Abstract
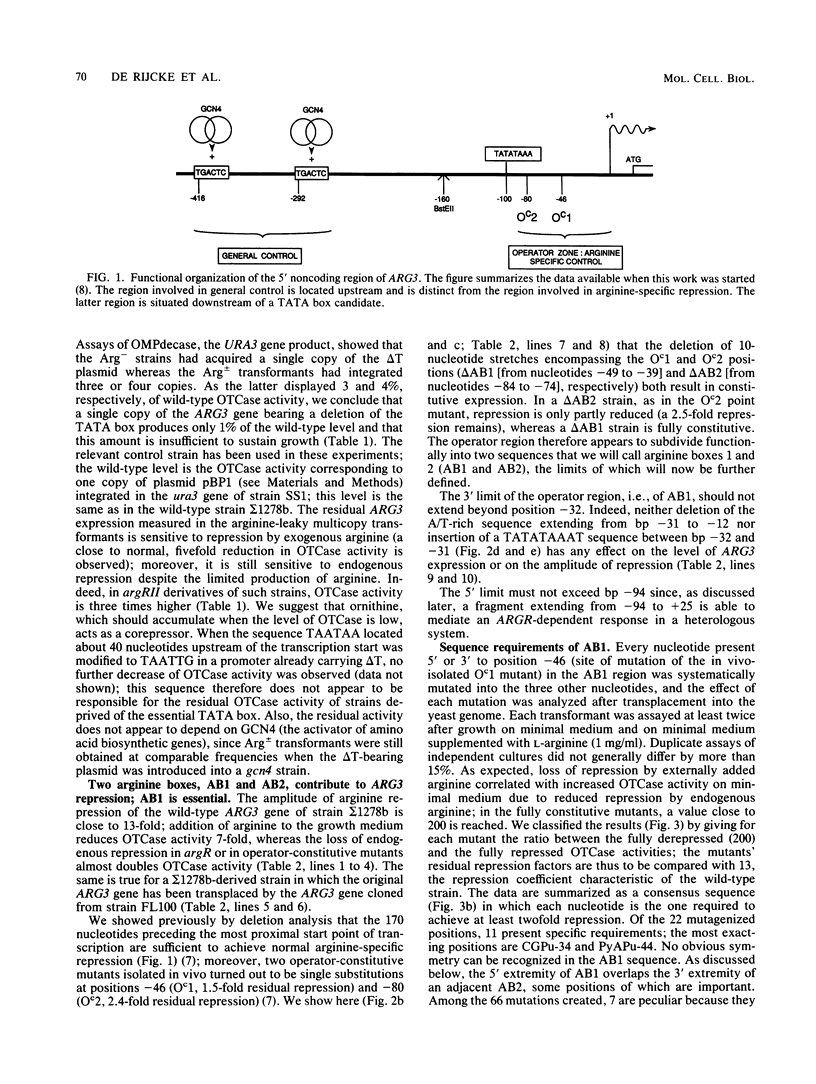
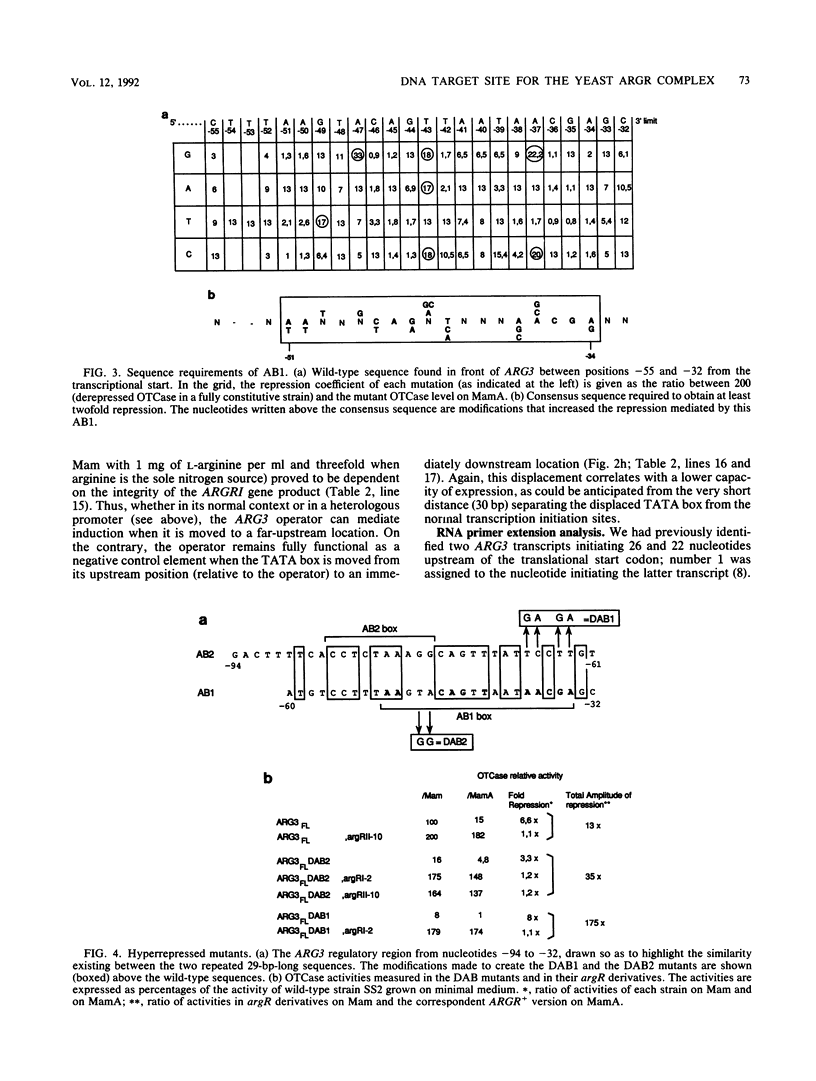
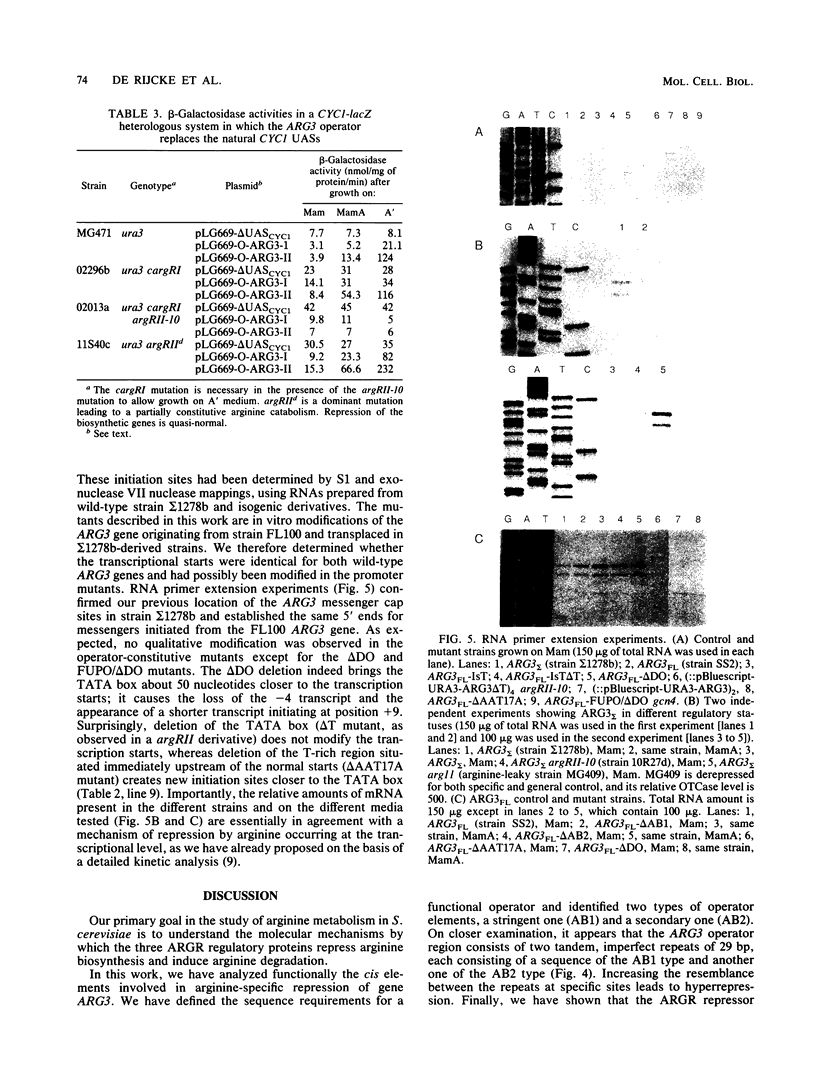
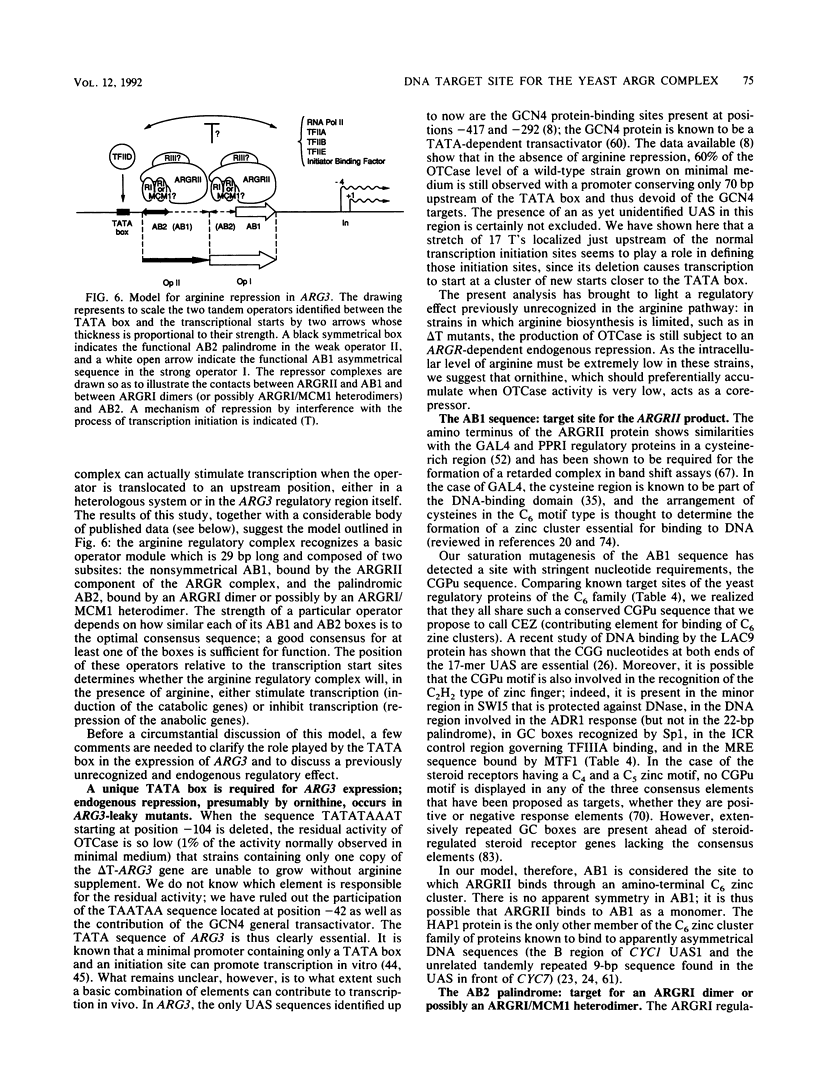
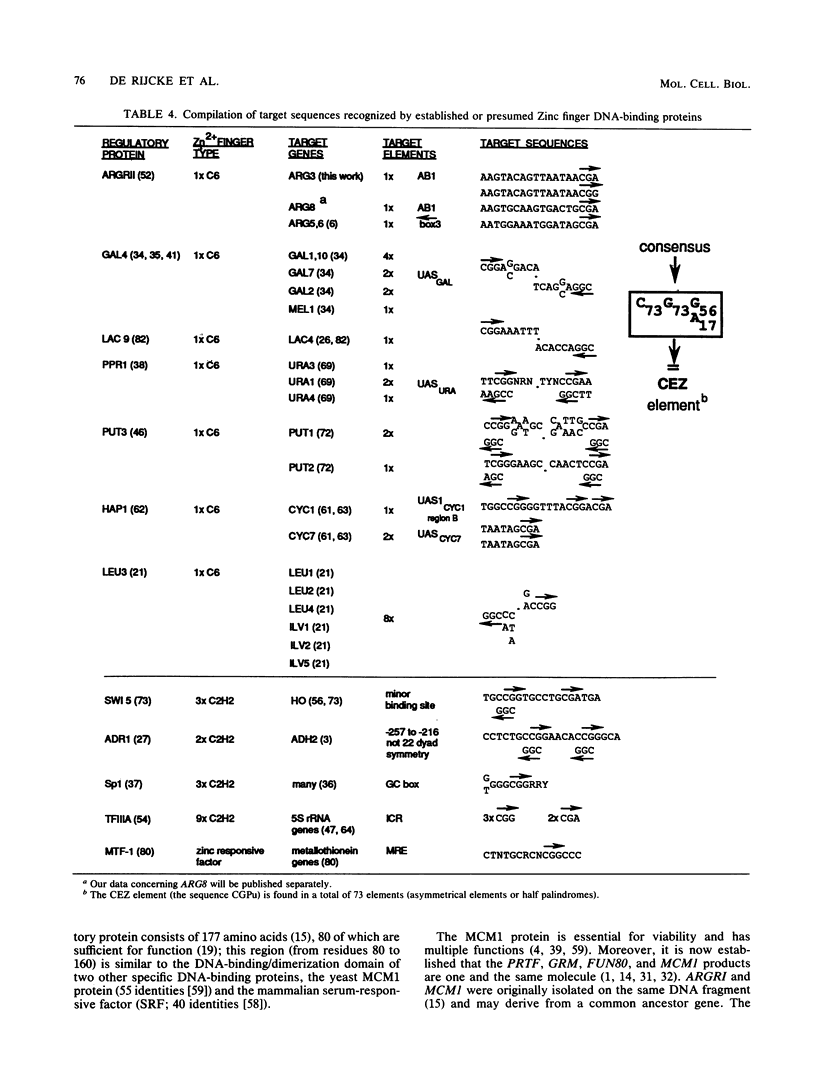
We have determined the sequences and positions of the cis elements required for proper functioning of the ARG3 promoter and proper arginine-specific control. A TATA box located 100 nucleotides upstream of the transcription start was shown to be essential for ARG3 transcription. Two sequences involved in normal arginine-mediated repression lie immediately downstream of the TATA box: an essential one (arginine box 1 [AB1]) and a secondary one (arginine box 2 [AB2]). AB1 was defined by saturation mutagenesis and is an asymmetrical sequence. A stringently required CGPu motif in AB1 is conserved in all known target sites of C6 zinc cluster DNA-binding proteins, leading us to propose that AB1 is the binding site of ARGRII, another member of the C6 family. The palindromic AB2 sequence is suggested, on the basis of published data, to be the binding site of ARGRI, possibly in heterodimerization with MCM1. AB2 and AB1 correspond respectively to the 5' and 3' halves of two adjacent similar sequences of 29 bp that appear to constitute tandem operators. Indeed, mutations increasing the similarity of the other halves with AB1 and AB2 cause hyperrepression. To mediate repression, the operator must be located close to the transcription initiation region. It remains functional if the TATA box is moved downstream of it but becomes inoperative in repression when displaced to a far-upstream position where it mediates an arginine and ARGR-dependent induction of gene expression. The ability of the ARG3 operator to act either as an operator or as an upstream activator sequence, depending on its location, and the functional organization of the anabolic and catabolic arginine genes suggest a simple model for arginine regulation in which an activator complex can turn into a repressor when able to interfere sterically with the process of transcription initiation.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ammerer G. Identification, purification, and cloning of a polypeptide (PRTF/GRM) that binds to mating-specific promoter elements in yeast. Genes Dev. 1990 Feb;4(2):299–312. doi: 10.1101/gad.4.2.299. [DOI] [PubMed] [Google Scholar]
- Bechet J., Greenson M., Wiame J. M. Mutations affecting the repressibility of arginine biosynthetic enzymes in Saccharomyces cerevisiae. Eur J Biochem. 1970 Jan;12(1):31–39. doi: 10.1111/j.1432-1033.1970.tb00817.x. [DOI] [PubMed] [Google Scholar]
