Abstract
AraC-like regulators play a key role in the expression of virulence factors in enteric pathogens, such as enteropathogenic Escherichia coli (EPEC), enterotoxigenic E. coli, enteroaggregative E. coli, and Citrobacter rodentium. Bioinformatic analysis of the genome of rabbit-specific EPEC (REPEC) strain E22 (O103:H2) revealed the presence of a gene encoding an AraC-like regulatory protein, RegR, which shares 71% identity to the global virulence regulator, RegA, of C. rodentium. Microarray analysis demonstrated that RegR exerts 25- to 400-fold activation on transcription of several genes encoding putative virulence-associated factors, including a fimbrial operon (SEF14), a serine protease, and an autotransporter adhesin. These observations were confirmed by proteomic analysis of secreted and heat-extracted surface-associated proteins. The mechanism of RegR-mediated activation was investigated by using its most highly upregulated gene target, sefA. Transcriptional analyses and electrophoretic mobility shift assays showed that RegR activates the expression of sefA by binding to a region upstream of the sefA promoter, thereby relieving gene silencing by the global regulatory protein H-NS. Moreover, RegR was found to contribute significantly to virulence in a rabbit infection experiment. Taken together, our findings indicate that RegR controls the expression of a series of accessory adhesins that significantly enhance the virulence of REPEC strain E22.
INTRODUCTION
Infectious diarrhea is an important cause of morbidity and death in children in developing countries. Enteropathogenic Escherichia coli (EPEC) is a leading cause of acute infantile diarrhea in this setting (1, 2). EPEC can be further categorized into two subgroups, typical EPEC (tEPEC) and atypical EPEC (aEPEC) (3), which differ from each other in terms of their genetic characteristics, serotypes, and virulence factors (4).
The central mechanism of EPEC pathogenesis is colonization of the intestine accompanied by the formation of characteristic “attaching-and-effacing” (A/E) lesions on the surface of intestinal epithelial cells (5, 6). A 35-kb chromosomal pathogenicity island, termed the locus of enterocyte effacement (LEE), contains all of the genes necessary for A/E lesion formation (7–10). All EPEC strains carry the LEE and are Shiga toxin negative, but tEPEC strains also carry an EPEC adherence factor plasmid (EAF), which encodes bundle-forming pili (BFP), an essential colonization factor, and PerA, which directly regulates the expression of these pili and indirectly controls expression of the LEE, via PerC (11). The EAF plasmid is absent from aEPEC.
Humans are the only host and reservoir of tEPEC. In contrast, aEPEC can cause disease in humans and other animals (4, 12, 13). Infection with rabbit-specific EPEC (REPEC) causes the same clinical and pathological features in baby rabbits that human-specific EPEC produces in children (14, 15). Thus, REPEC infection of rabbits serves as a valuable model of human EPEC infection and has been used to establish the contribution of LEE-encoded proteins to virulence (16).
Recent epidemiological studies indicate that aEPEC is more prevalent than tEPEC in both developed and developing countries (17) and is frequently associated with persistent diarrhea (18). The mechanism by which aEPEC infection causes diarrhea is incompletely understood. Although aEPEC strains lack the EAF plasmid, they are able to cause disease; unlike tEPEC strains, which become markedly attenuated when they are “cured” of this plasmid (19). These observations suggest that aEPEC must produce colonization factors that compensate for the absence of BFP and also for PerA, but the identities of these virulence factors are yet to be determined.
Transcriptional control of virulence genes is essential for bacterial survival in their hosts, as it enables bacteria to respond to changing environmental conditions, such as a change in temperature or pH, or to adapt to a niche environment (20, 21). The AraC superfamily is an important group of transcriptional regulators that play a central role in the expression of virulence factors in enteric pathogens (22). Members of this superfamily include PerA of EPEC (23), Rns of enterotoxigenic E. coli (ETEC) (24), AggR of enteroaggregative E. coli (EAEC) (25), and ToxT of Vibrio cholerae (26). In general, AraC-like proteins are composed of 200 to 300 amino acids, comprising two distinct domains: an N-terminal stretch, which can be highly variable, and a C-terminal region harboring a characteristic double helix-turn-helix (HTH) motif for DNA binding (27). Importantly, AraC-like regulators can directly sense environmental chemicals that are abundant at the sites where the bacterial pathogens colonize and damage their hosts (20).
We recently identified a non-LEE-encoded, AraC-like regulatory protein, RegA, which plays a key role in the ability of Citrobacter rodentium to colonize mouse intestine (28). C. rodentium is a mouse-specific pathogen that is similar to EPEC in that it possesses the LEE, which allows it to cause A/E lesions in mouse intestine (29, 30). C. rodentium infection of mice is also used as a convenient small-animal model to study EPEC virulence (31–33). RegA functions as a global transcriptional regulator in C. rodentium by activating the transcription of grlA (global regulator of LEE activator) and a number of genes encoding colonization factors, such as fimbriae (kfc) and an autotransporter adhesin (adcA), while repressing transcription of a series of housekeeping genes (28, 34). Importantly, RegA requires a gut-associated environmental signal, bicarbonate, to exert its effect on gene expression (35). In this study, we characterized an AraC-like transcriptional regulator, RegR, from REPEC strain E22 (O103:H2). Our in vitro and in vivo analyses demonstrate that RegR, which controls the expression of several putative virulence proteins, is an important virulence determinant of REPEC.
MATERIALS AND METHODS
Bacterial strains, plasmids, primers, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Unless stated otherwise, bacteria were grown at 37°C in Luria-Bertani broth (LB) or on Luria agar (LA) plates supplemented with the appropriate antibiotics at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 25 μg/ml; kanamycin, 50 μg/ml; and trimethoprim, 40 μg/ml. Primers used in this study are listed in Table 2.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| E22 | REPEC serotype O103:H2, Rifr | 14 |
| E22 ΔregR | E22 ΔregR, Rifr | This study |
| E22 Δ2 | E22 ΔregR ΔEcE22_5293, Kanr Rifr | This study |
| E22 ΔsefA::kan | E22 ΔsefA, Kanr Rifr | This study |
| MC4100 | F− araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR thiA | 66 |
| BL21(DE3) | F− ompT hsdSB(rB− mB−) gal dcm (DE3) | New England BioLabs |
| TOP10 | F− mcrA Δ(mrr− hsdRMS− mcrBC) ϕ80lacZΔM15 ΔlacX74 nupG recA1 araD139 Δ(ara-leu)7697 galE15 galK16 rpsL(Strr) endA1 λ− | Invitrogen |
| Plasmids | ||
| pGEM-T Easy | High-copy-no. vector, Apr | Promega |
| pCR2.1-TOPO | High-copy-no. vector, Apr Kanr | Invitrogen |
| pACYC184 | Medium-copy-no. vector, Cmr Tcr | 67 |
| pFT-A | Low-copy-no. vector, flp Apr | 68 |
| pKD4 | FRT-flanked kan gene, Kanr Apr | 36 |
| pKD46 | Low-copy-no. vector, PBAD-λred Apr | 36 |
| pMU2385 | Single-copy-no. transcriptional fusion vector, Tpr | 69 |
| pMAL-c2x | Expression vector for N-terminal MBP fusion proteins | New England Biolabs |
| pYS1 | pGEM-T Easy+ΔregR::kan | This study |
| pYS2 | pCR2.1-TOPO+regR | This study |
| pYS3 | pACYC184+regR | This study |
| pYS4 | pGEM-T Easy+sefA1 | This study |
| pYS5 | pGEM-T Easy+sefA2 | This study |
| pYS7 | pGEM-T Easy+sefA3 | This study |
| pYS8 | pGEM-T Easy+sefA4 | This study |
| pYS9 | pMU2385+sefA1 | This study |
| pYS10 | pMU2385+sefA2 | This study |
| pYS11 | pMU2385+sefA3 | This study |
| pYS12 | pMU2385+sefA4 | This study |
| pYS13 | pGEM-T Easy+regR | This study |
| pYS14 | pMAL-c2x+regR | This study |
| pYS15 | pGEM-T Easy+sefAprom | This study |
| pYS16 | pGEM-T Easy+sefAcontrol | This study |
| pYS17 | pGEM-T Easy+ΔsefA::kan | This study |
| pYS21 | pGEM-T Easy+ ΔEcE22_5293::kan | This study |
Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Kanr, kanamycin resistance; Tcr, tetracycline resistance; Tpr, trimethoprim resistance.
Table 2.
