Abstract
Tannerella forsythia is an anaerobic, Gram-negative bacterium involved in the so-called “red complex,” which is associated with severe and chronic periodontitis. The surface layer (S-layer) of T. forsythia is composed of cell surface glycoproteins, such as TfsA and TfsB, and is known to play a role in adhesion/invasion and suppression of proinflammatory cytokine expression. Here we investigated the association of this S-layer with serum resistance and coaggregation with other oral bacteria. The growth of the S-layer-deficient mutant in a bacterial medium containing more than 20% non-heat-inactivated calf serum (CS) or more than 40% non-heat-inactivated human serum was significantly suppressed relative to that of the wild type (WT). Next, we used confocal microscopy to perform quantitative analysis on the effect of serum. The survival ratio of the mutant exposed to 100% non-heat-inactivated CS (76% survival) was significantly lower than that of the WT (97% survival). Furthermore, significant C3b deposition was observed in the mutant but not in the WT. In a coaggregation assay, the mutant showed reduced coaggregation with Streptococcus sanguinis, Streptococcus salivarius, and Porphyromonas gingivalis but strong coaggregation with Fusobacterium nucleatum. These results indicated that the S-layer of T. forsythia plays multiple roles in virulence and may be associated with periodontitis.
INTRODUCTION
Periodontitis is an infectious disease caused by periodontopathogenic bacteria that colonize the tooth surface and gingival sulcus (1, 2). This disease is initiated by the formation of a biofilm, known as dental plaque, which consists of many oral bacteria. Currently, many periodontal pathogens, such as Porphyromonas gingivalis, Treponema denticola, Tannerella forsythia, Aggregatibacter actinomycetemcomitans, Fusobacterium nucleatum, and Prevotella intermedia, have been identified (3, 4). These pathogens produce several virulence factors and immune evasion factors, causing inflammation and destruction of periodontal tissues (5–8). T. forsythia is an anaerobic, Gram-negative bacterium isolated from the gingival sulci and periodontal pockets of patients with periodontitis (9, 10). Three pathogens (T. forsythia, P. gingivalis, and T. denticola), known collectively as the “red complex,” are strongly associated with the pathogenesis and progression of destructive forms of periodontitis (3, 9). Several T. forsythia virulence factors have been identified (10–12), including BspA, the S-layer, glycosidases, lipoproteins, and metalloproteinase.
The S-layer is the outermost cell envelope component of many bacteria, including archaea, Gram-positive bacteria, and Gram-negative bacteria (13, 14). Among Gram-negative bacteria, S-layers of Campylobacter rectus (15), Campylobacter fetus (16), and T. forsythia have been reported (17–19). The S-layer functions as a protective barrier and molecular sieve, and it plays a role in cell adhesion and surface recognition (11, 13, 14, 20–22). The S-layer of T. forsythia consists of two proteins, TfsA and TfsB, which are encoded by tfsA (TF2661 and TF2662) and tfsB (TF2663), respectively; these genes are located tandemly in an operon (17). Wild-type (WT) T. forsythia has a regularly arrayed structure at the outermost cell surface, while a mutant lacking both tfsA and tfsB completely lost its structure, and a mutant lacking either tfsA or tfsB retained smaller amounts of its structure (23). S-layer proteins are glycoproteins with apparent molecular sizes of 230 kDa (TfsA) and 270 kDa (TfsB). Recently, the characterization and structure of oligosaccharides in S-layer proteins have been reported (24). Oligosaccharides were O-glycosidically linked to three amino acid motifs, D(S/T)(A/I/L/M/T/V), on either of the two S-layer proteins (TfsA or TfsB) (24). These two proteins are specifically recognized in the sera of patients with periodontitis (25). Also, TfsA and TfsB may mediate adhesion to, and/or invasion of, human gingival epithelial cells and epidermal carcinoma cells of the mouth and are associated with hemagglutination (11, 23). Sekot et al. have demonstrated that T. forsythia lacking the S-layer induces significantly higher levels of proinflammatory cytokines than WT T. forsythia (26), which suggests that the S-layer attenuates the host immune response by evading innate immune recognition.
In this study, we investigated the associations of the S-layer of T. forsythia with serum resistance and coaggregation with Streptococcus sanguinis, Streptococcus salivarius, Streptococcus mutans, Streptococcus mitis, Aggregatibacter actinomycetemcomitans, F. nucleatum, and P. gingivalis. Serum contains a complement, including an innate immune factor present in gingival crevicular fluid (GCF) at concentrations as high as 70%, that can kill bacteria upon activation (27). Human complement can function through three activation pathways, including the classical pathway, an alternative pathway, and the lectin pathway (28, 29). Bacterial components such as lipopolysaccharides (LPS), peptidoglycan, and lipoproteins can induce complement activation. However, many bacteria, including periodontal pathogens, are resistant to human serum (16, 30–33). Coaggregation between different bacteria plays a key role in the colonization of the gingival crevice and the organization of biofilm formation by periodontal pathogens. Among the oral bacteria, F. nucleatum plays an important role in bacterial coaggregation. Previously, T. forsythia was reported to coaggregate with F. nucleatum and T. denticola (34, 35). Overall, we concluded that the S-layer is associated with serum resistance and coaggregation with other oral bacteria.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. A T. forsythia mutant lacking the S-layer (ΔtfsAB) was constructed as described previously (23). T. forsythia was grown in brain heart infusion (BHI) medium containing heat-inactivated calf serum (CS) (5%, vol/vol), yeast extract (5 g/liter), l-cysteine (1 g/liter), N-acetylmuramic acid (10 mg/liter), hemin (5 mg/liter), and menadione (0.5 mg/liter) (TF medium) for 3 to 7 days at 37°C under anaerobic conditions using the GasPak system (Mitsubishi Gas Chemical Company Inc., Tokyo, Japan). Chloramphenicol (10 μg/ml) was added when necessary. A. actinomycetemcomitans was grown in Todd-Hewitt medium containing yeast extract (10 g/liter) (THY) overnight at 37°C under 5% CO2. F. nucleatum was grown in BHI medium overnight at 37°C under anaerobic conditions. P. gingivalis was grown in BHI medium containing hemin (5 mg/liter) and menadione (0.5 mg/liter) for 2 days at 37°C under anaerobic conditions. Streptococcus mutans, S. sanguinis, S. salivarius, and S. mitis were grown in tryptic soy broth overnight at 37°C under 5% CO2. Escherichia coli strains were grown in Luria-Bertani (LB) broth overnight at 37°C.
Table 1.
Strains used in this study
| Strain | Descriptiona | Reference or sourceb |
|---|---|---|
| Tannerella forsythia | ||
| ATCC 43037 | Wild type | ATCC |
| ΔtfsAB mutant | TfsA and TfsB deletion mutant; Camr | 23 |
| Aggregatibacter actinomycetemcomitans | ||
| HK1651 | Wild type | ATCC |
| Y4 | Wild type | ATCC |
| Fusobacterium nucleatum ATCC 25586 | Wild type | ATCC |
| Porphyromonas gingivalis W83 | Wild type | ATCC |
| Streptococcus mutans UA159 | Wild type | ATCC |
| Streptococcus mitis GTC 495 | Wild type | GTC |
| Streptococcus salivarius GTC 215 | Wild type | GTC |
| Streptococcus sanguinis GTC 217 | Wild type | GTC |
| Escherichia coli | ||
| XL-II | endA1 supE44 thi-1 hsdR17 recA1 gyrA96 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr) Amy Camr] | Stratagene |
| RA31 | HB101 carrying empty vector (pGEM-T Easy); Ampr | 36 |
| RA11 | HB101 Omp100-expressing strain; Ampr | 36 |
Camr, chloramphenicol resistance; Tetr, tetracycline resistance; Ampr, ampicillin resistance.
ATCC, American Type Culture Collection; GTC, Gifu type culture.
