Abstract
Chromosomal double-strand breaks (DSBs) are resected by 5′-nucleases to form 3′ single-strand DNA (ssDNA) substrates for binding by homologous recombination and DNA damage checkpoint proteins. Two redundant pathways of extensive resection were described both in cells 1-3 and in vitro 4-6, one relying on Exo1 exonuclease and the other on Sgs1 helicase and Dna2 nuclease. However, it remains unknown how resection proceeds within the context of chromatin where histones and histone-bound proteins represent barriers for resection enzymes. Here, we have identified the yeast nucleosome remodeling enzyme Fun30 as novel factor promoting DSB end resection. Fun30 is the major nucleosome remodeler promoting extensive Exo1- and Sgs1-dependent resection of DSBs while the RSC and INO80 chromatin remodeling complexes play redundant roles with Fun30 in resection adjacent to DSB ends. ATPase and helicase domains of Fun30, which are needed for nucleosome remodeling 7, are also required for resection. Fun30 is robustly recruited to DNA breaks and spreads around the DSB coincident with resection. Fun30 becomes less important for resection in the absence of the histone-bound Rad9 checkpoint adaptor protein known to block 5′ strand processing 8 and in the absence of either histone H3 K79 methylation or γ-H2A, which mediate recruitment of the Rad9 9, 10. Together these data suggest that Fun30 helps to overcome the inhibitory effect of Rad9 on DNA resection.
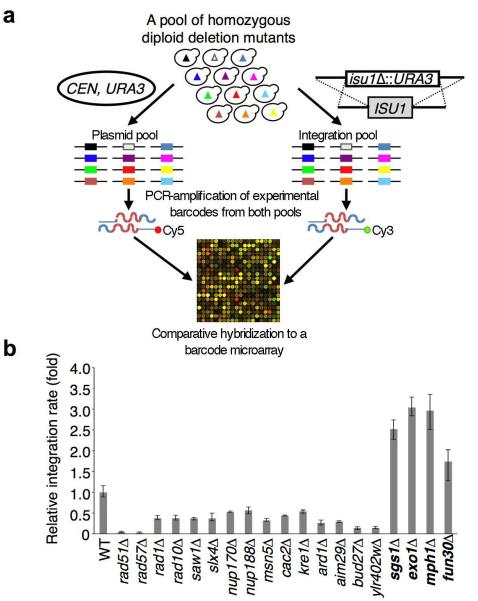
In order to identify novel proteins that play important roles in homologous recombination (HR), we screened a pool of 4,836 homozygous diploid yeast deletion mutants11 for those with altered rates of integration of a URA3 cassette at two independent loci (ISU1 and SOD2) (Fig. 1a). These screens identified most of the known as well as several novel genes that are essential or important in HR, including the SGS1 and EXO1 that are known to suppress gene targeting (Fig. 1b and Table S1). Higher rates of gene integration in the sgs1Δ and exo1Δ mutants are likely attributable to increased stability of the transformed linear DNA fragments12 as Sgs1 helicase and Exo1 nuclease enzymes are involved in the degradation of 5′ strands at DSB ends2, 3. The Mph1 helicase that was also identified in the screen may inhibit gene targeting by unwinding early recombination intermediates 13. Thus, degradation of a transformed integration cassette by resection enzymes and disruption of early recombination intermediates by DNA helicases likely prevent efficient integration in cells. In addition to the sgs1Δ, exo1Δ and mph1Δ mutants, gene targeting was significantly increased in the fun30Δ mutant (Fig. 1b), suggesting that Fun30 may also be involved in either DSB end resection or processing recombination intermediates. Fun30 is a member of the well-conserved yet poorly characterized Etl1 subfamily of Snf2 nucleosome remodeling factors14 that acts as a homodimeric ATP-dependent nucleosome remodeling enzyme7, and has also been implicated in gene silencing15, 16.
Figure 1. A genome-wide screen identifies novel genes that regulate gene integration in yeast.
a. A barcode microarray-based screen for mutants with altered frequency of gene integration. A mixed population of 4,836 homozygous diploid deletion mutant strains was transformed with an integrating cassette (i.e. isu1Δ∷URA3) or a centromeric plasmid (control). Molecular barcodes representing the deletion mutants were PCR-amplified from genomic DNA samples isolated from both transformed pools using Cy5- or Cy3-labeled primers and co-hybridized to a barcode microarray to reveal mutants with altered gene integration efficiency. b. Integration frequency in selected mutants. Error bars represent standard deviation, (n=3).