Primers used in this study
| Primer | Sequence (5′ → 3′) |
|---|---|
| RegR-F | GGGACCTGTTAAATGCATATGTAC |
| RegR-kanR | GAAGCAGCTCCAGCCTACACAGCATTTTGACTATTCATACAGTCACC |
| RegR-kanF | CTAAGGAGGATATTCATATGGCTTGAAAGTGAAAAGGAAATCATAATATG |
| RegR-R | CAGAACTCAATGGCGGCGATGTC |
| RegR-F1 | GGCCGAACCTCTGTATATGAG |
| RegR-R1 | CGTATCACCTGACACACTTCTC |
| RalR-F | GAGAAACGCAGAGCAGCCTAAA |
| RalR-kanR | GAAGCAGCTCCAGCCTACACAGCCCCTTCGCATTAGCCAAT |
| RalR-kanF | CTAAGGAGGATATTCATATGGGTCTGGGGTTGAATCAGTGCCT |
| RalR-R | CATTATCTGGCCATTTGCATCGTC |
| RalR-F1 | ACGCAGAGCAGCCTAAAACG |
| RalR-R1 | CAAGCCCCCCTCAAAAAT |
| RegR-BamHIF | CGGGATCCCGAGTTTACAGAGTGGATATCC |
| RegR-SalIR | CGGTCGACCAGTAAGAACTGCTTCTGCC |
| RegR-BamHIF | GGATCCATGAATAGTCAAAATGCTTTAAGTAATATGAGGCTTG |
| RegR-HindIIIR | AAGCTTTTATGATTTCCTTTTCACTTTCAAGGAAAACTGT |
| SefA-PE-F | CTCCCTAAATCCACTCATTCTTACTTA |
| SefA-PE-R | GCTGTTTCAGCGGCATTTGC |
| SefA_RegRB1_F | CCAATCAAAATAACAAAAAAACGCGCGTATACAAAACAA |
| SefA_RegRB1_R | TTGTTTTGTATACGCGCGTTTTTTTGTTATTTTGATTGG |
| SefA_RegRB2_F | CAAAACCAATAAAACACTCCACGCGCGTAAAAAAATATG |
| SefA_RegRB2_R | CATATTTTTTTACGCGCGTGGAGTGTTTTATTGGTTTTG |
| SefAP1-HindIII-F | GCCGAAGCTTTAGGCACTGACAATACGGCTGCAAACTT |
| SefAP-EcoRI-R | GCCGGAATTCTGGAGCCTGAACAGTAGCAACATCACCTAT |
| SefAFP2-HindIII-F | GCCGAAGCTTTAATGAAAATTCACTTAACCTAACGATTAAATTG |
| SefA-F | AAGTGTATGAGGTGCGTGGTCAGC |
| SefA-kanR | GAAGCAGCTCCAGCCTACACACCTGAACAGTAGCAACATCACCTATAGC |
| SefA-kanF | CTAAGGAGGATATTCATATGAGCAACATTCTACATTCAACAATATCAAGATTAA |
| SefA-R | TGGGGCTTTCGTCTTTTTATCG |
| SefA-F1 | CATCCATCCACAACTGGTTAG |
| SefA-R1 | CCAGACCGAAATCAGTTGAC |
| SefA-EMSA-F | TAGGCACTGACAATACGGCTGCAAACTT |
| SefA-EMSA-R | TTACAATGAAATAAAAACAATATAATTCACGCA |
| SefA-EMSAc-F | CTTATTCTTGCTTTGATGACGTGTG |
| SefA-EMSAc-R | ATCTCCCGTAACATAGACCCCTGC |
| pKD4F | TGTGTAGGCTGGAGCTGCTTC |
| pKD4R | CATATGAATATCCTCCTTAG |
| pKD4seqF | TGACGAGTTCTTCTGAGCGGGAC |
| pKD4seqR | TCTAGCTATCGCCATGTAAGCC |
Construction of EPEC E22 ΔregR, E22 Δ2 and E22 ΔsefA::kan nonpolar mutant strains.
The λ Red recombinase system was utilized to construct nonpolar deletion mutants of REPEC E22 (36). Phusion high-fidelity DNA polymerase (Finnzymes), which generates blunt-ended fragments, and primer pairs RegR-F/RegR-kanR and RegR-R/RegR-kanF were used to amplify the DNA sequences flanking the regR gene from the E22 chromosome, and primers pKD4F and pKD4R were used to amplify the kanamycin resistance (kan) gene, bordered by Flp recombination target (FRT) sites, from plasmid pKD4. The products of these three PCRs (100 ng each) served as templates in overlapping-extension PCR (37) using Platinum Taq DNA polymerase (Invitrogen) and primers RegR-F and RegR-R, to generate a DNA fragment carrying a kan gene flanked by ∼500-bp regions up- and downstream of regR. This DNA fragment was cloned into pGEM-T Easy (Promega), the recombinant plasmid (pYS1) was introduced into E. coli K-12 TOP10 cells (Invitrogen), and the insert was then confirmed by sequencing. pYS1 was used as a template in a PCR with primer pair RegR-F/RegR-R to amplify the linear allelic replacement DNA fragment, which was introduced into strain E22 expressing λ Red recombinase from plasmid pKD46. The resultant E22 ΔregR::kan mutant was confirmed by PCR using primer pairs in which one primer flanked the targeted region and the other primed within the kan gene (RegR-F1/pKD4seqR) and (RegR-R1/pKD4seqF). The kan gene was then excised from the E22 ΔregR::kan strain by using Flp recombinase encoded on plasmid pFT-A, generating a marker-free deletion mutant, E22 ΔregR, which was confirmed by PCR and sequencing using primers RegR-F1 and RegR-R1.
The same general protocol was employed to delete the EcE22_5293 gene from E22 ΔregR using the EcE22_5293-specific primer pairs RalR-F/RalR-kanR and RalR-R/RalR-kanF. The resultant 2.2-kb fragment was cloned into pGEM-T Easy to generate pYS21. The ΔEcE22_5293::kan mutation was confirmed by PCR using primer pairs RalR-F1/pKD4seqR and RalR-R1/pKD4seqF, and the PCR products were sequenced by using primers RalR-F1 and RalR-R1. The resultant double mutant strain, E22 ΔregR ΔEcE22_5293::kan, here referred to as E22 Δ2, was used as the parent strain in microarray analysis.
To generate the E22 ΔsefA::kan mutant, primer pairs SefA-F/SefA-kanR and SefA-R/SefA-KanF were used. The resultant 2.5-kb fragment was cloned into pGEM-T Easy to obtain pYS17. This construct was used as a template in a PCR with primer pair SefA-F/SefA-R to amplify the linear allelic replacement DNA fragment, which was introduced into strain E22 expressing λ Red recombinase from plasmid pKD46. The E22 ΔsefA::kan mutant was confirmed by PCR using primer pairs SefA-F1/pKD4seqR and SefA-R1/pKD4seqF, and the PCR products were sequenced using primers SefA-F1 and SefA-R1.
Construction of a trans-complementing plasmid, pYS3.
For trans complementation of the regR mutants of E22, a 950-bp fragment containing regR was amplified from genomic DNA of EPEC strain E22 using primer pair RegR-BamHIF/RegR-SalIR. The resultant fragment was cloned into pCR2.1-TOPO to yield pCR2.1-TOPO+regR (pYS2), which was then confirmed by sequencing. The fragment was excised by BamHI/SalI digestion and ligated into BamHI/SalI of pACYC184 to generate the plasmid pYS3 (pACYC184-regR).
Antisense EPEC E22 microarrays.
An antisense oligonucleotide microarray was custom designed using the Agilent eArray platform (Agilent Technologies). The array contained representative sequences from the open reading frames (ORFs), representing all gene predictions, for the E. coli E22 genome available at NCBI (GenBank accession number AAJV00000000). Each ORF was represented by at least three different oligonucleotides.
RNA isolation and labeling.
E. coli strains E22 Δ2(pYS3) and E22 Δ2(pACYC184) were cultivated in LB overnight at 37°C. Quadruplicate cultures of a 1:100 dilution were grown to an optical density at 600 nm (OD600) of 0.85. One volume of cells was incubated with two volumes of RNAprotect solution and processed according to the manufacturer's instructions (Qiagen). Cell lysis and RNA preparation were carried out by using the FastRNA Pro Blue kit (Qbiogene Inc.). After a 10-min treatment with RNase-free DNase I (Qiagen), the RNA was further purified utilizing the RNeasy MiniElute kit (Qiagen). A total of 5 μg of RNA was labeled either with Cy5-ULS or Cy3-ULS as described in the Kreatech ULS labeling manual (Kreatech Diagnostics). RNA quality and degree of labeling were determined with an Agilent 2100 bioanalyzer and an ND-1000 spectrophotometer (NanoDrop Technologies). A dye swap was performed for two of the four cultures to minimize the effects of any labeling artifacts.
Fragmentation, microarray hybridization, scanning, and analysis.
Fragmentation, hybridization, and scanning were performed at the Australian Genome Research Facility Ltd. (AGRF), Melbourne, Australia. Normalization and data analysis were performed using the limma package in Bioconductor (38–40). Genes were considered differentially expressed if they showed an average change of ≥2-fold with an adjusted P value of ≤0.05.
Primer extension.
Primer extension was performed as described previously (41). Briefly, total cellular RNA was purified from E. coli MC4100(pYS9, pYS3) and MC4100(pMU2385, pYS3). Cells were grown to mid-log phase (OD600 = 0.8), and RNA was isolated by using the FastRNA Pro kit. Primer SefA-PE-R was labeled at its 5′ end with 32P by T4 polynucleotide kinase and [γ-32P]ATP. The labeled primer was coprecipitated with 5 μg of total RNA. Hybridization was carried out at 45°C for 15 min in 10 μl of Tris-EDTA (TE) buffer containing 150 mM KCl. Primer extension reactions were started by the addition of 24 μl of extension solution (20 mM Tris-HCl [pH 8.4], 10 mM MgCl2, 10 mM dithiothreitol [DTT], 2 mM deoxynucleoside triphosphates [dNTPs], 1 U/ml avian myeloblastosis virus [AMV] reverse transcriptase) and were carried out at 42°C for 60 min. Samples were then precipitated and analyzed on a sequencing gel. A GA ladder was made by using the method of Maxam and Gilbert (42) to sequence a sefA fragment that was generated by PCR using primers SefA-PE-F and 32P-SefA-PE-R.
Construction of lacZ fusion plasmids.
The sefA-lacZ transcriptional fusions were constructed by PCR amplification of the regulatory region of sefA by using primer pairs SefAP1-HindIII-F/SefAP-EcoRI-R and SefAP2-HindIII-F/SefAP-EcoRI-R. The PCR fragments were cloned into pGEM-T Easy to generate plasmids pYS4 and pYS5, and these fragments were sequenced. The fragments were then excised from the pGEM-T Easy derivatives by HindIII/EcoRI digestion and cloned into the same sites of the single-copy transcriptional fusion vector pMU2385 to create the sefA-lacZ fusions sefA-lacZ1 (pYS9) and sefA-lacZ2 (pYS10).
sefA-lacZ3 (pYS11) carrying a RegR-Box1 mutation was generated by using a multiple-step overlapping PCR method. The first PCR involved the use of primer pairs SefAP1-HindIII-F/SefA_RegRB1_R and SefA_RegRB1_F/SefAP-EcoRI-R, template sefA-lacZ1 (pYS9), and Phusion high-fidelity DNA polymerase. The two resulting PCR fragments (100 ng each) were mixed together with Platinum Taq DNA polymerase master mix. After seven cycles of extension, primers SefAP1-HindIII-F and SefAP-EcoRI-R were added to the mixture. The sample was then subjected to 30 cycles of PCR, which resulted in the formation of a 700-bp DNA fragment. This fragment was cloned into pGEM-T Easy to generate pYS7, and the mutation was confirmed by sequencing. The mutant sefA fragment was excised by HindIII/EcoRI digestion and cloned into the same sites of the single-copy transcriptional fusion vector pMU2385 to create the sefA-lacZ3 (pYS11). sefA-lacZ4 (pYS12) was constructed in the same manner, except that primers pairs SefAP1-HindIII-F/SefA_RegRB2_R and SefA_RegRB2_F/SefAP-EcoRI-R were used in the Phusion PCR and the resultant fragment generated from the Platinum Taq PCR was cloned into pGEM-T Easy to generate pYS8.
