Abstract
Myogenin is a skeletal muscle-specific transcription factor that can activate myogenesis when introduced into a variety of nonmuscle cell types. Activation of the myogenic program by myogenin is dependent on its binding to a DNA sequence known as an E box, which is associated with numerous muscle-specific genes. Myogenin shares homology with MyoD and other myogenic regulatory factors within a basic region and a helix-loop-helix (HLH) motif that mediate DNA binding and dimerization, respectively. Here we show that the basic region-HLH motif of myogenin alone lacks transcriptional activity and is dependent on domains in the amino and carboxyl termini to activate transcription. Analysis of these N- and C-terminal domains through creation of chimeras with the DNA-binding domain of the Saccharomyces cerevisiae transcription factor GAL4 revealed that they act as strong transcriptional activators. These transcription activation domains are dependent for activity on a specific amino acid sequence within the basic region, referred to as the myogenic recognition motif (MRM), when an E box is the target for DNA binding. However, the activation domains function independent of the MRM when DNA binding is mediated through a heterologous DNA-binding domain. The activation domain of the acidic coactivator VP16 can substitute for the myogenin activation domains and restore strong myogenic activity to the basic region-HLH motif. Within a myogenin-VP16 chimera, however, the VP16 activation domain also relies on the MRM for activation of the myogenic program. These findings reveal that DNA binding and transcriptional activation are separable functions, encoded by different domains of myogenin, but that the activity of the transcriptional activation domains is influenced by the DNA-binding domain. Activation of muscle-specific transcription requires collaboration between the DNA-binding and activation domains of myogenin and is dependent on events in addition to DNA binding.
Full text
PDF






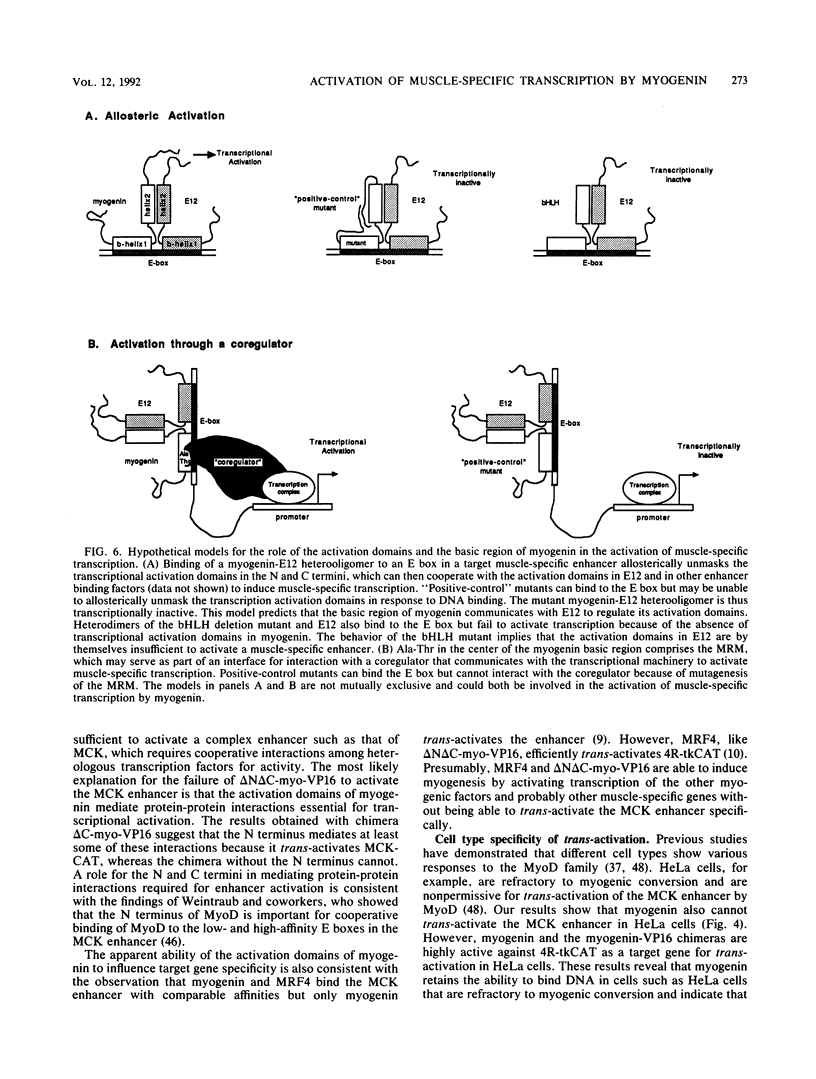


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Binétruy B., Smeal T., Karin M. Ha-Ras augments c-Jun activity and stimulates phosphorylation of its activation domain. Nature. 1991 May 9;351(6322):122–127. doi: 10.1038/351122a0. [DOI] [PubMed] [Google Scholar]
- Braun T., Bober E., Winter B., Rosenthal N., Arnold H. H. Myf-6, a new member of the human gene family of myogenic determination factors: evidence for a gene cluster on chromosome 12. EMBO J. 1990 Mar;9(3):821–831. doi: 10.1002/j.1460-2075.1990.tb08179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun T., Buschhausen-Denker G., Bober E., Tannich E., Arnold H. H. A novel human muscle factor related to but distinct from MyoD1 induces myogenic conversion in 10T1/2 fibroblasts. EMBO J. 1989 Mar;8(3):701–709. doi: 10.1002/j.1460-2075.1989.tb03429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun T., Winter B., Bober E., Arnold H. H. Transcriptional activation domain of the muscle-specific gene-regulatory protein myf5. Nature. 1990 Aug 16;346(6285):663–665. doi: 10.1038/346663a0. [DOI] [PubMed] [Google Scholar]