- Beier D. R., Sledziewski A., Young E. T. Deletion analysis identifies a region, upstream of the ADH2 gene of Saccharomyces cerevisiae, which is required for ADR1-mediated derepression. Mol Cell Biol. 1985 Jul;5(7):1743–1749. doi: 10.1128/mcb.5.7.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A., Sprague G. F., Jr MAT alpha 1 protein, a yeast transcription activator, binds synergistically with a second protein to a set of cell-type-specific genes. Cell. 1987 Aug 28;50(5):681–691. doi: 10.1016/0092-8674(87)90326-6. [DOI] [PubMed] [Google Scholar]
- Bercy J., Dubois E., Messenguy F. Regulation of arginine metabolism in Saccharomyces cerevisiae: expression of the three ARGR regulatory genes and cellular localization of their products. Gene. 1987;55(2-3):277–285. doi: 10.1016/0378-1119(87)90287-3. [DOI] [PubMed] [Google Scholar]
- Boonchird C., Messenguy F., Dubois E. Characterization of the yeast ARG5,6 gene: determination of the nucleotide sequence, analysis of the control region and of ARG5,6 transcript. Mol Gen Genet. 1991 Apr;226(1-2):154–166. doi: 10.1007/BF00273599. [DOI] [PubMed] [Google Scholar]
- Crabeel M., Huygen R., Cunin R., Glansdorff N. The promoter region of the arg3 gene in Saccharomyces cerevisiae: nucleotide sequence and regulation in an arg3-lacZ gene fusion. EMBO J. 1983;2(2):205–212. doi: 10.1002/j.1460-2075.1983.tb01406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabeel M., Huygen R., Verschueren K., Messenguy F., Tinel K., Cunin R., Glansdorff N. General amino acid control and specific arginine repression in Saccharomyces cerevisiae: physical study of the bifunctional regulatory region of the ARG3 gene. Mol Cell Biol. 1985 Nov;5(11):3139–3148. doi: 10.1128/mcb.5.11.3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabeel M., Lavalle R., Glansdorff N. Arginine-specific repression in Saccharomyces cerevisiae: kinetic data on ARG1 and ARG3 mRNA transcription and stability support a transcriptional control mechanism. Mol Cell Biol. 1990 Mar;10(3):1226–1233. doi: 10.1128/mcb.10.3.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabeel M., Messenguy F., Lacroute F., Glansdorff N. Cloning arg3, the gene for ornithine carbamoyltransferase from Saccharomyces cerevisiae: expression in Escherichia coli requires secondary mutations; production of plasmid beta-lactamase in yeast. Proc Natl Acad Sci U S A. 1981 Aug;78(8):5026–5030. doi: 10.1073/pnas.78.8.5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabeel M., Seneca S., Devos K., Glansdorff N. Arginine repression of the Saccharomyces cerevisiae ARG1 gene. Comparison of the ARG1 and ARG3 control regions. Curr Genet. 1988 Feb;13(2):113–124. doi: 10.1007/BF00365645. [DOI] [PubMed] [Google Scholar]