β-Galactosidase assay.
Bacteria were grown to mid-log phase (OD600 ∼ 0.6). β-Galactosidase activity was assayed as described by Miller (43), and the specific activity was expressed in Miller units (MU). The data shown are the results of at least three independent assays.
Expression and purification of MBP::RegR.
The coding sequence of regR was amplified from E22 genomic DNA by using primer pair RegR-BamHIF/RegR-HindIIIR and then cloned into pGEM-T Easy to obtain plasmid pYS13. regR was excised by BamHI/HindIII digestion and inserted into expression vector pMAL-c2 (New England BioLabs) for N-terminal fusion to malE. The resulting vector, pYS14, was transformed into E. coli strain BL21(DE3). Overnight cultures of transformants were diluted 1:100 into fresh LB and grown at 30°C with shaking at 200 oscillations per min to an OD600 of 0.9. Induction of gene expression was carried out for 19 h at 16°C by the addition of isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 0.1 mM. Afterwards, bacterial cells were harvested (15 min, 3,000 × g, 4°C) and disrupted by the addition of lysozyme (100 μg/ml) and subsequent sonication in column buffer (20 mM Tris-HCl [pH 7.4], 1 M NaCl, 1 mM EDTA). Purification of RegR was achieved through binding of the fusion protein to an amylose resin as recommended by the manufacturer (New England BioLabs). All steps were carried out at 4°C. The concentration and purity of eluted maltose-binding protein (MBP)::RegR were determined by using an ND-1000 spectrophotometer and by SDS-PAGE of the fusion protein.
EMSA.
Labeling of DNA fragments with 32P was performed as follows. Primers SefA-EMSA-F and SefA-EMSAc-F were labeled at their 5′ ends by using [γ-32P]ATP and T4 polynucleotide kinase. The DNA fragments to be analyzed for RegR binding were generated by PCR using primer pairs SefA-EMSA-F/SefA-EMSA-R (for fragment sefAprom) and SefA-EMSAc-F/SefA-EMSAc-R (for fragment sefAcontrol) from plasmids carrying either the entire promoter region of sefA (pYS15, for sefAprom) or part of the downstream coding sequences for the control experiment (pYS16, for sefAcontrol). Electrophoretic mobility shift assay (EMSA) was carried out as published previously (35). Briefly, each fragment was incubated with various amounts of purified MBP::RegR protein at 25°C for 30 min in binding buffer [10 mM Tris-HCl (pH 7.4), 100 mM KCl, 0.1 mM DTT, 0.01% (vol/vol) Triton X-100, 1 mM EDTA, and 100 μg/ml bovine serum albumin (BSA), 5 ng/μl poly(dI-dC), 10% (vol/vol) glycerol]. DNA and DNA-protein complexes were then separated on 5% native polyacrylamide gels (37.5:1) for approximately 12 h at 10 V/cm and 4°C.
Preparation of surface-associated proteins and immunoblotting.
Bacterial strains were grown overnight in 10 ml of LB with and without 45 mM sodium bicarbonate at 37°C. Cells were harvested by centrifugation at 3,000 × g for 10 min, resuspended in 160 μl of phosphate-buffered saline (PBS) (pH 7.4), vortexed at high speed for 1 min, and subsequently incubated at 60°C for 30 min with intermittent vortexing. The samples were then pelleted by centrifugation at 3,000 × g for 10 min, and the supernatant was transferred to a fresh tube, where it was mixed with NuPAGE lithium dodecyl sulfate (LDS) sample reducing buffer and boiled at 100°C for 5 min. The samples were separated by SDS-PAGE using 4 to 12% Bis-Tris NuPAGE gels (Invitrogen), and the proteins were stained with Coomassie brilliant blue R-250. For immunoblotting, proteins were transferred to a polyvinylidene difluoride (PVDF) membrane and blocked with 1% casein in PBS. The membrane was probed with 1-in-600 dilution of mouse anti-SefA serum followed by 1-in-10,000 goat anti-mouse horseradish peroxidase (HRP)-labeled secondary antibody (Bio-Rad). Immunopositive bands were developed with tetramethylbenzidine (TMB) substrate (KPL).
Preparation of secreted proteins.
Bacterial strains were grown in Dulbecco modified Eagle medium (DMEM) (Invitrogen). Cells were harvested by centrifugation (5,000 × g, 10 min), and the supernatant, containing the extracellular proteins, was passed through a 0.20-μm-pore-size filter (Sartorius). Proteins in the supernatants were precipitated with 10% (vol/vol) trichloroacetic acid on ice for 1 h, washed in 25% (vol/vol) acetone, separated by SDS-PAGE using 4 to 12% Bis-Tris NuPAGE gels, and stained with Coomassie brilliant blue R250.
Protein identification.
Tandem mass spectrometry was performed at the Walter and Eliza Hall Institute for Medical Research, Proteomics Laboratory, Melbourne, Australia.
Immunoelectron microscopy.
Immune serum was obtained by immunizing mice with purified SefA protein obtained from homogenized gel slices via three subcutaneous injections at 2-week intervals. Specific anti-SefA serum was obtained by absorbing the immune sera with strain E22 ΔsefA::kan bacteria. The absorbed serum was then diluted for use in Western blotting or immunogold labeling as required.
Strains were grown overnight at 37°C on LA supplemented with 45 mM bicarbonate and appropriate antibiotics. Colonies were suspended in PBS using a swab, and 10 μl of the suspension was applied onto 200-mesh carbon-coated copper grids, left for 5 min, and then negatively stained with 1% (wt/vol) ammonium molybdate for 10 s. For immunoelectron microscopy, the bacteria were reacted for 15 min with mouse anti-SefA serum (diluted 1:10 in PBS), followed by a 15-min incubation with goat anti-mouse IgG(Fc) conjugated to 10-nm gold particles (diluted 1:20 in PBS containing 1% bovine serum albumin) (Amersham). A Philips CM120 BioTwin transmission electron microscope operated at a voltage of 120 kV was used to examine the grids.
Infection of rabbits.
For in vivo assays of virulence, 4- to 5-week-old New Zealand White rabbits were inoculated with the wild-type REPEC strain, E22, or an isogenic mutant. Rabbits were weighed daily and examined for clinical signs of illness, including weight loss and evidence of diarrhea, as described previously (44). Fecal shedding of the infecting strains was determined by culture of rectal swabs on MacConkey agar supplemented with rifampin (50 μg/ml) to distinguish them from the rabbits' microbiota. Animals were euthanized if they lost more than 15% of their body weight or demonstrated severe diarrhea.
Statistical analysis.
All analyses of quantitative data were performed by using Student's t test. A two-tailed P value of <0.05 was taken to indicate statistical significance.
Microarray data accession number.
The microarray data have been submitted to the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE41093.
RESULTS
Identification of a putative virulence AraC-like regulator and its gene targets in E. coli E22.
We screened the entire genome of REPEC strain E22 (GenBank accession number AAJV02000000) for open reading frames (ORFs) homologous to the C. rodentium RegA protein by performing a BLASTp search (National Center for Biotechnology Information [http://www.ncbi.nlm.nih.gov/]). A protein encoded by ORF EcE22_5323 which exhibited 71% identity with the RegA protein was identified, and this ORF was designated regR. Interestingly, regR is located on an ∼62-kb, E22-specific genetic island (GenBank accession number AAJV02000028.1), which also includes an ORF, EcE22_5293, that encodes a protein homologous to the LdaA regulator (48% identity, 68% similarity) responsible for the expression of an afimbrial adhesin in EPEC (45).
To identify those genes whose expression is controlled by RegR, we carried out a microarray analysis. Because RegR and the LdaA-like protein share significant homology within their HTH DNA-binding motifs and to avoid any complication arising from possible redundancy and interplay between the two proteins in the microarray assay, we knocked out both loci in the E22 genome to generate strain E22 Δ2 (E22 ΔregR ΔEcE22_5293). This strain was used as the host strain in the microarray assay.
The microarray data showed that 270 genes represented by 663 probes were significantly differentially expressed (adjusted P value of <0.05) (see Table S1 in the supplemental material). However, only six of these genes, which are located adjacent to the regR locus, were strongly upregulated (>15-fold) by the RegR protein. Four of these genes (sefABCD) appear to form one operon, encoding proteins with similarity, ranging from 59 to 74%, to the fimbrins chaperone and usher of SEF14 fimbriae of Salmonella enterica serovar Enteritidis (Table 3) (46). Strikingly, transcription of sefA, the first gene of the sef operon, was activated 390-fold by RegR.
Table 3.
Summary of genes that were strongly differentially regulated by RegR as identified by microarray analysisa
| ORF | Gene | Predicted function | Avg fold regulationb | Adjusted P value < 0.05c |
|---|---|---|---|---|
| EcE22_2922 | glcA | Glycolate permease | −6.3 | 1.77E−10 |
| EcE22_2924 | glcB | Malate synthase G | −11.1 | 1.05E−10 |
| EcE22_2925 | glcG | Glyoxylate | −10.0 | 1.05E−10 |
| EcE22_2926 | glcF | Glycolate oxidase, iron-sulfur subunit | −8.9 | 1.57E−08 |
| EcE22_2927 | glcE | Glycolate oxidase, subunit | −10.8 | 8.69E−10 |
| EcE22_2928 | glcD | Glycolate oxidase, subunit | −12.9 | 8.51E−10 |
| EcE22_5321 | espC | Putative serine protease | 22.6 | 1.05E−10 |
| EcE22_5333 | sefA | Fimbrin subunit | 390.3 | 1.74E−07 |
| EcE22_5334 | sefB | Fimbrial chaperone | 39.4 | 1.53E−11 |
| EcE22_5335 | sefC | Fimbrial usher | 35.3 | 2.90E−11 |
| EcE22_5336 | sefD | Fimbrin subunit | 49.3 | 4.23E−10 |
| EcE22_5340 | adcA/tsh | Putative autotransporter adhesin | 17.3 | 5.76E−11 |
See Table S1 in the supplemental material for the complete list of ORFs regulated by RegR.
