Abstract
β-thalassemia is caused by mutations in the β-globin locus resulting in loss of, or reduced, hemoglobin A (adult hemoglobin, HbA, α2β2) production. Hydroxyurea treatment increases fetal γ-globin (fetal hemoglobin, HbF, α2γ2) expression in postnatal life substituting for the missing adult β-globin and is, therefore, an attractive therapeutic approach. Patients treated with hydroxyurea fall into three categories: i) ‘responders’ who increase hemoglobin to therapeutic levels; (ii) ‘moderate-responders’ who increase hemoglobin levels but still need transfusions at longer intervals; and (iii) ‘non-responders’ who do not reach adequate hemoglobin levels and remain transfusion-dependent. The mechanisms underlying these differential responses remain largely unclear. We generated RNA expression profiles from erythroblast progenitors of 8 responder and 8 non-responder β-thalassemia patients. These profiles revealed that hydroxyurea treatment induced differential expression of many genes in cells from non-responders while it had little impact on cells from responders. Part of the gene program up-regulated by hydroxyurea in non-responders was already highly expressed in responders before hydroxyurea treatment. Baseline HbF expression was low in non-responders, and hydroxyurea treatment induced significant cell death. We conclude that cells from responders have adapted well to constitutive stress conditions and display a propensity to proceed to the erythroid differentiation program.
Introduction
Hemoglobin disorders, particularly β-thalassemia and sickle cell disease (SCD), are the most common single gene disorders worldwide.1 They are caused by mutations in the β-globin locus resulting in abnormal or reduced rates of hemoglobin A (HbA) production. Clinical symptoms include anemia, infarction, bone marrow expansion and splenomegaly. The disease is lethal at a very early age, but patients receiving up to date treatment have a life expectancy of approximately four decades.2,3
In humans, fetal γ-globin and adult β-globin are the major β-like globins. They associate with α-globin chains to produce HbF (α2γ2) during the fetal period and HbA (α2β2) in adult life. This developmentally regulated globin gene expression pattern, known as globin switching, has been the subject of intense research during the last 30 years, mainly because reactivation of γ-globin expression would be beneficial to β-hemoglobinopathy patients. In β-thalassemia patients, γ-globin expression can reduce α-globin chain precipitation and compensate for the lack of β-globin chains through the formation of HbF. In SCD patients, high HbF reduces hemoglobin polymerization. This prevents sickling and improves the life span of the erythrocytes, thereby ameliorating disease symptoms.4
Several drugs can induce γ-globin gene expression resulting in increased HbF production and amelioration of the disease. Three well-known HbF-inducing agents are sodium butyrate (a histone deacetylase inhibitor),5 5-azacytidine (a DNA demethylating agent)6 and hydroxyurea (a ribonucleotide reductase inhibitor).7 Hydroxyurea (HU) is FDA approved for treatment of SCD patients and it is also widely used for β-thalassemia.8–12 How HU induces HbF production is poorly understood. Mechanisms proposed for the induction of HbF by HU include rapid erythroid regeneration, increased erythropoietin (EPO) production, apoptosis, nitric oxide (NO) production,13 increased guanylate cyclase activity13 and activation of the p38 MAPK pathway.14 Induction of HbF by HU in β-thalassemia patients was reported to be of similar magnitude as found in the cells of normal individuals (1.3- to 3.5-fold) and SCD patients (2- to 5-fold). In erythroid progenitor cells treated with HU in vitro, HbF induction was comparable to the increase of HbF in peripheral blood of SCD patients following HU therapy in vivo.15 The majority of patients increase HbF production upon HU treatment.4 However, HbF baseline and response magnitude among the patients is highly variable. The absolute HU response and the HbF baseline is likely dependent on genetic factors that modulate different regulatory pathways and trans-acting factors involved in γ-globin production.
Correlation of multiple single nucleotide polymorphisms (SNPs) with high HbF baseline has been reported in several studies.4 The XmnI (G)γ SNP at −158 (C>T) for instance is associated with high HbF baseline in SCD and β-thalassemia patients.16 The correlation between gene mutations and clinical response to HU has been investigated in many articles. While the β-globin mutations are not associated with HbF response to HU therapy, the Xmn I polymorphism has yielded inconsistent results. A SNP association study reported multiple significantly associated SNPs with HbF response to HU.10–12,17–21 However, a recent study in a large group of SCD children evaluated the effect of previously reported pharmacogenetic variables on HU response and found no significant association between any β-globin haplotypes (including XmnI) with HbF maximum HU tolerated dose after adjustment for HbF baseline.22 A total of 23 previously reported SNPs17 for high HbF baseline were also examined for their association with HU pharmacodynamics variables and only 2 SNPs (in the ARG1 and ARG2 genes) were found to be significantly associated with the change in HbF and maximum HU tolerated dose.22
Regulation of γ-globin gene expression is complex and can be influenced by different regulatory pathways, genetic- and environmental factors. The net outcome of these determines the response to HU.4 We hypothesized that some of the regulatory mechanisms may be deduced from the comparison of the expression profiles of cultured human erythroid progenitor cells (HEP) derived from HU ‘responder’ (R-HEP) and ‘non-responder’ (NR-HEP) patients. In addition, data from such an approach may help to understand the mechanism by which HU induces γ-globin expression. It may also help explaining the difference between ‘responders’ (R) and ‘non-responders’ (NR) regarding baseline levels of HbF and factors that are involved in γ-globin induction. Only a few such studies have been reported. One study reported expression profiling of peripheral blood mononuclear cells to characterize the role of circulating leukocytes in sickle cell pathogenesis.23 The expression profile of these cells is, therefore, not informative for the HbF response to HU in erythroid progenitor cells. Other studies on the effect of HU in SCD patients show decreased expression of adhesion molecules in vascular endothelial and red cells,24–25 its stimulating effect on proinflammatory gene expression,26 and several other pathways.27–28 To our knowledge, none of these studies address the differential HU response in β-thalassemia. Here we studied two groups of β-thalassemia patients: those who did not respond sufficiently to HU treatment and remained fully dependent on regular blood transfusions and those who become transfusion-independent upon HU treatment. We expanded HEP cells from peripheral blood of tesponder and non-responder patients and compared their proliferation, hemoglobin production and gene expression profiles in the presence and absence of HU. Our data shows that R-HEPs showed a relatively high baseline level of HbF compared to that of NR-HEPs. NR-HEPs change the expression pattern of a large number of genes upon HU treatment, while R-HEPs showed only minor changes after treatment. Differential gene expression profiles of these two groups indicated that high HbF was associated with a continuously activated stress response, and with genes that protect from stress-induced apoptosis. This suggests that HU is effective in R patients because their baseline HbF levels are relatively high and their erythroblasts have already activated a stress response program that protects them from the cytotoxic effects of HU. Moreover, R-HEPs displayed a propensity to proceed to terminal erythroid differentiation.
