Abstract
Bacterial transcription activators regulate transcription by making essential protein–protein interactions with RNA polymerase, for example, with region 4 of the σ70 subunit (σ70 R4). Rob, SoxS, and MarA comprise a closely related subset of members of the AraC/XylS family of transcription factors that activate transcription of both class I and class II promoters. Recently, we showed that interactions between SoxS and σ70 R4 occlude the binding of σ70 R4 to the −35 promoter element of class II promoters. Although Rob shares many similarities with SoxS, it contains a C-terminal domain (CTD) that the other paralogs do not. Thus, a goal of this study was to determine whether Rob makes protein–protein interactions with σ70 R4 at class II promoters and, if so, whether the interactions occlude the binding of σ70 R4 to the −35 hexamer despite the presence of the CTD. We found that although Rob makes fewer interactions with σ70 R4 than SoxS, the two proteins make the same, unusual, position-dependent interactions. Importantly, we found that Rob occludes σ70 R4 from binding the −35 hexamer, just as does SoxS. Thus, the CTD does not substantially alter the way Rob interacts with σ70 R4 at class II promoters. Moreover, in contrast to inferences drawn from the co-crystal structure of Rob bound to robbox DNA, which showed that only one of Rob’s dual helix–turn–helix (HTH) DNA binding motifs binds a recognition element of the promoter’s robbox, we determined that the two HTH motifs each bind a recognition element in vivo.
Keywords: SoxS, genetic epistasis, σ70 R4, prerecruitment, Rob−micF crystal structure
Introduction
In eubacteria, like Escherichia coli,1 transcription initiation and elongation are carried out by the holo-RNA polymerase (RNAP). RNAP is composed of a “core”, containing the α2, β, β′, and ω subunits, and a σ factor. Although the core alone can carry out transcription elongation, it does so only after binding nonspecifically to DNA. However, the addition of a σ factor to the core polymerase endows RNAP (α2, β, β′, ω, σ) with the capacity to recognize and bind specifically to promoter sites, thereby stimulating specific and efficient transcription from them. The specific binding of RNAP to promoters is mediated by the interaction of specific domains of the σ factor with the promoter’s key sequence elements. For example, during exponential growth, the predominant σ factor is σ70, whose regions 2.1 and 4.2 (σ70 R2.1 and σ70 R4.2) recognize the hexameric −10 and −35 sequence elements, respectively, which are centered at approximately −10 bp and −35 bp upstream of the start site of transcription of the “housekeeping” promoters.1,2 However, E. coli encodes six other σ factors that recognize different DNA-binding elements. The expression and/or activation of these alternative σ factors is one way in which the cell can adapt to different growth conditions or to provide a defense against potentially lethal environmental stresses, since they enhance the transcription of regulons encoding genes whose products mediate the respective responses.1-5
Environment-dependent enhancement of the transcription of specific genes or regulons is also brought about by the subclass of transcription factors called transcription activators. Most bacterial transcription activators function by first binding to a specific site in a target promoter; then, the DNA-bound activator recruits RNAP to the promoter by making adhesive protein–protein interactions with it.6,7 Once recruited to a promoter, RNAP then initiates transcription. The nature of the interactions between the transcription activator and RNAP depends on the position and orientation of the DNA-bound activator with respect to the location of the −35 promoter hexamer.8 Thus, at class I promoters, the activator binding site lies upstream of the −35 element, and the predominant interaction of the activator is with the “287” determinant of the C-terminal domain (CTD) of the α subunit of RNAP.9 On the other hand, at class II promoters, the activator binding site partially or completely overlaps the −35 element such that the DNA-bound activator is able to interact directly with region 4 of the σ70 subunit (σ70 R4).10-15
Rob, SoxS, MarA, and TetD form a highly paralogous subset of proteins16 within the AraC/XylS family of transcriptional activators, whose hallmark is the presence of two helix–turn–helix (HTH) DNA binding motifs per monomer.17 Extensive amino acid sequence identity is shared between Rob (55%), MarA (42%), and TetD (49%) over the l07-amino-acid length of SoxS, the shortest member of the subset.16 Accordingly, the paralogs share many characteristics. For example, Rob, SoxS, and MarA function as monomers,18 bind the same but highly degenerate target sequences,18-23 and activate transcription of a common set of ~100 genes,16,24-28 by the same, novel prerecruitment/DNA scanning mechanism,11,29-31 although each paralog differentially activates transcription of the target genes.16,32,33
Rob differs from its paralogs in several ways: it is synthesized constitutively34,35 rather than in response to an inducing signal, as are SoxS36 and MarA,37,38 and it contains a CTD.35,39 The CTD is both necessary and sufficient for sequestration–dispersal,40 the novel mechanism for turning off and turning on Rob’s ability to function as a transcription activator in the presence or absence, respectively, of an inducing signal, for example, bile salts.37 In addition, the co-crystal structure of Rob bound to the robbox in the micF promoter39 differs significantly from the co-crystal structure of MarA bound to the marbox in the mar promoter.41 (We note that even though the binding sites for MarA, Rob, and SoxS are the same in a given promoter, the site is given the name of the protein bound there in a specific situation.)
In the MarA–mar structure, the recognition helices of both the N-terminal HTH motif (helix-3) and the C-terminal HTH motif (helix-6) are bound to successive major grooves containing recognition elements 1 and 2 (RE1 and RE2), respectively.20,41 In contrast, the structure of the Rob–micF complex shows that the recognition helix of the N-terminal HTH motif (helix-3) binds RE1 of the robbox while that of the C-terminal HTH motif (helix-6) is not bound to DNA.39 Moreover, the DNA in the MarA–mar co-crystal structure is bent 35°,41 whereas the DNA in the Rob–micF co-crystal structure is straight,39 probably because Rob’s second HTH motif does not specifically contact base pairs in RE2. These qualitative differences in the co-crystal structures and supportive biochemical tests led Kwon et al.39 to propose that MarA and Rob bind DNA in two different ways.
Thus, the unusual mode of target DNA binding by Rob led us to ask whether the binding of Rob to the robbox of class II promoters occludes the binding of σ70 R4 to the −35 hexamer as does the binding of SoxS.42 The alternative question is whether the presence of the CTD, the absence of DNA bending, and the absence of RE2 binding by Rob’s C-terminal HTH motif such that the motif is not anchored to the DNA in proximity to σ70 R4 have required that Rob make different protein–protein interactions with σ70 R4 than SoxS during transcription activation of class II promoters. As a corollary to this question, we also asked whether PC (positive control) mutants of Rob deficient only in activation of class II promoters are substitutions of amino acids homologous to those of the class II PC mutants of SoxS.43
Accordingly, in this paper, we used alanine scanning libraries of σ70 to identify amino acid residues of σ70 that are required for Rob-dependent transcription activation of class II promoters.42 We also determined whether Rob and σ70 R4 can accommodate one another at class II promoters or whether the binding of Rob occludes the binding of σ70 R4, as is the case with SoxS.42 To investigate whether Rob’s CTD alters its mechanism of transcription activation at class II promoters, we used SoxS as a model to construct PC mutants of the class II surface of Rob that are defective in activating transcription of the class II fumC and micF promoters but bind DNA normally. Using these PC mutants in genetic epistasis tests, we identified amino acid residues in σ70 R4 that make specific protein–protein interactions with amino acids within the class II PC surface of Rob. Additionally, using Rob mutants defective in DNA binding and base-pair substitutions in the robbox of the fumC promoter, we identified specific Rob–robbox interactions in vivo. In turn, this allowed us to determine the orientation and precise position of Rob on the robbox of these class II promoters and to identify specific amino acid–base-pair interactions.
Results
The effect of single alanine substitutions of σ70 R4.2 at positions 590–613 on Rob-dependent transcription activation of class II promoters
We wished to determine whether any residues of σ70 from region 4.2 to the C-terminal tail, that is, positions 590–613, are important for Rob-dependent transcription activation of the class II fumC and micF promoters. To do so, we introduced a library of 17 single alanine substitutions of this region of σ70 carried on plasmid pGEX-2T-σ70 13 into derivatives of strain RA4468 [pBAD33-Rob] lysogenic for single-copy λ prophages carrying transcriptional fusions of lac to the fumC and micF promoters.33 In these strains, Rob expression is under control of the arabinose-inducible PBAD promoter while the wild-type and mutant forms of σ70 are expressed from a partially constitutive lac promoter. Accordingly, the effect of the σ70 mutants on Rob-dependent activation of the two class II promoters was determined by assaying β-galactosidase activity44 in the absence and presence of arabinose.
Figure 1 shows the effect of the single alanine substitutions on transcription activation of the fumC and micF promoters, expressed as a percentage of the amount of β-galactosidase activity produced by the lysogens carrying wild-type pGEX-2T-σ70. We note that Lonetto et al. determined that, even in the absence of full induction of the lac promoter by treatment with IPTG, the plasmid-encoded σ70 proteins are produced to a level about equal to that of the chromosomally encoded wild-type protein.13 As such, when an alanine substitution of plasmid-encoded σ70 reduces the total amount of activated transcription to a level that is <80% of that when both chromosome-encoded and plasmid-encoded σ70 are wild type, the contribution to the total activity by the mutant σ70 has been reduced by >40%. Accordingly, a >20% reduction in total activated transcription has commonly been considered to be significant.13,15,45 By this criterion, alanine substitutions K593A (76%) and R599A (59%) of σ70 clearly interfere with Rob-dependent transcription activation of the fumC promoter (Fig. 1a). Figure 1b shows the effect of members of the single alanine scanning library of σ70 at the micF promoter. In contrast to the reduction of Rob-dependent transcription activation at the fumC promoter, neither K593A (114%) nor R599A (99%) affected activation at the micF promoter. Instead, only R603A (73%) of σ70 reduced Rob-dependent activation of the micF promoter. Thus, whereas SoxS requires the same four amino acid residues of σ70 (K593, R596, R599, and R603) in its activation of the two class II promoters, Rob requires two of them at the fumC promoter (K593 and R599), a third (R603) at the micF promoter, and the fourth (R596) at neither. As discussed below, the smaller number of interactions between Rob and σ70 compared to the number of interactions between SoxS and σ70 are at least partially compensated for by interactions between Rob and the αCTD.
Fig. 1.