- Degols G. Functional analysis of the regulatory region adjacent to the cargB gene of Saccharomyces cerevisiae. Nucleotide sequence, gene fusion experiments and cis-dominant regulatory mutation analysis. Eur J Biochem. 1987 Nov 16;169(1):193–200. doi: 10.1111/j.1432-1033.1987.tb13597.x. [DOI] [PubMed] [Google Scholar]
- Dubois E., Bercy J., Descamps F., Messenguy F. Characterization of two new genes essential for vegetative growth in Saccharomyces cerevisiae: nucleotide sequence determination and chromosome mapping. Gene. 1987;55(2-3):265–275. doi: 10.1016/0378-1119(87)90286-1. [DOI] [PubMed] [Google Scholar]
- Dubois E., Bercy J., Messenguy F. Characterization of two genes, ARGRI and ARGRIII required for specific regulation of arginine metabolism in yeast. Mol Gen Genet. 1987 Apr;207(1):142–148. doi: 10.1007/BF00331501. [DOI] [PubMed] [Google Scholar]
- Dubois E., Hiernaux D., Grennon M., Wiame J. M. Specific induction of catabolism and its relation to repression of biosynthesis in arginine metabolism of Saccharomyces cerevisiae. J Mol Biol. 1978 Jul 15;122(4):383–406. doi: 10.1016/0022-2836(78)90417-5. [DOI] [PubMed] [Google Scholar]
- Dubois E., Messenguy F. Isolation and characterization of the yeast ARGRII gene involved in regulating both anabolism and catabolism of arginine. Mol Gen Genet. 1985;198(2):283–289. doi: 10.1007/BF00383008. [DOI] [PubMed] [Google Scholar]
- Evans R. M., Hollenberg S. M. Zinc fingers: gilt by association. Cell. 1988 Jan 15;52(1):1–3. doi: 10.1016/0092-8674(88)90522-3. [DOI] [PubMed] [Google Scholar]
- Friden P., Schimmel P. LEU3 of Saccharomyces cerevisiae encodes a factor for control of RNA levels of a group of leucine-specific genes. Mol Cell Biol. 1987 Aug;7(8):2708–2717. doi: 10.1128/mcb.7.8.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griggs D. W., Johnston M. Regulated expression of the GAL4 activator gene in yeast provides a sensitive genetic switch for glucose repression. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8597–8601. doi: 10.1073/pnas.88.19.8597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Ptashne M. Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2199–2203. doi: 10.1073/pnas.78.4.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters: positive and negative elements. Cell. 1984 Apr;36(4):799–800. doi: 10.1016/0092-8674(84)90028-x. [DOI] [PubMed] [Google Scholar]