Sera, factor H, and antibodies.
Human serum was obtained from healthy volunteers, with the approval of the ethics committee of Kagoshima University. CS was purchased from Gibco Life Technologies Japan (Tokyo, Japan). Serum was inactivated by heat (56°C for 30 min) when necessary. Human factor H was purchased from Complement Technology (Tyler, TX). Mouse monoclonal IgG against human C3 and a mouse monoclonal antibody against human factor H were obtained from BioPorto Diagnostics A/S (Gentofte, Denmark). A mouse polyclonal antibody against human C4b-binding protein (C4BP) was acquired from Abnova (Taipei, Taiwan). Horseradish peroxidase (HRP)-conjugated anti-mouse IgG(H+L) (Promega, Madison, WI) and Alexa Fluor 594-conjugated goat anti-mouse IgG(H+L) (Molecular Probes Inc., Eugene, OR) were used for detection.
Serum survival assay.
Two methods were used to evaluate susceptibility to serum. For the growth kinetics method, 105 cells of WT T. forsythia or the S-layer-deficient mutant grown for 5 days were added to 100 μl of modified TF medium (BHI medium containing yeast extract [5 g/liter], l-cysteine [1 g/liter], N-acetylmuramic acid [10 mg/liter], hemin [5 mg/liter], and menadione [0.5 mg/liter]) containing various concentrations of non-heat-inactivated and heat-inactivated serum (human serum at 80%, 60%, 40%, and 20%; CS at 60%, 40%, 20%, and 10%) on a 96-well plate. Then the plate was incubated for 10 days at 37°C under anaerobic conditions. The cell density (at 600 nm) was measured at 24-h intervals by a SpectraMax 340PC384 absorbance microplate reader (Molecular Devices Japan, Tokyo, Japan). For the quantitative method used to evaluate serum killing, 108 WT or S-layer-deficient mutant T. forsythia cells grown for 5 days in TF medium, and E. coli strain XL-II grown in LB medium overnight, were incubated with 10 ml of 100% CS for 2 h at 37°C under anaerobic conditions. Bacterial cells were pelleted by centrifugation and were washed with 5 ml of phosphate-buffered saline (PBS). After washing with PBS, the cells were stained using the BD Cell Viability kit (BD Biosciences, San Jose, CA), which enables visualization of dead and/or live cells. Confocal laser scanning microscopy (CLSM) was performed with a Carl Zeiss LSM700 microscope (Carl Zeiss Micro Imaging Co., Ltd., Tokyo, Japan). The microscope was equipped with detectors to monitor red (excitation wavelength, 555 nm; dichroic mirror wavelength, 585 nm) and green (excitation wavelength, 488 nm; dichroic mirror wavelength, 590 nm) fluorescence. Confocal images were obtained with a 40× oil lens (numerical aperture, 1.3) for an optical section thickness of approximately 0.7 μm. The experiment was performed three times independently. In each experiment, more than 10 microscopic fields were observed, and 3 images were randomly extracted. After three images of dead and live cells were obtained in each experiment, the areas occupied by dead and/or live cells in the CLSM images were analyzed by ImageJ 1.44i (National Institutes of Health, Bethesda, MD). Then the ratio of live bacteria to total bacteria was calculated.
Electron microscopic analysis.
For transmission electron microscopy (TEM), T. forsythia strains grown in TF medium were washed twice with PBS. TEM was used for ultrastructural characterization of the cell, as described previously (37–39). A total of 1010 bacterial cells were reacted with 40 ml of CS or PBS for 2 h at 37°C under anaerobic conditions. After centrifugation, the bacterial cells were washed twice with PBS. Next, the cells were fixed with 2.5% glutaraldehyde for 12 h. The samples were dehydrated in a series of ethanol concentrations and were then embedded in Spurr's resin for 2 days at 60°C (39). Thin sections were cut using an ultramicrotome (Ultracut R; Leica, Tokyo, Japan) with a diamond knife and were examined using a JEM-1400 transmission electron microscope (JEOL Ltd., Tokyo, Japan) at 80 kV.
Preparation of the outer membrane protein and SDS-PAGE.
Sarkosyl-insoluble outer membrane protein fractions were prepared as described previously (40). Briefly, WT and S-layer-deficient mutant T. forsythia cells grown in TF medium for 7 days were pelleted and suspended in PBS. The cells were disrupted by sonication, and the remaining undisrupted bacterial cells were removed by centrifugation. The outer and inner membrane protein fractions were collected as a pellet by centrifugation at 100,000 × g for 60 min at 4°C, and the pellet was redissolved in sodium lauryl sarcosinate (1%, wt/vol). The outer membrane protein fractions were collected as a pellet by centrifugation at 100,000 × g for 60 min at 4°C, and the pellet was redissolved in sodium dodecyl sulfate (SDS) (1%, wt/vol). The outer membrane protein fractions of the WT and mutant T. forsythia strains were separated by 12% SDS-polyacrylamide gel electrophoresis (PAGE), and the gels were stained with Coomassie brilliant blue R-250.
Detection of deposition of complement factor C3b on the bacterial surface.
WT T. forsythia and the S-layer-deficient mutant were grown for 5 days in TF medium. Then 2 × 108 cells were incubated in 0.5 ml of 30% human serum in PBS, or in PBS alone, for 30 min at 37°C under anaerobic conditions. The bacteria were pelleted by centrifugation; then the pellets were washed three times with 0.5 ml of PBS and were fixed with 0.5 ml of 2.5% paraformaldehyde for 10 min at room temperature. The bacteria were incubated with 0.5 ml of blocking solution (PBS containing 0.05% Tween 20 and 1% bovine serum albumin) for 30 min at 37°C. After blocking, the bacteria were treated with 1 ml of PBS-T (PBS containing 0.05% Tween 20) containing a mouse monoclonal antibody against human C3 (dilution, 1:10,000) and were incubated for 30 min at room temperature. The bacteria were washed three times with 0.5 ml of PBS, treated with 1 ml of PBS-T containing Alexa Fluor 594-conjugated goat anti-mouse IgG (1:250), and incubated for 30 min at room temperature. The bacteria were then washed three times with 0.5 ml of PBS, resuspended with 1 ml of PBS containing 4′,6-diamidino-2-phenylindole (DAPI) (5 μg/ml) (Molecular Probes Inc.), and incubated for 10 min at room temperature. After washing with PBS, the bacterial cells were suspended in 10 μl of PBS. CLSM was performed with a Carl Zeiss LSM700 microscope. The areas in CLSM images occupied by cells on which C3b was deposited or by whole cells were analyzed by ImageJ 1.44i (NIH).
Factor H and C4BP binding assays.