Design and Methods
Patients
The studies reported here were approved by the local medical ethical review committee and written informed consent was obtained from all the participating patients or their guardians. A large collection of β-thalassemic patients with milder phenotype, higher Hb level, less or no skeletal deformity, receiving a similar amount of blood transfusions on regular bases were monitored for their clinical manifestation and response to hydroxyurea (HU) treatment. Transfusions were temporarily stopped at the start point of HU treatment, and hematologic indexes were analyzed at different time points. Detailed clinical data, molecular analysis, hematologic indexes, transfusion and treatments are provided in the Online Supplementary Design and Methods and Online Supplementary Table S1. Two distinct groups were defined based on their response to HU: ‘responders (R)’ with a good response to HU treatment resulting in maintaining the mean hemoglobin level up to 8.5 g/dl (our threshold for transfusion) resulting in transfusion independency. ‘Non-responders (NR)’ with a poor response to HU treatment showing a mean Hb of less than 8.5 g/dl. This group of patients developed side effects to HU therapy, had discomfort using the drug, and no or very little change in transfusion intervals during the treatment period. Peripheral blood was collected from the patients at the same time for culturing of erythroid progenitors used for subsequent studies.
Mutations in the α- and β-globin genes and the −158 XmnI Gγ-globin gene (C>T SNP) were determined as described previously.29,30 Since positive correlation between α-globin deletions and good response to HU therapy is reported,12,19,31 patients with mutated or deleted α-globin alleles were excluded from the study. Although a high HbF base line is correlated with the XmnI polymorphism as well other SNPs, HbF response to HU therapy does not seem to be correlated with the Xmn I polymorphism as well as β-globin gene mutation.10,18–19 We, therefore, did not bias our study for these variables. We selected 8 patients from each group who comply with the above mentioned criteria for microarray analysis.
Hematologic analysis
A complete blood count was carried out on a hematology counter and standard methods were used for peripheral blood smear examination and reticulocyte count. The variant hemoglobin testing system was used for hemoglobin analysis.
Cell culture
HEPs were cultured essentially as published32 (see Online Supplementary Design and Methods for details). The cultures were expanded until Day 10 resulting in a homogenous population of erythroid progenitor cells.33 After Day 10, the cells were divided into control and HU treated (100 μM/mL HU) and the cultures were continued. RNA was isolated 48 h after HU treatment; the remaining cells were kept in culture for another three days to determine total hemoglobin, HbF and growth rates.
Cell morphology
Cell morphology was analyzed using cytospins stained with histological dyes and neutral benzidine.34 Pictures were taken with an Olympus BX40 microscope (40× objective, NA 0.65) equipped with an Olympus DP50 CCD camera and Viewfinder Lite 1.0 acquisition software.
Hemoglobin content
Aliquots of approximately 2×106 cells of the original cultures were removed and analyzed for hemoglobin content by photometry as described.35 The relative ratios of HbA, HbA2 and HbF were determined by HPLC (BioRad, Hercules, CA, USA).
RNA purification and quantitative RT-PCR analysis
Total RNA was extracted from cells using the TRI reagent (Sigma). For quantitative real-time polymerase chain reaction (RT-PCR) cDNA was synthesized from 1 μg of total RNA using random hexamers and Superscript II (Invitrogen, Carlsbad, CA, USA). RNase-free DNaseI (Invitrogen) was used to degrade contaminating DNA and primers were designed spanning an intron. Quantitative PCR was performed as described in the Online Supplementary Design and Methods.
Statistical analysis
P values were calculated by the Mann-Whitney method and ANOVA with Bonferroni’s correction using Stata11.0 (Stata Corp, College Station, TX, USA). At least 5 independent biological samples were analyzed in triplicate in each group.
Affymetrix microarrays
HEPs were lysed using the TRIzol Reagent (Invitrogen) and RNA was isolated. Initial RNA yield and quality of the labeled fragmented cRNA were determined using the 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). A total of 5 μg of cRNA was hybridized to U133 Plus 2.0 arrays, according to the manufacturer's protocols (Affymetrix, Santa Clara, CA, USA). Additional technical details and data analysis are described in the Online Supplementary Design and Methods.
Results
HU induces hemoglobinization and reduces cell proliferation
Human erythroid progenitor cells (HEPs) were expanded from peripheral blood mononuclear cells as described.32,33 We first titrated HU treatment to determine the concentration-dependent effects on cell survival, proliferation, HbA and HbF accumulation. We used cells from 2 unrelated healthy donors for these experiments. HEPs were expanded from peripheral blood mononuclear cells for ten days. HU was added at Day 10 in concentrations ranging from 0 to 400 μM and cell proliferation was monitored daily. Hemoglobin production, the percentage of HbA and HbF and cell morphology were analyzed after 18 days when the experiment was terminated (Online Supplementary Figure S1). Cell density was maintained between 1–2 million/mL by daily dilution and cumulative cell numbers were calculated. Increasing HU concentrations progressively decreased cell proliferation. Only 400 μM HU was immediately toxic to the cells (Online Supplementary Figure S1A). HU induced a concentration-dependent increase in total hemoglobin that reached a maximum at 100 μM HU (Online Supplementary Figure S1B). Analysis of hemoglobin subtypes by HPLC indicated that HU increased the HbF percentage concentration-dependent from 6% in non-HU treated to 9.7% in 200 μM HU-treated samples (Online Supplementary Figure S1C). Using total Hb levels and relative ratios of HbA, HbA2 and HbF, the accumulation of different hemoglobins can be calculated (Online Supplementary Figure S1D). This shows that both HbA and HbF were induced by HU. Finally, we analyzed hemoglobinization at the cellular level. Under proliferation conditions, addition of HU increased the percentage of hemoglobinized cells from 30% in control cells to 40% with 50 μM HU and up to 50% with 200 μM HU in the cultures (Online Supplementary Figure S1E). Based on these observations, and in agreement with previous publications,36–37 we conclude that HU induces erythroid differentiation and hemoglobinization. Since the best response with least cell toxicity was observed with 100 μM HU in the cultures, we used these conditions for all other experiments reported here. The selection of two groups of β-thalassemia patients was based on either the complete absence of response to HU treatment (NR) or a good response to HU treatment resulting in transfusion-independence (R). The XmnI (G)γ SNP (−158C>T) that has been associated with increased HbF baseline expression is present in 19% of the NR and in 69% of the R patient chromosomes. All of the patients were screened for α- and β-globin gene mutations. β-globin mutations were detected in all cases and no mutations in the α-globin genes were present in the selected patients (Online Supplementary Table S1).