The negative and positive values represent the degrees of down- and upregulation, respectively.
Only the most significant P values are shown.
The other two genes significantly upregulated by RegR encode a homolog of the autotransporters AdcA of C. rodentium (90% similarity) and Tsh of avian-pathogenic E. coli (APEC) (63% similarity) and a homolog of the serine protease EspC of enteropathogenic E. coli (75% similarity).
In addition, RegR downregulated the expression of six genes (>6-fold) (Table 3), including members of the glycolate and glyoxylate degradation pathway (glcDEFGB) and its associated glycolate permease gene, glcA.
Mapping of the sefA promoter.
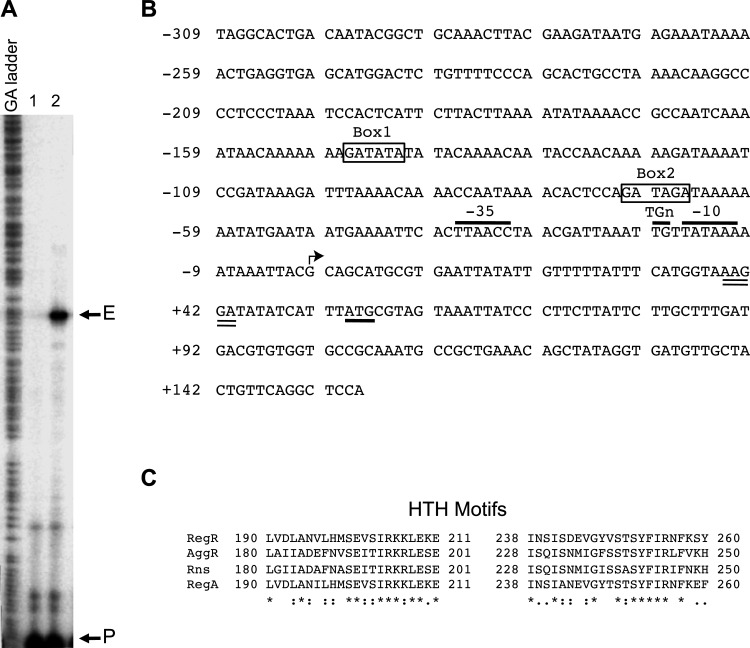
To characterize the molecular interaction of RegR with its gene targets, we investigated the sefA promoter, as sefA is the most upregulated gene target identified for RegR (Table 3). To identify the start site of transcription of sefA we performed a primer extension experiment as follows. A 32P-labeled primer (Sef14-PE-R) was hybridized with total RNA isolated from strain MC4100 carrying a pMU2385 derivative containing the sefA-lacZ1 transcriptional fusion (see below) and pACYC184+regR (pYS3). Only one major extension product was seen in the sample, indicating the presence of a single promoter for the sef operon (Fig. 1A). The start site of transcription was mapped to a guanine residue located 53 nucleotides (nt) upstream of the start codon for SefA (Fig. 1B). Based on this start site, a putative sefA promoter was identified (Fig. 1B). Its putative core elements included an extended −10 region (TGtTATAAA), a −35 region (TTAACC), and a spacer of 15 bp (Fig. 1B).
Fig 1.
The sefA regulatory region and the double helix-turn-helix DNA-binding motifs of RegR and its homologs. (A) The start site of transcription of the sefA promoter was mapped by primer extension using RNA isolated from E. coli MC4100 strains containing pACYC184-regR (pYS3) with either pMU2385 (control) or sefA-lacZ1 (pYS9). Lane 1, control experiment using RNA from E. coli MC4100 containing pACYC184-regR and pMU2385. Lane 2, experiment using RNA from E. coli MC4100 containing pACYC184-regR and sefA-lacZ1. The positions corresponding to the primer and the extension product are marked with P and E, respectively. (B) Nucleotide sequence of the sefA regulatory region. The numbering on the left of the sequence is relative to the transcriptional start site of sefA, which is indicated by an angled arrow. The putative −10 region, TGn motif, and −35 region are indicated and overlined. The putative start codon and ribosome-binding site are underlined and double underlined, respectively. The putative RegR-binding sites, Box1 and Box2, are boxed and labeled. (C) Alignment of helix-turn-helix (HTH) motifs of RegR, AggR, Rns, and RegA was done using the PS01124 profile defined in PROSITE (65). The numbers flanking the sequences indicate the positions of amino acid residues of the regulatory proteins. Identical and conserved amino acids are indicated with asterisks and colons, respectively.
Analysis of RegR-mediated activation of the sefA promoter.
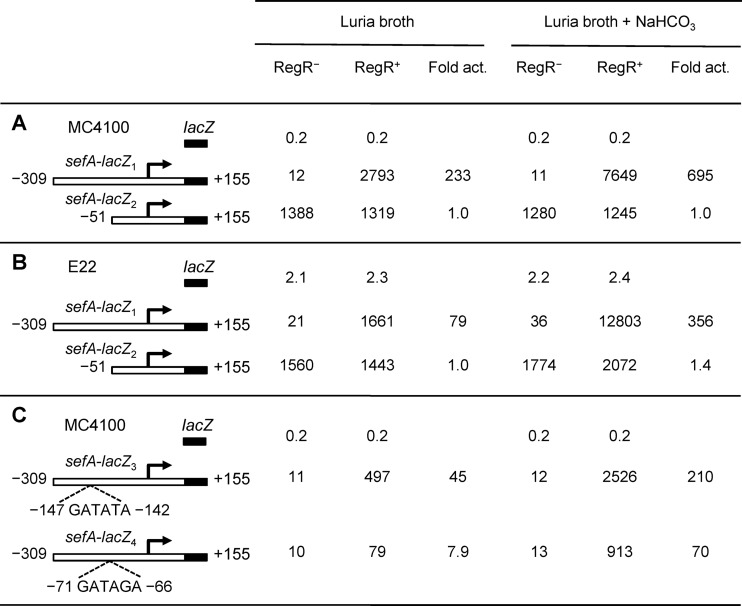
To examine RegR-mediated activation of sefA expression, we carried out transcriptional analyses using a lacZ reporter system. Two promoter-lacZ transcriptional fusions were constructed by cloning DNA fragments, encompassing positions −309 to +155 and −51 to +155 (relative to the transcriptional start site of sefA), in front of the lacZ structural gene of the single-copy vector pMU2385. The resulting plasmids, sefA-lacZ1 (pYS9) and sefA-lacZ2 (pYS10), were each transformed into E. coli MC4100 RegR+ [MC4100(pYS3)] and RegR− [MC4100(pACYC184)]. Because the RegA protein from C. rodentium (a homolog of RegR) requires sodium bicarbonate as a cofactor to exert maximal regulatory effects (35), we also tested whether this chemical is involved in RegR-mediated activation of sefA transcription. These strains were grown to mid-log phase in the absence or presence of 45 mM bicarbonate, and promoter activity was measured by assaying for β-galactosidase activity.
Although extremely low levels of β-galactosidase activities (11 to 12 MU) were detected for the sefA-lacZ1 construct in the RegR− background, high levels of expression were observed in the RegR+ background, where the sefA promoter activity increased from around 230 times in the absence of bicarbonate to around 700 times in the presence of bicarbonate, respectively (Fig. 2A). These data clearly demonstrate that RegR strongly activates sefA transcription and that bicarbonate positively influences the RegR-mediated effect.
Fig 2.
Expression of β-galactosidase by various sefA-lacZ fusions in both E. coli and E22 strains. The E. coli and E22 derivatives were grown in LB in the absence or presence of sodium bicarbonate (45 mM). The numbering of the sefA fragments is relative to the start site of transcription of sefA. The promoter activities of the various constructs are shown as specific activities of β-galactosidase (Miller units), which are the mean values from three independent assays, with variation of less than 15%. Fold activation (Fold act.) is the specific activity of β-galactosidase of the RegR+ strain divided by that of the RegR− strain. (A) The analysis of sefA-lacZ1 and sefA-lacZ2 was performed in E. coli RegR− [MC4100(pACYC184)] and RegR+ [MC4100(pYS3)] strains. (B) The analysis of sefA-lacZ1 and sefA-lacZ2 was performed in E22 RegR− [ΔregR(pACYC184)] and RegR+ [ΔregR(pYS3)] strains. (C) The analysis of sefA-lacZ3 and sefA-lacZ4 was performed in E. coli RegR− [MC4100(pACYC184)] and RegR+ [MC4100(pYS3)] strains.
For sefA-lacZ2, no difference of transcriptional activity was seen in the RegR− and RegR+ backgrounds with or without bicarbonate (Fig. 2A). Moreover, compared to that of sefA-lacZ1, the basal level of promoter activity of sefA-lacZ2 in the RegR− background increased approximately 100 times (Fig. 2A). The lack of RegR activation and the strong increase in basal-level transcription of this construct could be attributed to the absence of cis-acting elements required for the negative and positive controls of the sefA promoter.
The transcriptional analysis of the sefA-lacZ1 and sefA-lacZ2 constructs was also carried out in E22 using RegR− [ΔregR(pACYC834)] and RegR+ [ΔregR(pYS3)] strains. As shown in Fig. 2B, the transcriptional activity of sefA-lacZ1 was activated 79- and 356-fold by RegR in the absence and presence of sodium bicarbonate, respectively. In contrast, no significant effect of RegR on transcription of sefA-lacZ2 was seen. These results are in agreement with those observed in the E. coli MC4100 backgrounds, indicating that only sefA-lacZ1 carries an operator sequence responsible for RegR-mediated activation.
Identification of the RegR-binding sites.
The helix-turn-helix DNA-binding motif of RegR is highly homologous to that of RegA from C. rodentium (86.6% identity and 100% similarity) (Fig. 1C), suggesting that the two regulatory proteins may recognize similar DNA-binding sites. Indeed, a search using the RegA consensus sequence GATATA (47) identified two possible RegR-binding sites, GATATA (Box1) and GATAGA (Box2), which are centered at −144.5 and −68.5, respectively, relative to the start site of sefA transcription (Fig. 1B). To test whether these sequences are involved in the positive control of sefA by RegR, we made changes to bases within the two sites, from GATATA to CGCGCG and from GATAGA to CGCGCG (Fig. 1B). The two constructs, sefA-lacZ3 (pYS11) and sefA-lacZ4 (pYS12), containing the mutant sefA fragments were assessed for their ability to be activated by RegR. The data in Fig. 2C showed that while neither mutation had any effect on the basal levels of sefA promoter activities in the RegR− background, the mutations in Box1 and Box2 caused a significant and marked reduction in sefA expression in the RegR+ background (Fig. 2C). These results indicate that although both boxes are required for maximal levels of expression, Box2 plays a more significant role than Box1 in RegR-mediated activation of the sefA promoter.