Previously, we demonstrated that Omp100, one of the outer membrane proteins in A. actinomycetemcomitans, had binding affinity for factor H (36). We found that E. coli HB101 carrying the empty vector pGEM-T Easy (RA31) was highly susceptible to serum, while E. coli HB101 expressing Omp100 (RA11) and A. actinomycetemcomitans Y4 were resistant to serum. Therefore, we used these strains as controls in the factor H binding assay. WT T. forsythia and the S-layer-deficient mutant were grown for 5 days in TF medium. E. coli strains were grown in LB broth overnight, and A. actinomycetemcomitans was grown in THY overnight. Then 2 × 109 bacterial cells were incubated with 0.5 ml of 50% human serum in GVB++ (Veronal-buffered saline containing 0.1% gelatin, 0.15 mM CaCl2, and 0.5 mM MgCl2) or with GVB++ alone at 37°C for 30 min. The bacteria were pelleted by centrifugation, and the pellets were washed three times with 0.5 ml GVB++. The pellets were then treated with 75 μl of 0.1 M glycine-HCl (pH 2.0). After centrifugation, the supernatant was mixed with 5 μl of 1.5 M Tris-HCl (pH 8.8), and the eluted fraction was collected. Next, the eluted samples and human factor H (8 ng) were separated by 7.5% SDS-PAGE and were transferred to a nitrocellulose membrane. Immunoblotting was performed as described previously (41). The membrane was blocked with 2% skim milk in Tris-buffered saline (TBS) (20 mM Tris and 137 mM NaCl [pH 8.0]) containing 0.05% Tween 20 (TBS-T) at 4°C overnight and was then incubated with a mouse monoclonal antibody against human factor H (diluted 1:10,000 in 1% skim milk in TBS-T) at room temperature for 1 h. After washing with TBS-T, the membrane was incubated with HRP-conjugated goat anti-mouse IgG (1:2,500) at room temperature for 1 h. The membrane was washed with TBS-T, and factor H was detected using a chemiluminescence detection system (Perkin-Elmer Japan Co., Ltd., Kanagawa, Japan). The intensities of factor H binding were measured with Image Lab (Bio-Rad Laboratories, Hercules, CA) and were quantified relative to the intensity of binding to WT T. forsythia. Data represent means ± standard deviations (SD) of measurements performed in triplicate. We also investigated the binding of factor H or C4BP to T. forsythia cells by CLSM. We used the same method as that used to detect the deposition of complement factor C3b, except that an anti-factor H or anti-C4BP antibody was used.
Zymographic analysis of proteolytic activity.
Since karilysin is associated with serum resistance in T. forsythia (42), we compared the proteolytic activities of the WT and the S-layer-deficient mutant. WT T. forsythia and the S-layer-deficient mutant were grown for 7 days in 10 ml of serum-free TF medium. The bacterial supernatants were collected by centrifugation and filtration. The supernatants were then concentrated 10-fold using a Centriplus YM-30 centrifugal filter unit (Millipore, Billerica, MA). The concentrated supernatants (10 μl) were mixed with nonreducing sample buffer and were separated on a 7.5% polyacrylamide gel containing gelatin (1 mg/ml). After electrophoresis, the gel was washed twice with 2.5% Triton X-100 for 30 min at room temperature. After washing, the gel was incubated with developing buffer (50 mM Tris-HCl, 10 mM CaCl2 [pH 7.5]) at 37°C for 12 h. The gel was then stained with Coomassie brilliant blue R-250 and was destained for visualization of the proteolytic bands.
Coaggregation assay.
WT T. forsythia and the S-layer-deficient mutant were grown for 3 or 7 days in TF medium. After the bacterial cells were harvested by centrifugation, WT T. forsythia and the S-layer-deficient mutant were suspended in coaggregation buffer (CAB) (1 mM CaCl2, 1 mM MgCl2, 150 mM NaCl, and Tris-HCl [pH 8.0]), and the suspensions were adjusted to an optical density at 550 nm (OD550) of 1.0. Overnight cultures of A. actinomycetemcomitans HK1651, F. nucleatum ATCC 25586, S. mutans UA159, S. mitis GTC 495, S. salivarius GTC 215, and S. sanguinis GTC 217 and a 2-day culture of P. gingivalis W83 (stationary phase), used as coaggregation partner strains, were suspended with CAB and adjusted to an OD550 of 1.0. Suspensions of S. sanguinis and S. mutans during exponential phase (OD550, 0.4) were also prepared. As a preliminary, we investigated the optimal condition for the coaggregation assay. Three ratios (T. forsythia/partner strain ratios of 1:1, 1:2, and 2:1) using S. sanguinis and S. salivarius were investigated, and we found that a T. forsythia/partner strain ratio of 2:1 was most significant. Therefore, we used the T. forsythia/partner strain ratio of 2:1 for the coaggregation assay. WT T. forsythia or the S-layer-deficient mutant was mixed with the partner strain (2:1) in a disposable cuvette (12.5 by 12.5 by 45 mm), and the initial OD of the mixture was measured. Suspensions were incubated at 37°C and were measured at 15-min intervals using an Ultrospec 2000 UV/visible spectrophotometer (Pharmacia Biotech Ltd., Cambridge, United Kingdom). Decreases in absorbance were indicative of cell aggregates precipitating to the bottom of the cuvette. As controls, single bacterial cells were measured (the WT and S-layer-deficient mutant T. forsythia strains were adjusted to an OD550 of 0.66, while partner strain cells were adjusted to an OD550 of 0.33).
To observe the effects of sugars on coaggregation activity, 10 mM each sugar was added to the reaction mixture. We used N-acetyl-d-mannosamine, d-glucuronic acid sodium salt, l-(−)-fucose, d-(+)-xylose, and d-(+)-galactose, which were reported to be components of sugar moieties in the T. forsythia S-layer (24).
RESULTS
The S-layer contributes to survival in serum.
To evaluate the role of the S-layer in serum resistance, we investigated the growth of WT T. forsythia and the S-layer-deficient mutant in TF medium containing various concentrations of calf serum (Fig. 1). For the WT, growth was gradually delayed as the serum concentration was increased. In the presence of 20% CS, the growth of the mutant was delayed for 1 day relative to that of the WT. More than 40% CS significantly suppressed the growth of the mutant, while the growth rates of the two strains were similar in heat-inactivated CS (Fig. 1). Similar results were obtained when human serum was used (data not shown). The growth of the mutant was suppressed in the presence of >40% human serum, but the mutant could grow well in the presence of heat-inactivated serum. Furthermore, we investigated the killing activity of human serum incubated for 1 to 7 days at 37°C under anaerobic conditions, and we found that the killing activity of the serum against E. coli XL-II was retained after 7 days of incubation (data not shown).
Fig 1.
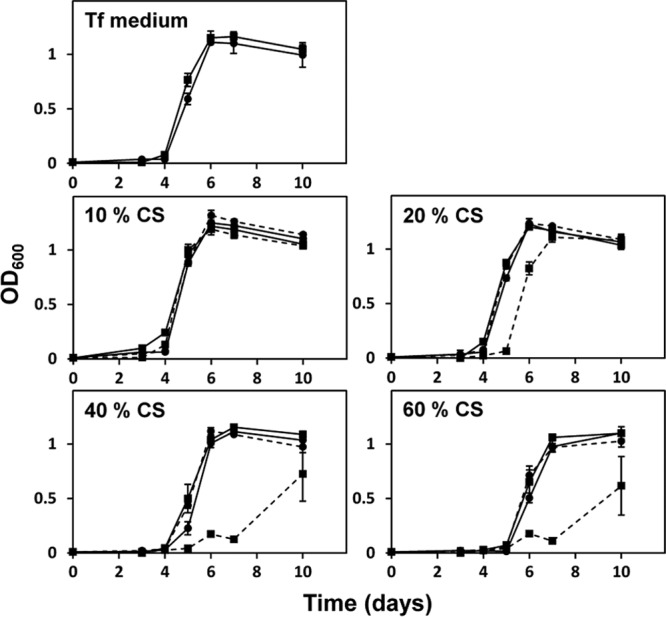
Growth kinetics of WT T. forsythia and the S-layer-deficient mutant in a medium containing serum. A total of 105 WT or mutant cells were inoculated into 100 μl of TF medium containing non-heat-inactivated serum on 96-well plates and were incubated for 10 days at 37°C under anaerobic conditions. The cell density (at 600 nm) was measured at 24-h intervals. Circles and squares indicate the growth of the WT and the mutant, respectively. Solid and dashed lines indicate the results with heat-inactivated CS and non-heat-inactivated CS, respectively. Data represent the means ± SD of measurements performed in triplicate.
Hence, we quantified the effect of serum on bacterial killing. Numerous dead cells were observed when the mutant was incubated with CS, but not when the WT cells were incubated with CS or when the mutant was incubated with heat-inactivated CS (Fig. 2A). The survival ratio of the mutant that was reacted with non-heat-inactivated CS (76% survival) was significantly lower than that of the WT (97% survival), and E. coli showed a lower survival ratio than the mutant (44% survival) (Fig. 2B). Also, the mutant treated with heat-inactivated serum contained only a few dead cells.
