SUMMARY
Telomerase is present in human cancer cells but absent in most somatic tissues. The mRNA of human telomerase (hTERT) is alternatively spliced into mostly non-functional products. We sought to understand splicing so we could decrease functional splice isoforms to reduce telomerase activity to complement direct enzyme inhibition. Unexpectedly, minigenes containing hTERT exons 5–10 flanked by 150–300bp intronic sequences did not produce alternative splicing. A 1.1kb region of 38bp repeats ~2kb from the exon 6/intron junction restored exclusion of exons 7/8. An element within intron 8, also >1kb from intron/exon junctions, modulated this effect. Transducing an oligonucleotide complementary to this second element increased non-functional hTERT mRNA from endogenous telomerase. These results demonstrate the potential of manipulating hTERT splicing for both chemotherapy and regenerative medicine, and provide the first specific sequences deep within introns that regulate alternative splicing in mammalian cells by mechanisms other than introducing cryptic splice sites.
INTRODUCTION
Chromosome ends are protected by repetitive non-coding TTAGGG sequences known as telomeres. Initially, each human chromosome is capped by 15–20kb of telomere repeats. DNA sequences are lost with every cell division due to incomplete replication of the DNA lagging strand (the end-replication problem) and end-processing events (Levy et al., 1992; Olovnikov, 1973; Watson, 1972). Telomeres shorten progressively until a critically shortened length is reached that triggers cellular senescence (Harley, 1991; Wright et al., 1996a). The limited proliferative capacity of replicative aging is thought to provide an initial barrier that prevents premalignant cells from progressing to malignant cancer cells (Harley, 1991).
To overcome this block to unlimited growth, cancer cells almost universally upregulate/reactivate telomerase. Telomerase is a ribonucleoprotein where the reverse transcriptase activity of the protein component (hTERT) catalyzes telomere elongation using the RNA template component (hTR) (Greider, 1990). Telomerase is normally expressed in proliferative stem and germline cells, but is repressed during development in most human tissues (Wright et al., 1996b). Re-expression of telomerase prevents the progressive shortening of telomeres and cellular senescence, and is almost universally the mechanism that cancer cells use to escape replicative senescence (Shay and Bacchetti, 1997). Indeed, expression of hTERT in telomerase-negative normal diploid cells results in cell immortalization (Bodnar et al, 1998) without progressing cells to cancer (Morales et al., 1999). Telomerase activity has been detected in the vast majority (~90%) of cancer cells (Kim et al., 1994; Shay and Bacchetti, 1997). A recombination-based mechanism named Alternative Lengthening of Telomeres (ALT) is used to maintain telomeres in some telomerase-negative tumors (Bryan et al., 1997).
Significant efforts have been expended to develop telomerase as a target for treating cancer. Imetelstat (GRN163L) is a modified oligonucleotide that binds to the template region of hTR, acting as an active site inhibitor (Marian et al., 2010). Imetelstat has shown successes in inhibiting telomerase activity in xenograft models (Joseph et al., 2010) and is currently in limited clinical trials. However, it remains unclear if Imetelstat as a single agent will achieve sufficient sustained inhibition in patients to drive cancer cells into crisis. Other types of telomerase inhibitors developed thus far (such as, antisense oligonucleotides, and ribozymes) with inhibitory effects on telomerase in model systems have not successfully progressed to clinical trials due to toxicity, low specificity, and/or inefficient delivery (Chen et al., 2009; Kole et al., 2012). Development of alternative telomerase inhibitors, used in combination or following standard cancer therapy, may enhance the potential of telomerase as a therapeutic target.
An in depth understanding of the regulation of telomerase in normal development and its aberrant expression in cancer cells may provide additional targets for the development of novel therapeutics. Increasing evidence suggests that both transcriptional and post-transcriptional processes are involved in regulating human telomerase (Daniel et al., 2012). In this study, alternative splicing of hTERT was examined as a post-transcriptional regulatory step that could potentially provide a new approach for inhibiting telomerase.
The telomerase transcript containing 16 exons can be spliced into multiple isoforms (Kilian et al., 1997). During kidney development, telomerase activity disappears before transcripts because of a dramatic shift in splicing patterns (Ulaner et al., 2001). The major isoforms studied in cultured cancer cells involve exons 6–9 that encode part of the reverse transcriptase domain. Of these, only the “full length” transcript (that includes all four intact exons) can be translated into an active enzyme. The minus alpha isoform is a dominant negative inhibitor of telomerase that lacks a portion of exon 6 (Yi et al., 2000). The major splice form (minus beta) skips exons 7 and 8 and contains a premature stop codon that is subject to nonsense mediated decay. The current best estimate for the number of catalytically active telomerase molecules/cell is 50–100 (Cohen et al., 2007), produced from ~20 mRNA molecules per cell (Yi et al., 2001). Only a small fraction of the hTERT transcripts are spliced into the full length form that can generate the catalytically active protein. A working hypothesis to explain these results is that the transcriptional machinery is unable to reduce transcription to a level that produces only a few mRNA molecules/cell, so that ~20 mRNA molecules per cell might represent the lower limit of transcriptional regulation. The cell then disposes of this excess transcription by alternatively splicing most into non-functional forms in order to produce the very low levels of protein needed to maintain telomere length in cancer cells. Almost nothing is currently known about the regulation of very low abundant proteins such as telomerase even though this gene was identified over 15 years ago.