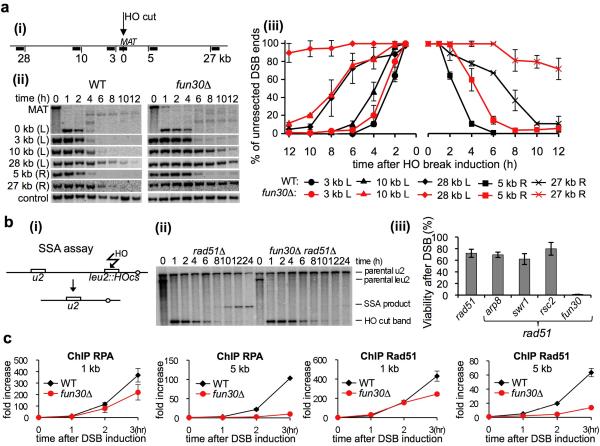
To assess a possible role of Fun30 in resection, we deleted FUN30 in a haploid strain where a DSB was induced by HO endonuclease at the MAT locus and the HML and HMR homologous sequences were deleted to prevent repair by HR. Southern blot analysis with multiple probes on both sides of the break allowed us to follow the loss of restriction fragments as a measure of 5′ strand resection at varying distances from the DSB3. Deletion of FUN30 only mildly affected the initial resection next to the DSB but severely delayed resection at regions 5, 10 and 27-28 kb from the break site (Fig. 2a). Consistently, fun30Δ mutants failed to repair DSBs by Single Strand Annealing (SSA) 17, a mechanism of DSB repair between two direct repeats that requires extensive resection followed by annealing of the repeats (Fig. 2b). In contrast, the mutants deficient in INO80, SWR1 or RSC complexes that were previously implicated in resection18-20, were proficient in SSA, suggesting a unique function of Fun30 in extensive resection (Fig. 2b). Consistent with defects in resection in fun30Δ mutants, loading of the ssDNA binding protein RPA, and the Rad51 recombinase, was moderately decreased within 1 kb from DSB but greatly diminished at 5 kb from the break site (Fig. 2c).
Figure 2. fun30Δ mutants are deficient in resection.
a. (i) Diagram of the MAT locus and probes used to follow resection kinetics are shown. Southern blot analysis (ii) and kinetics (iii) of DSB end resection in wild-type and fun30Δ mutants. Plotted in this and all following graphs are mean values of three independent experiments with error bars denoting standard deviation. b. (i) Schematic representation of SSA assay17 (ii) Southern blot analysis of SSA kinetics, and (iii) cell viability in response to a DSB repaired by SSA. c. ChIP analysis of Rad51 and RPA recruitment in wild-type and fun30Δ cells.
To test if Fun30 impacts on processing HR intermediates as Mph1 or Sgs1 we tested crossover frequency using an ectopic recombination assay involving MATa and MATa-inc sequences located on chromosomes V and III, respectively21. Crossover and noncrossover products can be distinguished based on restriction fragment size (Fig. S1). In this assay fun30Δ mutant displayed repair efficiency comparable to wild type cells and to other single mutants that affect extensive resection 3. However, a 2-fold increase in crossover frequency was found in fun30Δ cells but not in mutants of other tested remodelers (Fig. S1). This is the first evidence linking a nucleosome remodeling enzyme to recombination pathway choice. In this study we focused on the role of Fun30 in resection.
There are two pathways of extensive resection in yeast that depend on either Exo1 or Sgs11-3. To test whether Fun30 works in either one or both pathways we compared resection in exo1Δ, sgs1Δ and fun30Δ single mutants with that in sgs1Δ fun30Δ and exo1Δ fun30Δ double mutants. Both double mutants exhibited more severe resection defects and DNA damage sensitivities than either single mutant (Fig. S2), and the triple mutant sgs1Δ exo1Δ fun30Δ had the same pattern of resection as sgs1Δ exo1Δ double mutant cells (Fig. S2 and 3). Therefore we conclude that Fun30 contributes to the both Exo1 and Sgs1 resection pathways.