Fig 2.
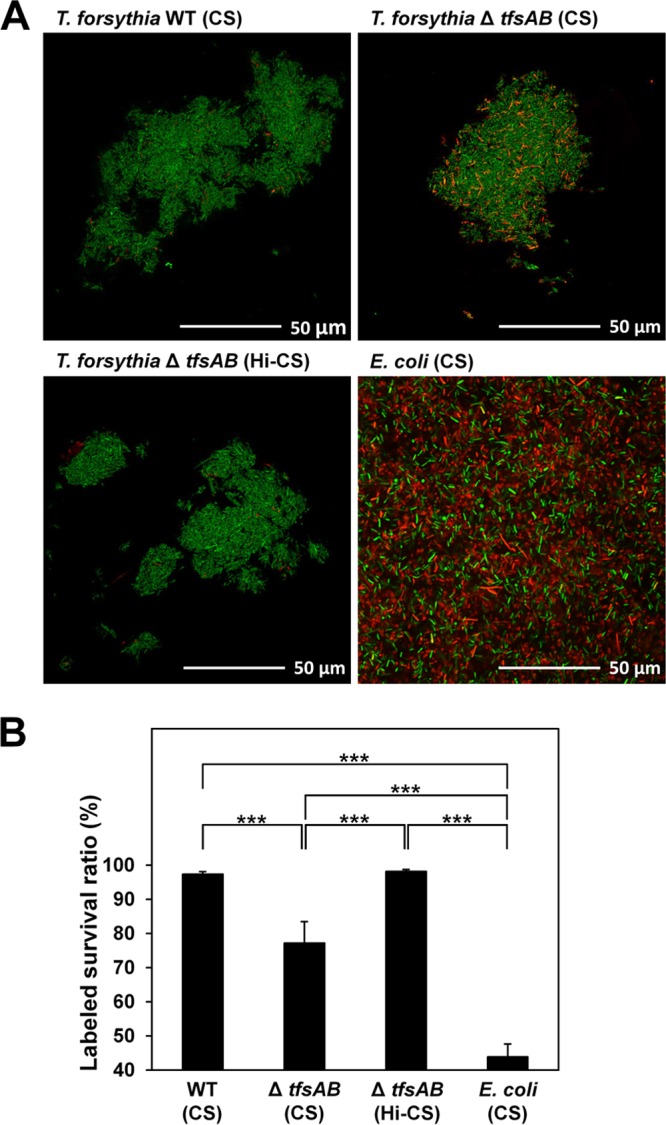
Effect of serum on the viability of T. forsythia. (A) WT T. forsythia, S-layer-deficient mutant, and E. coli XL-II cells were exposed to CS or heat-inactivated CS (Hi-CS). Details are given in Materials and Methods. In CLSM analysis, green and orange cells represent viable and dead cells, respectively. (B) The areas occupied by dead and/or live cells in the CLSM images were analyzed, and the ratio of live bacteria to total bacteria was calculated. Data represent means ± SD of measurements performed in triplicate. Asterisks indicate significant differences (***, P < 0.001) as determined by Tukey's honestly significant difference test. The y axis begins with 40%.
For TEM analysis, we found that the ratio of disrupted cells of the mutant treated with CS was significantly higher than that of the WT in the low-power field of electron microscopic observation (see Fig. S1 in the supplemental material). Disruption of the cytoplasmic membrane or the outer membrane was observed in both strains. Figure 3B shows the typical features of the T. forsythia mutant treated with CS. The outer membrane was disrupted (indicated by arrows), and cytoplasmic swelling was observed as a result. Also, in the WT, the S-layer was observed outside the outer membrane (Fig. 3A), while the mutant completely lost the S-layer at the cell surface, reducing the thickness of the cell surface relative to that of the WT (Fig. 3C). In addition, we confirmed the expression of TfsA and TfsB in the WT and the S-layer-deficient mutant. The WT expressed S-layer proteins, showing apparent molecular sizes of 230 kDa (TfsA) and 270 kDa (TfsB), while the mutant did not express either protein (Fig. 4).
Fig 3.
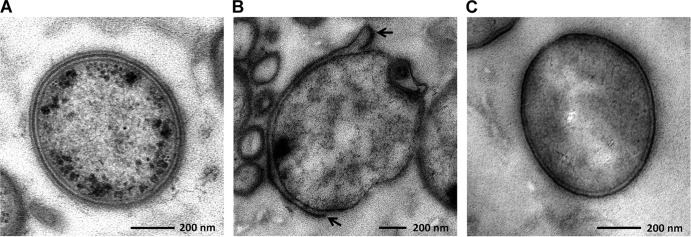
Electron micrographs of T. forsythia exposed to serum. (A and B) WT T. forsythia (A) and the S-layer-deficient mutant (B) were exposed to 100% CS at 37°C for 2 h. Then samples were prepared for observation by TEM. Arrows indicate the disrupted outer membrane. (C) As a control, the mutant treated with PBS was prepared.
Fig 4.
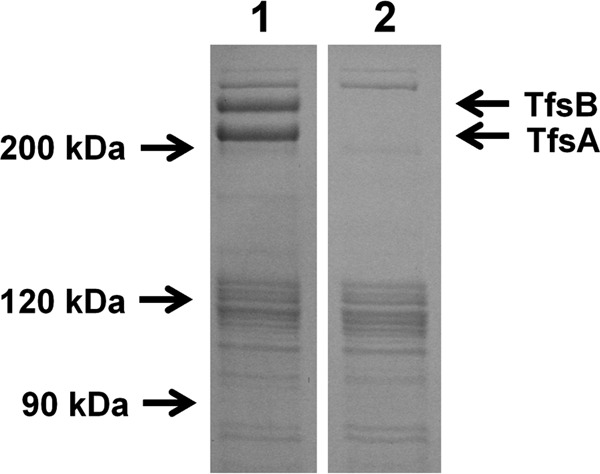
SDS-PAGE analysis of the outer membrane proteins in the WT and the S-layer-deficient mutant. The outer membrane protein fraction of WT T. forsythia and the S-layer-deficient mutant were prepared and separated on a 12.5% SDS-PAGE gel. Details are given in Materials and Methods. Lane 1, WT; lane 2, S-layer-deficient mutant.
Deposition of complement factor C3b is inhibited by the S-layer.
Because the S-layer-deficient mutant showed decreased serum resistance, we postulated that the S-layer affects the activation of complement in serum. Initially, we investigated the deposition of complement factor C3b, which accumulates on the bacterial cell surface by activating the complement pathway, resulting in assembly of the membrane attack complex (MAC) for bacterial membrane perforation. The level of C3b deposition was significantly higher for the mutant than for the WT (Fig. 5A). The area occupied by cells on which C3b was deposited versus the area occupied by whole cells was calculated using image analysis. The areas occupied by cells on which C3b was deposited were 2.9% and 34% of the total area of the image for the WT and the mutant, respectively (Fig. 5B). When the WT and the mutant were not treated with serum, C3b deposition was not observed on cells of either strain.
Fig 5.