Erythroblasts derived from responders are less sensitive to HU treatment and express higher HbF at baseline
First, we examined how HU sensitivity at the cellular level corresponds to the response of β-thalassemic patients. HEPs were expanded from blood mononuclear cells of the 16 selected β-thalassemic patients (Online Supplementary Table S1). After ten days of culture, the HEPs were further expanded in the presence or absence of 100 μM HU. Cell numbers were monitored daily. The cells displayed similar size distributions, indicative of similar differentiation stages. This was confirmed by the similar expression of differentiation markers (see below). Total hemoglobin production, ratio of hemoglobin types, and cell morphology were analyzed at Day 15 when the cultures were terminated. The first difference observed between cultures derived from NR-HEPs and R-HEPs was the relatively poor growth rate of R-HEPs (38.7×106vs. 10.6×106 cells, respectively) (Figure 1A and B and Online Supplementary Figure S2). Although proliferation of both NR- and R-HEPs was inhibited by HU (declining to 7.6×106vs. 4.8×106 cells, respectively) (Figure 1A-C), the growth rate of R-HEP cultures was affected much less than that of the NR-HEP cultures. We conclude that NR-HEPs are much more sensitive to HU treatment.
Figure 1.
Erythroblasts derived from responders are less sensitive to HU treatment and express higher HbF at baseline. NR-HEPs (A) grow faster than R-HEPs (B). After HU treatment, growth curves of NR-HEPs (A) decline more than those of R-HEPs (B). (C) HU sensitivity curves for NR-HEPs and R-HEPs, normalized by taking the ratio of cell proliferation of HU-treated cells over non-treated cells. (D) Hemoglobin induction by 100 μM HU (5 days) in NR-HEPs and R-HEPs; as in (C). (A-D) P<0.05 are indicated. (E) Representative cytospins of NR-HEPs and R-HEPs treated with HU for three days. Cytospins were stained with histological dyes and neutral benzidine. Hemoglobinized cells are stained brown. Pyknotic cells are indicated by arrows.
Total hemoglobin (in arbitrary units, a.u.) and the ratios of fetal (HbF) versus adult (HbA) hemoglobin expression were measured five days after the start of HU treatment (Day 15 of culture) and the absolute distribution of different hemoglobins was calculated (Figure 1D). HU treatment slightly altered the low expression of adult HbA in these thalassemic cells, while the expression of HbF increased significantly (NR from 8 to 33 a.u., R from 106 to 248 a.u.). Since the basal level of total hemoglobin is much higher in R-HEPs, these cells express the highest HbF levels upon HU treatment (Figure 1D). Analysis of HEPs treated with HU for three days, stained for hemoglobin in combination with histological dyes,34 showed that R-HEP cultures accumulated more hemoglobinized cells after HU treatment (25% in R-HEPs vs. 10% in NR-HEPs; Figure 1E). Cultures of NR-HEPs accumulated more pyknotic cells (20% in NR-HEPs vs. 10% in R-HEPs; Figure 1E).
In conclusion, when compared to R-HEPs, NR-HEPs are more likely to succumb to cell death in response to HU. Furthermore, although the fold-change in HbF levels upon HU treatment is higher in NR-HEPs than that observed in R-HEPs, the increase from the low baseline levels of HbF in NR-HEPs does not yield adequate HbF levels in these cells.
Responder HEPs constitutively express a stress-program that is induced by HU in non-responder HEPs
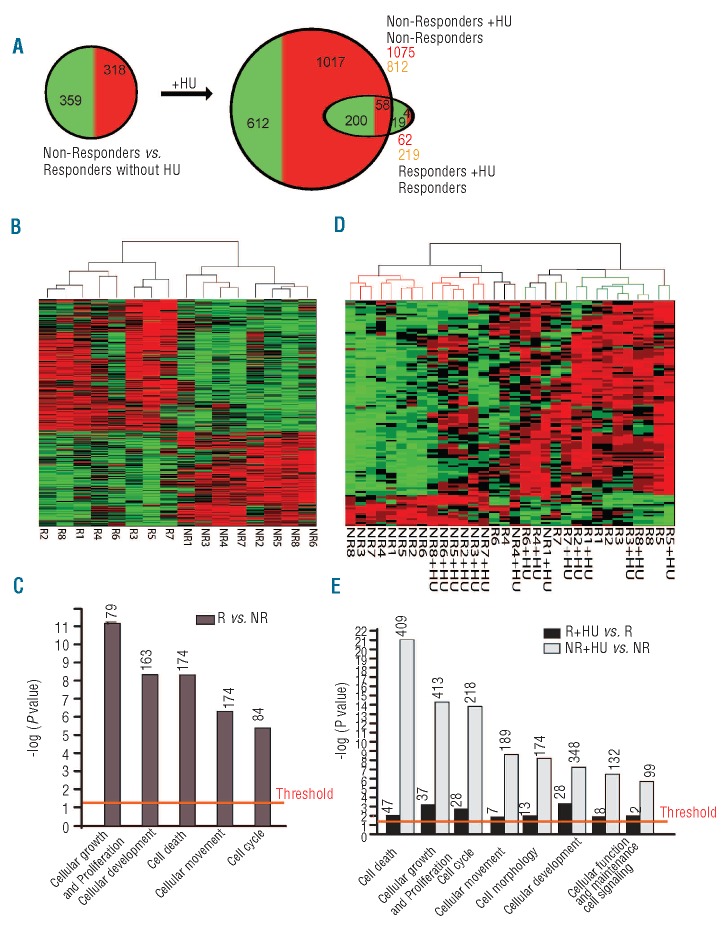
In order to carry out gene expression profiling analysis, HEPs were expanded for ten days and subsequently treated with 100 μM HU or solvent for two days. RNA expression profiles were compared between 8 R-HEP and 8 NR-HEP cultures. The expression profiles of the samples were compared using the SAM algorithm38 to identify HU response-associated genes. While the expression of CD71 and GPA were at similar levels indicative of similar stages of differentiation in all samples, comparison of the expression profiles between NR- and R-HEPs showed numerous differentially expressed genes in the absence of HU (677 genes, 1105 differentially expressed probe sets; Figure 2A and B). A striking difference between the NR and R profiles is their response to HU at the gene expression level. HU induced many changes in gene expression of NR-HEPs (1887 genes, 2664 differentially expressed probe sets) whereas far fewer changes were observed in R-HEPs (281 genes, 320 differentially expressed probe sets), the majority of which were shared with NR-HEPs (258 out of 281 genes; Figure 2A). Complete lists of probe sets are provided in Online Supplementary Table S2.
Figure 2.
Responder HEPs constitutively express a stress-program that is induced by HU in non-responders HEPs. (A) The number of differentially expressed genes between NR-and R-HEPs before and after HU treatment. (B and C) Supervised clustering of differentially expressed genes without (B) and with (C) HU treatment. Responder: R; non-responder: NR; HU treated: +HU. (D) Functional annotation of differentially expressed genes in R- versus NR-HEPs before HU treatment. (E) Functional annotation of differentially expressed genes in R- versus NR-HEPs after HU treatment. Functional annotation for biological processes was processed by Ingenuity pathway analysis (P< 0.05).