To address this deficiency, we constructed a hTERT minigene containing ~150–300 intronic sequences flanking hTERT exons 5–10. Surprisingly, it failed to produce significant amounts of alternatively spliced products. It was previously known that large long-lived mammals regulated telomerase differently from many small short-lived mammals (Friedman et al., 1999). We then identified several intronic sequences in the telomerase gene that were conserved in primates but not in other mammalian species. These included a 1100bp VNTR (variable number tandem repeats) composed of 38bp repeats in intron 6, and a ~260bp sequence that was present as a direct repeat in both introns 6 and 8 (flanking the exons removed in the major nonfunctional isoform that eliminates exons 7 and 8). Each of these elements was a kilobase or more from any splice junction. Including these elements in the minigene reconstituted alternative splicing. Exon-skipping oligonucleotides complementary to a region at the beginning of the 260nt direct repeat decreased the amount of functional splice forms transcribed from the endogenous gene, providing validation of this novel mechanism involved in splicing. Understanding the detailed mechanism(s) by which these regulatory sequences function to direct hTERT splicing choices may identify new telomerase inhibitor targets that could be used in combination with standard cancer chemotherapy and/or other telomerase inhibitors to reduce cancer recurrences (e.g. durable responses), as well as interventions to increase telomerase activity in stem cells for regenerative medicine.
RESULTS
Telomerase splicing is differentially regulated in different cancer cell lines
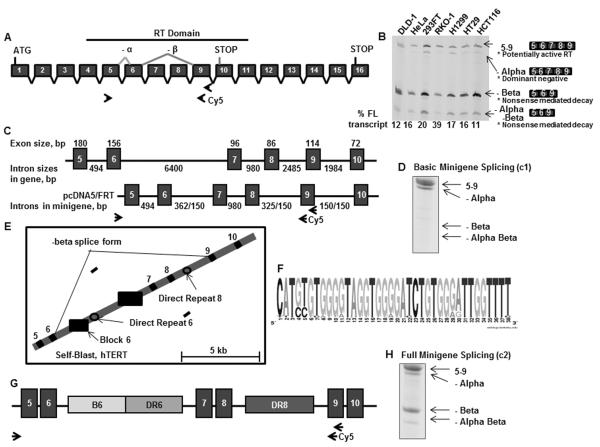
The 42kb telomerase (hTERT) gene on chromosome 5 contains 16 exons. Besides the full length transcript with all 16 exons, none of the identified alternative spliced forms has reverse transcriptase activity (Saeboe-Larssen et al., 2006; Yi et al., 2000). The alternatively spliced isoforms within the reverse transcriptase domain include minus alpha, minus beta, or both (minus alpha beta) (Fig. 1A). The inclusion of all five exons (5-6-7-8-9) is designated 5–9 (this would encode active telomerase in the absence of additional changes outside this region). The minus alpha splicing isoform uses an alternative 3' splice acceptor site 36bp into exon 6, resulting in an in-frame transcript that is translated into a dominant negative protein without reverse transcriptase activity (Yi et al., 2000). The minus beta splicing isoform skips exons 7 and 8, creating a frame-shift and encountering a pre-mature stop codon in exon 10. It is the major splicing isoform of hTERT transcripts in cancer cells and its steady state level is at least 5 times less than its rate of production since it is subject to degradation by non-sense mediated decay.
Figure 1.
Identification of long-range interactions required to produce minus beta splicing.