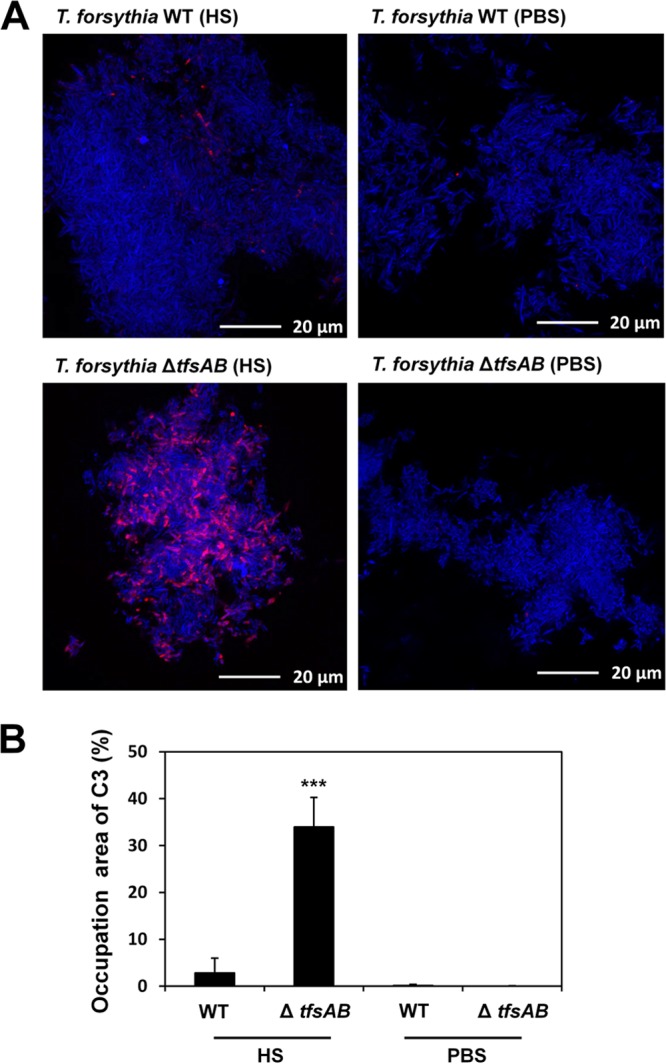
C3b deposition on the surfaces of WT T. forsythia and the S-layer-deficient mutant. (A) WT T. forsythia and the S-layer-deficient mutant were exposed either to 30% human serum (HS) in PBS or to PBS alone at 37°C for 30 min. After washing, the cells were reacted first with an anti-C3 antibody and then with Alexa Fluor 594-conjugated anti-mouse IgG. DAPI staining was performed as well. Blue and red cells represent whole cells and cells with C3b bound to the surface, respectively. (B) The efficiency of C3b deposition on the bacterial cell surface, as shown in the CLSM images, was analyzed. Details are given in Materials and Methods. Data represent means ± SD of measurements performed in triplicate. Significant changes were determined using Bonferroni's t test (***, P < 0.001).
Factor H and C4BP binding.
Inhibition of C3b deposition by recruiting the complement-controlling protein factor H or C4BP contributes to serum resistance (30, 32, 33). Factor H inhibits the alternative pathway in complement activation, while C4BP inhibits the classical and lectin pathways. Therefore, we investigated whether the S-layer has binding affinity with factor H or C4BP. E. coli strain RA11, expressing Omp100, showed factor H binding affinity, while E. coli RA31 showed almost no affinity. The binding affinity of RA11 with factor H was 11-fold higher than that of RA31. Also, A. actinomycetemcomitans Y4 possessed binding affinity. Both WT T. forsythia and the S-layer-deficient mutant had binding affinity for factor H, and the level of binding of the mutant was slightly higher (1.7-fold) than that of the WT (Fig. 6). By CLSM analysis, the level of binding of factor H with the mutant was slightly higher than that with the WT, although the binding affinity was weak in both strains (see Fig. S2 in the supplemental material). Also, C4BP binding affinity was weak in both strains, and no difference was observed between the two strains (see Fig. S3 in the supplemental material).
Fig 6.
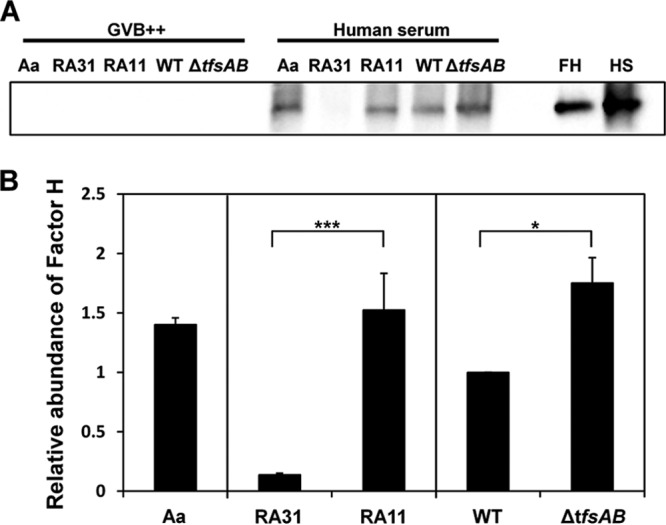
Binding of factor H to WT T. forsythia and the S-layer-deficient mutant. (A) WT T. forsythia, the S-layer-deficient mutant, an E. coli Omp100-expressing strain (RA11), E. coli RA31, and A. actinomycetemcomitans (Aa) were used. In total, 2 × 109 bacterial cells were used for the factor H binding assay, carried out as described in Materials and Methods. The samples, 8 ng of authentic human factor H (FH), and 0.1 μl of human serum (HS) were first separated by 7.5% SDS-PAGE and then transferred to a nitrocellulose membrane. Immunoblotting was performed as described in Materials and Methods. (B) The intensities of factor H binding were measured with Image Lab and were quantified relative to that for WT T. forsythia. Data represent means ± SD of measurements performed in triplicate. Significant differences between RA31 and RA11, and between WT T. forsythia and the S-layer-deficient mutant, were determined using Student's t test (***, P < 0.001; *, P < 0.05).
Proteolytic activities of WT T. forsythia and the S-layer-deficient mutant.
Proteolytic activity was analyzed using zymographic analysis. Two major bands, corresponding to molecular masses of 52 kDa and 48 kDa, were observed both in WT T. forsythia and in the mutant (Fig. 7). A previous report demonstrated that full-length karilysin undergoes autocatalytic processing to generate mature karilysin, showing a molecular mass of 48 kDa (43), suggesting that the proteolytic band of 48 kDa found in zymographic analysis is karilysin. The proteolytic activity of the mutant was similar to that of the WT (Fig. 7).
Fig 7.
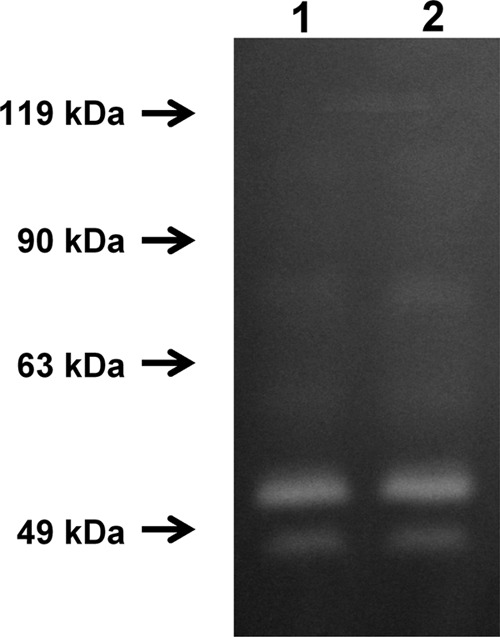
Zymographic analysis of WT T. forsythia and the S-layer-deficient mutant. The protease activities in the supernatants of WT and mutant T. forsythia cells were analyzed using a 7.5% polyacrylamide gel containing gelatin (1 mg/ml). Lane 1, WT; lane 2, S-layer-deficient mutant.
Association of the S-layer with coaggregation of T. forsythia with oral bacteria.