- Halvorsen Y. D., Nandabalan K., Dickson R. C. Identification of base and backbone contacts used for DNA sequence recognition and high-affinity binding by LAC9, a transcription activator containing a C6 zinc finger. Mol Cell Biol. 1991 Apr;11(4):1777–1784. doi: 10.1128/mcb.11.4.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartshorne T. A., Blumberg H., Young E. T. Sequence homology of the yeast regulatory protein ADR1 with Xenopus transcription factor TFIIIA. Nature. 1986 Mar 20;320(6059):283–287. doi: 10.1038/320283a0. [DOI] [PubMed] [Google Scholar]
- Heimberg H., Boyen A., Crabeel M., Glansdorff N. Escherichia coli and Saccharomyces cerevisiae acetylornithine aminotransferase: evolutionary relationship with ornithine aminotransferase. Gene. 1990 May 31;90(1):69–78. doi: 10.1016/0378-1119(90)90440-3. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs P., Jauniaux J. C., Grenson M. A cis-dominant regulatory mutation linked to the argB-argC gene cluster in Saccharomyces cerevisiae. J Mol Biol. 1980 Jun 5;139(4):691–704. doi: 10.1016/0022-2836(80)90055-8. [DOI] [PubMed] [Google Scholar]
- Jarvis E. E., Clark K. L., Sprague G. F., Jr The yeast transcription activator PRTF, a homolog of the mammalian serum response factor, is encoded by the MCM1 gene. Genes Dev. 1989 Jul;3(7):936–945. doi: 10.1101/gad.3.7.936. [DOI] [PubMed] [Google Scholar]
- Jarvis E. E., Hagen D. C., Sprague G. F., Jr Identification of a DNA segment that is necessary and sufficient for alpha-specific gene control in Saccharomyces cerevisiae: implications for regulation of alpha-specific and a-specific genes. Mol Cell Biol. 1988 Jan;8(1):309–320. doi: 10.1128/mcb.8.1.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauniaux J. C., Dubois E., Vissers S., Crabeel M., Wiame J. M. Molecular cloning, DNA structure, and RNA analysis of the arginase gene in Saccharomyces cerevisiae. A study of cis-dominant regulatory mutations. EMBO J. 1982;1(9):1125–1131. doi: 10.1002/j.1460-2075.1982.tb01307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol Rev. 1987 Dec;51(4):458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M., Dover J. Mutations that inactivate a yeast transcriptional regulatory protein cluster in an evolutionarily conserved DNA binding domain. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2401–2405. doi: 10.1073/pnas.84.8.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga J. T., Carner K. R., Masiarz F. R., Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987 Dec 24;51(6):1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- Kammerer B., Guyonvarch A., Hubert J. C. Yeast regulatory gene PPR1. I. Nucleotide sequence, restriction map and codon usage. J Mol Biol. 1984 Dec 5;180(2):239–250. doi: 10.1016/s0022-2836(84)80002-9. [DOI] [PubMed] [Google Scholar]