- Brennan T. J., Chakraborty T., Olson E. N. Mutagenesis of the myogenin basic region identifies an ancient protein motif critical for activation of myogenesis. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5675–5679. doi: 10.1073/pnas.88.13.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan T. J., Olson E. N. Myogenin resides in the nucleus and acquires high affinity for a conserved enhancer element on heterodimerization. Genes Dev. 1990 Apr;4(4):582–595. doi: 10.1101/gad.4.4.582. [DOI] [PubMed] [Google Scholar]
- Buskin J. N., Hauschka S. D. Identification of a myocyte nuclear factor that binds to the muscle-specific enhancer of the mouse muscle creatine kinase gene. Mol Cell Biol. 1989 Jun;9(6):2627–2640. doi: 10.1128/mcb.9.6.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty T., Brennan T. J., Li L., Edmondson D., Olson E. N. Inefficient homooligomerization contributes to the dependence of myogenin on E2A products for efficient DNA binding. Mol Cell Biol. 1991 Jul;11(7):3633–3641. doi: 10.1128/mcb.11.7.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty T., Brennan T., Olson E. Differential trans-activation of a muscle-specific enhancer by myogenic helix-loop-helix proteins is separable from DNA binding. J Biol Chem. 1991 Feb 15;266(5):2878–2882. [PubMed] [Google Scholar]
- Chakraborty T., Olson E. N. Domains outside of the DNA-binding domain impart target gene specificity to myogenin and MRF4. Mol Cell Biol. 1991 Dec;11(12):6103–6108. doi: 10.1128/mcb.11.12.6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cress W. D., Triezenberg S. J. Critical structural elements of the VP16 transcriptional activation domain. Science. 1991 Jan 4;251(4989):87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- Davis R. L., Cheng P. F., Lassar A. B., Weintraub H. The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell. 1990 Mar 9;60(5):733–746. doi: 10.1016/0092-8674(90)90088-v. [DOI] [PubMed] [Google Scholar]
- Davis R. L., Weintraub H., Lassar A. B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987 Dec 24;51(6):987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Edmondson D. G., Brennan T. J., Olson E. N. Mitogenic repression of myogenin autoregulation. J Biol Chem. 1991 Nov 15;266(32):21343–21346. [PubMed] [Google Scholar]
- Edmondson D. G., Olson E. N. A gene with homology to the myc similarity region of MyoD1 is expressed during myogenesis and is sufficient to activate the muscle differentiation program. Genes Dev. 1989 May;3(5):628–640. doi: 10.1101/gad.3.5.628. [DOI] [PubMed] [Google Scholar]
- Gonzalez G. A., Montminy M. R. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989 Nov 17;59(4):675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- Henthorn P., Kiledjian M., Kadesch T. Two distinct transcription factors that bind the immunoglobulin enhancer microE5/kappa 2 motif. Science. 1990 Jan 26;247(4941):467–470. doi: 10.1126/science.2105528. [DOI] [PubMed] [Google Scholar]
- Hochschild A., Irwin N., Ptashne M. Repressor structure and the mechanism of positive control. Cell. 1983 Feb;32(2):319–325. doi: 10.1016/0092-8674(83)90451-8. [DOI] [PubMed] [Google Scholar]
- Ingraham H. A., Flynn S. E., Voss J. W., Albert V. R., Kapiloff M. S., Wilson L., Rosenfeld M. G. The POU-specific domain of Pit-1 is essential for sequence-specific, high affinity DNA binding and DNA-dependent Pit-1-Pit-1 interactions. Cell. 1990 Jun 15;61(6):1021–1033. doi: 10.1016/0092-8674(90)90067-o. [DOI] [PubMed] [Google Scholar]