Since T. forsythia is known to coaggregate with oral bacteria (34, 35), we investigated a possible association between the S-layer and coaggregation. Figure 8 shows the coaggregation of T. forsythia strains grown for 7 days with stationary-phase cells of S. sanguinis over time. WT T. forsythia aggregated with S. sanguinis, while the S-layer-deficient mutant did not. The OD of the mixture of WT T. forsythia and S. sanguinis gradually decreased until 60 min, and finally a 35% reduction in the OD was observed. We further investigated the association of the S-layer with coaggregation by using stationary-phase cells of other bacteria. WT T. forsythia grown for 7 days coaggregated with S. salivarius, F. nucleatum, and P. gingivalis but not with S. mutans, S. mitis, or A. actinomycetemcomitans (Fig. 9A). For the mutant, no coaggregation with S. sanguinis or S. salivarius was observed, while coaggregation with F. nucleatum was increased over that of the WT. We investigated the coaggregation of T. forsythia grown for 3 days with stationary-phase cells of oral bacteria and found results similar (except for P. gingivalis) to those for T. forsythia grown for 7 days (Fig. 9B). Furthermore, we compared the coaggregation of T. forsythia (after 3 or 7 days of incubation) with S. sanguinis and S. mutans cells in the exponential phase (OD550, 0.4) and obtained results similar to those with stationary-phase cells (data not shown). Then we investigated the effects of sugars on coaggregation activity and found that N-acetylmannosamine suppressed the coaggregation of WT T. forsythia with S. sanguinis and S. salivarius, while other sugars did not. However, the coaggregation of WT or mutant T. forsythia with F. nucleatum was not inhibited by sugars (Fig. 10).
Fig 8.
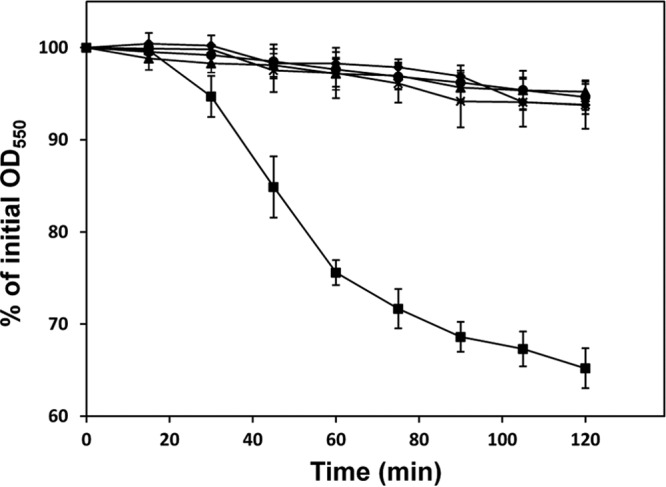
Coaggregation of T. forsythia with S. sanguinis. Coaggregation of WT T. forsythia and the S-layer-deficient mutant with S. sanguinis was investigated as described in Materials and Methods. A 7-day culture of WT T. forsythia or the mutant was mixed with an overnight culture of S. sanguinis (2:1) in a cuvette, and the mixture was incubated at 37°C. The OD550 of the upper phase in the cuvette was measured using a spectrophotometer at 15-min intervals. A decrease in absorbance was considered to indicate coaggregation. Squares indicate the coaggregation of WT T. forsythia with S. sanguinis. Diamonds indicate the coaggregation of the T. forsythia mutant with S. sanguinis. As controls, the densities of single cells of WT T. forsythia (circles), mutant T. forsythia (triangles), and S. sanguinis (crosses) were measured. Data represent means ± SD of measurements performed in triplicate.
Fig 9.
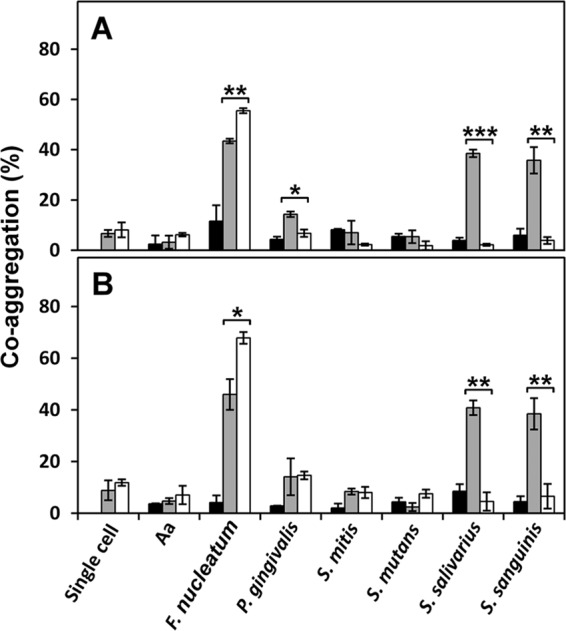
Coaggregation of T. forsythia with oral bacteria. Coaggregation of WT T. forsythia and the S-layer-deficient mutant with seven species of oral bacteria was investigated as described in Materials and Methods. WT T. forsythia (shaded bars) or the mutant (open bars) was mixed with oral bacteria (2:1) in a cuvette, and the mixture was incubated at 37°C for 60 min. The OD550 of the upper phase in the cuvette was measured using a spectrophotometer. The percentage of coaggregation was calculated as [(initial OD − postincubation OD)/initial OD] × 100. Filled bars indicate the percentage of autoaggregation of partner strains. Aa, A. actinomycetemcomitans. (A) Percentage of coaggregation of stationary-phase cells of T. forsythia (7-day culture) and stationary-phase cells of oral bacteria. (B) Percentage of coaggregation of exponential-phase cells of T. forsythia (3-day culture) and stationary-phase cells of oral bacteria. Data represent means ± SD of measurements performed in triplicate. Significant differences in coaggregation between the WT and mutant T. forsythia strains were determined using Student's t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Fig 10.
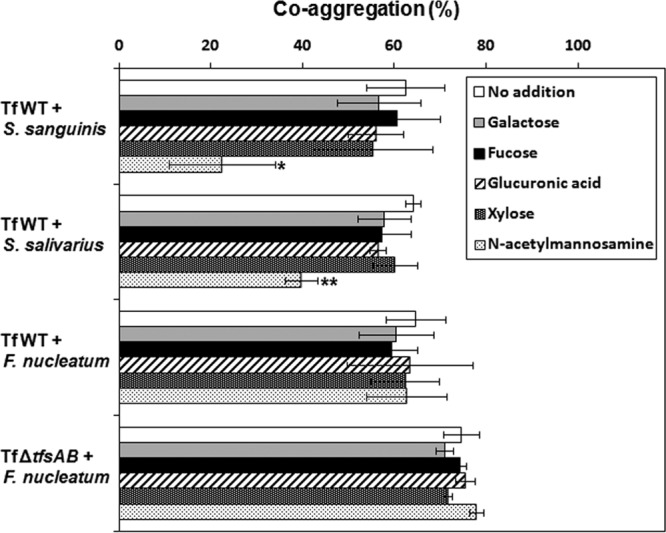
Effect of sugar on the coaggregation of T. forsythia with oral bacteria. To observe the effects of sugars on coaggregation activity, 10 mM each sugar was added to the reaction mixture. We used N-acetyl-d-mannosamine, d-glucuronic acid sodium salt, l-(−)-fucose, d-(+)-xylose, and d-(+)-galactose. Coaggregation activity (120 min) was investigated as described in Materials and Methods. The percentage of coaggregation was calculated as [(initial OD − postincubation OD)/initial OD] × 100. Data represent means ± SD of measurements performed in triplicate. Significant differences in coaggregation between bacteria with added sugar and bacteria with no sugar added were determined using Dunnett's test (*, P < 0.05; **, P < 0.01).