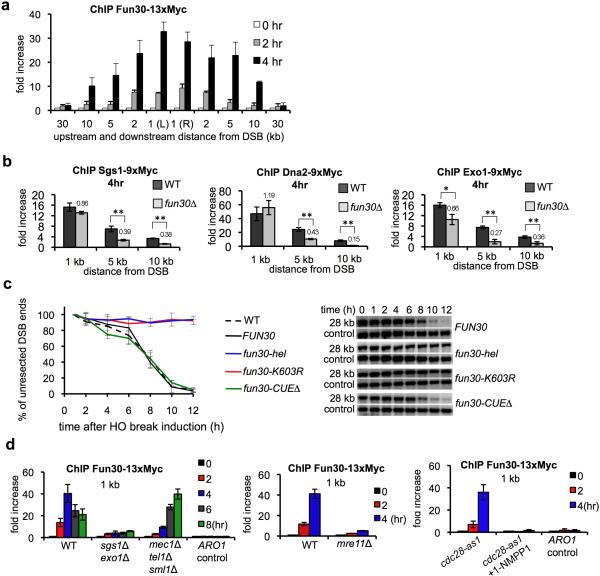
Fun30 likely regulates resection directly because we observed robust Fun30 recruitment to DSBs which spread in both directions around the DSB coincident with resection (Fig. 3a). Also, Fun30 co-immunoprecipitates with other resection proteins including Exo1, Dna2 and RPA (Fig. S4). Further evidence comes from the observation that in fun30Δ cells resection enzymes were recruited next to DSB ends, but failed to efficiently spread further from the break site (Fig. 3b), suggesting that Fun30 is important for the progression of resection. Finally, both helicase and ATPase domains needed for nucleosome remodeling by Fun30 7 are also required for efficient resection (Fig. 3c). It is unlikely that Fun30 plays indirect role in resection via changing general chromatin structure becasue the patterns of nucleosome positioning at multiple loci around an HO cut site in fun30Δ and the wild type cells is comparable (Fig. S3). Also microarray analysis of genome-wide gene expression showed that Fun30 deletion cause only modest changes in expression of fourteen genes unrelated to DNA damage response15, 16 (Table S2) and cellular levels of proteins involved in resection were comparable in wild-type and fun30Δ cells (Fig. S3).
Figure 3. Fun30 plays a direct role in DSB ends resection.
a. ChIP analysis of Fun30-13xMyc recruitment at DSB. b. Recruitment of resection enzymes analyzed by ChIP. Statistically significant differences in protein enrichment are indicated by “*” (P<0.05) or “**” (P<0.01, t-test). Relative enrichment in fun30Δ is shown. c. Analysis of resection kinetics in indicated FUN30 mutants. The level of mutant proteins and their recruitment to DSB were comparable to wild-type Fun30 (Fig. S5). d. ChIP analysis of Fun30-13xMyc recruitment in mutants deficient in resection or DNA damage checkpoint. Recruitment at the ARO1 locus was used as a control.
Chromatin remodeling at DSBs could occur independent of resection or in a coupled manner with resection. The second possibility is more likely because recruitment of Fun30 to DSBs is severely impaired in mutants defective in resection such as sgs1Δ exo1Δ, mre11Δ, or in the absence of active Cdk1 kinase22, 23 (Fig. 3d). This defect is unlikely due to a poor checkpoint activation because Fun30 was efficiently recruited in checkpoint-deficient cells (mec1Δ tel1Δ sml1Δ), albeit with a delay (Fig. 3d). Together with the observation that Fun30 spreads around the DSB coincident with resection and it is required for efficient spreading of resection enzymes away from the DSB we reason that Fun30-mediated chromatin remodeling and resection appear in a coupled manner. Consistent with this view there is no detectable change in histone H3 occupancy before resection as monitored by histone H3 ChIP. However histones remain bound to DSB ends longer in fun30Δ or sgs1Δ exo1Δ mutants, this can be attributed to slower resection (Fig. S6 and 24).
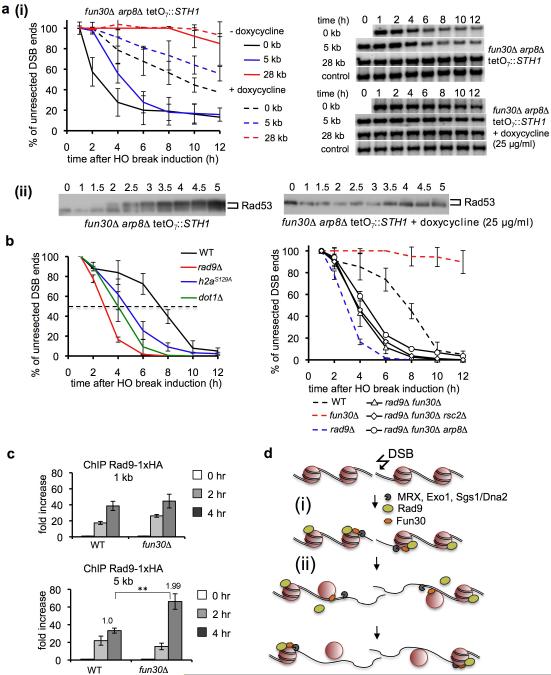
Previous studies indicated an important role for the INO8020, RSC19, and SWR118 ATP-dependent nucleosome remodeling complexes in resection. To examine possible redundant activities in resection we created single mutants lacking individual components of these and other complexes and the corresponding double mutants that also lack Fun30. No significant resection defect was observed in mutants deficient in CHD1, ISW1a/b or SWR1 activity alone, and in combination with fun30Δ the defect was comparable to that in fun30Δ single mutant (Fig. S7-S8). Also Rad54, a Snf2-like protein 25, has no role in resection. However the damage sensitivity of rad54Δ is epistatic with fun30Δ, providing further evidence for Fun30’s role in the HR pathway (Fig. S9). A very mild defect was observed in arp8Δ (INO80 complex) mutant cells while in rsc2Δ mutant initial resection was significantly delayed, a phenotype similar to but weaker than that observed in MRX mutants3. The arp8Δ fun30Δ and rsc2Δ fun30Δ double mutants showed more dramatic defects in resection and DNA damage response than the single mutants (Fig. S7-S8). Elimination of all three remodeling factors impaired resection and checkpoint activation very severely (Fig. 4a). Taken together these results indicate that Fun30 is the most important remodeler promoting extensive resection while initial resection close to DSBs is stimulated by RSC and to lesser extent by Fun30 and INO80.