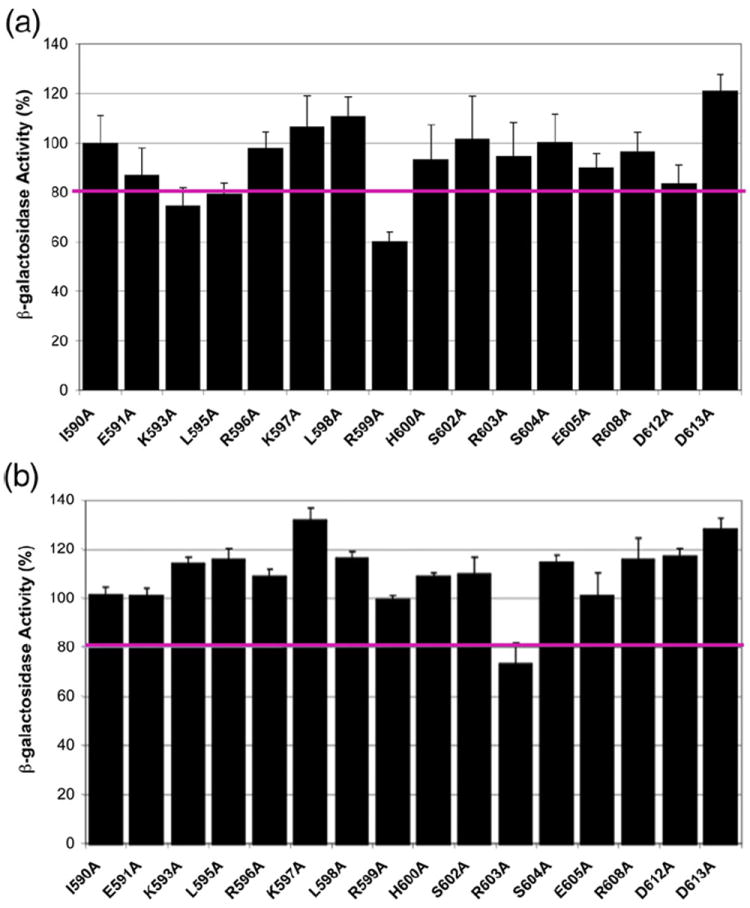
The effects of single alanine substitutions of σ70 on Rob-dependent transcription activation of the class II fumC and micF promoters. Derivatives of strain RA4468 containing single-copy λ prophages carrying transcriptional fusions of lac to the fumC (a) and micF (b) promoters and harboring plasmid pBAD33-Rob were transformed with compatible plasmids encoding wild-type σ70 or mutant alleles of σ70 with single alanine substitutions at the indicated positions. Determination of the activity of β-galactosidase in the respective strains was carried out by themethod of Griffith and Wolf,44 as described in Materials and Methods. The β-galactosidase activity produced by each mutant strain is given as a percentage of the activity produced by the strain expressing wild-type σ70. Activity values at or below 80% of the activity produced by wild-type σ70, denoted by the horizontal line, are considered to be significant, as discussed in the text.
Effects of a tri-alanine scanning library of σ70 R4 at positions 531–590 on Rob-dependent transcription activation
With a tri-alanine scanning library of amino acid residues of σ70 from positions 531–590 (residues 531–540 lie in the distal portion of σ70 R3.2, residues 541–570 lie in σ70 R4.1, and residues 571–590 lie in the N-terminal portion of σ70 R4.2), 1,46,47 we recently determined that none of the amino acid residues within this region are required for SoxS-dependent activation of transcription from class II promoters fumC and micF, including some substitutions that replace amino acids that are known to interact with nucleotides of the −35 hexamer.14,42 However, several of the tri-alanine substitutions reduced the basal level of transcription from these class II promoters.42 We inferred from these data that the binding of SoxS to the soxboxes of the class II promoters masks the effects of the tri-alanine substitutions that reduce basal transcription by occluding the binding there of σ70 R3.2–R4.2.42 Accordingly, we used this library to determine whether Rob and σ70 R3.2–R4.2 can accommodate one another during transcription activation of class II promoters. If so, then these data would support the hypothesis that the presence of Rob’s CTD has required Rob to use a different mechanism than SoxS in its activation of class II promoters; if not, then the hypothesis would be invalidated.
Figure 2 shows the results of this experiment. Again, with the use of the criterion for a significant effect of a substitution as one that reduces transcription to < 80% of that produced by wild-type σ70, none of the library’s substitutions significantly reduced Rob-dependent transcription activation of the three class II promoters (i.e., fumC, micF, and inaA) (Fig. 2a–c). In our previous work with the tri-alanine scanning library, we also determined that several members of the library reduced SoxS-dependent activation of transcription from the class I fpr promoter, as expected, because the SoxS binding site of class I promoters lies upstream of the −35 hexamer, which is therefore exposed and available for binding by σ70 R4.42 Here, we found that several tri-alanine substitutions of σ70 R4 also reduce Rob-dependent activation of the fpr promoter (Fig. 2d).
Fig. 2.
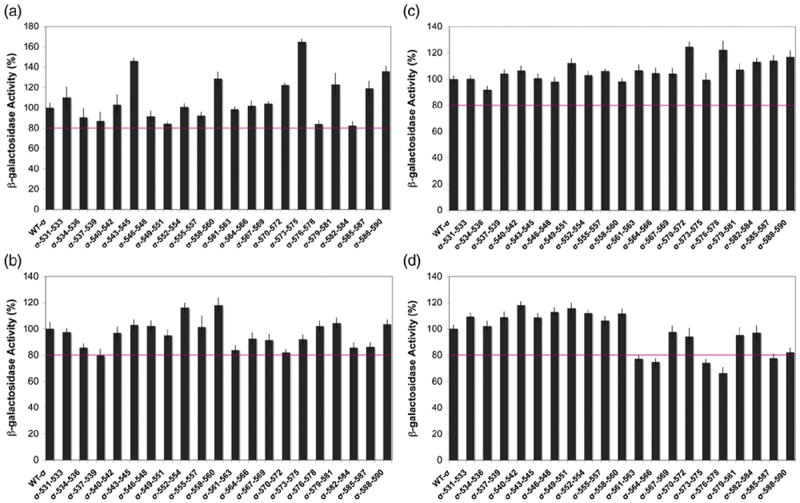
The effects of tri-alanine substitutions of σ70 on Rob-dependent transcription activation of one class I promoter and three class II promoters. Derivatives of strain N7840 Δ(araBAD)714 [pBAD33-his6-Rob] harboring a member of the library of tri-alanine substitutions of σ7042 carried on compatible plasmid pVR-σ and lysogenic for a single-copy λ prophage containing a transcriptional fusion of lac to a class II promoter, viz., fumC (a), inaA (b), and micF (c), or a class I promoter, viz., fpr (d), were grown as described in Materials and Methods. Rob was expressed from the arabinose-inducible PBAD promoter, while wild-type σ70 and σ70 R4 mutants were expressed constitutively from a truncated galP1 promoter. Wild-type σ70 was also constitutively expressed from chromosomal rpoD. Determination of the activity of β-galactosidase in the respective strains was carried out by the method of Griffith and Wolf,44 as described in Materials and Methods. Activity values at or below 80% of the activity produced by wild-type σ70, denoted by the horizontal line, are considered to be significant.
The experiments in Fig. 2a–c suggest that the binding of Rob to the robboxes of the fumC, micF, and inaA promoters occludes the binding of σ70 R4 to the −35 hexamer of these class II promoters, as is also the case during SoxS-dependent activation of the fumC and micF promoters.42 Thus, despite the presence of a CTD composed of ~167 amino acid residues,39 the binding of Rob to the three class II promoters is still able to prevent the binding of σ70 R4 to the −35 promoter element. In turn, the results of these experiments led us to analyze further the transcription activation of class II promoters by Rob, in order to determine whether the presence of the CTD has any effect on the underlying mechanism.
Orientation of Rob at class II promoters
As described above, the structures of the Rob–robbox and the MarA–marbox complexes are qualitatively different with respect to whether one (Rob) or both (MarA) recognition helices of the HTH motifs bind target DNA and whether the DNA is straight (Rob) or bent (MarA). Accordingly, for these reasons, and also because our previous work18 showed that the binding of Rob to the robboxes of the fumC and micF produces a 30° bend in the DNA, we thought that it was important to determine the position and orientation of Rob on the robbox of a class II promoter in vivo. To accomplish this objective, we carried out genetic epistasis tests as described previously.42,48
Epistasis occurs when one mutation masks the phenotype conferred by another mutation. The application of epistasis used here is to address the question of whether the side group of an amino acid residue within a recognition helix of Rob interacts specifically with a functional group of a given base pair in the corresponding recognition element in a robbox. If the side group of the wild-type amino acid makes a specific protein–DNA contact with a functional group of the wild-type base pair, then the effect on transcription activation of combining a single alanine substitution of that amino acid of Rob and a single base-pair substitution at that position of the robbox would not be significantly different from that conferred by the more severe of the two individual mutations; that is, the effect on activation conferred by one mutation would be masked by the effect conferred by the other, and thus, the mutations would be epistatic to one another. On the other hand, if the side group of a given wild-type amino acid residue does not make a specific protein–DNA contact with a functional group of a wild-type base pair, then the effect on transcription activation of combining a single alanine substitution of that amino acid of Rob and a single base-pair substitution at that position of the robbox would be significantly different from that conferred by the more severe of the two individual mutations; that is, the effect on activation conferred by one mutation would not be masked by the effect conferred by the other, and thus, the mutations would be non-epistatic to one another.42,48
According to the co-crystal structure of Rob bound to micF DNA (Fig. 3a), residue W36 in helix-3 of Rob’s N-terminal HTH motif makes a van der Waals contact with nucleotide C5 in RE1 (GCAC; C5 in boldface),39 the most highly conserved residue in the Rob/SoxS/MarA binding sites.20-22,39 Indeed, changing C5 to any of the other three nucleotides severely reduces SoxS-dependent transcription activation of the fpr and zwf promoters.20 Thus, for our epistasis experiment, we used site-directed mutagenesis to introduce a C5T substitution into the robbox of a micF-lac transcriptional fusion carried on plasmid pMLB1022 and to prepare a derivative of plasmid pBAD33-Rob carrying substitution W36A of Rob.
Fig. 3.