The expression data were used for cluster analysis clearly separated R-HEPs from NR-HEPs (Figure 2B). Differentially expressed gene enrichment analysis indicated that genes involved in cellular proliferation and apoptosis were already differentially expressed in R-HEPs compared to NR-HEPs before HU treatment (Figure 2C). The untreated NR samples clustered separately from those treated with HU. In contrast, the R samples clustered independent of HU treatment. HU treatment barely affected the gene expression profiles in samples R1, R3, R5, R7 and R8, and only moderately in samples R2, R4 and R6 (Figure 2D). Interestingly, upon HU treatment, the NR-HEP samples clustered more closely to the R-HEP samples (Figure 2D). Thus, while HU treatment had a major effect on the transcriptome of NR-HEPs, it caused much less change in that of R-HEPs. Differentially expressed gene enrichment analysis between the two groups indicated that HU treatment caused differential expression of genes involved in apoptosis and cell cycle regulation in NR-HEPs (Figure 2C and E). Collectively, our data show that R-HEPs are adopted to constitutively express a stress response program that is activated in NR-HEPs upon exposure to HU.
The INK4b-ARF-INK4a locus is differentially regulated between responders and non-responders
The INK4b-ARF-INK4a locus is known to be involved in hematopoietic differentiation, proliferation, and stress response.39–41 The probe sets on the microarrays do not discriminate between the overlapping ORFs of p14ARF and p16INK4a. Therefore, we analyzed expression of p15INK4b, p14ARF and p16INK4a, encoded by the INK4b-ARF-INK4a locus, in more detail (Online Supplementary Figure S3A). Expression of p16INK4a was increased upon HU treatment in both NR- and R-HEPs. Expression of p14ARF was at average 10-fold increased in NR-HEPs when compared to R-HEPs independent of HU-treatment. These data suggest that the effect of HU on the proliferation rate would be different between the two groups of patients.
Stress response genes
From the array data we also selected a number of genes with a role in the adaptation to stress that were consistently differentially expressed between NR- and R-HEPs (Online Supplementary Figure S3 and Table S3). Expression of these genes was analyzed using qRT-PCR. Forkhead box O3 (FOXO3) is a transcription factor inducing genes that enforce the oxidative firewall. Arginase 1 and 2 (ARG1, ARG2) compete with NO synthase (NOS) for the substrate L-arginine,42 and thereby protect against oxidative stress from NO. Homeodomain interacting protein kinase 2 (HIPK2) is involved in apoptosis, differentiation and also activation of CBP/P300.43,44 ARG2 and HIPK2 are known FOXO3 target genes.45 These genes were all consistently up-regulated in R-HEPs compared to NR-HEPs, and further up-regulated upon treatment with HU in both groups (Online Supplementary Figure S3B). Notably, expression levels of ARG1 and ARG2 were higher in the untreated R-HEP cultures than in HU-treated NR-HEP cultures and may have a role in protecting R-HEPs from HU-induced cell death. Kruppel like factor 10 (KLF10) was identified as a protein that protects stromal cells and acute lymphoblastic leukemia blasts against chemotherapy.46 KLF10 was expressed at elevated levels in R-HEPs compared to NR-HEPs, and reached similar levels in both groups upon HU treatment (Online Supplementary Figure S3D).
Apoptosis genes
BCLXL (BCL2L1) protects erythroblasts from apoptosis. Both in the absence and presence of HU, BCLXL levels were higher in R-HEPs. BCLXL is under control of STAT5B47 which was decreased in NR-HEPs upon HU treatment, although the difference was not as marked as that observed for BCLXL (Online Supplementary Figure S3C and data not shown). Whether BCL6 is directly involved in apoptosis is currently not clear. During VDJ rearrangement in B cells it is responsible for methylation of the ATR gene and thus prevents the activation of the DNA damage response during the rearrangement process.48 Expression of BCL6 is not increased by HU in both groups, but it is expressed almost 10-fold higher in R-HEPs than in NR-HEPs (Online Supplementary Figure S3C and Online Supplementary Table S3).
γ-globin expression and erythroid maturation
Whereas stress erythropoiesis induces γ-globin expression,49–51 several transcription factors have been reported to modify γ-globin expression, including KLF1, BCL11A and SOX6.52 Expression levels of SOX6 are approximately 3.5-fold higher in R-HEPs compared to NR-HEPs, whereas expression of SOX4 was slightly less (Online Supplementary Figure S3D and Online Supplementary Table S3). No significant differences were seen in BCL11A expression between the two groups. Expression of the γ-globin gene itself at the transcript level was 4-fold higher in R-HEPs compared to NR-HEPs before HU treatment (Online Supplementary Figure S3D). Finally genes important to maintain expansion of erythroblasts such as MYB,53 PRMT5,54 IKAROS55 and STAT56 remained constant (data not shown).
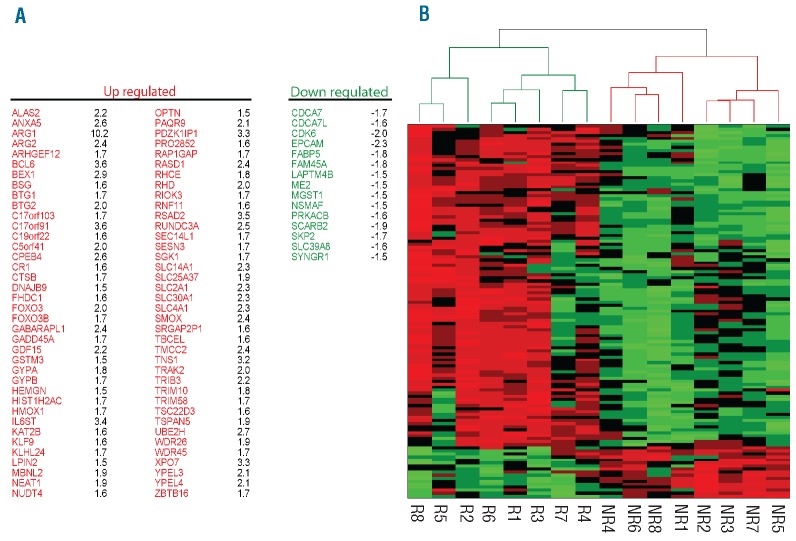
Merryweather-Clarke et al. reported global gene expression analysis of human erythroid progenitors.57 Comparison of differentially expressed genes associated with terminal erythroid differentiation (Online Supplementary Table S3)57 with the list of genes differentially expressed between untreated R-HEPs and NR-HEPs revealed a considerable number of overlapping genes (28%; 91 of 327 genes). Interestingly, the expression dynamics of these genes in R-HEPs was favoring terminal erythroid differentiation (Figure 3A). Clustering analysis with these 91 genes clustered R-HEPs and NR-HEPs in two distinct groups (Figure 3B). Collectively, these data are consistent with the notion that compared to NR-HEPs, R-HEPs have an intrinsically activated terminal erythroid differentiation program.
Figure 3.
Expression dynamics of substantial numbers of differentially expressed genes in responder HEPs are towards terminal erythroid differentiation. (A) Overlap of differentially expressed genes in R-HEPs (R) versus NR-HEPs (NR), with genes involved in the erythroid differentiation program (28%; 91 of 327).57 (B) Clustering analysis of genes involved in the erythroid differentiation program that are also differentially expressed between R and NR-HEPs.