DISCUSSION
The mechanism of complement resistance in bacteria functions through three main pathways: protease digestion of complement components; recruitment of factors such as factor H and C4BP, which inhibit complement activation, to the bacterial cell surface; and polysaccharide-mediated suppression of complement activation (16, 30–33). In periodontal bacteria, P. gingivalis possesses all three resistance systems. Gingipains, surface polysaccharides, and C4BP-binding proteins in P. gingivalis have been characterized in previous studies (32, 33). In T. denticola, FhbB has been reported to have factor H-binding activity (44). In the present study, we demonstrated that the S-layer was involved in serum resistance. First, we hypothesized that the S-layer has binding affinity for factor H or C4BP, both of which are involved in the conversion of C3b to an inactivated molecule. However, we failed to observe significant differences in factor H and C4BP binding between the WT and the S-layer-deficient mutant. Finally, we observed inhibition of C3b deposition in WT T. forsythia, but not in the mutant, suggesting that the S-layer in T. forsythia inhibits the binding of C3b to the cell surface. The S-layers of C. rectus and C. fetus have been reported to mediate serum resistance by inhibiting the binding of C3b to the cell surface through the S-layer (16). Recently, a metalloproteinase, karilysin, was reported to inhibit all pathways of the complement system, contributing to serum resistance in T. forsythia (42). In the present study, we observed no differences in protease activity between WT T. forsythia and the mutant (Fig. 7). Based on these results, we concluded that the S-layer mediates serum resistance by inhibiting the deposition of C3b onto the bacterial cell surface. In the growth experiment, the S-layer-deficient mutant grew more slowly than the WT in calf or human serum. Since complement activity in serum without bacteria did not decrease after 7 days of incubation at 37°C under anaerobic conditions (data not shown), the visible growth of both strains after 4 days was considered to result from the inactivation of complement by proteases. However, the expression of proteinases was similar for the WT and the mutant. Therefore, the slow growth of the mutant in serum was considered to be due to a significant reduction in the number of viable mutant cells by complement activity relative to the number of viable WT cells when the strains were initially exposed to serum.
Furthermore, although the S-layer-deficient T. forsythia mutant was more susceptible to serum than the WT, the mutant still showed a lower level of susceptibility than E. coli XL-II. As shown in Fig. 2, the mutant had a survival ratio almost 20% lower than that of the WT when exposed to serum, while the E. coli strain had a survival ratio lower than that of the mutant, suggesting that factors other than the S-layer are involved in serum resistance in T. forsythia. Based on the factor H binding assay, the mutant had factor H binding affinity, suggesting that a factor H binding protein(s) is localized to the cell surface. These results indicate that, as in P. gingivalis, several factors, including the S-layer, karilysin, and factor H-binding protein, play a role in the serum resistance of T. forsythia.
Bacterial coaggregation is important for the formation of a biofilm, which is involved in resistance to various immune factors derived from the host, antibacterial agents, and disinfectants. T. forsythia is commonly found in dental plaques (biofilms on the tooth surface) (3, 4). Based on our results, the S-layer is associated with binding to S. sanguinis, S. salivarius, P. gingivalis, and F. nucleatum. Moreover, F. nucleatum coaggregates strongly with the S-layer mutant, suggesting that other surface molecules (besides the S-layer) in T. forsythia play a role in binding to F. nucleatum, but not to the oral bacteria tested in this study. T. forsythia BspA may play a major role in coaggregation with F. nucleatum (35, 45). Because BspA is a cell-associated secreted protein and belongs to the leucine-rich repeat family, deletion of the S-layer may promote coaggregation with F. nucleatum through BspA binding. Recently, the characterization of an S-layer protein consisting of two glycoproteins has been reported (24). Oligosaccharides containing N-acetylmannosaminuronic acid, 5-acetimidol-7-N-glycolylpseudaminic acid, digitoxose, fucose, xylose, glucuronic acid, and galactose are O-glycosidically linked to three amino acid motifs, D(S/T)(A/I/L/M/T/V), at either TfsA or TfsB. We investigated the effects of monosaccharides, such as N-acetylmannosamine, fucose, xylose, glucuronic acid sodium salt, and galactose, on coaggregation activity and found that N-acetylmannosamine suppressed the coaggregation of WT T. forsythia with S. sanguinis and S. salivarius (Fig. 10). However, the coaggregation of the WT and mutant T. forsythia strains with F. nucleatum was not inhibited by sugars. Therefore, the binding site of TfsAB may differ for different bacterial species.
In conclusion, we demonstrated that the S-layer in T. forsythia contributes to serum resistance and coaggregation with other oral bacteria, suggesting that this layer has several virulence effects. Therefore, the S-layer in T. forsythia is important for colonization in the gingival sulcus and is associated with periodontal disease.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported in part by a Grant-in-Aid for Young Scientists (B) from the Ministry of Education, Culture, Sports, Sciences, and Technology of Japan.
We thank the Joint Research Laboratory, Kagoshima University Graduate School of Medical and Dental Sciences, for the use of their facilities.
Footnotes
Published ahead of print 28 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00983-12.
REFERENCES
- 1. Pihlstrom BL, Michalowicz BS, Johnson NW. 2005. Periodontal diseases. Lancet 366:1809–1820 [DOI] [PubMed] [Google Scholar]
- 2. Socransky SS, Haffajee AD. 2005. Periodontal microbial ecology. Periodontol. 2000 38:135–187 [DOI] [PubMed] [Google Scholar]
- 3. Holt SC, Ebersole JL. 2005. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex”, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol. 2000 38:72–122 [DOI] [PubMed] [Google Scholar]
- 4. Rylev M, Kilian M. 2008. Prevalence and distribution of principal periodontal pathogens worldwide. J. Clin. Periodontol. 35:346–361 [DOI] [PubMed] [Google Scholar]
- 5. Dashper SG, Seers CA, Tan KH, Reynolds EC. 2011. Virulence factors of the oral spirochete Treponema denticola. J. Dent. Res. 90:691–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Henderson B, Ward JM, Ready D. 2010. Aggregatibacter (Actinobacillus) actinomycetemcomitans: a triple A* periodontopathogen? Periodontol. 2000 54:78–105 [DOI] [PubMed] [Google Scholar]
- 7. Holt SC, Bramanti TE. 1991. Factors in virulence expression and their role in periodontal disease pathogenesis. Crit. Rev. Oral Biol. Med. 2:177–281 [DOI] [PubMed] [Google Scholar]
- 8. Nakayama K. 2003. Molecular genetics of Porphyromonas gingivalis: gingipains and other virulence factors. Curr. Protein Pept. Sci. 4:389–395 [DOI] [PubMed] [Google Scholar]
- 9. Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134–144 [DOI] [PubMed] [Google Scholar]
- 10. Tanner AC, Izard J. 2006. Tannerella forsythia, a periodontal pathogen entering the genomic era. Periodontol. 2000 42:88–113 [DOI] [PubMed] [Google Scholar]
- 11. Sharma A. 2010. Virulence mechanisms of Tannerella forsythia. Periodontol. 2000 54:106–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yoo JY, Kim HC, Zhu W, Kim SM, Sabet M, Handfield M, Hillman J, Progulske-Fox A, Lee SW. 2007. Identification of Tannerella forsythia antigens specifically expressed in patients with periodontal disease. FEMS Microbiol. Lett. 275:344–352 [DOI] [PubMed] [Google Scholar]