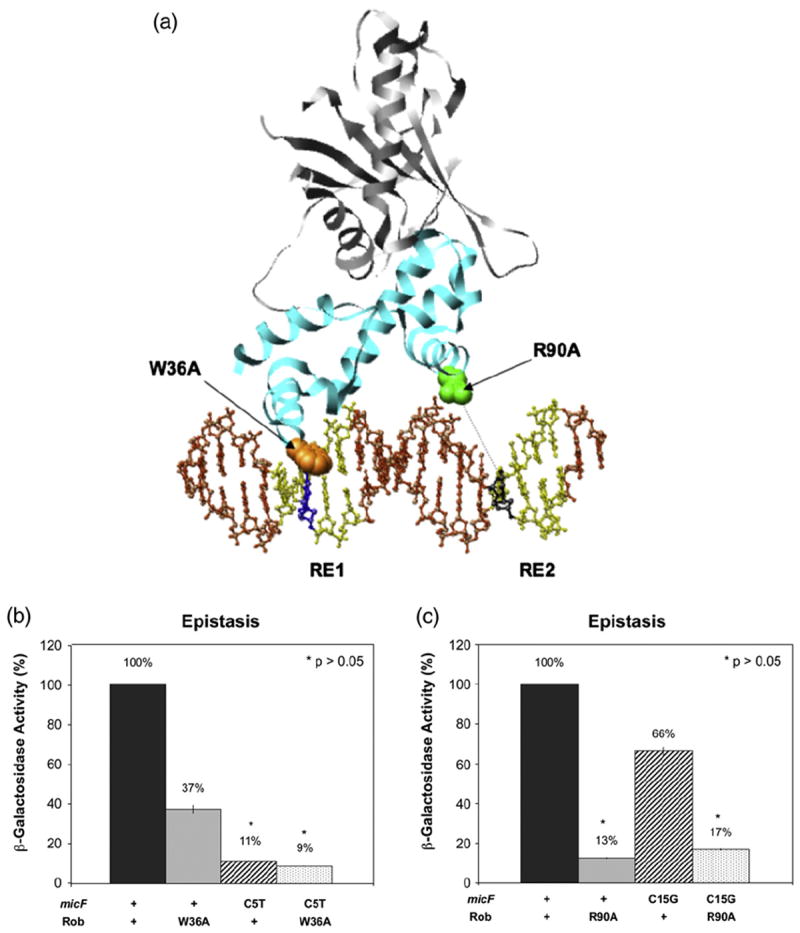
Genetic epistasis experiments determining the orientation of Rob and its interactions with RE1 and RE2 of the robbox of the micF promoter. (a) The crystal structure of the Rob–micF binary complex39 highlighting amino acid–nucleotide interactions. The NTD of Rob is shown in cyan, the CTD is gray, and RE1 and RE2 are colored yellow. Amino acid W36 of helix-3 of Rob, space-filled and colored orange, lies close to nucleotide C5 (blue) of RE1 of the micF robbox while amino acid R90 of helix-6, space-filled and colored green, lies above nucleotide C15 (black) of RE2. (b) Genetic epistasis test of alanine substitution W36A of Rob and nucleotide substitution C5T of RE1 in the robbox of the micF-lac transcriptional fusion. (c) Genetic epistasis test of alanine substitution R90A of Rob and nucleotide substitution C15G of RE2 in the robbox of the micF-lac transcriptional fusion. The β-galactosidase activity of each combination is given as a percentage of the activity produced by the combination of wild-type Rob and the wild-type robbox of the micF-lac fusion. (*) The p-values given in (b) and (c) denote the absence of a statistically significant difference between the β-galactosidase activity produced by the single mutant that confers the most severe defect and that produced by the corresponding double mutant (both identified with an asterisk), where the activity of the double mutant is not significantly less than the activity produced by either single mutant, that is, an epistatic interaction. The DNA sequence of the 20-bp robbox of the micF promoter is 5′-ACAGCACTGAATGTCAAAAC-3′, with bases C5 and C15 in boldface. The complete DNA sequence of the micF promoter is shown in Fig. 8.
Importantly, we also carried out a genetic epistasis experiment to determine whether the side group of an amino acid within helix-6 of Rob’s C-terminal HTH motif interacts specifically with a functional group of a base pair of RE2 in the robbox of the micF promoter, as do amino acids of helix-6 of MarA and base pair of RE2 in the marbox of the mar promoter.41 We note that the amino acid sequences of helix-6 of Rob and MarA are identical at 10 of 12 positions.16 To conduct the genetic epistasis test, we used site-directed mutagenesis to substitute amino acid R90 of pBAD33-Rob with alanine43 and to introduce a C15G substitution into the robbox of a micF-lac fusion carried on plasmid pMLB1022. We chose these substitutions because the co-crystal structure of MarA bound to the marbox of the mar promoter indicates that R90 of MarA makes a van der Waals contact with C15, a conserved residue in RE2 (CAAA; C15 in boldface).21,22
The pMLB1022 plasmids carrying the micF-lac transcriptional fusion with either the wild-type robbox or the robbox with the C5T substitution were introduced into strain RA4468 harboring either wild-type Rob or Rob W36A on the compatible pBAD33 plasmid. Similarly, pMLB1022 plasmids carrying the micF-lac fusion with either the wild-type robbox or the robbox with the C15G substitution were introduced into strain RA4468 harboring either wild-type Rob or Rob R90A on plasmid pBAD33. The effect of each combination of alleles on transcription activation was determined by assay of β-galactosidase activity after induction of Rob expression.
Figure 3b shows that single substitutions W36A of Rob and C5T of the robbox reduced transcription activation of the micF-lac fusion to 37% and 11% of that conferred by wild-type Rob or wild-type robbox, respectively. However, the combination of the W36A substitution of Rob and the C5T substitution of the robbox of the micF-lac fusion reduced transcription activation to 9% of wild type, statistically the same as the level of activation conferred by C5T alone. Thus, these data indicate that Rob mutant W36A is epistatic to robbox substitution C5T and that W36 interacts with C5 in vivo. Accordingly, since an amino acid located within Rob’s N-terminal HTH motif contacts a nucleotide of RE1 and since the binding of Rob to the micF promoter region protects the entire 20 bp robbox and one to two additional base pairs on either side from attack by DNase I,18 the position and orientation of Rob on the micF robbox in vivo is the same as that observed in the co-crystal structure.39
To determine if an epistatic interaction occurs between Rob’s C-terminal HTH motif and RE2, we compared the effects on transcription activation of the micF-lac fusion by wild-type Rob or substitution R90A in combination with either the wild-type robbox or its substitution C15G. Figure 3c shows that the single mutations R90A of Rob and C15G of the robbox reduced transcription activation of the micF-lac fusion to 13% and 66% of wild type, respectively. The double mutant reduced activation to 17% of wild type, which is statistically the same as the level of activation conferred by R90A alone. Therefore, an epistatic interaction occurs between R90A and C15G in RE2 of the micF promoter. Moreover, these data not only show that the orientation of Rob on the robbox of the micF promoter is the same as that in the co-crystal structure but also provide in vivo evidence that an amino acid within Rob’s C-terminal HTH motif specifically contacts a base pair within RE2.
Identification of PC mutations of Rob that reduce transcription activation of class II promoters
Recently, we showed that SoxS and σ70 R4 interact with one another in the yeast two-hybrid system and that the interaction is disrupted by mutations of the class I/II surface but is not disrupted by mutations of the class II surface.12 Thus, we concluded that the protein–protein interaction in yeast between SoxS and σ70 R4 can occur in solution, that is, “off DNA”, and requires amino acids of the class I/II surface but not amino acids of the class II surface.12 In turn, this indicates that the interactions in E. coli between amino acid residues of the class II surface of SoxS and amino acid residues of σ70 R4 occur “on DNA”.12 Given the data presented above indicating that Rob’s CTD does not interfere with the binding of Rob’s N-terminal domain (NTD) to the robbox of a class II promoter or its activation of several of them (Figs. 2 and 3), together with the high degree of amino acid sequence identity between the C-terminal HTH motifs of SoxS and Rob (9 of 16 amino acids match),16 we hypothesize here that, at class II promoters, SoxS and Rob interact with RNAP using similar functional domains. Accordingly, we used site-directed mutagenesis to introduce single alanine substitutions of Rob at the positions equivalent to the five amino acid residues of SoxS comprising its class II surface.43 As a control, we also prepared an alanine substitution of Rob at the position equivalent to that of K30A of SoxS, the amino acid substitution of its class I/II surface that confers the greatest reduction in transcription activation.43
Figure 4a and b shows the effects of the substitutions within Rob’s putative class II surface on transcription activation of the class II fumC and micF promoters, respectively. At the fumC promoter, substitutions L74A, D75A, L78A, and Q85 of the putative class II surface, as well as K30A of Rob’s putative class I/II surface, reduced transcription activation to 12–25% of that produced by wild-type Rob. In addition, at the micF promoter, substitution L74A reduced Rob-dependent transcription activation to 17% of the activation by wild-type Rob, while substitutions K30A, D75A, L78A, and Q85 had smaller effects than they did at the fumC promoter, reducing activation to 33–50% of that produced by wild-type Rob. Interestingly, substitution Q79A had no effect on Rob-dependent activation at either promoter. However, substituting alanine for the aspartic acid at the equivalent position in SoxS (i.e., D79A) reduces transcription activation at the fumC and micF promoters to 41% and 26%, respectively, of the wild-type level.43 Thus, the failure of Rob’s Q79 to play a role in activation of class II promoters is probably because it cannot make a contact with a basic residue of σ70, for example, K593, as is the case with SoxS.12
Fig. 4.
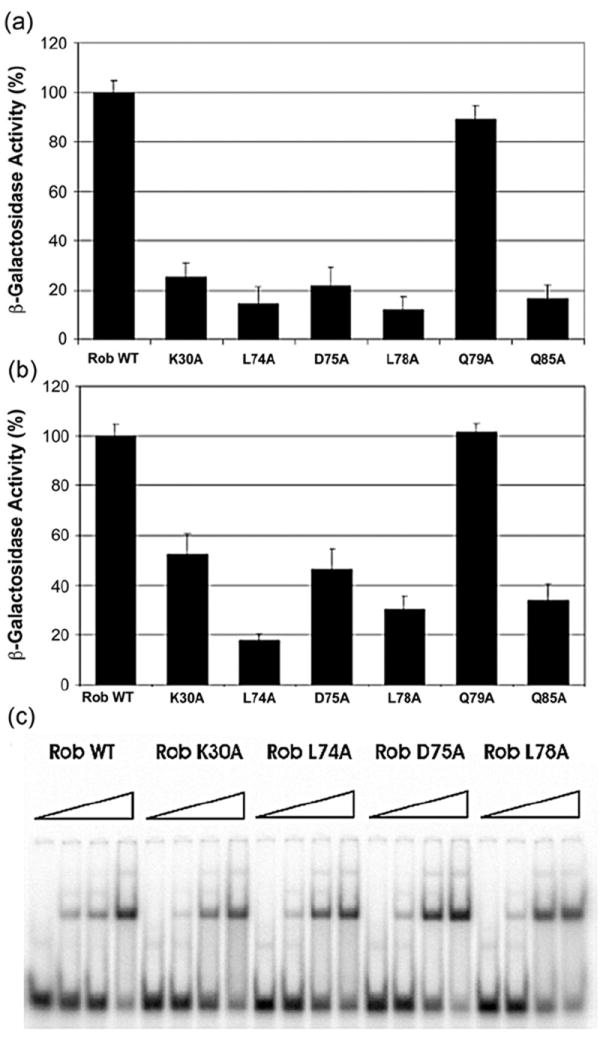
The identification of PC mutations of Rob that reduce transcription activation of class II promoters. Derivatives of strainRA4468 lysogenic for a single-copy λ prophage carrying a transcriptional fusion of lac to the fumC promoter (a) or to the micF promoter (b) were transformed with plasmid pBAD33-Rob or single alanine substitutions of it. The β-galactosidase activity produced by eachmutant strain is given as the percentage of the activity produced by the strain expressing wild-type Rob. (c) EMSAs showing the effect on binding fumC DNA of increasing concentrations (0, 8, 80, and 800 nM) of wild-type Rob and PCmutants K30A, L74A, D75A, and L78A were carried out as described in Materials and Methods.
Before concluding that the alanine substitutions that reduce Rob-dependent transcription activation of the fumC and micF promoters are truly PC mutations rather than general loss of function mutations, it was necessary to determine whether they retained the ability to bind robbox DNA. Accordingly, we placed a his6 tag on the N-terminus of Rob and used it to purify, by Ni-NTA affinity chromatography, the wild-type protein and mutant proteins with the K30A, L74A, D75A, L78A, and Q85A substitutions. Then, we used electrophoretic mobility shift assays (EMSA) to compare semi-quantitatively the DNA binding affinity of wild-type Rob to that of the putative PC mutants. Figure 4c shows EMSA for wild-type Rob and mutants K30A, L74A, D75A, and L78A (Q85; data not shown). Each of the tested mutants bound DNA with approximately the same affinity as wild-type Rob. Since the mutant proteins are severely defective in activating transcription from class II promoters but bind to robbox DNA with approximately wild-type affinity, they are, by definition, PC mutants of Rob’s class II surface.