Discussion
Increased γ-globin gene expression ameliorates β-thalassemia or SCD symptoms. However, γ-globin repression is well controlled and its pharmacological alleviation is difficult to achieve. Proliferative stress, such as induced by HU, can increase γ-globin expression, but not all patients increase HbF to a therapeutic level that decreases their dependency on regular transfusions. Here we show that cultured cells from patients that respond or do not respond to HU treatment display differential gene expression profiles already before HU treatment. We conclude from these data that cells from responders as opposed to non-responders have adapted to constitutive stress conditions and display a propensity to proceed to the erythroid differentiation program. We based these conclusions after characterization of the proliferation, differentiation kinetics and gene expression profiles of erythroblasts expanded from β-thalassemia patients at the extremes of clinical response to HU treatment. The subjects were selected from a large cohort of patients. All selected patients were on regular transfusion according to their age and body weight (Online Supplementary Table S1). The trial of the HU treatment protocol was over a period of 2–6 months at a dose of 5–20 mg/kg bodyweight per day with the aim of decreasing transfusion dependency and clinical severity of the disease. Two groups of patients were defined based on hemoglobin level and transfusion dependency after HU treatment protocol. At this stage, blood samples were drawn from all patients and their HEP cells expanded for ten days followed by 0 and 100 μM HU for an additional five days to compare proliferation, hemoglobin production and gene expression profiles between the two groups. When compared to R-HEPs, we observed that NR-HEPs were more sensitive to HU treatment and that their initial HbF levels were lower. HU treatment of NR-HEPs increased expression of a number of genes involved in cellular proliferation and apoptosis pathways, which is likely responsible for the observed increase in cell death. They also expressed more HbF in response to HU treatment. The fold-change in HbF is higher in NR-HEPs than in R-HEPs, but the final HbF levels remain lower in NR-HEPs due to their lower starting levels. The R-HEPs express relatively high initial γ-globin levels, and this increases further upon HU treatment. We observed that R-HEPs and NR-HEPs already display significantly different gene expression profiles at the start of the cultures. In contrast to the NR-HEPs, HU treatment had a relatively minor impact on the global gene expression profiles of R-HEPs. Based on these observations we suggest that in the cultured cells the responder phenotype is associated with gene expression signatures of terminal erythroid differentiation, an activated stress response and protection from stress-induced apoptosis. We suggest that these differential gene expression profiles could potentially be used to distinguish responders from non-responders in β-thalassemia patients who have not yet received HU treatment. Whether such a test could be performed directly on RNA isolated from peripheral blood, or requires derivation of HEP cultures first, must still be determined.
Factors regulating γ-globin expression
The −158 XmnI SNP in the promoter of the γ-globin gene has been linked to HbF expression.16 In our patient cohort, 11 of 16 (69% of chromosomes) of the responders carried the −158 XmnI C>T SNP. Among the non-responders, only 3 of 16 (19% of chromosomes) carried this SNP. Although there is a correlation between the presence of −158 XmnI SNP and base line HbF expression this SNP does not solely determine HbF and HU response in this patient group. It is very important to note that the most significant reported SNPs are correlated with high HbF base line while HbF responsiveness to HU is probably mediated by additional mechanisms that protect hematopoietic cells against stress-induced apoptosis and drives them to terminal erythroid differentiation.
The array data reveal more genes differentially expressed in R versus NR that may have an effect on the regulation of HbF, such as ID1 and HHEX (Online Supplementary Table S3). We note that many genes involved in erythroid differentiation are expressed at higher levels in R-HEPs (Figure 3).57 This suggests that R-HEPS have a propensity to enter the terminal differentiation pathway while NR-HEPs maintain a proliferative state. This difference may explain decreased survival of NR-HEPs upon HU treatment, whilst R-HEPs survive by undergoing terminal differentiation.
The role of stress factors in γ-globin expression
The low proliferation rate of the R-HEPs and their expression profiles suggest that these cells have adapted to permanent stress conditions. This is supported by the observation that HU treatment did not have a major impact on gene expression in the R samples, while 2664 probe sets were differentially expressed in response to HU in the NR samples. The most striking observation from the expression profiles is the expression of stress proteins in R-HEPs. An example validated by qRT-PCR is expression of FOXO3 (Online Supplementary Figure S3B and Online Supplementary Table S3). FOXO3 is up-regulated during erythroid differentiation35 and in response to various types of stress such as ROS and DNA damage.58 The increased FOXO3 expression in R-HEPs compared to NR-HEPs and the further upregulation in response to HU indicate increased levels of cellular stress. A number of FOXO3 target genes are also up-regulated, for example HIPK2 and BTG1 (Online Supplementary Table S3).45 Interestingly, the function of stress-induced FOXO3 protein is not to induce cell death, but to increase the potential of cells to prevent and repair oxidative damage and to slow down the cell cycle to allow DNA repair before replication.
The INK4b-ARF-INK4a locus is a master regulator of cellular senescence, differentiation and apoptosis programs governed by the Rb and p53 signaling networks. Genes of this locus are epigenetically silenced in hematopoietic stem cells but become poised for transcription as blood cells differentiate.41 Upregulation of p15INK4b and p16INK4a, but not p14ARF, causes G1 arrests followed by differentiation of hematopoietic progenitor cells.39,40 p14ARF is involved in activation of the p53 apoptotic pathway.41 Thus we propose that the increased expression of p16INK4a upon HU treatment drives the R-HEPs and NR-HEPs towards differentiation, but the relatively high expression of p14ARF in NR-HEPs, as well as other components of the p53 mediated apoptosis pathway such as CDKN1A and MDM2 (Online Supplementary Figure S3A and Online Supplementary Table S3), results in apoptosis rather than terminal differentiation. These two differential HU effects, one due to increased apoptosis and the other due to more differentiation, result in similar proliferation rates of NR-HEPs and R-HEPs (7.6×106vs. 4.8×106 cells, respectively, Figure 1A and B).
HU is a ribonucleotide reductase inhibitor that stalls cells in S phase.59 Stalled replication forks are potent inducers of senescence or apoptosis, but possibly also of HbF. Defects in the Fanconi anemia pathway result in increased replication fork stalling. It is therefore interesting to note that Fanconi anemia is associated with high HbF levels.60 Importantly, a stalled replication fork activates ATR kinase, which is crucial to all downstream events. Increased expression of BCLXL (BCL2L1, Online Supplementary Table S3) could also contribute to enhanced survival of R-HEPs in the presence of HU. In normal erythroid progenitors BCLXL is induced by EPO to maintain viability of erythroid cells during terminal maturation.61
ARG 1 and ARG 2
HU increases NO production through phosphorylation and activation of nitric oxide synthetase (NOS).62 Although NO has been implicated in upregulation of HbF through activation of γ-globin expression,63 NO also inhibits growth of erythroid primary cells and colony cultures.64 Arginase hydrolyzes L-arginine to urea and L-ornithine in the urea cycle and inhibits nitric oxide (NO) production via competition with NOS for the substrate L-arginine.42 During erythroid differentiation NO levels decrease significantly.65 NO is inhibited more by fetal RBCs when compared to adult RBCs suggesting that fetal RBCs have a higher level of NO scavengers.66 On the other hand, high NO concentrations promote apoptosis, while low NO concentrations result in resistance to apoptosis.