Figure 4. Fun30 chromatin remodeling factor promotes resection within Rad9-bound nucleosomes.
a. Analysis of DSB end resection (i) and Western blot showing Rad53 phosphorylation (ii). STH1 expression (component of RSC) was eliminated by adding doxycycline. b. Analysis of resection at 28 kb from a DSB. Southern blots are shown in Figure S10. c. ChIP analysis of Rad9-HA binding. d. Model of coupled resection and chromatin remodeling. (i) Nucleases and helicases resect DNA within a nucleosome-free region next to a break. Histones/histone-bound Rad9 impedes further resection. (ii) Fun30 is recruited to nucleosomes being resected and increases the access to nucleosomal DNA.
Besides tight histone-DNA interactions, the histone-bound checkpoint mediator protein Rad9 represents an additional barrier for resection. Rad9 is recruited to histone H2A phosphorylated at S129 (also called γ-H2A) and histone H3 methylated at K79 by Dot1 9, 10, and it inhibits resection both at DSBs and at telomeres8, 26, a phenomenon conserved in evolution 27, 28. We examined the possible role of nucleosome remodelers in resection within nucleosomes associated with Rad9. In rad9Δ cells extensive resection is dramatically increased, with an average rate of 10 kb/hr compared to 4 kb/hr in wild-type cells3 while in hta1/2-S129* or dot1Δ mutant cells resection is moderately increased (6-7 kb/hr) (Fig. 4b). Resection in fun30Δ rad9Δ cells was much faster than in fun30Δ cells and similarly Fun30 becomes partially dispensable for resection in the absence of γH2A or Dot1 (Fig. 4b; Fig. S10). Accordingly, deletion of DOT1 partially suppressed the DNA damage sensitivity of fun30Δ mutants, indicating that the observed sensitivity of fun30Δ cells is at least partially due to a resection defect (Fig. S10). Faster resection in fun30Δ rad9Δ cells was not due to increased INO80 or RSC activity within Rad9-free chromatin as resection kinetics in the fun30Δ rad9Δ rsc2Δ and fun30Δ rad9Δ arp8Δ triple mutants was still increased (Fig. 4b) relative to fun30Δ cells. These data suggest that Fun30 is particularly important for remodeling and resection within Rad9-bound chromatin. One possibility is that Fun30 helps to overcome the resection barrier formed by Rad9. Accordingly, more Rad9 was accumulated at DSB ends in fun30Δ than in wild-type cells (Fig. 4c, model in Fig. 4d).
In summary, we have identified Fun30 as a novel regulator of HR that promotes resection in the context of chromatin. Our findings in yeast shed light on the coordinated processes of DNA repair and chromatin remodeling and likely provide insights into possible roles of human homologue, SMARCAD129, in DNA repair.
Methods summary
All experiments were performed in Saccharomyces cerevisiae strains listed in Supplemental Table 3. Resection kinetics was analyzed by Southern blotting using probes specific for broken chromosome. Protein recruitment was followed by ChIP, proteins interactions were studied by co-immunoprecipitation and Western blotting. Detailed methods are presented in Supplementary Information.