(A) Diagram illustrating the exons and introns of human telomerase (not to scale). Functional telomerase is made from the full length transcript containing all 16 exons. The splicing isoforms within the RT domain (minus alpha and minus beta) are indicated. The minus beta splicing isoform creates a frame-shift and a premature stop codon as indicated in exon 10. The gene specific primer used to make the cDNA lies within exon 9 (arrow). PCR primers for endogenous splicing used a forward primer in exon 5 and a reverse cy5-labelled primer in exon 9 (arrows). (B) PCR analysis of endogenous hTERT splicing, showing a similar splicing pattern among cancer cell lines. 5–9 splicing includes the functional full length transcript (other potential isoforms might contain changes outside this assayed region). Minus beta is the dominant isoform in all cell lines, but variations in the distribution of each splice variant is cell line specific. (C) Diagram of human telomerase from exon 5 to exon 10 with exon sizes indicated above and intron sizes below (top) and the sequences included in the telomerase basic minigene (construct 1, c1)(bottom). All exonic sequences from exon 5 to exon 10 are included in the minigene, whereas only flanking intronic sequences are included. The values before and after the forward slash (/) indicate the 5' intronic and 3'intronic sequences adjacent to the exons used. The minigene was inserted into pcDNA5/FRT driven by a CMV promoter. The gene specific primer used to make cDNA and the reverse cy5-labelled primer for PCR is the same as the primers used for endogenous TERT analysis. However, the forward PCR primer lies within the pcDNA5/FRT vector just before exon 5, making it minigene-specific. (D) Splicing analysis of the basic minigene illustrated in (C) yielded only 5–9 and minus alpha products. (E) Self-blast of human telomerase showing a block of repeats in intron 6 (block 6), a direct repeat in intron 6 (DR6), and a direct repeat in intron 8 (DR8). (F) Sequence logo created by WebLogo (Parra et al., 2012) showing the strong 38 nucleotide consensus sequence of the 26 repeats in block 6. (G) Diagram of the full minigene (c2) including block 6 (B6), the direct repeat in intron 6 (DR6), and the direct repeat in intron 8 (DR8). (H) Splicing analysis of the full minigene illustrated in (E) showed minus beta formation. Data are from HeLa FRT clone 8 cells stably transfected with the minigenes.
Telomerase alternative splicing was examined in a variety of cancer cell lines (Fig. 1B). The ratio of transcripts spliced into each of the isoforms varied slightly between each line.
Minigene requires conserved distant intronic sequences to recapitulate normal splicing
We constructed a minigene containing exons 5 to 10 and 150–300 flanking nucleotides (Fig. 1C). The regions immediately downstream of exons 6 and 8 were unusually GU rich, so we included 325–362bp from these regions. This minigene containing 3669bp was inserted into the pcDNA/FRT vector, and stably transfected into a HeLa clone selected for a single FRT site introduced by Invitrogen's Flp-In system to reduce the variability arising from random genomic incorporation. Alternative splicing was determined using a minigene-specific forward primer. The basic minigene only produced 5–9 and minus alpha splicing, and therefore was unable to recapitulate the endogenous telomerase splicing pattern (Fig. 1D).
The human telomerase gene contains a block of 26 short repeats with a strong 38 nucleotide consensus sequence ~1900nt downstream of exon 6 (Fig. 1E, F). This block of repeats in intron 6 (B6, Block 6) is immediately followed by a 256 nucleotide direct repeat in intron 6 (DR6), which has 85% sequence similarity with a 258 nucleotide direct repeat in intron 8 (DR8). DR8 is ~1500nt downstream of exon 7. These elements are highly conserved among old world primates (Supplemental Fig. S1), suggesting a functional role in regulation. A separate and unrelated second block of repeats ~1270nt downstream from DR6 is also conserved among primates but the number of repeats varies greatly and therefore was not included in the minigene in this initial study. The endogenous hTERT splicing isoforms were restored when B6, DR6, and DR8 were incorporated into the minigene (“full” minigene) (Fig. 1G, H, construct c2).
Block 6 is necessary for minus beta splicing
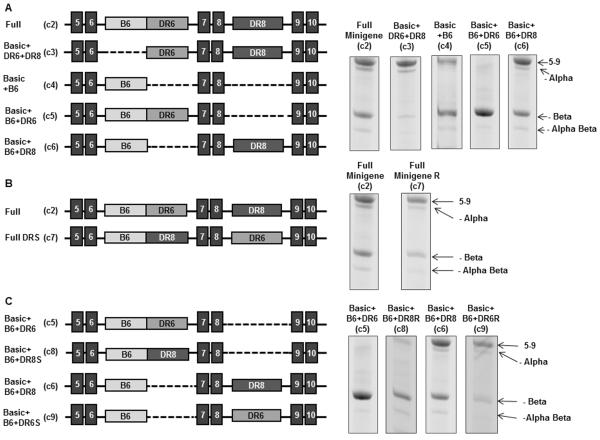
The addition of just the direct repeats in intron 6 and 8 to the basic minigene failed to produce significant amounts of minus beta splicing (Fig. 2A, construct c3). In contrast, the addition of B6 alone (Figure 2A, construct c4) was sufficient to enhance exon 7 and 8 skipping to produce a high proportion of minus beta splicing. The combination of B6 and the direct repeat in intron 6 produced exclusively minus beta splicing (Fig.2A, construct c5), while the combination of B6 and the direct repeat in intron 8 shifted splicing towards greater inclusion of all the exons (Fig. 2A, construct c6). Thus, while the direct repeats by themselves are not sufficient to produce the skipping of exons 7 and 8 (minus beta, construct c3), they modulate the ability of B6 to shift splicing towards non-functional forms.