- 13. Boot HJ, Pouwels PH. 1996. Expression, secretion and antigenic variation of bacterial S-layer proteins. Mol. Microbiol. 21:1117–1123 [DOI] [PubMed] [Google Scholar]
- 14. Sleytr UB, Beveridge TJ. 1999. Bacterial S-layers. Trends Microbiol. 7:253–260 [DOI] [PubMed] [Google Scholar]
- 15. Braun M, Kuhnert P, Nicolet J, Burnens AP, Frey J. 1999. Cloning and characterization of two bistructural S-layer-RTX proteins from Campylobacter rectus. J. Bacteriol. 181:2501–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thompson SA. 2002. Campylobacter surface-layers (S-layers) and immune evasion. Ann. Periodontol. 7:43–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee SW, Sabet M, Um HS, Yang J, Kim HC, Zhu W. 2006. Identification and characterization of the genes encoding a unique surface (S-) layer of Tannerella forsythia. Gene 371:102–111 [DOI] [PubMed] [Google Scholar]
- 18. Sabet M, Lee SW, Nauman RK, Sims T, Um HS. 2003. The surface (S-) layer is a virulence factor of Bacteroides forsythus. Microbiology 149:3617–3627 [DOI] [PubMed] [Google Scholar]
- 19. Sekot G, Posch G, Oh YJ, Zayni S, Mayer HF, Pum D, Messner P, Hinterdorfer P, Schäffer C. 2012. Analysis of the cell surface layer ultrastructure of the oral pathogen Tannerella forsythia. Arch. Microbiol. 194:525–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grogono-Thomas R, Blaser MJ, Ahmadi M, Newell DG. 2003. Role of S-layer protein antigenic diversity in the immune responses of sheep experimentally challenged with Campylobacter fetus subsp. fetus. Infect. Immun. 71:147–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Beveridge TJ, Pouwels PH, Sára M, Kotiranta A, Lounatmaa K, Kari K, Kerosuo E, Haapasalo M, Egelseer EM, Schocher I, Sleytr UB, Morelli L, Callegari ML, Nomellini JF, Bingle WH, Smit J, Leibovitz E, Lemaire M, Miras I, Salamitou S, Béguin P, Ohayon H, Gounon P, Matuschek M, Koval SF. 1997. Functions of S-layers. FEMS Microbiol. Rev. 20:99–149 [DOI] [PubMed] [Google Scholar]
- 22. Sleytr UB, Messner P. 1983. Crystalline surface layers on bacteria. Annu. Rev. Microbiol. 37:311–339 [DOI] [PubMed] [Google Scholar]
- 23. Sakakibara J, Nagano K, Murakami Y, Higuchi N, Nakamura H, Shimozato K, Yoshimura F. 2007. Loss of adherence ability to human gingival epithelial cells in S-layer protein-deficient mutants of Tannerella forsythensis. Microbiology 153:866–876 [DOI] [PubMed] [Google Scholar]
- 24. Posch G, Pabst M, Brecker L, Altmann F, Messner P, Schäffer C. 2011. Characterization and scope of S-layer protein O-glycosylation in Tannerella forsythia. J. Biol. Chem. 286:38714–38724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yoneda M, Hirofuji T, Motooka N, Nozoe K, Shigenaga K, Anan H, Miura M, Kabashima H, Matsumoto A, Maeda K. 2003. Humoral immune responses to S-layer-like proteins of Bacteroides forsythus. Clin. Diagn. Lab. Immunol. 10:383–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sekot G, Posch G, Messner P, Matejka M, Rausch-Fan X, Andrukhov O, Schäffer C. 2011. Potential of the Tannerella forsythia S-layer to delay the immune response. J. Dent. Res. 90:109–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hajishengallis G. 2010. Complement and periodontitis. Biochem. Pharmacol. 80:1992–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Walport MJ. 2001. Complement. First of two parts. N. Engl. J. Med. 344:1058–1066 [DOI] [PubMed] [Google Scholar]
- 29. Walport MJ. 2001. Complement. Second of two parts. N. Engl. J. Med. 344:1140–1144 [DOI] [PubMed] [Google Scholar]
- 30. McDowell JV, Frederick J, Miller DP, Goetting-Minesky MP, Goodman H, Fenno JC, Marconi RT. 2011. Identification of the primary mechanism of complement evasion by the periodontal pathogen, Treponema denticola. Mol. Oral Microbiol. 26:140–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ngampasutadol J, Tran C, Gulati S, Blom AM, Jerse EA, Ram S, Rice PA. 2008. Species-specificity of Neisseria gonorrhoeae infection: do human complement regulators contribute? Vaccine 26(Suppl. 8):I62–I66 [DOI] [PubMed] [Google Scholar]
- 32. Potempa M, Potempa J, Okroj M, Popadiak K, Eick S, Nguyen KA, Riesbeck K, Blom AM. 2008. Binding of complement inhibitor C4b-binding protein contributes to serum resistance of Porphyromonas gingivalis. J. Immunol. 181:5537–5544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Slaney JM, Gallagher A, Aduse-Opoku J, Pell K, Curtis MA. 2006. Mechanisms of resistance of Porphyromonas gingivalis to killing by serum complement. Infect. Immun. 74:5352–5361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bolstad AI, Jensen HB, Bakken V. 1996. Taxonomy, biology, and periodontal aspects of Fusobacterium nucleatum. Clin. Microbiol. Rev. 9:55–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ikegami A, Honma K, Sharma A, Kuramitsu HK. 2004. Multiple functions of the leucine-rich repeat protein LrrA of Treponema denticola. Infect. Immun. 72:4619–4627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Asakawa R, Komatsuzawa H, Kawai T, Yamada S, Goncalves RB, Izumi S, Fujiwara T, Nakano Y, Suzuki N, Uchida Y, Ouhara K, Shiba H, Taubman MA, Kurihara H, Sugai M. 2003. Outer membrane protein 100, a versatile virulence factor of Actinobacillus actinomycetemcomitans. Mol. Microbiol. 50:1125–1139 [DOI] [PubMed] [Google Scholar]
- 37. Yamada S, Sugai M, Komatsuzawa H, Matsumoto A. 2001. Suppressed localization of a major autolysin on Staphylococcus aureus treated with tetracycline. J. Electron Microsc. 30:359–364 [DOI] [PubMed] [Google Scholar]
- 38. Yamada S, Sugai M, Komatsuzawa H, Nakashima S, Oshida T, Matsumoto A, Suginaka H. 1996. An autolysin ring associated with cell separation of Staphylococcus aureus. J. Bacteriol. 178:1565–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yamada S, Matsumoto A. 1984. Localization of protein A on the cell surface of Staphylococcus aureus Cowan I and protein A-deficient strains. J. Electron Microsc. 33:172–174 [PubMed] [Google Scholar]
- 40. Komatsuzawa H, Asakawa R, Kawai T, Ochiai K, Fujiwara T, Taubman MA, Ohara M, Kurihara H, Sugai M. 2002. Identification of six major outer membrane proteins from Actinobacillus actinomycetemcomitans. Gene 288:195–201 [DOI] [PubMed] [Google Scholar]
- 41. Kawada-Matsuo M, Mazda Y, Oogai Y, Kajiya M, Kawai T, Yamada S, Miyawaki S, Oho T, Komatsuzawa H. 2012. GlmS and NagB regulate amino sugar metabolism in opposing directions and affect Streptococcus mutans virulence. PLoS One 7:e33382 doi:10.1371/journal.pone.0033382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jusko M, Potempa J, Karim AY, Ksiazek M, Riesbeck K, Garred P, Eick S, Blom AM. 2012. A metalloproteinase karilysin present in the majority of Tannerella forsythia isolates inhibits all pathways of the complement system. J. Immunol. 188:2338–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Karim AY, Kulczycka M, Kantyka T, Dubin G, Jabaiah A, Daugherty PS, Thogersen IB, Enghild JJ, Nguyen KA, Potempa J. 2010. A novel matrix metalloprotease-like enzyme (karilysin) of the periodontal pathogen Tannerella forsythia ATCC 43037. Biol. Chem. 391:105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Miller DP, Bell JK, McDowell JV, Conrad DH, Burgner JW, Héroux A, Marconi RT. 2012. Structure of factor H-binding protein B (FhbB) of the periopathogen, Treponema denticola: insights into progression of periodontal disease. J. Biol. Chem. 287:12715–12722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sharma A, Inagaki S, Sigurdson W, Kuramitsu HK. 2005. Synergy between Tannerella forsythia and Fusobacterium nucleatum in biofilm formation. Oral Microbiol. Immunol. 20:39–42 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


