dye-tagged ssODNs than the most-oxidized one (C/O = 1.1) and also has 30% more enhanced fluorescence quenching efficiency in water. At low target concentrations (10-50 nm), the detection sensitivity in serum for the least-oxidized graphene oxide, as measured by fluorescence intensity on a per-capture-DNA-strand basis, is over 30 fold that in Tris-HCl buffer, making this platform highly suitable for use in applications with biological fluids.
Notably, the finely tunable chemical compositions of graphene oxide allow it to be used as a flexible DNA sensing platform, as distinctly different DNA-detection performance profiles can be achieved in Tris-HCl buffer solution and serum by simply varying the C/O ratio of the graphene oxide nanosheets. As a versatile platform for developing a nanomaterial-based DNA sensor, graphene oxide nanosheets can offer a broad range of materials having different fluorescence quenching efficiencies and binding interactions with ssODNs, in contrast to other nanomaterials without such tunable features. Together with its other desirable properties, including excellent water solubility,[33,52] good biocompatibility,[53,54] facile surface modification,[25,26] and strong fluorescence quenching ability,[31,32] this newly found accessible versatility of graphene oxide as a DNA detection platform without target labeling should spur new efforts toward the application of graphene oxide in the development of a reliable and easy-to-use biomolecular detection assay tool.
Experimental Section
Characterization
Elemental analysis (EA) was carried out by Atlantic Microlabs (Norcross, GA). Karl Fischer titration of the water content in graphene oxide samples was carried out using a C20 Compact Karl Fischer Coulometer (Mettler Toledo, Columbus, OH). The resulting water content of the graphene oxide is then used to eliminate the contribution of water to the EA-determined C/O ratio (see Tables S1 and S2 and the accompanying discussion in the SI). UV-vis absorption and fluorescence emission spectra were obtained on a CARY 300 Bio UV-vis spectrophotometer (Varian Medical Systems, Inc., Palo Alto, CA) and a Jobin Yvon Fluorolog fluorometer (λex = 647, 546, and 546 and λem = 664, 563, and 569 nm for Cy5, Cy3, and Alexa Fluor 546, respectively; slit width = 3 nm), respectively. Details for tapping-mode AFM experiments, solid-state 13C MAS NMR spectra collection, and calculation of the C/O ratios of the graphene oxide samples are available in the SI.
Preparation of Graphene Oxide Nanosheets
Graphite was oxidized to graphite oxide using a modification of the Hummer's method.[48] A suspension of graphite oxide in water was exfoliated into an aqueous dispersion with ultrasonication (Vibra-Cell™ VC 505 (500 watts), Sonics & Materials, Inc.). Details for preparing graphene oxide nanosheets are available in the SI.
Preparation of dye-tagged ssODN-graphene oxide complex
An aliquot of dye-tagged ssODN solution (10 μL of a 200 μm solution) was added to an aliquot of graphene oxide solution (490 μL of a 0.102 mg mL-1 solution) prior to brief vortexing (S/P® Vortex Mixer (Charlotte, NC)). After 2 h incubation in the dark at room temperature, the solution was mixed with aqueous NaCl (500 μL of a 2 N solution) and further incubated for 30 min in the dark at room temperature to precipitate dye-tagged ssODN-graphene oxide complexes. The solution was centrifuged at 14,000 g for 30 min and the supernatant was discarded to remove unbound dye-tagged ssODNs. The collected ssODN-graphene oxide complexes were washed twice by the following cycle: dissolving in aqueous NaCl (500 μL of a 2 N solution), centrifuging at 14,000 g for 30 min, and discarding the supernatant. The isolated ssODN-graphene oxide complexes were then dissolved in ultrapure deionized water (400 μL) and purified again with an Amicon® Ultra centrifugal filter (100K MWCO, Millipore) via centrifugation at 14,000 g for 30 min to remove NaCl and residual unbound ssODNs. The materials remain on the filter was then washed twice with ultrapure deionized water (2 × 400 μL) via centrifugation at 14,000 g for 30 min. The purified ssODN-graphene oxide complexes were isolated from the filter and redispersed in ultrapure deionized water (1 mL) for further analysis with UV-vis and fluorescence spectroscopy. Details for UV-vis absorption and fluorescence measurements and their analysis are available in the SI.