Figure 2.
Effects of distal elements B6, DR6 and DR8 on alternative splicing.
(A) DR6 and DR8 (construct c3) do not generate significant exon skipping in the absence of B6 (construct c2). B6 without DR6 and DR8 is sufficient to produce minus beta splicing (c5). Adding DR6 to B6 (c5) modifies its activity and virtually eliminates all 5–9 products that include all of the exons. Adding only DR8 to B6 (c6) shifts splicing in the opposite direction and increases the amount of 5–9 splicing. (B) Switching the position of DR6 and DR8 in the presence of B6 does not change the ratio of 5–9 to minus beta splicing (c7 vs c2). (C) DR6 and DR8 produce similar effects when positioned as the direct repeat only in intron 6 (constructs 5 vs 8) or only in intron 8 (constructs 6 vs 9). It is thus the position of the direct repeats rather than their sequence difference that influence splice choice. All constructs were analyzed in HeLa FRT clone 8 cells.
The effects of the direct repeats on the alternative splicing of telomerase is position dependent
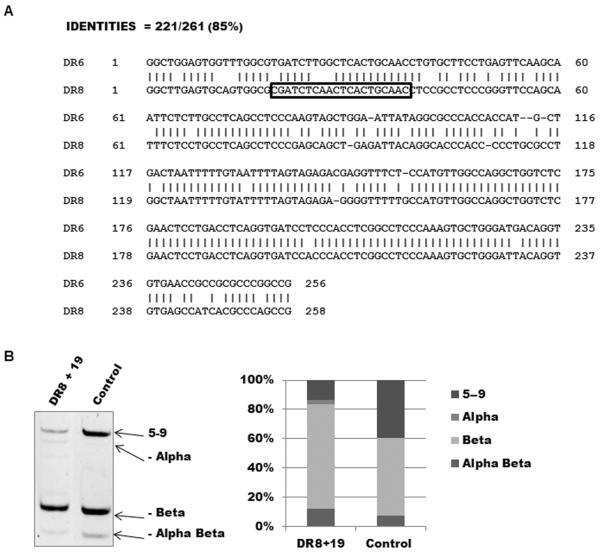
Switching the positions of direct repeats 6 and 8 did not change splicing of the hTERT minigene (Fig. 2B, constructs c2 vs c7). Similarly, there was little difference when the direct repeats were substituted for each other. Replacing DR6 with DR8 (Fig. 2C, constructs c5 vs c8) or replacing DR8 with DR6 (Fig 2C, constructs c6 vs c9) did not significantly change the splicing pattern. This suggests that it is the intronic location rather than the sequence differences between DR6 and DR8 (Fig. 3A) that influences their effect on splicing.
Figure 3.
DR8 directed oligonucleotides alter the splicing of endogenous telomerase.
(A) The direct repeats DR6 and DR8 are 85% identical and are members of the J1 family of Alu repeats. The sequence of the 20bp 2-O-methyl oligonucleotide containing 50% thiophosphate linkages complementary to the region starting at nt19 is boxed. (B) Introduction of the oligonucleotide DR8+19 dramatically shifts splicing of the endogenous telomerase mRNA towards the production of the non-functional minus beta isoform. Cells were treated with 100nM DR8+19 oligonucleotide or vehicle control. Right side shows the quantification.
Antisense oligonucleotide supports the effect of the direct repeat in intron 8 established by the minigene model
Because the direct repeat in intron 8 suppressed the minus beta promoting effects of the direct repeat in intron 6 in the minigene, we tested whether interfering with its structure might change the splicing of the endogenous TERT gene. Figure 3B demonstrates that an antisense oligonucleotide (DR8+19, a 20-mer beginning at nt19 of the DR8 sequence) directed against DR8 shifted the splicing of endogenous hTERT to mostly minus beta splicing. This confirms that the behavior of the minigene is recapitulating important aspects of endogenous regulation.
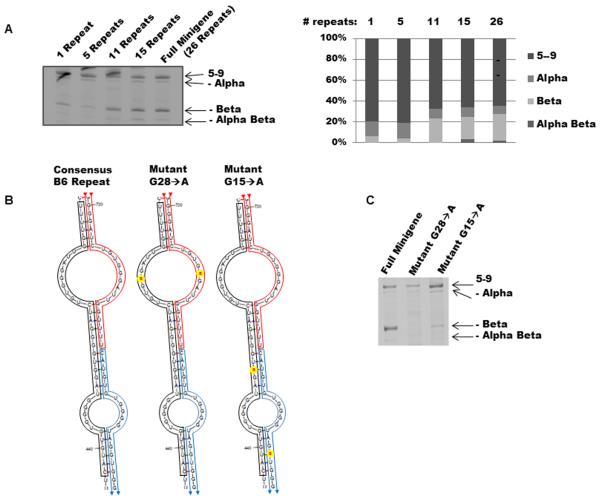
A minimum number of consensus repeats are required for the minus beta splicing effects of block 6
The minimum number of repeats present in the primate genomes examined was 9 repeats (bonobo, Supplementary Fig. S1). The B6 sequence in the full minigene (c2) was replaced with varying number of the consensus repeat sequence in order to determine the minimum number required to increase minus beta splicing. With 1 or 5 repeats, the minigenes did not induce minus beta splicing. However, 11 or more repeats did restore the effect (Fig. 4A).