Genetic evidence for protein–protein interactions between Rob and σ70 R4 at the fumC and micF promoters
We screened for specific interactions between amino acids of Rob and of σ70 R4 with two sets of genetic epistasis experiments similar to those described previously.12,49 In one set, plasmids expressing wild-type Rob or its four class II PC mutants and plasmids expressing wild-type σ70 or its derivatives K593A and R599A were co-transformed into strain RA4468 (rob∷kan) carrying a fumC-lacZ fusion. In the second set, plasmids expressing the Rob alleles and plasmids expressing wild-type σ70 or substitution R603A were introduced into RA4468 carrying the micF-lac fusion. Then, the effects on transcription activation of the two promoters by the respective substitutions alone and in combination with each other were determined by assay of the amount of β-galactosidase activity produced by each combination. For these experiments, we applied the same logic as was used in the epistasis tests aimed at identifying interactions between amino acids of Rob and base pair of the robbox (Fig. 3), except that, here, the purpose of the test is to determine whether the R groups of a pair of amino acid residues, one residing on Rob and the other residing on σ70 R4, interact with one another.
In the first set of genetic epistasis tests, we found that D75A of Rob is epistatic to R599A of σ70 during activation of the fumC-lac fusion, but none of the other three PC mutants of Rob’s class II surface are epistatic to R599A (Fig. 5). Moreover, none of the PC mutants are epistatic to K593A of σ70 (Fig. 5). We offer three explanations for why none of the class II substitutions are epistatic to K593A: (i) K593 interacts with the keto group of a peptide bond, and hence, the interaction is unaffected by any amino acid substitution at that position; (ii) K593 interacts with an undiscovered amino acid residue of Rob that confers a PC mutant phenotype when substituted with alanine; or (iii) the activation defect conferred by K593A results from a negative effect (direct or indirect) of the mutation on Rob’s ability to make a protein–DNA interaction with the robbox of the fumC promoter.
Fig. 5.
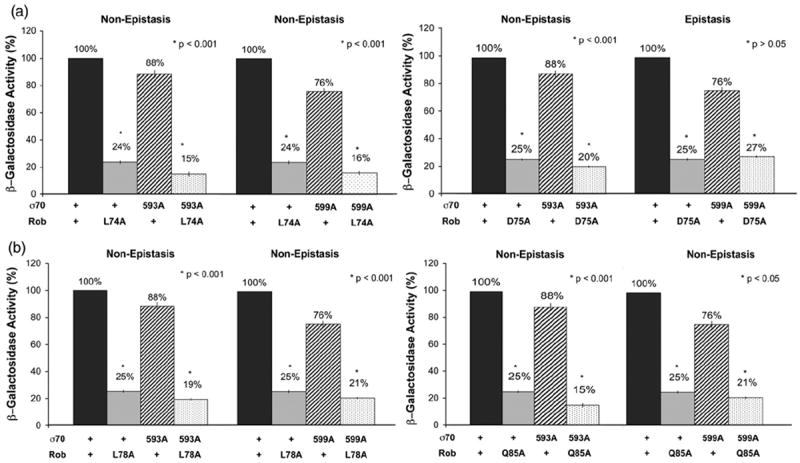
Genetic epistasis tests at the fumC promoter between PC mutants of Rob’s class II surface and mutant alleles of σ70. Plasmid pBAD33-Rob carrying the gene for wild-type Rob or PC mutant alleles of it (L74A, D75A, L78A, or Q85A) and compatible plasmid pVR-σ70 carrying the gene for wild-type σ70 or mutant alleles of it (K593A or R599A) in all possible combinations were introduced into strain RA4468 carrying a transcriptional fusion of lac to the fumC promoter on a single-copy λ prophage. The β-galactosidase activity of the strain containing wild-type σ70 in combination with wild-type Rob is set to 100%, and the activity produced by each mutant gene or combination of mutant genes is presented as a percentage of the wild-type activity. (a) The left panel shows that K593A and R599A are not epistatic to L74A, while the right panel shows that K593A is not epistatic to D75A but R599A is epistatic to D75A. (b) The left panel shows that K593A and R599A are not epistatic to L78A, while the right panel shows that K593A and R599A are not epistatic to Q85A. (*) The p-values given in the panels denote a statistically significant difference (or no statistically significant difference) between the β-galactosidase activity produced by the single mutant that confers the most severe defect and that produced by the corresponding double mutant (both identified with an asterisk), where the activity of the double mutant is significantly less (is not significantly less) than the activity produced by either single mutant (i.e., a non-epistatic interaction or an epistatic interaction, respectively).
In the second set of experiments, we found that Rob D75A is epistatic to σ70 R603A at the micF promoter and that no other epistasis was observed (Fig. 6). Thus, a single amino acid residue of Rob, D75, interacts with different residues of σ70 R4, depending on the promoter, that is, with R599 at the fumC promoter and with R603 at the micF promoter. A preliminary molecular modeling of the complexes of Rob and σ70 R4 bound to the −35 regions of the fumC and micF promoters (data not shown) suggests that the reason that Rob D75 interacts with σ70 R599 at the fumC promoter and with R603 at the micF promoter is because the extent of overlap of the −35 element by the robbox differs in the two promoters: the position of the robbox of the micF promoter with respect to the −35 element is 4 bp upstream of the relative position of the robbox of the fumC promoter.
Fig. 6.
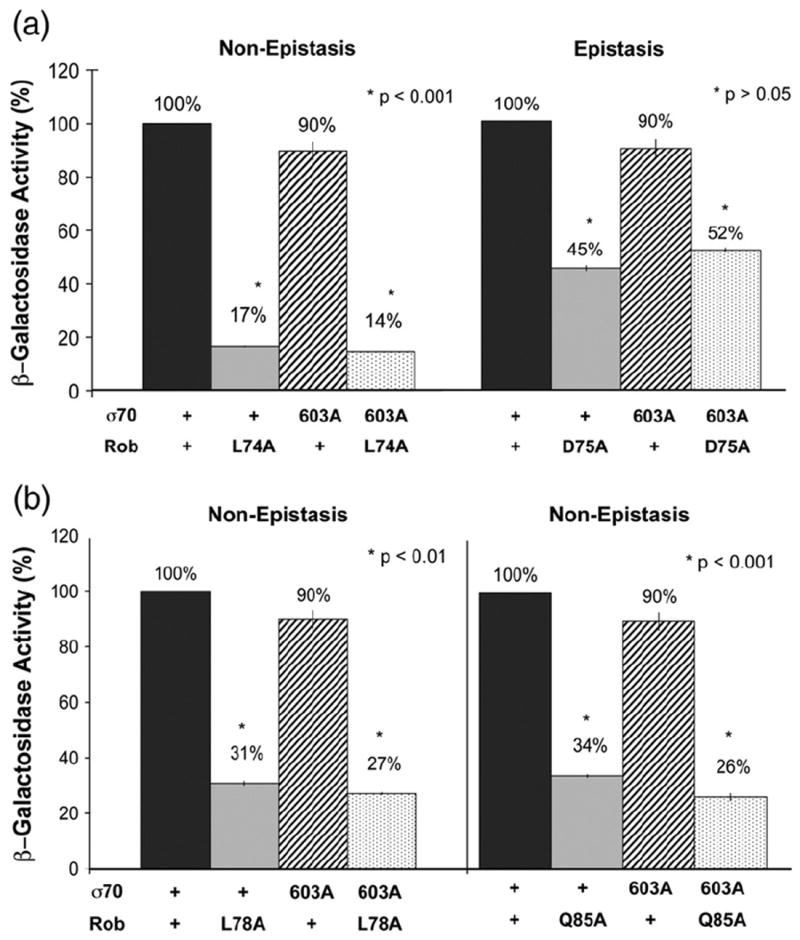
Genetic epistasis tests at the micF promoter between PC mutants of Rob’s class II surface and substitution R603A of σ70. Plasmid pBAD33-Rob carrying the gene for wild-type Rob or PC mutant alleles of it (L74A, D75A, L78A, or Q85A) and compatible plasmid pVR-σ70 carrying the gene for wild-type σ70 or substitution R603A of it were introduced into strain RA4468 carrying a transcriptional fusion of lac to the micF promoter on a single-copy λ prophage. The β-galactosidase activity of the strain containing wild-type σ70 in combination with wild-type Rob is set to 100%, and the activity produced by each mutant gene or combination of mutant genes is presented as a percentage of the wild-type activity. (a) (Left) Nonepistatic interaction between R603A and L74A. (Right) Epistatic interaction between R603A and D75A. (b) (Left) Non-epistatic interaction between R603A and L78A. (Right) Non-epistatic interaction between R603A and Q85A. (*) The p-values given in the panels denote a statistically significant difference (or no statistically significant difference) between the β-galactosidase activity produced by the single mutant that confers the most severe defect and that produced by the corresponding double mutant (both identified with an asterisk), where the activity of the double mutant is significantly less (is not significantly less) than the activity produced by either single mutant (i.e., a non-epistatic or an epistatic interaction, respectively).
Figure 7 shows that amino acid K30 of Rob, whose K30 homolog in SoxS is part of its class I/II surface,43 does not interact with R599 of σ70 R4 at the fumC promoter nor with R603 at the micF promoter, presumably because Rob’s K30 is too far from σ70 R4 at the two promoters to interact with it. On the other hand, Rob’s class II surface must be close enough to σ70 R4 to allow these Rob–σ70 R4 protein interactions to occur.
Fig. 7.
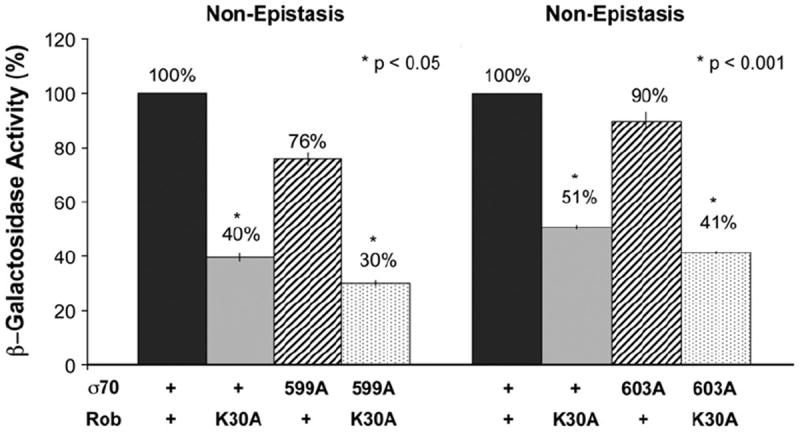
Genetic epistasis tests at the fumC and micF promoters between a PC mutant of Rob’s class I/II surface and two mutant alleles of σ70. Plasmid pBAD33-Rob carrying the gene for wild-type Rob or PC mutant K30A and compatible plasmid pVR-σ70 carrying the gene for wild-type σ70 or mutant alleles of it (R599A or R603A) were introduced into derivatives of strain RA4468 carrying a transcriptional fusion of lac to either the fumC (left) or the micF (right) promoter on a singlecopy λ prophage. The β-galactosidase activity of wild-type σ70 in combination with wild-type Rob is set to 100%, and the activity produced by each mutant gene or combination of mutant genes is presented as a percentage of the wild-type activity. (*) The p-values denote a statistically significant difference in the β-galactosidase activity produced by the single mutant that confers the most severe defect and that produced by the corresponding double mutant (both identified with an asterisk), where the activity of the double mutant is significantly less than the activity produced by either single mutant, that is, a non-epistatic interaction.