The Arginase 1, ARG1, Arginase 2, ARG2, and argininosuccinate synthetase 1, ASS1 genes are differentially expressed between NR-HEPs and R-HEPs. Their expression is 10.2-, 2.4- and 1.9-fold increased in R-HEPs compared to NR-HEPs (Online Supplementary Figure S3B and Online Supplementary Table S3). In an SNP association study, ARG1/ARG2, ASS1, NOS1 and NOS2A were reported to be significantly associated with response to HU treatment.17 Moreover, ARG1 and ARG2 SNPs were also found to be significantly associated with the change in HbF and maximum HU tolerated dose in SCD patients.22 This suggests that high expression of ARG1, ARG2 and ASS1 protects hematopoietic progenitor cells against excessive amounts of NO after HU treatment to prevent apoptosis, and scavenge the extra NO at later stages of differentiation.
In conclusion, the biological and molecular analysis of erythroblast cultures of β-thalassemia patients suggests that several mechanisms are involved in high HbF expression and HU responsiveness. Although the basal HbF level is correlated with polymorphisms such as the XmnI (Gγ) SNP (−158C>T), cell survival, stress response and propensity towards terminal erythroid differentiation are also playing important roles in β-thalassemia patients HU response. R-HEPs, compared to NR-HEPs, proliferate at a slower rate, are intrinsically poised to undergo terminal erythroid differentiation, and have a constitutively activated stress response. Thus, our data indicate that drugs activating the stress response may increase HbF, provided that cell survival and differentiation are not compromised.
Acknowledgments
The authors would like to acknowledge the patients and their families for their contribution to this study, Dr. Yuri Mochkine from the Department of Biochemistry, Erasmus MC, Rotterdam and Dr. George Garinis from the Department of Genetics, Erasmus MC, Rotterdam for helpful advice on microarray analysis, Kirsten van Lom from the Department of Hematology, Erasmus MC, Rotterdam for HPLC analysis of hemoglobin subtypes, and Teus van Gent from the Department of Cell Biology, Erasmus MC, Rotterdam for statistical analysis.
Footnotes
The online version of this article has a Supplementary Appendix.
Funding
The work was supported by the NIH (RO1 HL 73455), NGI, NWO (ZonMW 912-07-019) and Landsteiner Foundation for Blood Transfusion Research (1040).
Authorship and Disclosures
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Weatherall DJ, Clegg JB, Higgs DR, Wood WG. The hemoglobinopathies. In: Scriver CR, ed. The Metabolic and Molecular Bases of Inherited Disease. New York, NY: McGraw-Hill; 2001:4571–636 [Google Scholar]
- 2.Borgna-Pignatti C, Gamberini MR. Complications of thalassemia major and their treatment. Expert Rev Hematol. 2011; 4(3):353–66 [DOI] [PubMed] [Google Scholar]
- 3.Borgna-Pignatti C, Rugolotto S, De Stefano P, Zhao H, Cappellini MD, Del Vecchio GC, et al. Survival and complications in patients with thalassemia major treated with transfusion and deferoxamine. Haematologica. 2004;89(10):1187–93 [PubMed] [Google Scholar]
- 4.Akinsheye I, Alsultan A, Solovieff N, Ngo D, Baldwin CT, Sebastiani P, et al. Fetal hemoglobin in sickle cell anemia. Blood. 2011;118(1):19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perrine SP, Miller BA, Greene MF, Cohen RA, Cook N, Shackleton C, et al. Butryic acid analogues augment gamma globin gene expression in neonatal erythroid progenitors. Bioche Biophys Res Commun. 1987;148(2):694–700 [DOI] [PubMed] [Google Scholar]
- 6.Ley TJ, DeSimone J, Anagnou NP, Keller GH, Humphries RK, Turner PH, et al. 5-azacytidine selectively increases γ-globin synthesis in a patient with β thalassemia. N Engl J Med. 1982;307(24):1469–75 [DOI] [PubMed] [Google Scholar]
- 7.Veith R, Galanello R, Papayannopoulou T, Stamatoyannopoulos G. Stimulation of F-cell production in patients with sickle-cell anemia treated with cytarabine or hydroxyurea. N Engl J Med. 1985;313(25):1571–5 [DOI] [PubMed] [Google Scholar]
- 8.Dixit A, Chatterjee TC, Mishra P, Choudhry DR, Mahapatra M, Tyagi S, et al. Hydroxyurea in thalassemia intermedia--a promising therapy. Ann Hematol. 2005; 84(7):441–6 [DOI] [PubMed] [Google Scholar]
- 9.Ansari SH, Shamsi TS, Ashraf M, Perveen K, Farzana T, Bohray M, et al. Efficacy of hydroxyurea in providing transfusion independence in beta-thalassemia. J Pediatr Hematol Oncol. 2011;33(5):339–43 [DOI] [PubMed] [Google Scholar]
- 10.Karimi M, Haghpanah S, Farhadi A, Yavarian M. Genotype-phenotype relationship of patients with beta-thalassemia taking hydroxyurea: a 13-year experience in Iran. Int J Hematol. 2012;95(1):51–6 [DOI] [PubMed] [Google Scholar]
- 11.Koren A, Levin C, Dgany O, Kransnov T, Elhasid R, Zalman L, et al. Response to hydroxyurea therapy in β-thalassemia. Am J Hematol. 2008;83(5):366–70 [DOI] [PubMed] [Google Scholar]
- 12.Italia KY, Jijina FJ, Merchant R, Panjwani S, Nadkarni AH, Sawant PM, et al. Response to hydroxyurea in β thalassemia major and intermedia: experience in western India. Clin Chim Acta. 2009;407(1–2):10–5 [DOI] [PubMed] [Google Scholar]
- 13.Cokic VP, Andric SA, Stojilkovic SS, Noguchi CT, Schechter AN. Hydroxyurea nitrosylates and activates soluble guanylyl cyclase in human erythroid cells. Blood. 2008;111(3):1117–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park JI, Choi HS, Jeong JS, Han JY, Kim IH. Involvement of p38 Kinase in Hydroxyurea-induced Differentiation of K562 Cells. Cell Growth Differ. 2001; 12(9):481–6 [PubMed] [Google Scholar]
- 15.Yang YM, Pace B, Kitchens D, Ballas SK, Shah A, Baliga BS. BFU-E colony growth in response to hydroxyurea: Correlation between in vitro and in vivo fetal hemoglobin induction. Am J Hematol. 1997; 56(4):252–8 [DOI] [PubMed] [Google Scholar]
- 16.Panigrahi I, Marwaha RK, Kulkarni K. The expanding spectrum of thalassemia intermedia. Hematology. 2009;14(6):311–4 [DOI] [PubMed] [Google Scholar]
- 17.Ma Q, Wyszynski DF, Farrell JJ, Kutlar A, Farrer LA, Baldwin CT, et al. Fetal hemoglobin in sickle cell anemia: genetic determinants of response to hydroxyurea. Pharmacogenomics J. 2007;7(6):386–94 [DOI] [PubMed] [Google Scholar]