Methods
Yeast strains and plasmids
All strains used are listed in Supplemental Table 3. The fun30 ATPase dead, helicase domain deletion, and Cue domain deletion mutants were first constructed using fusion PCR with a high fidelity Phusion DNA polymerase (NEB). In each case, the mutation or deletion was introduced via a pair of overlapping PCR primers. The PCR products were subsequently cloned onto pXP735, a derivative of the plasmid pFA6a-kanMX6 30 with a TRP1 promoter inserted between the pTEF and kanMX coding sequences. Wild-type FUN30 was similarly cloned into pXP735. In each case, the FUN30 gene, including 490 bp of the 5’ untranslated region (5’UTR), the coding sequence, and 300 bp of the 3’UTR was inserted between the pTEF and TRP1 promoter sequences on the vector and verified with DNA sequencing. The resultant plasmid was then used as the template to amplify an integration cassette containing the 5’UTR, coding region, and 3’UTR of FUN30, the TRP1 promoter that directs expression of the kanMX gene and a tTEF fragment. The PCR products were transformed into a fun30Δ∷natMX mutant. The 5’UTR of FUN30 and the tTEF sequences at both ends of the PCR products direct their integration into the genome by replacing the existing fun30Δ∷natMX cassette via homologous recombination. As a result, the wild-type or mutant alleles were integrated at the endogenous FUN30 locus, with its expression controlled by the endogenous regulatory elements. Integrants were selected for G418 resistance and Clonat sensitivity, PCR amplified, and verified with DNA sequencing. The sequences of all oligonucleotide primers used for constructing and sequencing these alleles are available upon request.
Genome-wide screens for mutants that regulate gene integration efficiency
Both isu1Δ∷URA3 and sod2Δ∷URA3 gene knockout cassettes containing ~1.5 kb of flanking sequences on each side of the endogenous ISU1 or SOD2 locus were separately transformed into a pool of 4,836 homozygous diploid deletion mutants. About 8×105 independent Ura+ transformants were obtained for each cassette. As a control, the centromeric circular plasmid pRS416 (URA3), which is maintained in yeast cells ectopically, was also transformed into the pool. For each transformation reaction, all Ura+ transformants were pooled and a genomic DNA sample was extracted. An integration sample and the control sample were analyzed with the barcode microarray to identify deletion mutants with altered integration frequency. Results of both the isu1Δ∷URA3 and sod2Δ∷URA3 screens were combined to compile a list of mutants with reduced or increased integration efficiency (by >=2-fold). These mutants were then individually validated for relative integration efficiency using a third integration cassette can1Δ∷URA3 in a well-controlled manner. Briefly, both the can1Δ∷URA3 cassette and a LEU2 plasmid (pRS415) were transformed into each strain within the same transformation reaction. Cells from such a reaction were split and plated on both SC-Ura plates to select for transformants resulting from integration and SC-Leu plates to select for cells containing the plasmid. The numbers of Ura+ and Leu+ transformants obtained from each reaction were compared to calculate the relative integration rate for each strain, with that of a wild-type strain arbitrarily set to 1.0 as a reference. Four such independent transformation reactions were carried out with each strain and the results were averaged.
Microarray analysis of gene expression
Both the wild-type BY4741 and an isogenic fun30Δ mutant were grown in 10 ml of synthetic complete medium until log phase. Cells were collected and resuspended in 400 μl of TES solution (10 mM Tris-HCl pH 7.5, 10 mM EDTA, 0.5% SDS). Cells were lysed by vortexing with glass beads after adding 400 μl of acidic phenol to the cell suspension. Cell lysates were clarified by centrifugation at 13,000 rpm for 10 min at 4°C. Protein in the supernatant was extracted once with an equal volume of phenol:chloroform (1:1) solution, followed by extraction once with an equal volume of chloroform. The resulting supernatant was mixed with three volumes of 100% ethanol and 0.1 volume of 3M NaOAc (pH 5.2) and incubated at −80°C for 2 hrs. Total RNA was precipitated by centrifugation at 13,000 rpm for 20 min at 4°C followed by washing once with 70% ethanol. After a brief air dry (5 min), RNA was dissolved in 50 μl of DEPC-treated water and analyzed with an Affimatrix microarray. Two independent experiments were carried out for each strain and the results were combined.
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation assays were carried out as previously described 31, 32 with modification. Briefly, cultures were grown to a density of about 1 × 107 cells/ml in pre-induction medium (YEP-Raffinose) and the HO endonuclease was induced by adding 2% galactose. Protein-DNA complexes were crosslinked with 1% (final concentration) formaldehyde and incubated for 10 min at room temperature with rotation. The reaction was quenched by adding glycine to a final concentration of 125 mM and incubated at RT for 5 min with rotation. Cells were lysed with glass beads in lysis buffer (50 mM HEPES pH 7.5, 1 mM EDTA, 140 mM NaCl, 1% Triton X-100, 0.1% NaDOC, 1 mg/ml Bacitracin, 1 mM Benzamidine, and 1 mM PMSF) supplemented with Roche protease inhibitor tablets. Cell extracts were sonicated to shear the DNA to an average size of 0.5 kb. IP samples were incubated with affinity-purified anti-Myc (Sigma M4439), anti-FLAG (Sigma M2), anti-Rad51 (gift from Dr. Patrick Sung), or anti-Rfa2 (gift from Dr. Wolf-Dietrich Heyer) antibodies overnight at 4°C, followed by incubation with protein-G agarose beads for 4 hr at 4°C. The protein bound beads were washed twice with lysis buffer used above, twice with lysis buffer containing 500 mM NaCl, twice in wash buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA, 0.25 M LiCl, 0.5% NP-40 substitute and 0.5% NaDOC), and twice in 1x TE. Protein/DNA complexes were eluted with elution buffer (10 mM Tris-HCl pH 8.0, 10 mM EDTA pH 8.0, 1%SDS) and incubated at 65°C overnight to reverse crosslinking. Samples were digested with proteinase K at 37°C for 6 hr. DNA was purified by phenol extraction and ethanol precipitation. Purified DNA samples were analyzed by real-time quantitative PCR (ABI 7900HT) with primers that specifically anneal to DNA sequences located at indicated distances from the DSB using the following conditions: 95°C for 10 min; 40 cycles of 95°C for 15 sec, 60°C for 1 min, and 72°C for 30 sec. Histone loss was measured according to the method described by Chen et al. 24.