Figure 4.
Length and mutational analysis of Block 6.
(A) Left side shows splicing products of HeLa FRT clone 8 cells stably transfected with minigenes containing varying numbers of block 6 consensus repeats. Right side shows the quantification. <11 repeats failed to produce significant amounts of minus beta splicing. (B) Secondary RNA folding structures predicted by Mfold for B6 self-interactions for the consensus sequence and two mutants. The structures with the lowest free energies are shown. The consensus 38bp repeat can self-fold into a highly structured staggered secondary structure. One complete (black) and two half consensus repeats (red and blue) are highlighted. Neither mutant G28→ A nor mutant G15→ A altered this structure and both self-folded similarly to the consensus repeat. The mutated nucleotides are in red and highlighted in yellow. (C) Mutations that do not alter the secondary structure of self-folding abolish minus beta splicing.
Single mutations disrupt block 6 function
When various numbers of 38nt repeats were analyzed using the Mfold web server (Zuker, 2003), the majority of the putative structures were dominated by the staggered overlap shown in Fig 4B, in which one repeat interacted with two adjacent half-repeats. We introduced two different single mutations into the 38bp repeat, G28→A (predicted to be found in the top loop of the self-complementary repeat structure) and G15→A (predicted to be located in two different double stranded regions, disrupting neither of them). These mutations completely or almost completely abolished the ability of 13 repeats to produce minus beta splicing (Figure 4C).
DISCUSSION
We used a minigene system to identify the pre-mRNA sequence elements required for in vivo hTERT (human telomerase) splicing regulation. In contrast to most alternative splicing events that have been studied, the splicing of hTERT is regulated by long-range interactions rather than by the intronic/exonic elements adjacent to the splice sites. We identified three elements that were highly conserved among primates that were embedded in introns 6 and 8. These elements surround the alternative exons 7 and 8, where the elimination of these exons results in the major non-functional minus beta telomerase isoform. Only when these elements were included in the telomerase minigene did we observe the various splicing isoforms seen with the endogenous gene. CHIP-seq data indicate many potential splicing factor binding sites at long distances from intron/exon junctions (Yeo et al., 2009), but few specific examples of regulation-at-a-distance have been previously reported. These elements in the telomerase gene represent the first examples in mammals of specific distant sequences that influence splice choice by a mechanism other than introducing cryptic splice sites (Dominski and Kole, 1993; Friedman et al., 1999; Parra et al., 2012; Pros et al., 2009; Salem et al., 2012).
The block of repeats in intron 6 is necessary for the enhancement of minus beta splicing of telomerase and a minimal number of repeats is required for its function, The number of B6 repeats varies greatly from 18–38 repeats amongst individuals (Leem et al., 2002). Yet, the mechanism of how B6 repeats influence hTERT splicing remains unknown. The importance of repeats can be seen in the alternative splicing of the Tau gene. Intronic mutations in Tau destabilize a stem-loop structure, thus increasing inclusion of exon 10 and this correlates with frontotemporal dementia and Parkinson's (Varani et al., 1999). Another example is seen in myotonic dystrophy type 1 (DM1), where an expansion of (CUG)n repeats in DM protein kinase (DMPK) sequesters Mbnl1 and increases the levels of CUGBP1, leading to the aberrant splicing of genes governed by these splicing factors (Mulders et al., 2009). The implication of altered repeats resulting in diseases suggests that a possible link between the B6 repeats of telomerase and cancer development warrants further investigation.
The direct repeat in introns 6 and 8 have 85% sequence similarity, but their effects on telomerase splicing is very different. When these elements were switched, the position of the element rather than the nucleotide sequence governed its effect on telomerase splicing. This suggests the direct repeats may function by recruiting similar RNA binding proteins based on their sequences and the interactions of these binding proteins with other neighboring proteins may dictate the splicing decision. This is similar to studies demonstrating that the effects of SR and hnRNP binding are highly dependent on their position with respect to splice sites (Llorian et al., 2010; Yeo et al., 2009). The mechanisms and factors involved in telomere maintenance throughout evolution are extraordinarily diverse, indicating the lack of constraints to evolving multiple specific mechanisms for protecting the ends of the chromosomes from degradation (Fisette et al., 2010). Even among mammals there is considerable diversity in the size and use of telomeres (Gomes et al., 2011). The conservation among higher primates, but not other mammals of the elements is consistent with the evolutionary diversity of known telomere maintenance mechanisms.