- Jaynes J. B., Johnson J. E., Buskin J. N., Gartside C. L., Hauschka S. D. The muscle creatine kinase gene is regulated by multiple upstream elements, including a muscle-specific enhancer. Mol Cell Biol. 1988 Jan;8(1):62–70. doi: 10.1128/mcb.8.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. S., Guarente L. Mutations that alter transcriptional activation but not DNA binding in the zinc finger of yeast activator HAPI. Nature. 1989 Nov 9;342(6246):200–203. doi: 10.1038/342200a0. [DOI] [PubMed] [Google Scholar]
- Lassar A. B., Buskin J. N., Lockshon D., Davis R. L., Apone S., Hauschka S. D., Weintraub H. MyoD is a sequence-specific DNA binding protein requiring a region of myc homology to bind to the muscle creatine kinase enhancer. Cell. 1989 Sep 8;58(5):823–831. doi: 10.1016/0092-8674(89)90935-5. [DOI] [PubMed] [Google Scholar]
- Lassar A. B., Davis R. L., Wright W. E., Kadesch T., Murre C., Voronova A., Baltimore D., Weintraub H. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell. 1991 Jul 26;66(2):305–315. doi: 10.1016/0092-8674(91)90620-e. [DOI] [PubMed] [Google Scholar]
- Lewin B. Commitment and activation at pol II promoters: a tail of protein-protein interactions. Cell. 1990 Jun 29;61(7):1161–1164. doi: 10.1016/0092-8674(90)90675-5. [DOI] [PubMed] [Google Scholar]
- Lillie J. W., Green M. R. Transcription activation by the adenovirus E1a protein. Nature. 1989 Mar 2;338(6210):39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- Lin H., Yutzey K. E., Konieczny S. F. Muscle-specific expression of the troponin I gene requires interactions between helix-loop-helix muscle regulatory factors and ubiquitous transcription factors. Mol Cell Biol. 1991 Jan;11(1):267–280. doi: 10.1128/mcb.11.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Ptashne M. A new class of yeast transcriptional activators. Cell. 1987 Oct 9;51(1):113–119. doi: 10.1016/0092-8674(87)90015-8. [DOI] [PubMed] [Google Scholar]
- Ma J., Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987 Mar 13;48(5):847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- Miner J. H., Wold B. Herculin, a fourth member of the MyoD family of myogenic regulatory genes. Proc Natl Acad Sci U S A. 1990 Feb;87(3):1089–1093. doi: 10.1073/pnas.87.3.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989 Mar 10;56(5):777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Vaessin H., Caudy M., Jan L. Y., Jan Y. N., Cabrera C. V., Buskin J. N., Hauschka S. D., Lassar A. B. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989 Aug 11;58(3):537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- Olson E. N. MyoD family: a paradigm for development? Genes Dev. 1990 Sep;4(9):1454–1461. doi: 10.1101/gad.4.9.1454. [DOI] [PubMed] [Google Scholar]
- Piette J., Bessereau J. L., Huchet M., Changeux J. P. Two adjacent MyoD1-binding sites regulate expression of the acetylcholine receptor alpha-subunit gene. Nature. 1990 May 24;345(6273):353–355. doi: 10.1038/345353a0. [DOI] [PubMed] [Google Scholar]
- Rhodes S. J., Konieczny S. F. Identification of MRF4: a new member of the muscle regulatory factor gene family. Genes Dev. 1989 Dec;3(12B):2050–2061. doi: 10.1101/gad.3.12b.2050. [DOI] [PubMed] [Google Scholar]
- Sartorelli V., Webster K. A., Kedes L. Muscle-specific expression of the cardiac alpha-actin gene requires MyoD1, CArG-box binding factor, and Sp1. Genes Dev. 1990 Oct;4(10):1811–1822. doi: 10.1101/gad.4.10.1811. [DOI] [PubMed] [Google Scholar]