Co-Immunoprecipitation(Co-IP)
To examine the potential interaction between Fun30 and resection enzymes, co-immunoprecipitation was performed as described by Lambert et al 33 with modification. Yeast cells (A600 ~ 1.0) grown in YEPD medium were treated with 0.1% methyl methanesulfonate (MMS) for 90 min. Treated cells or untreated control cells were lysed on a bead beater in lysis buffer (100 mM HEPES pH 8.0, 20 mM Mg(Ac)2, 150 mM NaCl, 10% glycerol (v/v), 0.4% Nonidet P-40, 10 mM EGTA, 0.1mM EDTA, plus protease and phosphatase inhibitors). The extracts were gently clarified by centrifugation at 1800 × g for 10 min at 4°C. The supernatant for each sample was collected and split into two aliquots. 1000 units of Benzonase (Sigma) were added to one of the aliquot (~1.5 ml) to digest genomic DNA and RNA. The supernatant was then incubated with protein G-agarose beads for 3 hr at 4°C to capture the proteins or protein-DNA complexes that bind non-specifically to the beads. Beads were discarded after centrifugation at 1800 × g for 5 min. The supernatant was incubated with 10 μg of anti-Myc or anti-FLAG antibody at 4°C overnight with agitation. In parallel, a mock immunoprecipitation was established for each set of Co-IP reactions by incubating one aliquot of MMS-treated sample with an appropriate IgG. Protein G-agarose beads were added to each sample and the mixtures were incubated for another 3 hr at 4°C. Then the beads were subjected to extensive washing at 4°C with wash buffer (100 mM HEPES pH 7.4, 20 mM Mg(Ac)2, 150 mM NaCl, 10% glycerol (v/v), 0.5% Nonidet P-40, 10 mM EGTA, 0.1mM EDTA, plus protease and phosphatase inhibitors). Immunoprecipitated proteins were eluted by boiling beads in 2x SDS loading buffer for 5 min.
Western Blotting
Whole-cell extracts (WCE) were prepared as previously described 31. Yeast cells grown overnight in rich medium (YEPD) or pre-induction medium (YEP-raffinose: 1% yeast extract, 2% peptone, 2% raffinose) either before or at different times following HO break induction were collected. WCEs were prepared using a trichloroacetic acid (TCA) method. Pelleted cells from 5 ml of culture were washed once with water and resuspended in 10% TCA. The cells were lysed by vortexing with acid-washed glass beads and the protein lysates were pelleted by centrifugation at 20,000 × g for 15 min. The pellets were washed with ice-cold 80% acetone and proteins were dissolved in 2x SDS sample loading buffer by boiling for 5 min. Samples were centrifuged for 5 min at 13,200 × g in a microcentrifuge and the supernatant was retained as the protein extract. WCEs or immunoprecipitated samples were resolved on a 7.5% or 8.5% SDS-PAGE gel and transferred onto a polyvinylidene difluoride membrane (Immobilon-P; Millipore). To examine the expression levels of Sgs1-9xMyc, Exo1-9xMyc, Mre11-13xMyc or Dna2-9xMyc in WT and fun30 cells, the membrane was probed with mouse monoclonal anti-Myc antibody (M4439, Sigma). To monitor checkpoint activation during DSB repair, the membrane was probed with anti-Rad53 antibody (gift from Dr. Achille Pellicioli). Anti-mouse and anti-rabbit IgG HRP-conjugated secondary antibodies were obtained from Santa Cruz Biotechnology. Blots were developed using the ECL plusWestern Blotting System (GE Healthcare).