There are large numbers of different avenues for exploiting these observations to develop therapeutic interventions. For example, screening for small molecules or siRNAs that alter the splicing pattern is likely to be the best long-term approach. There are many recently discovered “telomeropathies” due to mutations in telomerase or one of its components (Garcia et al., 2007), and increasing evidence suggests that telomere shortening in stem cells may contribute to the failure to adequately maintain tissues in late life. There may be many minor or previously uncharacterized spliceosome factors that influence these long-range interactions in different directions, where some factors would decrease full length functional telomerase and would be important for the treatment of cancer while others would increase functional telomerase and would provide therapeutics for regenerative medicine. Increasing our understanding of the regulation of telomerase at the level of splicing may provide additional pathways for manipulating the function of this critical ribonucleoprotein enzyme that influences human health at multiple levels.
EXPERIMENTAL PROCEDURES
Cell culture
HeLa cervical carcinoma, H1299 lung adenocarcinoma, 293FT embryonic kidney cells, DLD-1 colon carcinoma, RKO-1 colon carcinoma, HT29 colon adenocarcinoma, and HCT116 colorectal carcinoma were cultured at 37°C in 5% CO2 in 4:1 DMEM:Medium 199, containing 10% calf serum (HyClone, Logan, UT).
Generation of cells containing FRT site
The Flp-In System (Invitrogen, Carlsbad, CA) was used to generate HeLa cells with a stably integrated FRT siteFRT stably integrated clones were screened for single insertion by southern blotting and adequate expression by beta galactosidase staining. The data from a single clone (HeLa Clone#8) is shown in Figures 2–4. Additional clones with different FRT insertion sites were used to confirm several of the results observed in HeLa#8.
Construction of minigene plasmids
Human telomerase sequences were inserted in the pcDNA5/FRT expression vector (Invitrogen, Carlsbad, CA) using a variety of restriction sites. The sequence of the minigene is available upon request.
RT-PCR and splicing analysis
RNA was extracted from stable minigene expressing HeLa cells using the RNeasy Plus Mini Kit (Qiagen, Valencia, CA). cDNA was made by combining 1ug of RNA, 1ng telomerase specific primer (5'-CGCAAACAGCTTGTTCTCCATGTC-3'), and hybridization buffer (1.5M NaCl, 50mM Tris pH 7.5, 10mM EDTA). The mixture was heated to 90°C. Temperature was decreased by 1°C every 20 seconds until 43°C, then Moloney murine leukemia virus reverse transcriptase (M-MLV RT) (Promega, Madison, WI) was added with 1.25× RT mix (1.25mM dNTP, 12.5mM DTT, 12.5mM Tris pH 8.0, 7.5mM MgCl2). The reaction was incubated at 43°C for 30 minutes and then heated to 94°C for 5 minutes. To amplify minigene specific telomerase splicing isoforms, the forward primer 5'-CTGGCTAACTAGAGAACCCACTGC-3' and the cy5-labelled reverse primer 5'- AGGCTGCAGAGCAGCGTGGAGAGG-3' were used with Taq polymerase PCR (Promega, Madison, WI) in RT-PCR buffer (0.5M KCl, 0.1M Tris pH 8.3, 15mM MgCl2, 0.01% gelatin). The reaction was initially denatured at 94°C for 3 minutes, then denatured at 94°C for 30 seconds, annealed at 61°C for 30 seconds, and extended at 72°C for 1 minute for 30 cycles, with a final extension at 72°C for 10 minutes. The PCR product was resolved on a 5% denaturing polyacrylamide gel and visualized at 650nm.
Annealing oligonucleotides to generate consensus and mutant block 6 repeats
In separate reactions, equal molar ratios of the top and bottom strands of the terminating ends containing restriction sites and the overlapping mutant repeats were annealed using 1× annealing buffer (10mM Tris pH 8.0, 50mM NaCl, 1mM EDTA), heated to 95°C for 5 minutes and then gradually cooled to room temperature. The sequence and strategy are shown in the Supplementary Methods. After purifying the annealed DNA using the QIAquick Gel Extraction Kit (Qiagen, Valencia, CA), the terminating ends were combined with the overlapping consensus or mutant repeats at a1:40:1 ratio. After ligating the mixture overnight at room temperature using T4 DNA ligase (Fermentas, Canada), ligation products were separated on a 1% agarose gel. Gel purified size classes were then digested with restriction enzymes, and ligated to the minigene vector.
Antisense oligonucleotide
The 2-O-methyl oligonucleotide DR8+19 oligonucleotide (5'– CGAUCUCAACUCACUGCAAC – 3') containing 50% phosphothioate linkages was synthesized by Integrated DNA Technologies, Inc. The oligonucleotide (100nM) was introduced into the cells by reverse transfection using Lipofectamine RNAiMAX (Life Technologies Corporation, CA). Transfected cells were collected after 48 hours and analyzed by RT-PCR.