Comparison of the epistatic interactions between PC mutants of Rob and alanine substitutions of σ70 R4 at class II promoters to the corresponding interactions between PC mutants of SoxS and substitutions of σ70 R4
The data in Table 1 allow two basic comparisons to be made with respect to the requirements for transcription activation by SoxS12 and Rob at the fumC and micF promoters: the identity of alanine substitutions of σ70 that reduce transcription activation by each activator at each promoter and the epistatic interactions at each promoter between PC mutants of a given activator and alanine substitutions of σ70. Interestingly, all σ70 substitutions that reduce Rob-dependent transcription activation at a given promoter also reduce transcription activation by SoxS12 at that promoter; however, the converse is not true. Thus, (i) K593A, R596A, R599A, and R603A reduce SoxS-dependent activation at the fumC and micF promoters, but R596A has no effect on Rob-dependent transcription activation at either promoter; (ii) K593A and R599A have no effect on Rob-dependent activation of the micF promoter; and (iii) R603A has no effect on Rob activation of the fumC promoter. Accordingly, a total of eight substitutions of σ70 reduce SoxS-dependent activation of the two class II promoters,12 but a total of only three σ70 substitutions reduce Rob activation of the two promoters (Table 1).
Table 1.
Epistatic interactions between alanine substitutions of σ70 R4 and alanine substitutions of the class II PC surface of Rob and SoxS at the fumC and micF promoters
| σ70 R4 substitution | Epistasis between alanine substitutions of σ70 R4 and alanine substitutions of the class II PC surface of Rob and SoxS at two promoters
|
|||
|---|---|---|---|---|
|
fumC promoter
|
micF promoter
|
|||
| Rob | SoxS | Rob | SoxS | |
| K593A | None | D75A, M78A | N.A. | M78A, D79A |
| R596A | N.A. | None | N.A. | None |
| R599A | D75A | D75A | N.A. | None |
| R603A | N.A. | None | D75A | D75A, D79A |
All amino acids of σ70 R4 that confer a defect in Rob-dependent transcription activation (Fig. 1) or SoxS-dependent transcription activation12 of the fumC and/or micF promoters when substituted with alanine are listed even if no interacting partners have been identified. The alanine substitutions within the class II PC surface of Rob (Figs. 5 and 6) and SoxS12 that are epistatic partners of the listed alanine substitutions of σ70 R4 are shown according to the promoter at which the epistasis was observed. N.A., not applicable because this substitution of σ70 R4 has no effect on the activity of the activator; none, no epistasis occurs between these combinations of the activator and the σ70 R4 substitutions.
With respect to epistatic interactions (Table 1), (i) PC mutants of SoxS make a total of four epistatic interactions with K593A at the two promoters, while Rob PC mutants make none; (ii) no PC mutants of either SoxS or Rob are epistatic to R596A at either promoter; (iii) the same PC mutant (D75A) of SoxS and Rob is epistatic to R599A at the fumC promoter, but no epistasis occurs at the micF promoter between R599A and a PC mutant of either activator; and (iv) at the fumC promoter, no epistasis occurs between R603A and a PC mutant of either activator, whereas at the micF promoter, both D75A and D79A of SoxS are epistatic to R603A while only D75A of Rob is epistatic to R603A. Accordingly, the PC mutants of the class II surface of SoxS make seven epistatic interactions with the four alanine substitutions of σ70 R4 that reduce SoxS activation of the two class II promoters,12 while the PC mutants of Rob’s class II surface make only two. Thus, the sum of the identity of alanine substitutions of σ70 R4 that reduce transcription activation at the two promoters by SoxS (8) or Rob (3) and the number of epistatic interactions made by Sox (7) and Rob (2) at the two promoters are three times greater for SoxS (15) than for Rob (5). This significant difference between the two activators could be because the CTD of Rob alters the conformation of Rob’s class II surface such that it is less able to interact with σ70 R4.
Despite the significant differences cited above (Table 1) between the epistatic interactions observed with alanine substitutions of σ70 R4 and PC mutants of SoxS or Rob, the interactions between the activators and σ70 R4 are strikingly similar in one unusual respect: D75A of Rob reduces transcription activation of the fumC and micF promoters, but it is epistatic to two different alanine substitutions of σ70 R4, R599A at the fumC promoter and R603A at the micF promoter (Table 1). Importantly, these two protein–protein interactions are correlated with the position of the robbox in the two promoters with respect to their −35 hexamers (Fig. 8) and consistent with our preliminary molecular modeling mentioned above. Moreover, given our hypothesis that Rob’s CTD affects the protein–protein interactions between Rob’s NTD and σ70 R4 and the evidence cited above that supports it, it is remarkable that Rob makes the same pair of position-dependent interactions with σ70 R4 as SoxS (Table 1). 12 Although we cannot explain all of the differences between the interactions of SoxS and Rob with σ70 R4 at the two promoters, we think that Rob would not make the same position-dependent interactions as SoxS if Rob’s CTD affected the conformation of the class II surface.
Fig. 8.
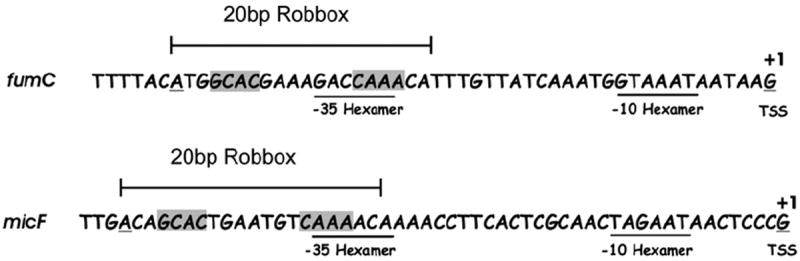
Comparison of the extent to which the Rob binding site (robbox) in the fumC and micF promoters overlaps the −35 promoter hexamer in the fumC and micF promoters. The sequences are aligned by their −35 promoter hexamers, which are underlined. The 20-bp sequences that are protected by MalE–SoxS and Rob from DNase I attack are bracketed.18,19 Important elements of the sequences are highlighted: the invariant “A” is underlined, RE1 and RE2 are shown in gray, and the promoter hexamers and the transcription start sites (TSS) are underlined. The −35 promoter elements were chosen as the hexamer located 17 bp upstream of the −10 hexamer of the promoter, identified as described previously.19 The robbox in the fumC promoter covers the −35 hexamer and extends 3 bp beyond62 while the binding site in the micF promoter only covers 5 bp of the hexamer.18,63
Discussion
As described above, Rob, SoxS, and MarA share many physiological and genetic properties, which have enabled us to address the question of whether Rob’s CTD interferes with Rob’s ability to interact with σ70 R4 at class II promoters: the premise upon which the hypothesis that Rob’s CTD interferes with the ability of Rob’s NTD to make protein–protein interactions with σ70 R4 was based in part on the co-crystal structures of MarA bound to mar41 and of Rob bound to micF DNA,39 which indicate that Rob and MarA bind DNA differently. In particular, recognition helix-3 and helix-6 of MarA’s dual HTH motifs bind RE1 and RE2, respectively, of mar’s robbox, while helix-3 within Rob’s N-terminal HTH motif binds RE1 of micF’s robbox, but helix-6 does not bind RE2. We used genetic epistasis experiments to determine whether helix-6 of Rob actually binds RE2 in vivo. Moreover, as a positive control, we also conducted a genetic epistasis test to determine whether Rob’s helix-3 binds RE1 in vivo, as it does in the co-crystal structure. In previous work,42 we found that epistatic and non-epistatic interactions between amino acid residues of SoxS and base pairs of soxbox DNA agreed with the predictions based on the co-crystal structure of MarA bound to marbox DNA.
Figure 3b shows that W36A within helix-3 of Rob is epistatic to C5T within RE1 of the micF robbox, indicating that the side group of W36 specifically contacts a functional group of C5 in vivo, just as it does in the co-crystal structure. In turn, this experiment provides a proof of principle, since it shows that a genetic epistasis test can determine whether an interaction observed in a crystal structure is an accurate representation of whether the interaction occurs in vivo and vice versa.
Figure 3c shows that R90A, within helix-6 of Rob, is epistatic to C15G within RE2 of the micF robbox. Thus, the data in Fig. 3b and Fig. 3c not only show that in vivo Rob binds to robbox DNA in the same orientation as in the co-crystal structure of MarA bound to the marbox of the mar promoter but also, more importantly, show that amino acids of helix-3 and helix-6 of Rob are bound in vivo to RE1 and RE2, respectively. Accordingly, the co-crystal structure of Rob bound to the micF robbox does not represent the Rob–micF interactions that occur in vivo. In turn, this indicates that Rob and MarA do not bind DNA differently and that in vivo Rob’s CTD does not cause it to bind robbox DNA differently than MarA binds marbox DNA in vitro41 or than SoxS binds soxbox DNA in vivo.42 We also note that the in vitro binding of Rob, SoxS, and MarA to a robbox/soxbox/marbox bends the DNA by about 30°;50 however, although mar DNA is bent by about 35° in the co-crystal structure with MarA,41 the micF DNA is straight in the co-crystal structure with Rob.39 In addition, the boundaries of the regions protected from DNase I digestion by the binding of purified Rob or MalE–SoxS to DNA containing the zwf and fumC promoters were the same;50 this would not be the case unless Rob’s C-terminal HTH motif was bound to RE2 of these promoters, as occurs with MalE–SoxS.51 Together, these data suggest that the absence of DNA binding by helix-6 and the linearity of DNA in the Rob–micF co-crystal structure do not accurately represent the DNA binding properties of Rob. Accordingly, we examined protein–protein interactions of Rob and RNAP to investigate further the DNA binding and transcriptional activation properties of Rob as compared to SoxS.
Importantly, since the interactions between SoxS and σ70 at class II promoters have been well characterized,12,42 we were able to determine whether Rob’s CTD affects the interactions between its NTD and σ70 by comparing them to those observed with SoxS. An unusual property of SoxS that has not been previously investigated with Rob or MarA is that the ability of σ70 R4 to bind to the −35 promoter elements of the class II fumC, inaA, and micF promoters in vivo is blocked by the binding of SoxS to the soxboxes of these promoters, which overlap the respective −35 hexamers.42 Accordingly, determination of whether the binding of Rob and σ70 R4 can accommodate one another at class II promoters in vivo would be a good test of whether Rob’s CTD interferes with Rob’s ability to interact with σ70 R4 at class II promoters. Using the same tri-alanine scanning library of amino acid residues 530–590 of σ70 R4 as was used in the experiments with SoxS,42 we found that none of the substitutions reduce Rob-dependent transcription activation of the fumC, inaA, and micF promoters (Fig. 2a–c), as was also the case with SoxS.42 Thus, presuming that occlusion requires a specific conformation of the DNA-bound activator, Rob’s CTD does not affect Rob’s ability to carry out this unusual function.