Analysis of resection at DSB ends
Resection of DSB ends was analyzed at an HO endonuclease-induced DSB at the MAT locus on chromosome III using Southern blots as previously described 3. Briefly, genomic DNA was isolated by glass bead disruption using a standard phenol extraction method. Purified DNA was digested with EcoRI and separated on a 0.8% agarose gel. Southern blotting and hybridization with radiolabeled DNA probes specific for the MAT locus was performed as described previously 34. The band signal intensities were analyzed with ImageQuant TL software (Amersham Biosciences) and normalized to the TRA1 probe. DSB end resection beyond each EcoRI site for each time point was estimated as the percentage of the band signal intensity corresponding to the EcoRI fragment of interest at 1 h after break induction.
Single-strand annealing (SSA) and ectopic recombination assays
Repair by SSA between two partial LEU2 gene repeats was followed by Southern blot analysis using a leu2 sequence probe as previously described (Vaze et al., 2002). Experiments are done in a rad51Δ background to prevent repair by break-induced replication (BIR). Repair by ectopic recombination between chromosome V and chromosome III was followed by Southern blot analysis using a MATa sequence probe. Crossing-over was measured at an 8 h time point as previously described 21. To test cell viability in response to a DSB repaired by SSA or by ectopic recombination, cells were grown in pre-induction medium (YEP-Raffinose) overnight to an early log phase. Cells were then diluted to a concentration of ~1×103 cells/ml and equal amounts of cells were plated on YEPD and YEP-Gal plates. Plates were incubated at 30°C for 3 days. Viability was calculated by dividing the number of colonies grown on YEP-Gal by the number of colonies grown on YEPD multiplied by 100%. Three independent experiments were performed for each strain.
Spot assays for analyzing sensitivity to DNA-damaging agents
Yeast cells were grown in YEPD rich medium overnight to saturation. Undiluted and 1/10 serial dilutions of each cell culture were spotted onto plates containing camptothecin or phleomycin DNA-damaging agent at the indicated concentrations. Plates were incubated at 30°C for two to three days before being photographed.
Micrococcal nuclease digestion
Yeast cells from 80 ml of culture (OD600 ~ 0.8-1.0) were harvested by centrifugation at 4000 × g and washed twice with distilled deionized H2O. The cell pellet was resuspended in 900 μl of sorbitol solution (1M Sorbitol, 50mM Tris-Cl pH 7.5). 0.56 μl of 14.3M β-mercaptoethanol and 100 μl of Zymolase 20T stock (dissolved in sorbitol solution, 25 mg/ml) were added to each sample. Samples were then incubated at 30°C with rotation for 25 min to digest cell membranes. Spheroplasts were harvested by centrifugation at 14,000 × g for 2 min and washed twice with ice-cold sorbitol solution. The pellet was resuspended with 1.3 ml of lysis buffer (0.5mM spermidine, 1mM β-mercaptoethanol, 0.075% Nonidet P-40, 50 mM NaCl, 10mM Tris-HCl pH8.0, 5mM MgCl2, 5mM CaCl2, plus protease inhibitors). Micrococcal nuclease (Roche Applied Science) was added to each sample to a final concentration of 100 U/ml. After mixing, 200 μl of nuclei suspension was immediately taken out as an undigested control. The remaining samples were subjected to MNase digestion at 37°C. Aliquots of nuclei suspension were taken out at 2, 4, 8, and 16 min, and the reactions were immediately stopped by addition of 20 mM EDTA pH 8.0 and 1% SDS. The supernatants were collected by centrifugation at 14,000 × g for 10 min, followed by Proteinase K digestion (0.1 mg/ml) at 50°C for 3 hr. The resultants were purified by phenol-chloroform extraction, DNA was recovered by ethanol precipitation. DNA pellets were washed once with 70% ethanol, and then dissolved in 1x TE. After RNase treatment, samples were separated on a 1.4% agarose gel. Nucleosome mapping at specific loci was examined by Southern blotting and hybridization with DNA probes specific for sequences located at the HO cleavage site or 5, 10 or 27 kb from the DSB.
Supplementary Material
Acknowledgements
We thank Drs. Patrick Sung, Wolf-Dietrich Heyer, and Achille Pellicioli for antibodies; Sang-Eun Lee, Steve Kron and Mary Osley for strains; Bertrand Llorente for sharing unpublished data; and Jessica Tyler, Patrick Sung and Sang Eun Lee for critical comments on the manuscript. This work was supported by the National Institutes of Health (NIH) grants GM080600 to G.I. and HG004840 to X.P.
Footnotes
Primary Accessions
Microarray data were deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE38601.
Author contributions
X.C. constructed most of the strains and analyzed chromatin structure, protein interactions, protein recruitment to DSBs, histone loss at DSBs, activation of the DNA damage checkpoint and DNA damage sensitivity. X.Z. and K. C. performed microarray analysis of gene expression. D.C., J.T., and X.P. performed the genetic screen and constructed fun30 point mutants. A.P. and C-D.C analyzed resection and crossover frequency. X.C., X.P. and G.I. designed the experiments, discussed the data and wrote the manuscript.