Supplementary Material
Highlights
Alternative splicing is a new target for telomerase inhibition/activation
Elements deep within introns regulate human telomerase splicing
These intronic elements contain both unusual short repeats and direct repeats
A direct repeat oligonucleotide modifies splicing of endogenous telomerase
ACKNOWLEDGMENTS
This work was supported by a pre-doctoral fellowship (BC100756 to M.S.W.), The UT Southwestern Medical Student Research Program and NIDDK T-35-DK066141 (R.K); CA154805 (W.E.W.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med. 1997;3:1271–1274. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- Chen H, Li Y, Tollefsbol TO. Strategies targeting telomerase inhibition. Molecular biotechnology. 2009;41:194–199. doi: 10.1007/s12033-008-9117-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SB, Graham ME, Lovrecz GO, Bache N, Robinson PJ, Reddel RR. Protein composition of catalytically active human telomerase from immortal cells. Science. 2007;315:1850–1853. doi: 10.1126/science.1138596. [DOI] [PubMed] [Google Scholar]
- Daniel M, Peek GW, Tollefsbol TO. Regulation of the human catalytic subunit of telomerase (hTERT) Gene. 2012;498:135–146. doi: 10.1016/j.gene.2012.01.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Kole R. Restoration of correct splicing in thalassemic pre-mRNA by antisense oligonucleotides. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:8673–8677. doi: 10.1073/pnas.90.18.8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisette JF, Toutant J, Dugre-Brisson S, Desgroseillers L, Chabot B. hnRNP A1 and hnRNP H can collaborate to modulate 5' splice site selection. RNA. 2010;16:228–238. doi: 10.1261/rna.1890310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman KJ, Kole J, Cohn JA, Knowles MR, Silverman LM, Kole R. Correction of aberrant splicing of the cystic fibrosis transmembrane conductance regulator (CFTR) gene by antisense oligonucleotides. The Journal of biological chemistry. 1999;274:36193–36199. doi: 10.1074/jbc.274.51.36193. [DOI] [PubMed] [Google Scholar]
- Garcia CK, Wright WE, Shay JW. Human diseases of telomerase dysfunction: insights into tissue aging. Nucleic Acids Res. 2007;35:7406–7416. doi: 10.1093/nar/gkm644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes NM, Ryder OA, Houck ML, Charter SJ, Walker W, Forsyth NR, Austad SN, Venditti C, Pagel M, Shay JW, et al. Comparative biology of mammalian telomeres: hypotheses on ancestral states and the roles of telomeres in longevity determination. Aging Cell. 2011;10:761–768. doi: 10.1111/j.1474-9726.2011.00718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider CW. Telomeres, telomerase and senescence. Bioessays. 1990;12:363–369. doi: 10.1002/bies.950120803. [DOI] [PubMed] [Google Scholar]
- Harley CB. Telomere loss: mitotic clock or genetic time bomb? Mutat Res. 1991;256:271–282. doi: 10.1016/0921-8734(91)90018-7. [DOI] [PubMed] [Google Scholar]
- Joseph I, Tressler R, Bassett E, Harley C, Buseman CM, Pattamatta P, Wright WE, Shay JW, Go NF. The telomerase inhibitor imetelstat depletes cancer stem cells in breast and pancreatic cancer cell lines. Cancer Res. 2010;70:9494–9504. doi: 10.1158/0008-5472.CAN-10-0233. [DOI] [PubMed] [Google Scholar]
- Kilian A, Bowtell DD, Abud HE, Hime GR, Venter DJ, Keese PK, Duncan EL, Reddel RR, Jefferson RA. Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum Mol Genet. 1997;6:2011–2019. doi: 10.1093/hmg/6.12.2011. [DOI] [PubMed] [Google Scholar]
- Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- Kole R, Krainer AR, Altman S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nature reviews Drug discovery. 2012;11:125–140. doi: 10.1038/nrd3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leem SH, Londono-Vallejo JA, Kim JH, Bui H, Tubacher E, Solomon G, Park JE, Horikawa I, Kouprina N, Barrett JC, et al. The human telomerase gene: complete genomic sequence and analysis of tandem repeat polymorphisms in intronic regions. Oncogene. 2002;21:769–777. doi: 10.1038/sj.onc.1205122. [DOI] [PubMed] [Google Scholar]
- Levy MZ, Allsopp RC, Futcher AB, Greider CW, Harley CB. Telomere end-replication problem and cell aging. J Mol Biol. 1992;225:951–960. doi: 10.1016/0022-2836(92)90096-3. [DOI] [PubMed] [Google Scholar]