A second property of Rob that would be different from SoxS if Rob’s CTD prevents Rob’s NTD from making the same protein–protein interactions with σ70 R4 as SoxS would be the location of the amino acids of Rob and of σ70 R4 that make the interactions. Table 1 shows that two amino acid residues of σ70 R4 are required for Rob-dependent activation of fumC and that only one is required for activation of micF. In contrast, SoxS-dependent activation of these class II promoters requires four amino acid residues of σ70 R4 at each promoter. This difference could be due to the presence of Rob’s CTD. Although SoxS-dependent activation of fumC and micF requires a total of eight amino acid residues of σ70 R412 (Table 1), while Rob-dependent activation requires only three (Table 1), the reduced number of interactions is still sufficient for activation by Rob. Further evidence that Rob’s CTD has not altered the ability of Rob’s NTD to activate transcription of class II promoters is that the amino acid residues of Rob required only for activation of class II promoters, that is, the class II surface, reside at four of the five positions as those of the class II surface of SoxS (Fig. 4).43 Thus, although transcription activation of class II promoters by Rob requires fewer residues of σ70 R4 than SoxS, the positions of the amino acids of Rob that are required for activation are essentially the same, that is, the presence of Rob’s CTD has not led to a change in the position of the class II surface.
Another unusual property of SoxS is that the same amino acid residue of its class II surface (D75) contacts two different amino acids of σ70 R4 during transcription activation of fumC (R599) and micF (R603).12 Remarkably, the genetic epistasis experiments presented in Figs. 5d and 6b show that D75 of Rob, like D75 of SoxS, contacts R599 of σ70 R4 at the fumC promoter and R603 at the micF promoter. Using the same amino acid of Rob (SoxS) to contact two different amino acids of σ70 R4 at the fumC and micF promoters is most likely due to differences in the extent of overlap of the −35 hexamer by the robbox (soxbox) at the two promoters, that is, the robbox (soxbox) in the fumC promoter covers all of the −35 element and extends 3 bp further downstream, whereas the robbox (soxbox) in the micF promoter only covers 5 bp of the hexamer (Fig. 8). Indeed, preliminary molecular modeling (data not shown) suggests that this unusual pair of protein–protein interactions indeed emanates from the relative position of the robbox in a given promoter with respect to the position of the −35 element in that promoter. The conservation of these position-dependent interactions between D75 of the activators and two different amino acids of σ70 R4 at two different promoters would not occur if Rob’s CTD had a significant effect on the interaction between Rob’s NTD and σ70 R4.
In this work, we observed that Rob makes only one protein–protein interaction with σ70 R4 at the fumC (D75–R599) and micF (D75–R603) promoters, respectively (Table 1), while SoxS makes three at fumC and four at micF.12 In addition, substitution K593A is the only other substitution of σ70 R4 that reduces Rob-dependent transcription activation, and it does so only at the fumC but not at the micF promoter (Table 1); since no epistatic interaction with it was identified, we presume that the side group of K593 interacts with Rob’s polypeptide backbone or with DNA.52 In contrast, R596A and R599A of σ70 R4 reduce SoxS activation of both promoters, but none of the four potential epistatic interactions were found; as such, their side groups are presumed to interact with DNA or with the polypeptide backbone of Rob. Accordingly, at the two promoters, Rob makes no more than three protein–protein interactions with σ70 R4, whereas SoxS may make as many as 11.
These data raise the interesting question of how Rob is able to interact well enough with RNAP to activate transcription from class II promoters. The answer is that σ70 R4 is not the only surface of RNAP with which Rob makes protein–protein interactions: like many other transcriptional activators, Rob requires the CTD of the RNAP α subunit for activation of transcription from both class I and class II promoters.31 However, alanine scanning mutagenesis of the αCTD (amino acid residues 255–329) showed that, unlike most other transcription activators that make protein–protein interactions with the αCTD through interaction with the 261, 265, and/or 287 determinants,6,8,53-55 Rob-dependent transcription activation of class II promoters fumC, micF, and inaA requires the side groups of amino acid residues within and near the “265 determinant”.31 Thus, with Rob-dependent transcription activation of fumC, micF, and inaA employing the side groups of seven, six, and seven amino acids of the αCTD, respectively, the potential problem of too little protein–protein interaction specificity and too little binding affinity is non-existent.31
Previously, we used the alanine scanning library of the αCTD to identify amino acid residues that are required for SoxS-dependent transcription activation of both class I and class II promoters56 (M. A. Zafar, K. L. Griffith, and R.E.W., unpublished results). Eight amino acids of the αCTD are required for activation of fumC, and of these, six are within or near the 265 determinant. Similarly, six amino acids of the αCTD are required for activation of micF, and of these, five are within or near the 265 determinant. Thus, a total of 14 amino acid residues of the αCTD are required for SoxS-dependent activation of fumC and micF, while a total of 13 are required for Rob-dependent activation of the two promoters. Accordingly, Rob’s interactions with the αCTD in activation of class II promoters help compensate for the dearth of interactions with σ70 R4.
Materials and Methods
Bacterial strains
Strain RA4468 (ΔlacU169 rob∷kan rpsL)35 harboring single-copy λ prophages carrying transcriptional fusions of lac to the fumC and micF promoters33 was used to determine the effect of single alanine substitutions of σ70 at positions 590–613 on Rob-dependent transcription activation of these promoters. Strain RA4468 is a derivative of strain GC4468 (ΔlacU169 rpsL).57 Strain N7840 (ΔlacU169 Δmar rpsL)58 harboring single-copy λ prophages carrying transcriptional fusions of lac to the fpr, fumC, inaA, and micF promoters was used to determine the effect of the tri-alanine substitutions of σ70 at positions 530–590 on Rob-dependent transcription activation of these promoters. This set of lysogens also carries the Δ(araBAD)714 deletion of the arabinose operon.43 Plasmid pMLB1022-micF (micF′-′trpA-′lacOZY from −121 to +85) carrying the micF promoter controlling expression of the ′trpA-lacOZYA fusion was used in the genetic epitasis tests between Rob and the robbox within the micF promoter. Plasmids pMLB1022-micF-C5T and pMLB1022-micF-C15G carrying the substitutions at C5 and C15 of the robboxes were prepared by the QuikChange method of site-directed mutagenesis (Stratagene).
Cloning of Rob
A two-step procedure was used (K. L. Griffith and R.E.W., unpublished results) to clone the rob gene into the expression vector pBAD33.59 The rob gene was amplified by polymerase chain reaction (PCR) from the chromosome of strain HB301 (W3110 ΔlacU169) using the following pair of oligonucleotides: 5′-Rob (5′-CTAGCATCTAGAAGGAGATATACATATGGATCA GGCCGGCAT-3′) and 3′-Rob (5′-AGATTCAAGCTTGGATCCTCATTAACGACGGATCG-3′), where the restriction sites are underlined and the stop codons are in boldface. Conditions used for PCR were as follows: 30 cycles at 95 °C for 1 min, 55 °C for 30 s, and 72 °C for 1 min. The PCR products were purified by agarose gel electrophoresis, digested with restriction enzymes NdeI and BamHI, and ligated into plasmid pET21a (Novagen), which was digested with the same two enzymes. An aliquot of the ligation mixture was transformed into chemically competent strain DH5α and plated onto LB agar medium containing ampicillin (50 μg/ml). Plasmid DNA was isolated by the alkaline lysis method and purified using QIAprep spin mini-prep columns, as described by the manufacturer (Qiagen). The resulting clones were screened by determining the DNA sequence using fluorescent dideoxy sequencing (University of Maryland, Baltimore County core facility). A clone with the correct DNA sequence and no secondary mutations was selected for subcloning into pBAD18 and pBAD33.
To place the rob gene under the control of the arabinose-inducible promoter, PBAD, we digested plasmid pET21a carrying rob with XbaI and HindIII to remove the DNA fragment containing the complete coding sequence of rob along with its ribosome binding site. The appropriate restriction fragment was purified by agarose gel electrophoresis and ligated into plasmids pBAD18 and pBAD33, which had also been digested with the same two enzymes. An aliquot of the ligation mixture was transformed into chemically competent cells of strain DH5α and plated onto LB agar medium supplemented with ampicillin (50 μg/ml) to select for pBAD18 and with chloramphenicol (20 μg/ml) to select for pBAD33. Plasmid DNA was isolated from the resulting transformants and screened for molecules containing the desired insertion by digestion with XbaI and HindIII followed by gel electrophoresis. The resulting plasmids were named pBAD18-Rob and pBAD33-Rob. The addition of his6 tags to create plasmid pBAD-his6-Rob and derivatives carrying single alanine substitutions K30A, L74A, D75A, L78A, and Q85A, the constructs used for protein purification, were created using add-on PCR to introduce six histidine codons between the first and the second codons of rob of the parental plasmids. The PCR products were digested with XbaI and HindIII and ligated into pBAD18 digested with the same enzymes as described above.
Preparation of single alanine substitutions of Rob
Single alanine substitutions of Rob were introduced into the rob gene of plasmids pBAD33-Rob and pBAD18-his6-Rob by the QuikChange method (Stratagene) of site-directed mutagenesis. To prepare the mutations, we linearly amplified segments of plasmids pBAD18 and pBAD33 with Pfu Turbo DNA Polymerase (Stratagene) and a pair of complementary oligonucleotides (Invitrogen) carrying a portion of rob and nucleotide sequences required to change the targeted amino acid of Rob to an alanine (codon GCN). A list of the oligonucleotide sequences used in this study is presented in Table S1. An aliquot of the PCR reactions was transformed into chemically competent cells of strain DH5α and plated onto LB agar medium supplemented with ampicillin (50 μg/ml) or chloramphenicol (20 μg/ml). Plasmid DNA was isolated from the transformants as described above and screened by DNA sequencing. Clones containing the introduced alanine substitution and no additional secondary mutations were then transformed into the strains described below.
Library of single alanine substitutions of σ70 590–613 (R4.2 and C-terminal tail)
The library encoding 17 single alanine substitutions of σ70 was carried on plasmid pGEX-2T and was a gift from C. A. Gross via S. M. Egan.13,45,60 The C-terminal region of each σ70 derivative was sequenced to confirm that the plasmid contained the correct alanine substitution and no secondary mutations. In the process of screening the library, we discovered that several of the mutant alleles had reverted to wild type. Dr. V. A. Rhodius kindly provided replacements. As mentioned above, the library was transformed into derivatives of strain RA4468 harboring plasmid pBAD33-Rob and lysogenic for single-copy λ prophages carrying transcriptional fusions of lacZ to the fumC and micF promoters. Rob expression was under control of the arabinose-inducible PBAD promoter carried on the compatible, low- to medium-copy pBAD33 plasmid,59 while the wild-type and mutant forms of σ70 were expressed from a constitutive lac promoter on plasmid pGEX-2T-σ70.13 The cultures were grown to A600=0.1 in LB medium supplemented with ampicillin (100 μg/ml) and chloramphenicol (20 μg/ml), divided into two parts, with half being treated with 0.2% arabinose and the other half being the uninduced control. After 1 h of continued incubation, the cells were collected and assayed for β-galactosidase activity by our high-throughput method44 as described below.