- Keleher C. A., Goutte C., Johnson A. D. The yeast cell-type-specific repressor alpha 2 acts cooperatively with a non-cell-type-specific protein. Cell. 1988 Jun 17;53(6):927–936. doi: 10.1016/s0092-8674(88)90449-7. [DOI] [PubMed] [Google Scholar]
- Kovari L., Sumrada R., Kovari I., Cooper T. G. Multiple positive and negative cis-acting elements mediate induced arginase (CAR1) gene expression in Saccharomyces cerevisiae. Mol Cell Biol. 1990 Oct;10(10):5087–5097. doi: 10.1128/mcb.10.10.5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laughon A., Gesteland R. F. Primary structure of the Saccharomyces cerevisiae GAL4 gene. Mol Cell Biol. 1984 Feb;4(2):260–267. doi: 10.1128/mcb.4.2.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luche R. M., Sumrada R., Cooper T. G. A cis-acting element present in multiple genes serves as a repressor protein binding site for the yeast CAR1 gene. Mol Cell Biol. 1990 Aug;10(8):3884–3895. doi: 10.1128/mcb.10.8.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lue N. F., Buchman A. R., Kornberg R. D. Activation of yeast RNA polymerase II transcription by a thymidine-rich upstream element in vitro. Proc Natl Acad Sci U S A. 1989 Jan;86(2):486–490. doi: 10.1073/pnas.86.2.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lue N. F., Kornberg R. D. Accurate initiation at RNA polymerase II promoters in extracts from Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8839–8843. doi: 10.1073/pnas.84.24.8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marczak J. E., Brandriss M. C. Isolation of constitutive mutations affecting the proline utilization pathway in Saccharomyces cerevisiae and molecular analysis of the PUT3 transcriptional activator. Mol Cell Biol. 1989 Nov;9(11):4696–4705. doi: 10.1128/mcb.9.11.4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConkey G. A., Bogenhagen D. F. Transition mutations within the Xenopus borealis somatic 5S RNA gene can have independent effects on transcription and TFIIIA binding. Mol Cell Biol. 1987 Jan;7(1):486–494. doi: 10.1128/mcb.7.1.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenguy F., Dubois E., Boonchird C. Determination of the DNA-binding sequences of ARGR proteins to arginine anabolic and catabolic promoters. Mol Cell Biol. 1991 May;11(5):2852–2863. doi: 10.1128/mcb.11.5.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenguy F., Dubois E., Descamps F. Nucleotide sequence of the ARGRII regulatory gene and amino acid sequence homologies between ARGRII PPRI and GAL4 regulatory proteins. Eur J Biochem. 1986 May 15;157(1):77–81. doi: 10.1111/j.1432-1033.1986.tb09640.x. [DOI] [PubMed] [Google Scholar]