References
- 1.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases dna2 and exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cejka P, et al. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010;467:112–116. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicolette ML, et al. Mre11-Rad50-Xrs2 and Sae2 promote 5’ strand resection of DNA double-strand breaks. Nat Struct Mol Biol. 2010;17:1478–1485. doi: 10.1038/nsmb.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niu H, et al. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature. 2010;467:108–111. doi: 10.1038/nature09318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Awad S, Ryan D, Prochasson P, Owen-Hughes T, Hassan AH. The Snf2 homolog Fun30 acts as a homodimeric ATP-dependent chromatin-remodeling enzyme. J Biol Chem. 2010;285:9477–9484. doi: 10.1074/jbc.M109.082149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lazzaro F, et al. Histone methyltransferase Dot1 and Rad9 inhibit single-stranded DNA accumulation at DSBs and uncapped telomeres. Embo J. 2008;27:1502–1512. doi: 10.1038/emboj.2008.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammet A, Magill C, Heierhorst J, Jackson SP. Rad9 BRCT domain interaction with phosphorylated H2AX regulates the G1 checkpoint in budding yeast. EMBO Rep. 2007;8:851–857. doi: 10.1038/sj.embor.7401036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toh GW, et al. Histone H2A phosphorylation and H3 methylation are required for a novel Rad9 DSB repair function following checkpoint activation. DNA Repair (Amst) 2006;5:693–703. doi: 10.1016/j.dnarep.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 11.Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 12.Chung WH, Zhu Z, Papusha A, Malkova A, Ira G. Defective resection at DNA double-strand breaks leads to de novo telomere formation and enhances gene targeting. PLoS Genet. 2010;6:e1000948. doi: 10.1371/journal.pgen.1000948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prakash R, et al. Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev. 2009;23:67–79. doi: 10.1101/gad.1737809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34:2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neves-Costa A, Will WR, Vetter AT, Miller JR, Varga-Weisz P. The SNF2-family member Fun30 promotes gene silencing in heterochromatic loci. PLoS One. 2009;4:e8111. doi: 10.1371/journal.pone.0008111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu Q, Zhang X, Bi X. Roles of chromatin remodeling factors in the formation and maintenance of heterochromatin structure. J Biol Chem. 2011;286:14659–14669. doi: 10.1074/jbc.M110.183269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaze M, et al. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires srs2 helicase. Mol Cell. 2002;10:373–385. doi: 10.1016/s1097-2765(02)00593-2. [DOI] [PubMed] [Google Scholar]
- 18.Kalocsay M, Hiller NJ, Jentsch S. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol Cell. 2009;33:335–343. doi: 10.1016/j.molcel.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 19.Shim EY, et al. RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol Cell Biol. 2007;27:1602–1613. doi: 10.1128/MCB.01956-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Attikum H, Fritsch O, Hohn B, Gasser SM. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell. 2004;119:777–788. doi: 10.1016/j.cell.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 21.Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ira G, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivanov EL, Sugawara N, White CI, Fabre F, Haber JE. Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:3414–3425. doi: 10.1128/mcb.14.5.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen CC, et al. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell. 2008;134:231–243. doi: 10.1016/j.cell.2008.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alexeev A, Mazin A, Kowalczykowski SC. Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51-ssDNA nucleoprotein filament. Nat Struct Biol. 2003;10:182–186. doi: 10.1038/nsb901. [DOI] [PubMed] [Google Scholar]
- 26.Lydall D, Weinert T. Yeast checkpoint genes in DNA damage processing: implications for repair and arrest. Science. 1995;270:1488–1491. doi: 10.1126/science.270.5241.1488. [DOI] [PubMed] [Google Scholar]
- 27.Bothmer A, et al. Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol Cell. 2011;42:319–329. doi: 10.1016/j.molcel.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bunting SF, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okazaki N, et al. The novel protein complex with SMARCAD1/KIAA1122 binds to the vicinity of TSS. J Mol Biol. 2008;382:257–265. doi: 10.1016/j.jmb.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 30.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 31.Chen X, et al. Cell cycle regulation of DNA double-strand break end resection by Cdk1-dependent Dna2 phosphorylation. Nat Struct Mol Biol. 2011;18:1015–1019. doi: 10.1038/nsmb.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugawara N, Wang X, Haber JE. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol Cell. 2003;12:209–219. doi: 10.1016/s1097-2765(03)00269-7. [DOI] [PubMed] [Google Scholar]
- 33.Lambert JP, et al. Defining the budding yeast chromatin-associated interactome. Mol Syst Biol. 2010;6:448. doi: 10.1038/msb.2010.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.