- Schena M., Freedman L. P., Yamamoto K. R. Mutations in the glucocorticoid receptor zinc finger region that distinguish interdigitated DNA binding and transcriptional enhancement activities. Genes Dev. 1989 Oct;3(10):1590–1601. doi: 10.1101/gad.3.10.1590. [DOI] [PubMed] [Google Scholar]
- Schäfer B. W., Blakely B. T., Darlington G. J., Blau H. M. Effect of cell history on response to helix-loop-helix family of myogenic regulators. Nature. 1990 Mar 29;344(6265):454–458. doi: 10.1038/344454a0. [DOI] [PubMed] [Google Scholar]
- Stern S., Tanaka M., Herr W. The Oct-1 homoeodomain directs formation of a multiprotein-DNA complex with the HSV transactivator VP16. Nature. 1989 Oct 19;341(6243):624–630. doi: 10.1038/341624a0. [DOI] [PubMed] [Google Scholar]
- Sternberg E. A., Spizz G., Perry W. M., Vizard D., Weil T., Olson E. N. Identification of upstream and intragenic regulatory elements that confer cell-type-restricted and differentiation-specific expression on the muscle creatine kinase gene. Mol Cell Biol. 1988 Jul;8(7):2896–2909. doi: 10.1128/mcb.8.7.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S., Richmond T. J. DNA binding-induced conformational change of the yeast transcriptional activator PRTF. Cell. 1990 Jul 27;62(2):367–377. doi: 10.1016/0092-8674(90)90373-m. [DOI] [PubMed] [Google Scholar]
- Tapscott S. J., Davis R. L., Thayer M. J., Cheng P. F., Weintraub H., Lassar A. B. MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science. 1988 Oct 21;242(4877):405–411. doi: 10.1126/science.3175662. [DOI] [PubMed] [Google Scholar]
- Tapscott S. J., Weintraub H. MyoD and the regulation of myogenesis by helix-loop-helix proteins. J Clin Invest. 1991 Apr;87(4):1133–1138. doi: 10.1172/JCI115109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer M. J., Tapscott S. J., Davis R. L., Wright W. E., Lassar A. B., Weintraub H. Positive autoregulation of the myogenic determination gene MyoD1. Cell. 1989 Jul 28;58(2):241–248. doi: 10.1016/0092-8674(89)90838-6. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Davis R., Lockshon D., Lassar A. MyoD binds cooperatively to two sites in a target enhancer sequence: occupancy of two sites is required for activation. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5623–5627. doi: 10.1073/pnas.87.15.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Dwarki V. J., Verma I., Davis R., Hollenberg S., Snider L., Lassar A., Tapscott S. J. Muscle-specific transcriptional activation by MyoD. Genes Dev. 1991 Aug;5(8):1377–1386. doi: 10.1101/gad.5.8.1377. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Tapscott S. J., Davis R. L., Thayer M. J., Adam M. A., Lassar A. B., Miller A. D. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5434–5438. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentworth B. M., Donoghue M., Engert J. C., Berglund E. B., Rosenthal N. Paired MyoD-binding sites regulate myosin light chain gene expression. Proc Natl Acad Sci U S A. 1991 Feb 15;88(4):1242–1246. doi: 10.1073/pnas.88.4.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright W. E., Sassoon D. A., Lin V. K. Myogenin, a factor regulating myogenesis, has a domain homologous to MyoD. Cell. 1989 Feb 24;56(4):607–617. doi: 10.1016/0092-8674(89)90583-7. [DOI] [PubMed] [Google Scholar]