- Messenguy F., Dubois E. Participation of transcriptional and post-transcriptional regulatory mechanisms in the control of arginine metabolism in yeast. Mol Gen Genet. 1983;189(1):148–156. doi: 10.1007/BF00326068. [DOI] [PubMed] [Google Scholar]
- Messenguy F., Dubois E. The yeast ARGRII regulatory protein has homology with various RNases and DNA binding proteins. Mol Gen Genet. 1988 Jan;211(1):102–105. doi: 10.1007/BF00338399. [DOI] [PubMed] [Google Scholar]
- Messenguy F. Regulation of arginine biosynthesis in Saccharomyces cerevisiae: isolation of a cis-dominant, constitutive mutant for ornithine carbamoyltransferase synthesis. J Bacteriol. 1976 Oct;128(1):49–55. doi: 10.1128/jb.128.1.49-55.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J., McLachlan A. D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minty A., Kedes L. Upstream regions of the human cardiac actin gene that modulate its transcription in muscle cells: presence of an evolutionarily conserved repeated motif. Mol Cell Biol. 1986 Jun;6(6):2125–2136. doi: 10.1128/mcb.6.6.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai K., Nakaseko Y., Nasmyth K., Rhodes D. Zinc-finger motifs expressed in E. coli and folded in vitro direct specific binding to DNA. Nature. 1988 Mar 17;332(6161):284–286. doi: 10.1038/332284a0. [DOI] [PubMed] [Google Scholar]
- Nakamaye K. L., Eckstein F. Inhibition of restriction endonuclease Nci I cleavage by phosphorothioate groups and its application to oligonucleotide-directed mutagenesis. Nucleic Acids Res. 1986 Dec 22;14(24):9679–9698. doi: 10.1093/nar/14.24.9679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman C., Runswick M., Pollock R., Treisman R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell. 1988 Dec 23;55(6):989–1003. doi: 10.1016/0092-8674(88)90244-9. [DOI] [PubMed] [Google Scholar]
- Passmore S., Maine G. T., Elble R., Christ C., Tye B. K. Saccharomyces cerevisiae protein involved in plasmid maintenance is necessary for mating of MAT alpha cells. J Mol Biol. 1988 Dec 5;204(3):593–606. doi: 10.1016/0022-2836(88)90358-0. [DOI] [PubMed] [Google Scholar]
- Pellman D., McLaughlin M. E., Fink G. R. TATA-dependent and TATA-independent transcription at the HIS4 gene of yeast. Nature. 1990 Nov 1;348(6296):82–85. doi: 10.1038/348082a0. [DOI] [PubMed] [Google Scholar]
- Pfeifer K., Arcangioli B., Guarente L. Yeast HAP1 activator competes with the factor RC2 for binding to the upstream activation site UAS1 of the CYC1 gene. Cell. 1987 Apr 10;49(1):9–18. doi: 10.1016/0092-8674(87)90750-1. [DOI] [PubMed] [Google Scholar]