Library of tri-alanine substitutions of σ70 530–590 (R3.2, R4.1, and R4.2)
Members of the library of tri-alanine substitutions of σ7042 carried on plasmid pVR-σ were transformed by electroporation into strain N7840 harboring compatible plasmid pBAD33-Rob or plasmids with mutant rob alleles and carrying transcriptional fusions of lac to the class II fumC, micF, and inaA promoters. In pVR-σ, members of the library are under the control of a truncated galP1 promoter that constitutively expresses the wild-type and mutant σ70 proteins. Strain N7840 carries wild-type rpoD in the chromosome. Cultures were grown in LB medium supplemented with kanamycin (50 μg/ml) and chloramphenicol (20 μg/ml). When A600 of the cultures reached 0.1–0.2, Rob expression in half of them was induced with 0.2% arabinose and the other half was uninduced. Assay of β-galactosidase activity was carried out as described below.
Assay of β-galactosidase activity
Assays of β-galactosidase activity61 were carried as described by Griffith and Wolf,44 except that the reactions were not stopped with chloramphenicol. Independent determinations of β-galactosidase activity were carried out on at least three different days. Activity values of 80% of wild-type or less were considered to be significant. Only assays with a standard deviation of less than 12% of the mean were retained. Two-tailed t tests were used to determine the significance of differences in the effects of the mutations on transcriptional activation as described previously.12
Purification of Rob
Wild-type and mutant Rob proteins were expressed from pBAD-his6-Rob and purified by nickel affinity chromatography utilizing the BioLogic DuoFlow protein purification system (Bio-Rad). The protocol was modified from a method described previously.37 Cultures were grown at 37 °C in LB medium containing ampicillin (100 μg/ml) to A600=0.5 at which point Rob expression was induced with 0.2% arabinose. After 3 h, cells were harvested by centrifugation, resuspended in binding buffer [20 mM Tris–Cl (pH 8.0), 0.5 M NaCl, and 5 mM imidazole], and disrupted by passage through a French pressure cell at 4 °C. The extract was centrifuged at 13,000 rpm for 30 min at 4 °C, and the supernatant was applied to a Ni-NTA column pre-equilibrated with binding buffer. The column was then washed with 10 column volumes of the binding buffer, followed by 6 volumes of wash buffer [20 mM Tris–Cl (pH 8.0), 0.5 M NaCl, and 30 mM imidazole]. Rob was eluted from the column with 6 column volumes of elution buffer [20 mM Tris–Cl (pH 8.0), 0.5 M NaCl, and 450 mM imidazole]. The purity of the Rob preparations was estimated to be >95% by SDS-PAGE (data not shown). Samples were concentrated by dialysis into Rob dialysis buffer [20 mM Tris–Cl (pH 8.0), 0.3 M NaCl, and 50% glycerol] and stored at −20 °C.
Electrophoretic mobility shift assays
We used EMSAs to compare the DNA binding affinity of wild-type Rob to that of mutant Rob proteins containing single alanine substitutions. Complementary 25-mer oligonucleotides containing the fumC robbox DNA sequence were synthesized (Invitrogen). One oligonucleotide of the pair was labeled on its 5′ end with bacteriophage T4 polynucleotide kinase (Fermentas) and [γ-32P]ATP. The labeled oligonucleotide was annealed to a twofold molar excess of the complementary strand, and the duplex was purified with a G-25 column (Eppendorf) to remove excess label and primers. The EMSAs were carried out as previously described.19,20,43 Binding reactions contained 5 mM Tris, 24 mM Hepes (pH 7.9), 50 mM potassium glutamate, 20 mM NaCl, 1.4 mM ethylenediaminetetraacetic acid, 0.4 mg/ml bovine serum albumin, 9% glycerol, 0.5 μg poly(dIdC), ~3 fmol labeled DNA, and increasing concentrations of protein in a final volume of 25 μl. Rob–DNA complexes were separated by electrophoresis in 12% (w/v) polyacrylamide gels under non-denaturing conditions.
Acknowledgments
We thank Carol Gross, Virgil Rhodius, and Susan Egan for providing the library of single alanine substitutions of σ70 at positions 590–613. Thanks also go to Ammar Zafar for critical reading of the manuscript, to Belinda Jackson for determining the sequence of σ70 in the σ70 590–613 plasmids, and to Jason Constantineau for help in purifying Rob and the mutant derivatives of it. We gratefully acknowledge Kevin Griffith for preparation of the pBAD plasmids carrying Rob and his6-Rob. This work was supported by Public Health Service grant GM27113 from the National Institutes of Health and from the Designated Research Initiative Fund of University of Maryland, Baltimore County, both of which were awarded to R.E.W. Funds supporting N.S.-A. were provided by the Fundació Cellex.
Abbreviations used
- RNAP
RNA polymerase
- σ70 R4
region 4 of the σ70 subunit
- CTD
C-terminal domain
- HTH
helix–turn–helix
- NTD
N-terminal domain
- EMSA
electrophoretic mobility shift assay
References
- 1.Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- 2.Gross CA, Chan C, Dombroski A, Gruber T, Sharp M, Tupy J, Young B. The functional and regulatory roles of sigma factors in transcription. Cold Spring Harbor Symp Quant Biol. 1998;63:141–155. doi: 10.1101/sqb.1998.63.141. [DOI] [PubMed] [Google Scholar]
- 3.Borukhov S, Nudler E. RNA polymerase holoenzyme: structure, function and biological implications. Curr Opin Microbiol. 2003;6:93–100. doi: 10.1016/s1369-5274(03)00036-5. [DOI] [PubMed] [Google Scholar]
- 4.Wade JT, Roa DC, Grainger DC, Hurd D, Busby SJ, Struhl K, Nudler E. Extensive functional overlap between sigma factors in Escherichia coli. Nat Struct Mol Biol. 2006;13:806–814. doi: 10.1038/nsmb1130. [DOI] [PubMed] [Google Scholar]
- 5.Hengge-Aronis R. Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase. Microbiol Mol Biol Rev. 2002;66:373–395. doi: 10.1128/MMBR.66.3.373-395.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benoff B, Yang H, Lawson CL, Parkinson G, Liu J, Blatter E, et al. Structural basis of transcription activation: the CAP-alpha CTD-DNA complex. Science. 2002;297:1562–1566. doi: 10.1126/science.1076376. [DOI] [PubMed] [Google Scholar]
- 7.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 8.Busby S, Ebright RH. Transcription activation by catabolite activator protein (CAP) J Mol Biol. 1999;293:199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- 9.Ebright RH. Transcription activation at class I CAP-dependent promoters. Mol Microbiol. 1993;8:797–802. doi: 10.1111/j.1365-2958.1993.tb01626.x. [DOI] [PubMed] [Google Scholar]
- 10.Busby S, Ebright RH. Transcription activation at class II CAP-dependent promoters. Mol Microbiol. 1997;23:853–859. doi: 10.1046/j.1365-2958.1997.2771641.x. [DOI] [PubMed] [Google Scholar]
- 11.Martin RG, Gillette WK, Martin NI, Rosner JL. Complex formation between activator and RNA polymerase as the basis for transcription activation by MarA and SoxS in Escherichia coli. Mol Microbiol. 2002;43:355–370. doi: 10.1046/j.1365-2958.2002.02748.x. [DOI] [PubMed] [Google Scholar]
- 12.Zafar MA, Shah IM, Wolf RE., Jr Protein–protein interactions between sigma(70) region 4 of RNA polymerase and Escherichia coli SoxS, a transcription activator that functions by the prerecruitment mechanism: evidence for “off-DNA” and “on-DNA” interactions. J Mol Biol. 2010;401:13–32. doi: 10.1016/j.jmb.2010.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lonetto MA, Rhodius V, Lamberg K, Kiley P, Busby S, Gross C. Identification of a contact site for different transcription activators in region 4 of the Escherichia coli RNA polymerase sigma(70) subunit. J Mol Biol. 1998;284:1353–1365. doi: 10.1006/jmbi.1998.2268. [DOI] [PubMed] [Google Scholar]
- 14.Dove SL, Darst SA, Hochschild A. Region 4 of sigma as a target for transcription regulation. Mol Microbiol. 2003;48:863–874. doi: 10.1046/j.1365-2958.2003.03467.x. [DOI] [PubMed] [Google Scholar]
- 15.Grainger DC, Webster CL, Belyaeva TA, Hyde EI, Busby SJW. Transcription activation at the Esherichia coli melAB promoter: interactions of MelR with its DNA target site and with domain 4 of the RNA polymerase sigma subunit. Mol Microbiol. 2004;51:1297–1310. doi: 10.1111/j.1365-2958.2003.03929.x. [DOI] [PubMed] [Google Scholar]
- 16.Griffith KL, Becker SM, Wolf RE., Jr Characterization of TetD as a transcriptional activator of a subset of genes of the Escherichia coli SoxS/Mar-A/Rob regulon. Mol Microbiol. 2005;56:1103–1117. doi: 10.1111/j.1365-2958.2005.04599.x. [DOI] [PubMed] [Google Scholar]
- 17.Gallegos MT, Schleif R, Bairoch A, Hofmann K, Ramos JL. AraC/XylS family of transcriptional regulators. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jair KW, Yu X, Skarstad K, Thöny B, Fujita N, Ishihama A, Wolf RE., Jr Transcriptional activation of promoters of the superoxide and multiple antibiotic resistance regulons by Rob, a binding protein of the Escherichia coli origin of chromosomal replication. J Bacteriol. 1996;178:2507–2513. doi: 10.1128/jb.178.9.2507-2513.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fawcett WP, Wolf RE., Jr Purification of a MalE–SoxS fusion protein and identification of the control sites of Escherichia coli superoxide-inducible genes. Mol Microbiol. 1994;14:669–679. doi: 10.1111/j.1365-2958.1994.tb01305.x. [DOI] [PubMed] [Google Scholar]
- 20.Griffith KL, Wolf RE., Jr Systematic mutagenesis of the DNA binding sites for SoxS in the Escherichia coli zwf and fpr promoters: identifying nucleotides required for DNA binding and transcription activation. Mol Microbiol. 2001;40:1141–1154. doi: 10.1046/j.1365-2958.2001.02456.x. Erratum Mol. Microbiol. 42:571. [DOI] [PubMed] [Google Scholar]
- 21.Martin RG, Gillette WK, Rhee S, Rosner JL. Structural requirements for marbox function in transcriptional activation of mar/sox/rob regulon promoters in Escherichia coli: sequence, orientation and spatial relationship to the core promoter. Mol Microbiol. 1999;34:431–441. doi: 10.1046/j.1365-2958.1999.01599.x. [DOI] [PubMed] [Google Scholar]
- 22.Wood TI, Griffith KL, Fawcett WP, Jair KW, Schneider TD, Wolf RE., Jr Interdependence of the position and orientation of SoxS binding sites in the transcriptional activation of the class I subset of Escherichia coli superoxide-inducible promoters. Mol Microbiol. 1999;34:414–430. doi: 10.1046/j.1365-2958.1999.01598.x. [DOI] [PubMed] [Google Scholar]