- 18.Bradai M, Abad MT, Pissard S, Lamraoui F, Skopinski L, de Montalembert M. Hydroxyurea can eliminate transfusion requirements in children with severe β-thalassemia. Blood. 2003;102(4):1529–30 [DOI] [PubMed] [Google Scholar]
- 19.Dixit A, Chatterjee TC, Mishra P, Choudhry DR, Mahapatra M, Tyagi S, et al. Hydroxyurea in thalassemia intermedia - a promising therapy. Ann Hematol. 2005; 84(7):441–6 [DOI] [PubMed] [Google Scholar]
- 20.Yavarian M, Karimi M, Bakker E, Harteveld CL, Giordano PC. Response to hydroxyurea treatment in Iranian transfusion-dependent β-thalassemia patients. Haematologica. 2004;89(10):1172–8 [PubMed] [Google Scholar]
- 21.Bradai M, Pissard S, Abad MT, Dechartres A, Ribeil JA, Landais P, et al. Decreased transfusion needs associated with hydroxyurea therapy in Algerian patients with thalassemia major or intermedia. Transfusion. 2007;47(10):1830–6 [DOI] [PubMed] [Google Scholar]
- 22.Ware RE, Despotovic JM, Mortier NA, Flanagan JM, He J, Smeltzer MP, et al. Pharmacokinetics, pharmacodynamics, and pharmacogenetics of hydroxyurea treatment for children with sickle cell anemia. Blood. 2011;118(18):4985–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jison ML, Munson PJ, Barb JJ, Suffredini AF, Talwar S, Logun C, et al. Blood mononuclear cell gene expression profiles characterize the oxidant, hemolytic, and inflammatory stress of sickle cell disease. Blood. 2004;104(1):270–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laurance S, Lansiaux P, Pellay FX, Hauchecorne M, Benecke A, Elion J, et al. Differential modulation of adhesion molecule expression by hydroxycarbamide in human endothelial cells from the micro-and macrocirculation: potential implications in sickle cell disease vasoocclusive events. Haematologica. 2011;96(4):534–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gambero S, Canalli AA, Traina F, Albuquerque DM, Saad ST, Costa FF, et al. Therapy with hydroxyurea is associated with reduced adhesion molecule gene and protein expression in sickle red cells with a concomitant reduction in adhesive proper-ties. Eur J Haematol. 2007;78(2):144–51 [DOI] [PubMed] [Google Scholar]
- 26.Laurance S, Pellay FX, Dossou-Yovo OP, Verger E, Krishnamoorthy R, Lapoumeroulie C, et al. Hydroxycarbamide stimulates the production of proinflammatory cytokines by endothelial cells: relevance to sickle cell disease. Pharmacogenet Genomics. 2010;20(4):257–68 [DOI] [PubMed] [Google Scholar]
- 27.Costa FC, da Cunha AF, Fattori A, de Sousa Peres T, Costa GG, Machado TF, et al. Gene expression profiles of erythroid precursors characterise several mechanisms of the action of hydroxycarbamide in sickle cell anaemia. Br J Haematol. 2007;136(2):333–42 [DOI] [PubMed] [Google Scholar]
- 28.Flanagan JM, Steward S, Howard TA, Mortier NA, Kimble AC, Aygun B, et al. Hydroxycarbamide alters erythroid gene expression in children with sickle cell anaemia. Br J Haematol. 2012;157(2):240–8 [DOI] [PubMed] [Google Scholar]
- 29.Najmabadi H, Karimi-Nejad R, Sahebjam S, Pourfarzad F, Teimourian S, Sahebjam F, et al. The β-thalassemia mutation spectrum in the Iranian population. Hemoglobin. 2001; 25(3):285–96 [DOI] [PubMed] [Google Scholar]
- 30.Sutton M, Bouhassira EE, Nagel RL. Polymerase chain reaction amplification applied to the determination of β-like globin gene cluster haplotypes. Am J Hematol. 1989;32(1):66–9 [DOI] [PubMed] [Google Scholar]
- 31.Panigrahi I, Dixit A, Arora S, Kabra M, Mahapatra M, Choudhry VP, et al. Do alpha deletions influence hydroxyurea response in thalassemia intermedia? Hematology. 2005;10(1):61–3 [DOI] [PubMed] [Google Scholar]
- 32.Leberbauer C, Boulme F, Unfried G, Huber J, Beug H, Mullner EW. Different steroids co-regulate long-term expansion versus terminal differentiation in primary human erythroid progenitors. Blood. 2005; 105(1):85–94 [DOI] [PubMed] [Google Scholar]
- 33.van den Akker E, Satchwell TJ, Pellegrin S, Daniels G, Toye AM. The majority of the in vitro erythroid expansion potential resides in CD34(−) cells, outweighing the contribution of CD34(+) cells and significantly increasing the erythroblast yield from peripheral blood samples. Haematologica. 2010;95(9):1594–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beug H, Palmieri S, Freudenstein C, Zentgraf H, Graf T. Hormone-dependent terminal differentiation in vitro of chicken erythroleukemia cells transformed by ts mutants of avian erythroblastosis virus. Cell. 1982;28(4):907–19 [DOI] [PubMed] [Google Scholar]
- 35.Bakker WJ, Blazquez-Domingo M, Kolbus A, Besooyen J, Steinlein P, Beug H, et al. FoxO3a regulates erythroid differentiation and induces BTG1, an activator of protein arginine methyl transferase 1. J Cell Biol. 2004;164(2):175–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Budzowska M, Jaspers I, Essers J, de Waard H, van Drunen E, Hanada K, et al. Mutation of the mouse Rad17 gene leads to embryonic lethality and reveals a role in DNA damage-dependent recombination. EMBO J. 2004;23(17):3548–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fibach E, Prasanna P, Rodgers GP, Samid D. Enhanced fetal hemoglobin production by phenylacetate and 4-phenylbutyrate in erythroid precursors derived from normal donors and patients with sickle cell anemia and β-thalassemia. Blood. 1993; 82(7):2203–9 [PubMed] [Google Scholar]
- 38.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98(9):5116–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minami R, Muta K, Umemura T, Motomura S, Abe Y, Nishimura J, et al. p16(INK4a) induces differentiation and apoptosis in erythroid lineage cells. Exp Hematol. 2003;31(5):355–62 [DOI] [PubMed] [Google Scholar]
- 40.Kheradmand Kia S, Solaimani Kartalaei P, Farahbakhshian E, Pourfarzad F, von Lindern M, Verrijzer CP. EZH2-dependent chromatin looping controls INK4a and INK4b, but not ARF, during human progenitor cell differentiation and cellular senescence. Epigenetics Chromatin. 2009; 2(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams RT, Sherr CJ. The INK4-ARF (CDKN2A/B) locus in hematopoiesis and BCR-ABL-induced leukemias. Cold Spring Harb Symp Quant Biol. 2008;73:461–7 [DOI] [PubMed] [Google Scholar]