DNA detection assay
The as-prepared complex of Alexa Fluor 546-tagged ssODNs and graphene oxide sheets (5 μg) was dissolved in 20 mm Tris-HCl buffer solution (1 mL, pH 7.4, containing 100 mm NaCl) and FBS, respectively. After 1 h incubation to achieve a constant background fluorescence signal (see Figure S7 in the SI), appropriate amounts of complementary target ssODNs were added to the solutions, followed by 1 h incubation in the dark at room temperature prior to fluorescence measurement.
Supplementary Material
Acknowledgements
This work is financially supported by the NIH (NCI Center of Cancer Nanotechnology Excellence Grant C54CA151880 and Core Grant P30CA060553 to the Robert H. Lurie Comprehensive Cancer Center of Northwestern University). We thank the Initiative for Sustainability and Energy at Northwestern (ISEN) for funding the purchase of the ultrasonicator and the Karl Fischer Coulometer. OCC was an ACC-NSF fellow (CHE-0936924). ZA is supported by the ARO (Award # W991NF-09-1-0541).
Footnotes
Supporting Information is available on the WWW under http://www.small-journal.com or from the author.
References
- 1.Heller MJ. Annu. Rev. Biomed. Eng. 2002;4:129. doi: 10.1146/annurev.bioeng.4.020702.153438. [DOI] [PubMed] [Google Scholar]
- 2.McGlennen RC. Clin. Chem. 2001;47:393. [PubMed] [Google Scholar]
- 3.Tecon R, van der Meer JR. Sensors. 2008;8:4062. doi: 10.3390/s8074062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spier CR, Vadas GG, Kaattari SL, Unger MA. Environ. Toxicol. Chem. 2011;30:1557. doi: 10.1002/etc.546. [DOI] [PubMed] [Google Scholar]
- 5.Cooper MA. Nat. Rev. Drug. Discovery. 2002;1:515. doi: 10.1038/nrd838. [DOI] [PubMed] [Google Scholar]
- 6.Walt DR, Franz DR. Anal. Chem. 2000;72:738A. doi: 10.1021/ac003002a. [DOI] [PubMed] [Google Scholar]
- 7.Schena M, Shalon D, Davis RW, Brown PO. Science. 1995;270:467. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 8.Hong BJ, Oh SJ, Youn TO, Kwon SH, Park JW. Langmuir. 2005;21:4257. doi: 10.1021/la046951y. [DOI] [PubMed] [Google Scholar]
- 9.Espina V, Woodhouse EC, Wulfkuhle J, Asmussen HD, Petricoin EF, III, Liotta LA. J. Immunol. Methods. 2004;290:121. doi: 10.1016/j.jim.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 10.Lequin RM. Clin. Chem. 2005;51:2415. doi: 10.1373/clinchem.2005.051532. [DOI] [PubMed] [Google Scholar]
- 11.Sendroiu IE, Gifford LK, Lupták A, Corn RM. J. Am. Chem. Soc. 2011;133:4271. doi: 10.1021/ja2005576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fritz J, Baller MK, Lang HP, Rothuizen H, Vettiger P, Meyer E, Güntherodt H-J, Gerber C, Gimzewski JK. Science. 2000;288:316. doi: 10.1126/science.288.5464.316. [DOI] [PubMed] [Google Scholar]