- 23.Jair KW, Martin RG, Rosner JL, Fujita N, Ishihama A, Wolf RE., Jr Purification and regulatory properties of MarA protein, a transcriptional activator of Escherichia coli multiple antibiotic and superoxide resistance promoters. J Bacteriol. 1995;177:7100–7104. doi: 10.1128/jb.177.24.7100-7104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blanchard JL, Wholey WY, Conlon EM, Pomposiello PJ. Rapid changes in gene expression dynamics in response to superoxide reveal SoxRS-dependent and independent transcriptional networks. PLoS One. 2007;2:e1186. doi: 10.1371/journal.pone.0001186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ariza RR, Cohen SP, Bachhawat N, Levy SB, Demple B. Repressor mutations in the marRAB operon that activate oxidative stress genes and multiple antibiotic resistance in Escherichia coli. J Bacteriol. 1994;176:143–148. doi: 10.1128/jb.176.1.143-148.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barbosa TM, Levy SB. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J Bacteriol. 2000;182:3467–3474. doi: 10.1128/jb.182.12.3467-3474.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin RG, Nyantakyi PS, Rosner JL. Regulation of the multiple antibiotic resistance (mar) regulon by marORA sequences in Escherichia coli. J Bacteriol. 1995;177:4176–4178. doi: 10.1128/jb.177.14.4176-4178.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pomposiello PJ, Bennik MH, Demple B. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J Bacteriol. 2001;183:3890–3902. doi: 10.1128/JB.183.13.3890-3902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Griffith KL, Shah IM, Myers TE, O’Neill MC, Wolf RE., Jr Evidence for “pre-recruitment” as a new mechanism of transcription activation in Escherichia coli: the large excess of SoxS binding sites per cell relative to the number of SoxS molecules per cell. Biochem Biophys Res Commun. 2002;291:979–986. doi: 10.1006/bbrc.2002.6559. [DOI] [PubMed] [Google Scholar]
- 30.Griffith KL, Wolf RE., Jr Genetic evidence for pre-recruitment as the mechanism of transcription activation by SoxS of Escherichia coli: the dominance of DNA binding mutations of SoxS. J Mol Biol. 2004;344:1–10. doi: 10.1016/j.jmb.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 31.Keen EF., III . Doctoral dissertation. University of Maryland; Baltimore County: 2005. Molecular characterization of the Escherichia coli Rob protein: identifying the protein–protein interactions with RNA polymerase and determining the domains required for transcription activation and DNA binding. [Google Scholar]
- 32.Ariza RR, Li Z, Ringstad N, Demple B. Activation of multiple antibiotic resistance and binding of stress-inducible promoters by Escherichia coli Rob protein. J Bacteriol. 1995;177:1655–1661. doi: 10.1128/jb.177.7.1655-1661.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin RG, Gillette WK, Rosner JL. Promoter discrimination by the related transcriptional activators MarA and SoxS: differential regulation by differential binding. Mol Microbiol. 2000;35:623–634. doi: 10.1046/j.1365-2958.2000.01732.x. [DOI] [PubMed] [Google Scholar]
- 34.Azam TA, Ishihama A. Twelve species of the nucleoid-associated protein from Escherichia coli. Sequence recognition specificity and DNA binding affinity. J Biol Chem. 1999;274:33105–33113. doi: 10.1074/jbc.274.46.33105. [DOI] [PubMed] [Google Scholar]
- 35.Skarstad K, Thony B, Hwang DS, Kornberg A. A novel binding protein of the origin of the Escherichia coli chromosome. J Biol Chem. 1993;268:5365–5370. [PubMed] [Google Scholar]
- 36.Gu M, Imlay JA. The SoxRS response of Escherichia coli is directly activated by redox-cycling drugs rather than by superoxide. Mol Microbiol. 2011;79:1136–1150. doi: 10.1111/j.1365-2958.2010.07520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenberg EY, Bertenthal D, Nilles ML, Bertrand KP, Nikaido H. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol Microbiol. 2003;48:1609–1619. doi: 10.1046/j.1365-2958.2003.03531.x. [DOI] [PubMed] [Google Scholar]
- 38.Rosner JL, Dangi B, Gronenborn AM, Martin RG. Posttranscriptional activation of the transcriptional activator Rob by dipyridyl in Escherichia coli. J Bacteriol. 2002;184:1407–1416. doi: 10.1128/JB.184.5.1407-1416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwon HJ, Bennik MHJ, Demple B, Ellenberger T. Crystal structure of the Escherichia coli Rob transcription factor in complex with DNA. Nat Struct Biol. 2000;7:424–430. doi: 10.1038/75213. [DOI] [PubMed] [Google Scholar]
- 40.Griffith KL, Fitzpatrick MM, Keen EF, III, Wolf RE., Jr Two functions of the C-terminal domain of Escherichia coli Rob: mediating “sequestration–dispersal” as a novel off–on switch for regulating Rob’s activity as a transcription activator and preventing degradation of Rob by Lon protease. J Mol Biol. 2009;388:415–430. doi: 10.1016/j.jmb.2009.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rhee S, Martin RG, Rosner JL, Davies DR. A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc Natl Acad Sci USA. 1998;95:10413–10418. doi: 10.1073/pnas.95.18.10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zafar MA, Sanchez-Alberola N, Wolf RE., Jr Genetic evidence for a novel interaction between transcriptional activator SoxS and region 4 of the sigma(70) subunit of RNA polymerase at class II SoxS-dependent promoters in Escherichia coli. J Mol Biol. 2011;407:333–353. doi: 10.1016/j.jmb.2010.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Griffith KL, Wolf RE., Jr A comprehensive alanine scanning mutagenesis of the Escherichia coli transcriptional activator SoxS: identifying amino acids important for DNA binding and transcription activation. J Mol Biol. 2002;322:237–257. doi: 10.1016/s0022-2836(02)00782-9. [DOI] [PubMed] [Google Scholar]
- 44.Griffith KL, Wolf RE., Jr Measuring beta-galactosidase activity in bacteria: cell growth, permeabilization, and enzyme assays in 96-well arrays. Biochem Biophys Res Commun. 2002;290:397–402. doi: 10.1006/bbrc.2001.6152. [DOI] [PubMed] [Google Scholar]
- 45.Bhende PM, Egan SM. Genetic evidence that transcription activation by RhaS involves specific amino acid contacts with sigma 70. J Bacteriol. 2000;182:4959–4969. doi: 10.1128/jb.182.17.4959-4969.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lonetto M, Gribskov M, Gross CA. The sigma 70 family: sequence conservation and evolutionary relationships. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 48.Bhende PM, Egan SM. Amino acid–DNA contacts by RhaS: an AraC family transcription activator. J Bacteriol. 1999;181:5185–5192. doi: 10.1128/jb.181.17.5185-5192.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wickstrum JR, Egan SM. Amino acid contacts between sigma 70 domain 4 and the transcription activators RhaS and RhaR. J Bacteriol. 2004;186:6277–6285. doi: 10.1128/JB.186.18.6277-6285.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jair KW, Wolf RE., Jr . Characterization of transcriptional activator proteins SoxS, MarA, and Rob in Escherichia coli. University of Maryland; Baltimore County: 1996. [Google Scholar]
- 51.Fawcett WP, Wolf RE., Jr Genetic definition of the Escherichia coli zwf “soxbox,” the DNA binding site for SoxS-mediated induction of glucose 6-phosphate dehydrogenase in response to superoxide. J Bacteriol. 1995;177:1742–1750. doi: 10.1128/jb.177.7.1742-1750.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nickels BD, Dove SL, Murakami KS, Darst SA, Hochschild A. Protein–protein and protein–DNA interactions of sigma 70 region 4 involved in transcription activation by lambda cI. J Mol Biol. 2002;324:17–34. doi: 10.1016/s0022-2836(02)01043-4. [DOI] [PubMed] [Google Scholar]
- 53.Busby S, Ebright RH. Promoter structure, promoter recognition, and transcription activation in prokaryotes. Cell. 1994;79:743–746. doi: 10.1016/0092-8674(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 54.Savery NJ, Lloyd GS, Busby SJW, Thomas MS, Ebright RH, Gourse RL. Determinants of the C-terminal domain of the Escherichia coli RNA polymerase alpha subunit important for transcription at class I cyclic AMP receptor protein-dependent promoters. J Bacteriol. 2002;184:2273–2280. doi: 10.1128/JB.184.8.2273-2280.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Savery NJ, Lloyd GS, Kainz M, Gaal T, Ross W, Ebright RH, et al. Transcription activation at class II CRP-dependent promoters: identification of determinants in the C-terminal domain of the RNA polymerase alpha subunit. EMBO J. 1998;17:3439–3447. doi: 10.1093/emboj/17.12.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shah IM, Wolf RE., Jr Novel protein–protein interaction between Escherichia coli SoxS and the DNA binding determinant of the RNA polymerase alpha subunit: SoxS functions as a co-sigma factor and redeploys RNA polymerase from UP-element-containing promoters to SoxS-dependent promoters during oxidative stress. J Mol Biol. 2004;343:513–532. doi: 10.1016/j.jmb.2004.08.057. [DOI] [PubMed] [Google Scholar]
- 57.Greenberg JT, Monach P, Chou JT, Demple B. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc Natl Acad Sci USA. 1990;87:6181–6185. doi: 10.1073/pnas.87.16.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martin R, Rosner J. Fis, an accessorial factor for transcriptional activation of the mar (multiple antibiotic resistance) promoter of Escherichia coli in the presence of the activator MarA, SoxS, or Rob. J Bacteriol. 1997;179:7410–7419. doi: 10.1128/jb.179.23.7410-7419.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guzman LM, Belin D, Carson MJ, Beckwith JR. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dombroski AJ, Walter WA, Record MT, Jr, Siegele DA, Gross CA. Polypeptides containing highly conserved regions of transcription initiation factor sigma 70 exhibit specificity of binding to promoter DNA. Cell. 1992;70:501–512. doi: 10.1016/0092-8674(92)90174-b. [DOI] [PubMed] [Google Scholar]
- 61.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1972. [Google Scholar]
- 62.Jair KW, Fawcett WP, Fujita N, Ishihama A, Wolf RE., Jr Ambidextrous transcriptional activation by SoxS: requirement for the C-terminal domain of the RNA polymerase alpha subunit in a subset of Escherichia coli superoxide-inducible genes. Mol Microbiol. 1996;19:307–317. doi: 10.1046/j.1365-2958.1996.368893.x. [DOI] [PubMed] [Google Scholar]
- 63.Andersen J, Delihas N, Ikenaka K, Green PJ, Pines O, Ilercil O, Inouye M. The isolation and characterization of RNA coded by the micF gene in Escherichia coli. Nucleic Acids Res. 1987;15:2089–2101. doi: 10.1093/nar/15.5.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]