- 42.Durante W, Johnson FK, Johnson RA. Arginase: a critical regulator of nitric oxide synthesis and vascular function. Clin Exp Pharmacol Physiol. 2007;34(9):906–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steinmann S, Schulte K, Beck K, Chachra S, Bujnicki T, Klempnauer KH. v-Myc inhibits C/EBP[β] activity by preventing C/EBP[β]-induced phosphorylation of the co-activator p300. Oncogene. 2009;28(26):2446–55 [DOI] [PubMed] [Google Scholar]
- 44.Yoshida H, Kitabayashi I. Chromatin regulation by AML1 complex. Int J Hematol. 2008;87(1):19–24 [DOI] [PubMed] [Google Scholar]
- 45.Bakker WJ, van Dijk TB, Parren-van Amelsvoort M, Kolbus A, Yamamoto K, Steinlein P, et al. Differential Regulation of Foxo3a Target Genes in Erythropoiesis. Mol Cell Biol. 2007;27(10):3839–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Døsen-Dahl G, Munthe E, Nygren MK, Stubberud H, Hystad ME, Rian E. Bone marrow stroma cells regulate TIEG1 expression in acute lymphoblastic leukemia cells: role of TGFβ/BMP-6 and TIEG1 in chemotherapy escape. Int J Cancer. 2008;123(12):2759–66 [DOI] [PubMed] [Google Scholar]
- 47.Moucadel V, Constantinescu SN. Differential STAT5 signaling by ligand-dependent and constitutively active cytokine receptors. J Biol Chem. 2005; 280(14):13364–73 [DOI] [PubMed] [Google Scholar]
- 48.Ranuncolo SM, Polo JM, Dierov J, Singer M, Kuo T, Greally J, et al. Bcl-6 mediates the germinal center B cell phenotype and lymphomagenesis through transcriptional repression of the DNA-damage sensor ATR. Nat Immunol. 2007;8(7):705–14 [DOI] [PubMed] [Google Scholar]
- 49.Blau CA, Constantoulakis P, al-Khatti A, Spadaccino E, Goldwasser E, Papayannopoulou T, et al. Fetal hemoglobin in acute and chronic states of erythroid expansion. Blood. 1993;81(1):227–33 [PubMed] [Google Scholar]
- 50.Papayannopoulou T, Vichinsky E, Stamatoyannopoulos G. Fetal Hb production during acute erythroid expansion. I. Observations in patients with transient erythroblastopenia and post-phlebotomy. Br J Haematol. 1980;44(4):535–46 [DOI] [PubMed] [Google Scholar]
- 51.Stamatoyannopoulos G, Veith R, Al-Khatti A, Fritsch EF, Goldwasser E, Papayannopoulou T. On the induction of fetal hemoglobin in the adult; stress erythropoiesis, cell cycle-specific drugs, and recombinant erythropoietin. Prog Clin Biol Res. 1987;251:443–53 [PubMed] [Google Scholar]
- 52.Sankaran VG, Lettre G, Orkin SH, Hirschhorn JN. Modifier genes in Mendelian disorders: the example of hemoglobin disorders. Ann N Y Acad Sci. 2010; 1214:47–56 [DOI] [PubMed] [Google Scholar]
- 53.Emambokus N, Vegiopoulos A, Harman B, Jenkinson E, Anderson G, Frampton J. Progression through key stages of haemopoiesis is dependent on distinct threshold levels of c-Myb. EMBO J. 2003; 22(17):4478–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rank G, Cerruti L, Simpson RJ, Moritz RL, Jane SM, Zhao Q. Identification of a PRMT5-dependent repressor complex linked to silencing of human fetal globin gene expression. Blood. 2010;116(9):1585–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lopez RA, Schoetz S, DeAngelis K, O'Neill D, Bank A. Multiple hematopoietic defects and delayed globin switching in Ikaros null mice. Proc Natl Acad Sci USA. 2002; 99(2):602–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boosalis MS, Bandyopadhyay R, Bresnick EH, Pace BS, Van DeMark K, Zhang B, et al. Short-chain fatty acid derivatives stimulate cell proliferation and induce STAT-5 activation. Blood. 2001;97(10):3259–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Merryweather-Clarke AT, Atzberger A, Soneji S, Gray N, Clark K, Waugh C, et al. Global gene expression analysis of human erythroid progenitors. Blood. 2011; 117(13):e96–108 [DOI] [PubMed] [Google Scholar]
- 58.Hattangadi SM, Lodish HF. Regulation of erythrocyte lifespan: do reactive oxygen species set the clock? J Clin Invest. 2007; 117(8):2075–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Szekeres T, Fritzer-Szekeres M, Elford HL, Jayaram HM. The Enzyme Ribonucleotide Reductase: Target for Antitumor and Anti-HIV Therapy. Crit Rev Clin Lab Sci. 1997;34(6):503–28 [DOI] [PubMed] [Google Scholar]
- 60.Gumruk F, Tavil B, Balta G, Unal S, Gurgey A. Significance of fetal hemoglobin values in detection of heterozygotes in fanconi anemia: reevaluation of fetal hemoglobin values by a sensitive method. J Pediatr Hematol Oncol. 2008;30(12):896–9 [DOI] [PubMed] [Google Scholar]
- 61.Motoyama N, Kimura T, Takahashi T, Watanabe T, Nakano T. bcl-x Prevents Apoptotic Cell Death of Both Primitive and Definitive Erythrocytes at the End of Maturation. J Exp Med 1999;189(11):1691–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cokic VP, Beleslin-Cokic BB, Tomic M, Stojilkovic SS, Noguchi CT, Schechter AN. Hydroxyurea induces the eNOS-cGMP pathway in endothelial cells. Blood. 2006; 108(1):184–91 [DOI] [PubMed] [Google Scholar]
- 63.Lou T, Singh M, Mackie A, Li W, Pace B. Hydroxyurea generates nitric oxide in human erythroid cells: mechanisms for gamma-globin gene activation. Exp Biol Med (Maywood). 2009;234(11):1374–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maciejewski JP, Selleri C, Sato T, Cho HJ, Keefer LK, Nathan CF, et al. Nitric oxide suppression of human hematopoiesis in vitro. Contribution to inhibitory action of interferon-γ and tumor necrosis α. J Clin Invest. 1995;96(2):1085–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kucukkaya B, Arslan DO, Kan B. Role of G proteins and ERK activation in hemin-induced erythroid differentiation of K562 cells. Life Sci. 2006;78(11):1217–24 [DOI] [PubMed] [Google Scholar]
- 66.Calatayud S, Beltrán B, Brines J, Moncada S, Esplugues JV. Foetal erythrocytes exhibit an increased ability to scavenge for nitric oxide. Eur J Pharmacol. 1998;347(2–3):363–6 [DOI] [PubMed] [Google Scholar]