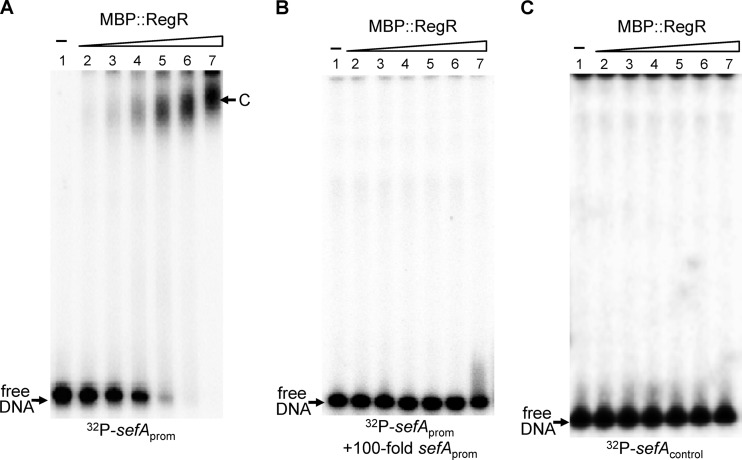
RegR binds directly to the sefA promoter region.
To provide further evidence that RegR interacts directly with the sefA regulatory region, we performed an electrophoretic mobility shift assay (EMSA). For this assay we used a purified fusion protein (MBP::RegR), because RegR, like many other AraC-like proteins, is insoluble in water. A 348-bp sefA promoter fragment (sefAprom) which spanned positions −309 to +38, relative to the start site of transcription, was end-labeled with 32P and incubated with various amounts of MBP::RegR. In the absence of MBP::RegR, the sefAprom fragment migrated to the bottom of the gel during electrophoresis to form a free DNA band (Fig. 3A, lane 1). However, after the addition of increasing amounts of MBP::RegR to the reaction mixture, this band gradually shifted (Fig. 3A, lanes 2 to 7). A major retarded band (C), representing a protein-DNA complex, was seen when MBP::RegR was added at final concentrations of between 65 and 500 nM (Fig. 3A, lanes 4 to 7). However, addition of excess amounts of cold sefA promoter fragment (sefAprom) into the reaction mixes outcompeted the binding of RegR to the labeled probe (Fig. 3B). An additional control experiment using a 32P-labeled DNA fragment (sefAcontrol) covering a sequence within the coding region of the sefA gene (between positions +75 and +302, relative to the start site of transcription) was also carried out. As shown in Fig. 3C, MBP::RegR failed to bind to this DNA fragment, further confirming the binding specificity of RegR for the sefA regulatory region.
Fig 3.
Electrophoretic mobility shift analysis of the binding of the recombinant RegR protein to the sefA regulatory region. The 32P-labeled PCR fragments containing the sefA regulatory region, sefAprom (A and B), and the coding region, sefAcontrol (C), were each mixed with 0, 15.5, 31.3, 62.5, 125, 250, and 500 nM MBP::RegR protein (lanes 1 to 7, respectively) in the presence of 45 mM sodium bicarbonate. A 100-fold molar excess of the unlabeled sefAprom fragment was added into the reaction mixture to demonstrate RegR specificity (B). Following incubation at 30°C for 20 min, the samples were analyzed on native polyacrylamide gels. The unbound DNA (free DNA) and protein-DNA complexes (band C) are marked.
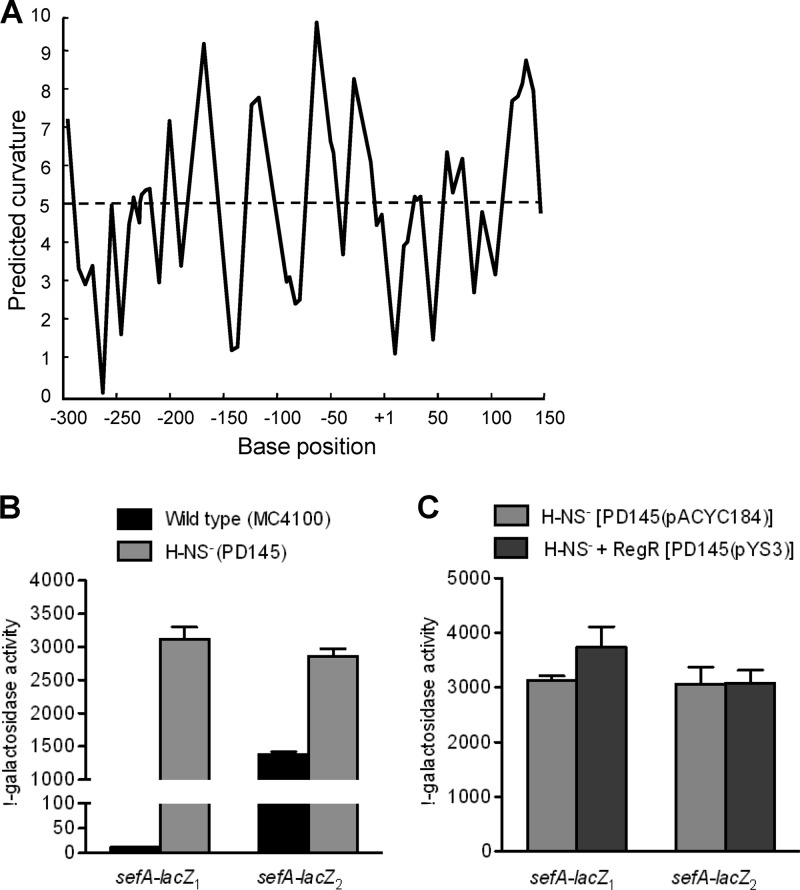
Effect of H-NS on expression of the sefA promoter.
As shown in the previous section, the transcription of the sefA promoter is subject to negative regulation by an unknown protein. As the DNA sequence surrounding the sefA promoter region is highly AT rich and is predicted by the bend.it program (http://hydra.icgeb.trieste.it/dna/index.php) to be highly curved (Fig. 4A), we hypothesized that H-NS is responsible for the repression. To test this, sefA-lacZ1 and sefA-lacZ2, were analyzed in strains MC4100 (wild type) and PD145 (hns). Relative to the promoter activities in MC4100, both constructs showed enhanced sefA expression in PD145 (hns) (Fig. 4B), confirming the involvement of H-NS in the negative regulation of sefA expression. Furthermore, the fact that deletion of the upstream sequence in the sefA-lacZ2 construct resulted in a large increase in promoter activity in E. coli MC4100 is consistent with the possibility that the region involved in H-NS interaction is partially removed (Fig. 4B).
Fig 4.
Analysis of H-NS-mediated repression of the sefA promoter. (A) In silico analysis of intrinsic curvature of the sefA regulatory region, using the bend.it program (http://hydra.icgeb.trieste.it/dna/index.php). The regions with >5 degrees per helical turn of DNA (dashed line) represent curved sequences. The base position is relative to the transcriptional start site of sefA. (B) Effects of H-NS on the expression of sefA. E. coli strains MC4100 (H-NS+) and PD145 (H-NS−) which contained either sefA-lacZ1 (−309 to +155) or sefA-lacZ2 (−51 to +155) were assayed for β-galactosidase activity. (C) Effects of RegR on the expression of sefA in the H-NS− background. E. coli strains PD145(pACYC184) (RegR−) and PD145(pYS3) (RegR+) which contained either sefA-lacZ1 (−309 to +155) or sefA-lacZ2 (−51 to +155) were assayed for β-galactosidase activity. The β-galactosidase activities (Miller units) shown are the means (± standard deviations [SD]) of results from three independent experiments.
We next analyzed the effect of RegR on transcription of the sefA-lacZ1 and sefA-lacZ2 constructs in the H-NS− backgrounds PD145(pACYC184) and PD145(pYS3). The results in Fig. 4C showed that in the absence of H-NS, the transcriptional activities of both constructs were essentially the same either with or without the expression of RegR. This suggests that the primary function of RegR at the sefA promoter is to overcome H-NS-mediated repression.
Effect of RegR on the synthesis of surface-associated and secreted proteins.
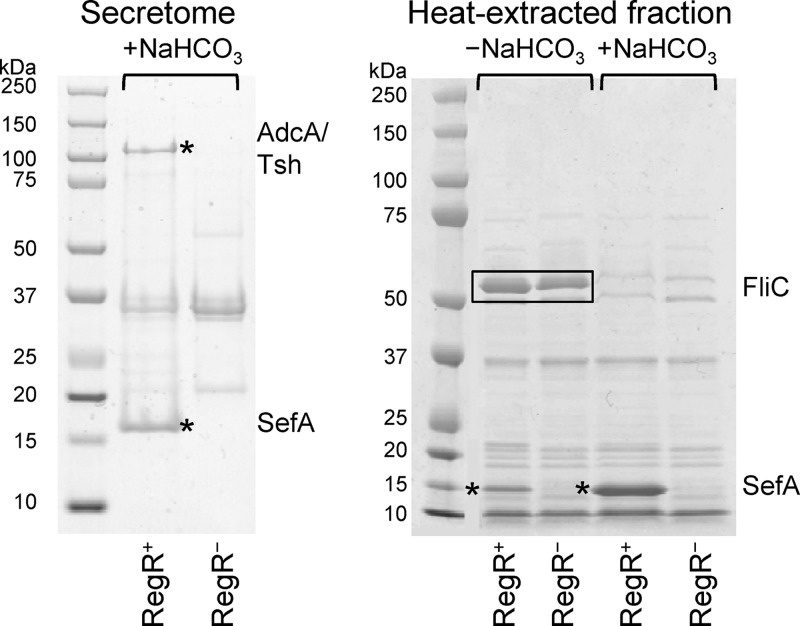
To identify secreted and surface-associated proteins whose synthesis is upregulated by RegR we carried out proteomic analyses of the secretomes and heat-extracted fractions of RegR+ [E22 ΔregR(pYS3)] and RegR− (E22 ΔregR) E22 strains using SDS-PAGE. Bacterial strains were grown in the presence and absence of bicarbonate, as our transcriptional analysis of RegR showed that bicarbonate enhances its activity. Comparison of the protein profiles revealed two protein bands in the secretome (∼135 and ∼15 kDa) and two in the heat-extracted fractions (∼17 kDa) of the RegR+ strain that were absent from the RegR− strain (Fig. 5). Tandem mass spectrometric analysis showed that the high-molecular-weight protein band consisted of two proteins: EcE22_5321, a homolog of EspC, and EcE22_5340, a homolog of AdcA. The low-molecular-weight proteins were SefA (EcE22_5333). A surface-associated protein of ∼ 52 kDa that was identified as FliC was seen only in the absence of bicarbonate, either with or without RegR. Because the expression of this protein is not under RegR regulation, it was not studied further.