- 13.Shen Z, Huang M, Xiao C, Zhang Y, Zeng X, Wang PG. Anal. Chem. 2007;79:2312. doi: 10.1021/ac061986j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tyagi S, Kramer FR. Nat. Biotechnol. 1996;14:303. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 15.Hong BJ, Sunkara V, Park JW. Nucleic Acids Res. 2005;33:e106. doi: 10.1093/nar/gni109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Satishkumar BC, Brown LO, Gao Y, Wang C-C, Wang H-L, Doorn SK. Nat. Nanotechnol. 2007;2:560. doi: 10.1038/nnano.2007.261. [DOI] [PubMed] [Google Scholar]
- 17.Yang R, Jin J, Chen Y, Shao N, Kang H, Xiao Z, Tang Z, Wu Y, Zhu Z, Tan W. J. Am. Chem. Soc. 2008;130:8351. doi: 10.1021/ja800604z. [DOI] [PubMed] [Google Scholar]
- 18.Rosi NL, Mirkin CA. Chem. Rev. 2005;105:1547. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 19.Li H, Rothberg L. Proc. Natl. Acad. Sci. U. S. A. 2004;101:14036. doi: 10.1073/pnas.0406115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu C-H, Yang H-H, Zhu C-L, Chen X, Chen G-N. Angew. Chem., Int. Ed. 2009;48:4785. doi: 10.1002/anie.200901479. [DOI] [PubMed] [Google Scholar]
- 21.Lu C-H, Li J, Liu J-J, Yang H-H, Chen X, Chen G-N. Chem.–Eur. J. 2010;16:4889. doi: 10.1002/chem.200903071. [DOI] [PubMed] [Google Scholar]
- 22.He S, Song B, Li D, Zhu C, Qi W, Wen Y, Wang L, Song S, Fang H, Fan C. Adv. Funct. Mater. 2010;20:453. [Google Scholar]
- 23.Lerf A, He H, Forster M, Klinowski J. J. Phys. Chem. B. 1998;102:4477. [Google Scholar]
- 24.Cai W, Piner RD, Stadermann FJ, Park S, Shaibat MA, Ishii Y, Yang D, Velamakanni A, An SJ, Stoller M, An J, Chen D, Ruoff RS. Science. 2008;321:1815. doi: 10.1126/science.1162369. [DOI] [PubMed] [Google Scholar]
- 25.Stankovich S, Piner RD, Nguyen ST, Ruoff RS. Carbon. 2006;44:3342. [Google Scholar]
- 26.Niyogi S, Bekyarova E, Itkis ME, McWilliams JL, Hamon MA, Haddon RC. J. Am. Chem. Soc. 2006;128:7720. doi: 10.1021/ja060680r. [DOI] [PubMed] [Google Scholar]
- 27.Erickson K, Erni R, Lee Z, Alem N, Gannett W, Zettl A. Adv. Mater. 2010;22:4467. doi: 10.1002/adma.201000732. [DOI] [PubMed] [Google Scholar]
- 28.Cote LJ, Kim F, Huang J. J. Am. Chem. Soc. 2009;131:1043. doi: 10.1021/ja806262m. [DOI] [PubMed] [Google Scholar]
- 29.Becerril HA, Mao J, Liu Z, Stoltenberg RM, Bao Z, Chen Y. ACS Nano. 2008;2:463. doi: 10.1021/nn700375n. [DOI] [PubMed] [Google Scholar]
- 30.Compton OC, Nguyen ST. Small. 2010;6:711. doi: 10.1002/smll.200901934. [DOI] [PubMed] [Google Scholar]
- 31.Kim J, Cote LJ, Kim F, Huang J. J. Am. Chem. Soc. 2010;132:260. doi: 10.1021/ja906730d. [DOI] [PubMed] [Google Scholar]
- 32.Treossi E, Melucci M, Liscio A, Gazzano M, Samori P, Palermo V. J. Am. Chem. Soc. 2009;131:15576. doi: 10.1021/ja9055382. [DOI] [PubMed] [Google Scholar]