- Pfeifer K., Kim K. S., Kogan S., Guarente L. Functional dissection and sequence of yeast HAP1 activator. Cell. 1989 Jan 27;56(2):291–301. doi: 10.1016/0092-8674(89)90903-3. [DOI] [PubMed] [Google Scholar]
- Pfeifer K., Prezant T., Guarente L. Yeast HAP1 activator binds to two upstream activation sites of different sequence. Cell. 1987 Apr 10;49(1):19–27. doi: 10.1016/0092-8674(87)90751-3. [DOI] [PubMed] [Google Scholar]
- Pieler T., Hamm J., Roeder R. G. The 5S gene internal control region is composed of three distinct sequence elements, organized as two functional domains with variable spacing. Cell. 1987 Jan 16;48(1):91–100. doi: 10.1016/0092-8674(87)90359-x. [DOI] [PubMed] [Google Scholar]
- Pollock R., Treisman R. A sensitive method for the determination of protein-DNA binding specificities. Nucleic Acids Res. 1990 Nov 11;18(21):6197–6204. doi: 10.1093/nar/18.21.6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H. F., Dubois E., Broën P., Messenguy F. Functional analysis of ARGRI and ARGRIII regulatory proteins involved in the regulation of arginine metabolism in Saccharomyces cerevisiae. Mol Gen Genet. 1990 Jul;222(2-3):192–200. doi: 10.1007/BF00633817. [DOI] [PubMed] [Google Scholar]
- Qui H. F., Dubois E., Messenguy F. Dissection of the bifunctional ARGRII protein involved in the regulation of arginine anabolic and catabolic pathways. Mol Cell Biol. 1991 Apr;11(4):2169–2179. doi: 10.1128/mcb.11.4.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qui H. F., Dubois E., Messenguy F. Dissection of the bifunctional ARGRII protein involved in the regulation of arginine anabolic and catabolic pathways. Mol Cell Biol. 1991 Apr;11(4):2169–2179. doi: 10.1128/mcb.11.4.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos F., Thuriaux P., Wiame J. M., Bechet J. The participation of ornithine and citrulline in the regulation of arginine metabolism in Saccharomyces cerevisiae. Eur J Biochem. 1970 Jan;12(1):40–47. doi: 10.1111/j.1432-1033.1970.tb00818.x. [DOI] [PubMed] [Google Scholar]
- Roy A., Exinger F., Losson R. cis- and trans-acting regulatory elements of the yeast URA3 promoter. Mol Cell Biol. 1990 Oct;10(10):5257–5270. doi: 10.1128/mcb.10.10.5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai D. D., Helms S., Carlstedt-Duke J., Gustafsson J. A., Rottman F. M., Yamamoto K. R. Hormone-mediated repression: a negative glucocorticoid response element from the bovine prolactin gene. Genes Dev. 1988 Sep;2(9):1144–1154. doi: 10.1101/gad.2.9.1144. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui A. H., Brandriss M. C. The Saccharomyces cerevisiae PUT3 activator protein associates with proline-specific upstream activation sequences. Mol Cell Biol. 1989 Nov;9(11):4706–4712. doi: 10.1128/mcb.9.11.4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman D. J., Bankier A. T., Seddon A., Groenhout E. G., Nasmyth K. A. Characterization of a transcription factor involved in mother cell specific transcription of the yeast HO gene. EMBO J. 1988 Feb;7(2):485–494. doi: 10.1002/j.1460-2075.1988.tb02836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Helix-turn-helix, zinc-finger, and leucine-zipper motifs for eukaryotic transcriptional regulatory proteins. Trends Biochem Sci. 1989 Apr;14(4):137–140. doi: 10.1016/0968-0004(89)90145-X. [DOI] [PubMed] [Google Scholar]
- Sumrada R. A., Cooper T. G. Nucleotide sequence of the Saccharomyces cerevisiae arginase gene (CAR1) and its transcription under various physiological conditions. J Bacteriol. 1984 Dec;160(3):1078–1087. doi: 10.1128/jb.160.3.1078-1087.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumrada R. A., Cooper T. G. Point mutation generates constitutive expression of an inducible eukaryotic gene. Proc Natl Acad Sci U S A. 1985 Feb;82(3):643–647. doi: 10.1073/pnas.82.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumrada R. A., Cooper T. G. Ubiquitous upstream repression sequences control activation of the inducible arginase gene in yeast. Proc Natl Acad Sci U S A. 1987 Jun;84(12):3997–4001. doi: 10.1073/pnas.84.12.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuriaux P., Ramos F., Wiame J. M., Grenson M., Béchet J. Sur l'existence de gènes régulateurs affectant simultanément la synthèse des enzymes biosynthétiques et cataboliques de l'arginine chez "Saccharomyces cerevisiae". Arch Int Physiol Biochim. 1968 Dec;76(5):955–956. [PubMed] [Google Scholar]
- Westin G., Schaffner W. A zinc-responsive factor interacts with a metal-regulated enhancer element (MRE) of the mouse metallothionein-I gene. EMBO J. 1988 Dec 1;7(12):3763–3770. doi: 10.1002/j.1460-2075.1988.tb03260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray L. V., Jr, Witte M. M., Dickson R. C., Riley M. I. Characterization of a positive regulatory gene, LAC9, that controls induction of the lactose-galactose regulon of Kluyveromyces lactis: structural and functional relationships to GAL4 of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Mar;7(3):1111–1121. doi: 10.1128/mcb.7.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong J., Ashraf J., Thompson E. B. The promoter and first, untranslated exon of the human glucocorticoid receptor gene are GC rich but lack consensus glucocorticoid receptor element sites. Mol Cell Biol. 1990 Oct;10(10):5580–5585. doi: 10.1128/mcb.10.10.5580. [DOI] [PMC free article] [PubMed] [Google Scholar]