Fig 5.
Effects of RegR on the expression of secreted and surface-associated proteins. RegR+ [E22 ΔregR(pYS3)] and RegR− (E22 ΔregR) strains of E22 were grown in DMEM (for secretome analysis) or in Luria broth in the absence or presence of 45 mM bicarbonate (for analysis of heat-extracted fractions). Proteins were separated by SDS-PAGE and stained with Coomassie brilliant blue R250. Four protein bands (two from the secretome fractions and two from the heat-extracted fractions) which were present in the RegR+ but not in the RegR− strain (indicated by asterisks) were excised and analyzed by tandem mass spectrometry. In the heat-extracted fractions, two bands which are abundant in the absence of bicarbonate (boxed) were analyzed by tandem mass spectrometry. The identities of these proteins are shown at the right of the gels.
Visualization of SefA on the surface of E22 by immunoelectron microscopy.
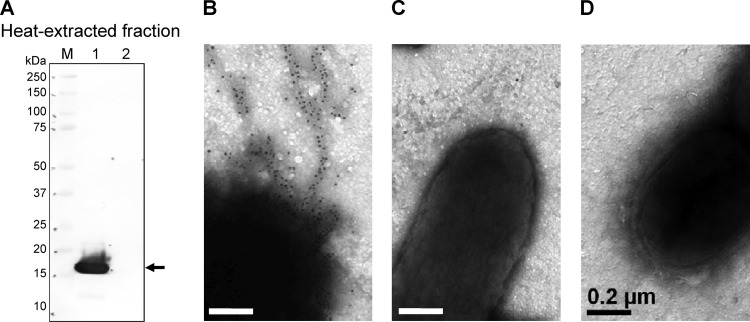
To confirm the presence of SefA on the surface of E22, we generated a sefA mutant, E22 ΔsefA::kan, and carried out electron microscopic examination of immunogold-labeled SefA+ [E22(pYS3)], SefA− [E22 ΔsefA::kan(pYS3)] and SefA+ RegR− (E22 ΔregR) strains. To ascertain the specificity of the antiserum, we probed heat-extracted fractions of the strains by immunoblotting using the SefA antiserum. This analysis revealed a protein of approximately 17 kDa in extracts of the sefA-expressing strain but not in the sefA mutant (Fig. 6A). When used for immunogold electron microscopy, the same antiserum revealed fimbria-like structures on the surface of the SefA+ strain (Fig. 6B), but not on the SefA− strain (Fig. 6C). Fimbria-like structures on the surface of the regR mutant strain also were not recognized by the antiserum (Fig. 6D), demonstrating that RegR is required for SefA expression and its presence on the cell surface.
Fig 6.
Localization of SefA on the cell surface. (A) Immunoblot of heat-extracted fractions from SefA+ [E22(pYS3)] and SefA− [E22 ΔsefA::kan(pYS3)] strains probed with mouse anti-SefA serum. The arrow indicates a protein of approximately 17 kDa produced by the SefA+ strain which was absent from the SefA− strain. Lanes: 1, E22(pYS3); 2, E22 ΔsefA(pYS3). M, protein molecular mass standards. (B to D) Immunoelectron microscopy of SefA+ [E22(pYS3)] (B), SefA− [E22 ΔsefA::kan(pYS3)] (C), and SefA+ RegR− (E22 ΔregR) (D) strains probed with mouse anti-SefA serum visualized with goat anti-mouse IgG(Fc) conjugated to 10-nm gold particles. Bars represent 0.2 μm.
RegR is a virulence determinant of REPEC strain E22.
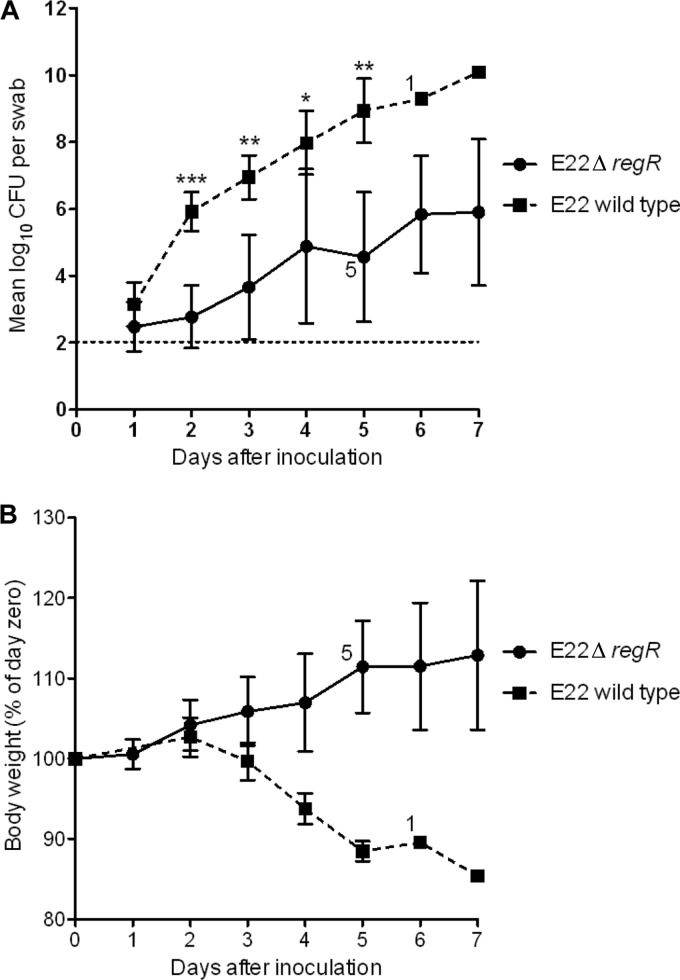
To investigate the effect of RegR on the virulence of EPEC, we infected infant rabbits with wild-type REPEC strain E22 or its isogenic regR mutant, E22 ΔregR. Following inoculation of six rabbits with 5 × 105 CFU of wild-type E22, large numbers of REPEC organisms were shed in stools, such that >105 CFU were recovered from rectal swabs from all rabbits within 2 days of inoculation (Fig. 7A). Loss of body weight began within 48 h of infection and continued until animals were euthanized (Fig. 7B). All of the rabbits developed clinical illness, characterized by diarrhea with weight loss, requiring euthanasia. Five rabbits were euthanized on day 5 and one on day 7. These results were consistent with previous reports of infection of rabbits with E22 (14). Following inoculation with 8 × 105 CFU of the regR mutant strain, rabbits shed the challenge strain from the second day after inoculation to 7 days postinoculation but yielded significantly lower numbers of bacteria in rectal swabs (P < 0.0005 on day 2, P < 0.005 on days 3 and 5, and P < 0.05 on day 4) (Fig. 7A). One rabbit inoculated with the regR mutant strain began to lose weight on day 2, developed diarrhea on day 3, and was subsequently euthanized on day 4.
Fig 7.
Effect of regR on virulence of REPEC strain E22. Six infant rabbits were inoculated with 5 × 105 CFU of wild-type E22 or 8 × 105 CFU of its isogenic regR mutant (E22 ΔregR). When some rabbits in a group were euthanized, due to loss of body weight of greater than 15% or the presence of severe diarrhea, the surviving numbers of rabbits are shown adjacent to the data points. (A) Colonization of rabbits by REPEC strain E22 and the regR mutant as measured by quantitative culture of rectal swabs. Data are the geometric mean CFU per swab (***, P < 0.0005; **, P < 0.005; and *, P < 0.05). The detection limit of the culture method is indicated by the horizontal line. (B) Body weight of rabbits. Values are the mean (± SD) for each group expressed as a percentage of the weight on the day of infection (day zero).
Body weight is another sensitive indicator of illness in the REPEC/rabbit infection model (16, 48), and during the course of infection, rabbits infected with the mutant strain showed weight gain, in contrast to those infected with the wild type (Fig. 7B). Taken together, these results indicated that RegR is required for the full virulence of REPEC strain E22.
DISCUSSION
In this study, we demonstrated that REPEC strain E22 possesses a RegR regulon, which is required for efficient infection of rabbits. The RegR regulon includes the master regulatory gene regR and its cognate gene targets adcA/tsh, espC, and sefABCD. Although these genes are carried on separate operons, they are colocated in the E22 genome, and their encoded proteins could function cooperatively in pathogenesis. The adcA and espC loci encode homologs of the autotransporter proteins AdcA/Tsh and EspC, respectively. These proteins are members of the SPATE (serine protease autotransporters of Enterobacteriaceae) subfamily of proteins (49, 50). The AdcA protein of C. rodentium is responsible for bacterial aggregation as well as for binding to cultured epithelial cells (28). Interestingly, adcA is also a member of the RegA regulon of C. rodentium, which is required for the intestinal colonization of mice. The Tsh protein of avian-pathogenic E. coli (APEC) has agglutinin and mucinolytic activities, which may facilitate colonization of the avian mucus by APEC (51). Other studies have shown that Tsh contributes to the pathogenesis of avian colibacillosis and the early stages of APEC infection of chickens (52, 53).
The EspC protein from EPEC mediates cell adhesiveness and causes cytoskeletal damage to epithelial cells following its internalization by the host cells (54, 55). Both the type III secretion system, which is located on the LEE, and type V secretion system are involved in secretion and translocation of EspC (56).