- Llorian M, Schwartz S, Clark TA, Hollander D, Tan LY, Spellman R, Gordon A, Schweitzer AC, de la Grange P, Ast G, et al. Position-dependent alternative splicing activity revealed by global profiling of alternative splicing events regulated by PTB. Nat Struct Mol Biol. 2010;17:1114–1123. doi: 10.1038/nsmb.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marian CO, Cho SK, McEllin BM, Maher EA, Hatanpaa KJ, Madden CJ, Mickey BE, Wright WE, Shay JW, Bachoo RM. The telomerase antagonist, imetelstat, efficiently targets glioblastoma tumor-initiating cells leading to decreased proliferation and tumor growth. Clin Cancer Res. 2010;16:154–163. doi: 10.1158/1078-0432.CCR-09-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales CP, Holt SE, Ouellette M, Kaur KJ, Yan Y, Wilson KS, White MA, Wright WE, Shay JW. Absence of cancer-associated changes in human fibroblasts immortalized with telomerase. Nature genetics. 1999;21:115–118. doi: 10.1038/5063. [DOI] [PubMed] [Google Scholar]
- Mulders SA, van den Broek WJ, Wheeler TM, Croes HJ, van Kuik-Romeijn P, de Kimpe SJ, Furling D, Platenburg GJ, Gourdon G, Thornton CA, et al. Triplet-repeat oligonucleotide-mediated reversal of RNA toxicity in myotonic dystrophy. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:13915–13920. doi: 10.1073/pnas.0905780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olovnikov AM. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J Theor Biol. 1973;41:181–190. doi: 10.1016/0022-5193(73)90198-7. [DOI] [PubMed] [Google Scholar]
- Parra MK, Gallagher TL, Amacher SL, Mohandas N, Conboy JG. Deep intron elements mediate nested splicing events at consecutive AG dinucleotides to regulate alternative 3' splice site choice in vertebrate 4.1 genes. Molecular and cellular biology. 2012;32:2044–2053. doi: 10.1128/MCB.05716-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pros E, Fernandez-Rodriguez J, Canet B, Benito L, Sanchez A, Benavides A, Ramos FJ, Lopez-Ariztegui MA, Capella G, Blanco I, et al. Antisense therapeutics for neurofibromatosis type 1 caused by deep intronic mutations. Human mutation. 2009;30:454–462. doi: 10.1002/humu.20933. [DOI] [PubMed] [Google Scholar]
- Saeboe-Larssen S, Fossberg E, Gaudernack G. Characterization of novel alternative splicing sites in human telomerase reverse transcriptase (hTERT): analysis of expression and mutual correlation in mRNA isoforms from normal and tumour tissues. BMC Mol Biol. 2006;7:26. doi: 10.1186/1471-2199-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem IH, Hsairi I, Mezghani N, Kenoun H, Triki C, Fakhfakh F. CAPN3 mRNA processing alteration caused by splicing mutation associated with novel genomic rearrangement of Alu elements. Journal of human genetics. 2012;57:92–100. doi: 10.1038/jhg.2011.129. [DOI] [PubMed] [Google Scholar]
- Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer. 1997;33:787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- Ulaner GA, Hu JF, Vu TH, Giudice LC, Hoffman AR. Tissue-specific alternate splicing of human telomerase reverse transcriptase (hTERT) influences telomere lengths during human development. Int J Cancer. 2001;91:644–649. [PubMed] [Google Scholar]
- Varani L, Hasegawa M, Spillantini MG, Smith MJ, Murrell JR, Ghetti B, Klug A, Goedert M, Varani G. Structure of tau exon 10 splicing regulatory element RNA and destabilization by mutations of frontotemporal dementia and parkinsonism linked to chromosome 17. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:8229–8234. doi: 10.1073/pnas.96.14.8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD. Origin of concatameric T4 DNA. Nature. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- Wright WE, Brasiskyte D, Piatyszek MA, Shay JW. Experimental elongation of telomeres extends the lifespan of immortal x normal cell hybrids. EMBO J. 1996a;15:1734–1741. [PMC free article] [PubMed] [Google Scholar]
- Wright WE, Piatyszek MA, Rainey WE, Byrd W, Shay JW. Telomerase activity in human germline and embryonic tissues and cells. Dev Genet. 1996b;18:173–179. doi: 10.1002/(SICI)1520-6408(1996)18:2<173::AID-DVG10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X, Shay JW, Wright WE. Quantitation of telomerase components and hTERT mRNA splicing patterns in immortal human cells. Nucleic Acids Res. 2001;29:4818–4825. doi: 10.1093/nar/29.23.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X, White DM, Aisner DL, Baur JA, Wright WE, Shay JW. An alternate splicing variant of the human telomerase catalytic subunit inhibits telomerase activity. Neoplasia. 2000;2:433–440. doi: 10.1038/sj.neo.7900113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.