- 33.Hong BJ, Compton OC, An Z, Eryazici I, Nguyen ST. ACS Nano. 2012;6:63. doi: 10.1021/nn202355p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swathi RS, Sebastian KL. J. Chem. Phys. 2008;129:054703. doi: 10.1063/1.2956498. [DOI] [PubMed] [Google Scholar]
- 35.Swathi RS, Sebastian KL. J. Chem. Phys. 2009;130:086101. doi: 10.1063/1.3077292. [DOI] [PubMed] [Google Scholar]
- 36.Qu L, Martin RB, Huang W, Fu K, Zweifel D, Lin Y, Sun Y-P, Bunker CE, Harruff BA, Gord JR, Allard LF. J. Chem. Phys. 2002;117:8089. [Google Scholar]
- 37.Yang R, Tang Z, Yan J, Kang H, Kim Y, Zhu Z, Tan W. Anal. Chem. 2008;80:7408. doi: 10.1021/ac801118p. [DOI] [PubMed] [Google Scholar]
- 38.Chang H, Tang L, Wang Y, Jiang J, Li J. Anal. Chem. 2010;82:2341. doi: 10.1021/ac9025384. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, Li Z, Hu D, Lin C-T, Li J, Lin Y. J. Am. Chem. Soc. 2010;132:9274. doi: 10.1021/ja103169v. [DOI] [PubMed] [Google Scholar]
- 40.Jang H, Kim Y-K, Kwon H-M, Yeo W-S, Kim D-E, Min D-H. Angew. Chem., Int. Ed. 2010;49:5703. doi: 10.1002/anie.201001332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cui L, Lin X, Lin N, Song Y, Zhu Z, Chen X, Yang CJ. Chem. Commun. 2012;48:194. doi: 10.1039/c1cc15412e. [DOI] [PubMed] [Google Scholar]
- 42.Eda G, Chhowalla M. Adv. Mater. 2010;22:2392. doi: 10.1002/adma.200903689. [DOI] [PubMed] [Google Scholar]
- 43.Ambrosi A, Bonanni A, Sofer Z, Cross JS, Pumera M. Chem.–Eur. J. 2011;17:10763. doi: 10.1002/chem.201101117. [DOI] [PubMed] [Google Scholar]
- 44.Loo AH, Bonanni A, Pumera M. Nanoscale. 2012;4:143. doi: 10.1039/c1nr10966a. [DOI] [PubMed] [Google Scholar]
- 45.Buglione L, Chng ELK, Ambrosi A, Sofer Z, Pumera M. Electrochem. Commun. 2012;14:5. [Google Scholar]
- 46.Xu Y, Zhao L, Bai H, Hong W, Li C, Shi G. J. Am. Chem. Soc. 2009;131:13490. doi: 10.1021/ja905032g. [DOI] [PubMed] [Google Scholar]
- 47.Huang WT, Shi Y, Xie WY, Luo HQ, Li NB. Chem. Commun. 2011;47:7800. doi: 10.1039/c1cc11631b. [DOI] [PubMed] [Google Scholar]
- 48.Hummers WS, Offeman RE. J. Am. Chem. Soc. 1958;80:1339. [Google Scholar]
- 49.Hontoria-Lucas C, López-Peinado AJ, López-González JDD, Rojas-Cervantes ML, Martín-Aranda RM. Carbon. 1995;33:1585. [Google Scholar]
- 50.Szabó T, Berkesi O, Forgó P, Josepovits K, Sanakis Y, Petridis D, Dékány I. Chem. Mater. 2006;18:2740. [Google Scholar]
- 51.Gao H, Kong Y. Annu. Rev. Mater. Res. 2004;34:123. [Google Scholar]
- 52.Li D, Müller MB, Gilje S, Kaner RB, Wallace GG. Nat. Nanotechnol. 2008;3:101. doi: 10.1038/nnano.2007.451. [DOI] [PubMed] [Google Scholar]
- 53.Liu Z, Robinson JT, Sun X, Dai H. J. Am. Chem. Soc. 2008;130:10876. doi: 10.1021/ja803688x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang K, Ruan J, Song H, Zhang J, Wo Y, Guo S, Cui D. Nanoscale Res. Lett. 2011;6 doi: 10.1007/s11671-010-9751-6. DOI:10.1007/s11671. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.