The E22 sefABCD cluster is homologous to that found in group D Salmonella. It consists of four cotranscribed genes encoding the major subunit (SefA), chaperone (SefB), usher (SefC), and minor subunit (SefD) of the SEF14 fimbriae (46). The E22 fimbria-like structures that we identified by immunogold electron microscopy using anti-SefA serum have a morphology similar to that of SEF14 fimbriae produced by S. Enteritidis, which play a role in colonization of epithelial cell surfaces (57). SEF14 fimbriae are required for binding of S. Enteritidis to macrophages and are a virulence determinant of Salmonella (58). Interestingly, the expression of Salmonella sefABCD is subject to positive control by the AraC-like transcriptional activator SefR (46).
Our microarray analysis showed that expression of the three operons adcA/tsh, espC, and sefABCD was strongly upregulated by RegR (Table 3). This finding was supported by proteomic analyses which revealed the abundant presence of the products of all three operons, either in secretomes or on the bacterial cell surface, in the RegR+ but not the RegR− background (Fig. 5). RegR-mediated activation was enhanced in the presence of the gut-specific environmental factor bicarbonate, which is also the cofactor for the virulence regulator RegA of C. rodentium and ToxT of V. cholerae (20, 59).
The mechanism of RegR-mediated activation was examined by using the promoter region of the sefA gene. A primer extension experiment indicated that transcription of the E22 sefABCD operon is driven by a single σ70 promoter which contains a well conserved −10 region (TATAAA versus the consensus TATAAT), a TGn motif, a less conserved −35 sequence (TTAACC versus the consensus TTGACA), and a shorter spacer (15 bp versus 17 bp) (Fig. 1B). Data from transcriptional assays using lacZ reporters indicated that all of the cis elements required for the control of the sefA promoter are contained within a region of 464 bp between positions −309 and +155, relative to the start site of transcription (Fig. 1B), as the construct sefA-lacZ1 exhibited a degree of maximal activation by RegR similar to that seen in the microarray assay (Fig. 2 and Table 3). The extremely high levels of regulatory outcome of sefA transcription are determined by the global negative regulatory protein H-NS and the REPEC-specific activator RegR, which exert opposing effects on sefA expression (Fig. 2). The entire regulatory region of sefA is highly AT rich and was predicted to form strong DNA curvatures, a general signature of H-NS-binding sites (Fig. 4A). Indeed, deletion of a region between positions −309 and −52 resulted in a pronounced loss of H-NS-mediated repression (Fig. 4).
Two putative RegR-binding sites were identified upstream of the promoter core sequences (Fig. 1B). These sites are similar to the operator sequences of the virulence operons controlled by the Rns, AggR, and RegA proteins (47, 60, 61). This homology is most likely due to the high degrees of similarity between the HTH DNA-binding motifs of RegR, Rns, AggR, and RegA (Fig. 1C), suggesting that these regulatory systems may have evolved from the same ancestry.
In addition to its ability to activate virulence gene expression, RegR also repressed expression of the glcDEFGBA operon, whose products are involved in the transport and utilization of glycolate (Table 3). Transcription of the glcDEFGBA operon is activated by GlcC (62), but the mode of action of RegR at the glcD promoter is unknown.
Although all EPEC strains carry the outer membrane adhesin protein intimin, which is encoded on the LEE, they require other adhesins for their initial attachment to the host intestinal epithelium (63) and regulators that control the expression of these adhesins (11). For tEPEC, these additional virulence determinants are BFP and its associated regulator, PerA (11). The virulence of aEPEC strains suggests that these pathogens also express one or more surface-located factors that compensate for the absence of BFP and a protein which regulates their expression. Our findings that aEPEC strain E22 carries the three operons adcA/tsh, espC, and sefABCD, which are regulated by RegR (a homolog of PerA), were highly induced and expressed under conditions similar to those found in the gut, and were required for full virulence in the rabbit model of infection (Fig. 7), support our hypothesis.
We have previously shown that aEPEC strains are a heterogeneous group of bacteria that exhibit considerable variations in terms of the serotype, pattern of adherence to mammalian cells, and carriage of known virulence determinants (64). This group of A/E pathogens carry the LEE and most likely carry AraC-like regulators, such as RegR of E22 and RegA of C. rodentium, which are able to sense the environment and regulate the expression of a variety of adherence factors that act cooperatively with intimin and are required for virulence. These AraC-like regulators and accessory adherence factors may have been acquired together via horizontal gene transfer. Alternatively, the regulatory protein may have been acquired independently and subsequently adapted to regulate existing adherence factors.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Australian National Health & Medical Research Council (NHMRC), the NHMRC Independent Research Institutes Infrastructure Support Scheme, and the Victorian State Government Operational Infrastructure Support Program to the Murdoch Childrens Research Institute. M.T. and Y.S. were recipients of an NHMRC Peter Doherty Australian Biomedical Fellowship.
Footnotes
Published ahead of print 22 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01325-12.
REFERENCES
- 1. Fagundes-Neto U, Scaletsky IC. 2000. The gut at war: the consequences of enteropathogenic Escherichia coli infection as a factor of diarrhea and malnutrition. Sao Paulo Med. J. 118:21–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Robins-Browne RM. 1987. Traditional enteropathogenic Escherichia coli of infantile diarrhea. Rev. Infect. Dis. 9:28–53 [DOI] [PubMed] [Google Scholar]
- 3. Kaper JB. 1996. Defining EPEC. Rev. Microbiol. Sao Paulo. 27:130–133 [Google Scholar]
- 4. Trabulsi LR, Keller R, Tardelli Gomes TA. 2002. Typical and atypical enteropathogenic Escherichia coli. Emerg. Infect. Dis. 8:508–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Finlay BB, Rosenshine I, Donnenberg MS, Kaper JB. 1992. Cytoskeletal composition of attaching and effacing lesions associated with enteropathogenic Escherichia coli adherence to HeLa cells. Infect. Immun. 60:2541–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Frankel G, Phillips AD, Rosenshine I, Dougan G, Kaper JB, Knutton S. 1998. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol. Microbiol. 30:911–921 [DOI] [PubMed] [Google Scholar]
- 7. Elliott SJ, Sperandio V, Giron JA, Shin S, Mellies JL, Wainwright L, Hutcheson SW, McDaniel TK, Kaper JB. 2000. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 68:6115–6126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McDaniel TK, Kaper JB. 1997. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol. Microbiol. 23:399–407 [DOI] [PubMed] [Google Scholar]
- 9. Tauschek M, Strugnell RA, Robins-Browne RM. 2002. Characterization and evidence of mobilization of the LEE pathogenicity island of rabbit-specific strains of enteropathogenic Escherichia coli. Mol. Microbiol. 44:1533–1550 [DOI] [PubMed] [Google Scholar]
- 10. Wong AR, Pearson JS, Bright MD, Munera D, Robinson KS, Lee SF, Frankel G, Hartland EL. 2011. Enteropathogenic and enterohaemorrhagic Escherichia coli: even more subversive elements. Mol. Microbiol. 80:1420–1438 [DOI] [PubMed] [Google Scholar]
- 11. Bieber D, Ramer SW, Wu CY, Murray WJ, Tobe T, Fernandez R, Schoolnik GK. 1998. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 280:2114–2118 [DOI] [PubMed] [Google Scholar]
- 12. Moura RA, Sircili MP, Leomil L, Matte MH, Trabulsi LR, Elias WP, Irino K, Pestana de Castro AF. 2009. Clonal relationship among atypical enteropathogenic Escherichia coli strains isolated from different animal species and humans. Appl. Environ. Microbiol. 75:7399–7408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Johnson TJ, Nolan LK. 2009. Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol. Mol. Biol. Rev. 73:750–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marches O, Nougayrede JP, Boullier S, Mainil J, Charlier G, Raymond I, Pohl P, Boury M, De Rycke J, Milon A, Oswald E. 2000. Role of Tir and intimin in the virulence of rabbit enteropathogenic Escherichia coli serotype O103:H2. Infect. Immun. 68:2171–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Milon A, Oswald E, De Rycke J. 1999. Rabbit EPEC: a model for the study of enteropathogenic Escherichia coli. Vet. Res. 30:203–219 [PubMed] [Google Scholar]
- 16. Abe A, Heczko U, Hegele RG, Brett Finlay B. 1998. Two enteropathogenic Escherichia coli type III secreted proteins, EspA and EspB, are virulence factors. J. Exp. Med. 188:1907–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ochoa TJ, Contreras CA. 2011. Enteropathogenic Escherichia coli infection in children. Curr. Opin. Infect. Dis. 24:478–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nguyen RN, Taylor LS, Tauschek M, Robins-Browne RM. 2006. Atypical enteropathogenic Escherichia coli infection and prolonged diarrhea in children. Emerg. Infect. Dis. 12:597–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Levine MM, Nataro JP, Karch H, Baldini MM, Kaper JB, Black RE, Clements ML, O'Brien AD. 1985. The diarrheal response of humans to some classic serotypes of enteropathogenic Escherichia coli is dependent on a plasmid encoding an enteroadhesiveness factor. J. Infect. Dis. 152:550–559 [DOI] [PubMed] [Google Scholar]
- 20. Yang J, Tauschek M, Robins-Browne RM. 2011. Control of bacterial virulence by AraC-like regulators that respond to chemical signals. Trends Microbiol. 19:128–135 [DOI] [PubMed] [Google Scholar]
- 21. Mahan MJ, Slauch JM, Mekalanos JJ. 1996. Environmental regulation of virulence gene expression in Escherichia, Salmonella, and Shigella spp., p 2803–2815 In Neidhardt FC, et al. (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed ASM Press, Washington, DC [Google Scholar]
- 22. Stoebel DM, Free A, Dorman CJ. 2008. Anti-silencing: overcoming H-NS-mediated repression of transcription in Gram-negative enteric bacteria. Microbiology 154:2533–2545 [DOI] [PubMed] [Google Scholar]
- 23. Gomez-Duarte OG, Kaper JB. 1995. A plasmid-encoded regulatory region activates chromosomal eaeA expression in enteropathogenic Escherichia coli. Infect. Immun. 63:1767–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Murphree D, Froehlich B, Scott JR. 1997. Transcriptional control of genes encoding CS1 pili: negative regulation by a silencer and positive regulation by Rns. J. Bacteriol. 179:5736–5743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nataro JP, Yikang D, Yingkang D, Walker K. 1994. AggR, a transcriptional activator of aggregative adherence fimbria I expression in enteroaggregative Escherichia coli. J. Bacteriol. 176:4691–4699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. DiRita VJ, Parsot C, Jander G, Mekalanos JJ. 1991. Regulatory cascade controls virulence in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 88:5403–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gallegos MT, Schleif R, Bairoch A, Hofmann K, Ramos JL. 1997. Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hart E, Yang J, Tauschek M, Kelly M, Wakefield MJ, Frankel G, Hartland EL, Robins-Browne RM. 2008. RegA, an AraC-like protein, is a global transcriptional regulator that controls virulence gene expression in Citrobacter rodentium. Infect. Immun. 76:5247–5256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Deng W, Li Y, Vallance BA, Finlay BB. 2001. Locus of enterocyte effacement from Citrobacter rodentium: sequence analysis and evidence for horizontal transfer among attaching and effacing pathogens. Infect. Immun. 69:6323–6335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schauer DB, Falkow S. 1993. Attaching and effacing locus of a Citrobacter freundii biotype that causes transmissible murine colonic hyperplasia. Infect. Immun. 61:2486–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Deng W, Puente JL, Gruenheid S, Li Y, Vallance BA, Vazquez A, Barba J, Ibarra JA, O'Donnell P, Metalnikov P, Ashman K, Lee S, Goode D, Pawson T, Finlay BB. 2004. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. U. S. A. 101:3597–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mundy R, MacDonald TT, Dougan G, Frankel G, Wiles S. 2005. Citrobacter rodentium of mice and man. Cell. Microbiol. 7:1697–1706 [DOI] [PubMed] [Google Scholar]
- 33. Frankel G, Phillips AD, Novakova M, Field H, Candy DC, Schauer DB, Douce G, Dougan G. 1996. Intimin from enteropathogenic Escherichia coli restores murine virulence to a Citrobacter rodentium eaeA mutant: induction of an immunoglobulin A response to intimin and EspB. Infect. Immun. 64:5315–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tauschek M, Yang J, Hocking D, Azzopardi K, Tan A, Hart E, Praszkier J, Robins-Browne RM. 2010. Transcriptional analysis of the grlRA virulence operon from Citrobacter rodentium. J. Bacteriol. 192:3722–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang J, Hart E, Tauschek M, Price GD, Hartland EL, Strugnell RA, Robins-Browne RM. 2008. Bicarbonate-mediated transcriptional activation of divergent operons by the virulence regulatory protein, RegA, from Citrobacter rodentium. Mol. Microbiol. 68:314–327 [DOI] [PubMed] [Google Scholar]
- 36. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chalker AF, Minehart HW, Hughes NJ, Koretke KK, Lonetto MA, Brinkman KK, Warren PV, Lupas A, Stanhope MJ, Brown JR, Hoffman PS. 2001. Systematic identification of selective essential genes in Helicobacter pylori by genome prioritization and allelic replacement mutagenesis. J. Bacteriol. 183:1259–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Smyth G. 2005. limma: linear models for microarray data, p 397–420 In Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S. (ed), Bioinformatics and computational biology solutions using R and Bioconductor. Springer, New York, NY [Google Scholar]
- 39. Smyth GK. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:Article 3 [DOI] [PubMed] [Google Scholar]
- 40. Smyth GK, Speed T. 2003. Normalization of cDNA microarray data. Methods 31:265–273 [DOI] [PubMed] [Google Scholar]
- 41. Yang J, Tauschek M, Strugnell R, Robins-Browne RM. 2005. The H-NS protein represses transcription of the eltAB operon, which encodes heat-labile enterotoxin in enterotoxigenic Escherichia coli, by binding to regions downstream of the promoter. Microbiology 151:1199–1208 [DOI] [PubMed] [Google Scholar]
- 42. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 43. Miller JH. 1974. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Plainview, NY [Google Scholar]
- 44. Adams LM, Simmons CP, Rezmann L, Strugnell RA, Robins-Browne RM. 1997. Identification and characterization of a K88- and CS31A-like operon of a rabbit enteropathogenic Escherichia coli strain which encodes fimbriae involved in the colonization of rabbit intestine. Infect. Immun. 65:5222–5230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Scaletsky IC, Michalski J, Torres AG, Dulguer MV, Kaper JB. 2005. Identification and characterization of the locus for diffuse adherence, which encodes a novel afimbrial adhesin found in atypical enteropathogenic Escherichia coli. Infect. Immun. 73:4753–4765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Edwards RA, Matlock BC, Heffernan BJ, Maloy SR. 2001. Genomic analysis and growth-phase-dependent regulation of the SEF14 fimbriae of Salmonella enterica serovar Enteritidis. Microbiology 147:2705–2715 [DOI] [PubMed] [Google Scholar]
- 47. Tan A, Yang J, Tauschek M, Praszkier J, Robins-Browne RM. 2011. Autogenous transcriptional regulation of the regA gene, encoding an AraC-like, essential virulence regulator in Citrobacter rodentium. J. Bacteriol. 193:1777–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhu C, Feng S, Thate TE, Kaper JB, Boedeker EC. 2006. Towards a vaccine for attaching/effacing Escherichia coli: a LEE encoded regulator (ler) mutant of rabbit enteropathogenic Escherichia coli is attenuated, immunogenic, and protects rabbits from lethal challenge with the wild-type virulent strain. Vaccine 24:3845–3855 [DOI] [PubMed] [Google Scholar]
- 49. Henderson IR, Navarro-Garcia F, Nataro JP. 1998. The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 6:370–378 [DOI] [PubMed] [Google Scholar]
- 50. Yen YT, Kostakioti M, Henderson IR, Stathopoulos C. 2008. Common themes and variations in serine protease autotransporters. Trends Microbiol. 16:370–379 [DOI] [PubMed] [Google Scholar]
- 51. Kobayashi RK, Gaziri LC, Vidotto MC. 2010. Functional activities of the Tsh protein from avian pathogenic Escherichia coli (APEC) strains. J. Vet. Sci. 11:315–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Skyberg JA, Johnson TJ, Johnson JR, Clabots C, Logue CM, Nolan LK. 2006. Acquisition of avian pathogenic Escherichia coli plasmids by a commensal E. coli isolate enhances its abilities to kill chicken embryos, grow in human urine, and colonize the murine kidney. Infect. Immun. 74:6287–6292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dozois CM, Dho-Moulin M, Bree A, Fairbrother JM, Desautels C, Curtiss R., 3rd 2000. Relationship between the Tsh autotransporter and pathogenicity of avian Escherichia coli and localization and analysis of the Tsh genetic region. Infect. Immun. 68:4145–4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Navarro-Garcia F, Canizalez-Roman A, Sui BQ, Nataro JP, Azamar Y. 2004. The serine protease motif of EspC from enteropathogenic Escherichia coli produces epithelial damage by a mechanism different from that of Pet toxin from enteroaggregative E. coli. Infect. Immun. 72:3609–3621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Xicohtencatl-Cortes J, Saldana Z, Deng W, Castaneda E, Freer E, Tarr PI, Finlay BB, Puente JL, Giron JA. 2010. Bacterial macroscopic rope-like fibers with cytopathic and adhesive properties. J. Biol. Chem. 285:32336–32342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Vidal JE, Navarro-Garcia F. 2008. EspC translocation into epithelial cells by enteropathogenic Escherichia coli requires a concerted participation of type V and III secretion systems. Cell Microbiol. 10:1975–1986 [DOI] [PubMed] [Google Scholar]
- 57. Ogunniyi AD, Kotlarski I, Morona R, Manning PA. 1997. Role of SefA subunit protein of SEF14 fimbriae in the pathogenesis of Salmonella enterica serovar Enteritidis. Infect. Immun. 65:708–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Edwards RA, Schifferli DM, Maloy SR. 2000. A role for Salmonella fimbriae in intraperitoneal infections. Proc. Natl. Acad. Sci. U. S. A. 97:1258–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Abuaita BH, Withey JH. 2009. Bicarbonate Induces Vibrio cholerae virulence gene expression by enhancing ToxT activity. Infect. Immun. 77:4111–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Morin N, Tirling C, Ivison SM, Kaur AP, Nataro JP, Steiner TS. 2010. Autoactivation of the AggR regulator of enteroaggregative Escherichia coli in vitro and in vivo. FEMS Immunol. Med. Microbiol. 58:344–355 [DOI] [PubMed] [Google Scholar]
- 61. Munson GP, Holcomb LG, Scott JR. 2001. Novel group of virulence activators within the AraC family that are not restricted to upstream binding sites. Infect. Immun. 69:186–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nunez MF, Pellicer MT, Badia J, Aguilar J, Baldoma L. 2001. The gene yghK linked to the glc operon of Escherichia coli encodes a permease for glycolate that is structurally and functionally similar to L-lactate permease. Microbiology 147:1069–1077 [DOI] [PubMed] [Google Scholar]
- 63. Krejany EO, Grant TH, Bennett-Wood V, Adams LM, Robins-Browne RM. 2000. Contribution of plasmid-encoded fimbriae and intimin to capacity of rabbit-specific enteropathogenic Escherichia coli to attach to and colonize rabbit intestine. Infect. Immun. 68:6472–6477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tennant SM, Tauschek M, Azzopardi K, Bigham A, Bennett-Wood V, Hartland EL, Qi W, Whittam TS, Robins-Browne RM. 2009. Characterisation of atypical enteropathogenic E. coli strains of clinical origin. BMC Microbiol. 9:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tobes R, Ramos JL. 2002. AraC-XylS database: a family of positive transcriptional regulators in bacteria. Nucleic Acids Res. 30:318–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Casadaban MJ. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541–555 [DOI] [PubMed] [Google Scholar]
- 67. Chang AC, Cohen SN. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Posfai G, Koob MD, Kirkpatrick HA, Blattner FR. 1997. Versatile insertion plasmids for targeted genome manipulations in bacteria: isolation, deletion, and rescue of the pathogenicity island LEE of the Escherichia coli O157:H7 genome. J. Bacteriol. 179:4426–4428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yang J, Baldi DL, Tauschek M, Strugnell RA, Robins-Browne RM. 2007. Transcriptional regulation of the yghJ-pppA-yghG-gspCDEFGHIJKLM cluster, encoding the type II secretion pathway in enterotoxigenic Escherichia coli. J. Bacteriol. 189:142–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.