Abstract
White blood cell count (WBC) is unique among identified inflammatory predictors of chronic disease in that it is routinely measured in asymptomatic patients in the course of routine patient care. We led a genome-wide association analysis to identify variants associated with WBC levels in 13,923 subjects in the electronic Medical Records and Genomics (eMERGE) Network. We identified two regions of interest that were each unique to subjects of genetically determined ancestry to the African continent (AA) or to the European continent (EA). WBC varies among different ancestry groups. Despite being ancestry specific, these regions were identifiable in the combined analysis. In AA subjects, the region surrounding the Duffy antigen/chemokine receptor gene (DARC) on 1q21 exhibited significant association (p value = 6.71e–55). These results validate the previously reported association between WBC and of the regulatory variant rs2814778 in the promoter region, which causes the Duffy negative phenotype (Fy−/−). A second missense variant (rs12075) is responsible for the two principal antigens, Fya and Fyb of the Duffy blood group system. The two variants, consisting of four alleles, act in concert to produce five antigens and subsequent phenotypes. We were able to identify the marginal and novel interaction effects of these two variants on WBC. In the EA subjects, we identified significantly associated SNPs tagging three separate genes in the 17q21 region: (1) GSDMA, (2) MED24, and (3) PSMD3. Variants in this region have been reported to be associated with WBC, neutrophil count, and inflammatory diseases including asthma and Crohn’s disease.
Introduction
White blood cell count (WBC) is an important marker of the health of the immune system. It is also emerging as a risk factor for chronic diseases. Elevated total WBC and its differential cell line levels have been identified as risk factors for multiple diseases including asthma (Gudbjartsson et al. 2009), coronary artery disease (Madjid et al.2004; Weijenberg et al. 1996), myocardial infarction (Gudbjartsson et al. 2009), and cancer (Nalls et al. 2008; Shankar et al. 2007). Genome wide association studies of total WBC (leukocyte count) and its constitutional cell lines (neutrophils, basophils, eosinophils, monocytes, and lymphocytes) are expected to identify genes that predict WBC variability as the end result of a variety of inflammatory pathways.
WBC varies with clinical events, which is why it is often gathered in the course of clinical care. High levels are associated with infections or leukemias, while low levels are associated with immune failure or diseased bone marrow. Within the normal range, however, several studies have suggested associations with chronic WBC levels and a variety of conditions, likely because the WBC is an indicator of overall inflammatory activity levels. While environmental influences (infection, cancer, etc.) likely are responsible for the majority of variability in acute WBC levels, chronic “resting state” WBC levels are likely to be influenced by genetic variation. We thus sought to use a genome wide association study (GWAS) of “resting state” WBC to identify genetic loci, with the ultimate goal of furthering our understanding of genetic influences on resting state inflammation.
WBC is also known to vary among groups of different ancestry (Bach 2002) and understanding these biological mechanisms may be useful in understanding the etiology of inflammation and ultimate effects on chronic inflammatory diseases. A lower neutrophil count, the major class of WBC in human peripheral blood, is reported in subjects with African ancestry (Nalls et al. 2008; Reich et al. 2009). This lower WBC in those of African ancestry has been attributed, in part, to an association with the Duffy antigen/chemokine receptor gene (DARC). The 336 amino acid Duffy glycoprotein is a receptor that binds cytokines released during inflammation.
Two SNPs (rs12075 and rs2814778) determine the Duffy antigen phenotype. The Duffy Fya and Fyb phenotypes differ at a single amino acid glycine → aspartic acid (G42D), due to a 125 G → A substitution (rs12075) (Dean 2005). The rs12075 variant has been shown to be associated with levels of monocyte chemo attractant protein-1 (MCP-1) and other inflammatory mediators (Schnabel et al. 2010). The null phenotype variant rs2814778 is located in the erythroid promoter region and is one of 34 SNPs used to infer ancestral origin by the SNPforID Consortium (Phillips et al. 2007). Individuals with the FYB allele who are homozygous for rs2814778 (−33T → C) have the phenotype Fy(a−b−) and do not express Duffy antigens on their red blood cells (RBCs) (Dean 2005). RBCs that lack the Duffy Fya and Fyb antigens are resistant to invasion of the malaria parasites Plasmodium vivax and Plasmodium knowlesi, a phenotype posited to result in positive selection in African populations. Lower WBC in subjects of African ancestry has been attributed to this variant ([MIM 613665]) (http://www.omim.org).
We performed a pooled and genetically determined ancestry (GDA)-stratified analysis of total WBC identified through electronic medical records (EMRs) among 13,923 subjects in the electronic Medical Records and Genomics (eMERGE) Network to assess influence of genetic/genomic variations on WBC level (http://www.gwas.net). An advantage of our study sample from all participating eMERGE sites was that most of the 13,923 subjects have longitudinal measurements over multiple years as well as clinical covariates, such as medications, to identify and eliminate WBC observations that may have reflected acute conditions such as cancer or infection.
Methods
Selection and description of participants
The eMERGE Network is a consortium of five U.S. cohorts linked to EMR data for conducting large-scale, high-throughput genetic research (McCarty et al. 2011). Participating sites include the following: (1) Group Health Cooperative, University of Washington and Fred Hutchinson Cancer Research Center partnership, Seattle, WA, (2) Marshfield Clinic, Marshfield, WI, (3) Mayo Clinic, Rochester, MN, (4) Northwestern University, Evanston, IL, and (5) Vanderbilt University, Nashville, TN (Roden et al.2008). The network brings together researchers with a wide range of expertise in genomics, statistics, ethics, informatics, and clinical medicine from leading medical research institutions across the country (McCarty et al.2011). Each center participating in the consortium, organized by the National Human Genome Research Institute (NHGRI), proposed studying the relationship between genetic variation and one or more common human traits, using the technique of genome-wide association analysis. The Center for Inherited Disease Research (CIDR) at Johns Hopkins University and The Broad Institute of MIT and Harvard served as the genotyping centers for the network.
The WBC data extraction algorithm for EMRs was developed and validated by Group Health and the University of Washington. Group Health, Marshfield Clinic and Mayo Clinic reported >98% of subjects as having WBC records. Northwestern University reported 92% and Vanderbilt University reported 91%. Visit and subject level exclusion criteria are listed in Table 1. In general, we excluded any visit and/or subject whose values were possibly reflecting something other than resting state WBC levels. The subjects were removed if there was a record of dialysis or HIV infection at any time. Visit-level exclusions included inpatient and emergency room visits. Other visit-level exclusions included the following: (1) active infections, (2) medications affecting WBC levels, (3) chemotherapy window of 6 months prior and 3 months post index visit, and (4) a prior diagnosis of Alzheimer’s disease. The algorithm was presented to all participating sites and data were returned to Group Health for harmonization. Once harmonized, the data were transferred to the University of Washington for the association analyses.
Table 1.
White blood cell count subject/visit algorithm using electronic medical records for the five eMERGE participating sites
| Subject-level exclusion criteria |
| Any indication at any time of HIV |
| Dialysis at any time |
| Visit-level exclusion criteria |
| Inpatient or emergency visit |
| Splenectomy record prior to lab |
| Prior diagnosis of myelodysplastic syndrome |
| Medications with minor impacts on WBC (aspirin at high doses). |
| Strongly immune affecting medications (oral or IV steroids chemotherapeutic agents such as methotrexate) |
| Indication of concurrent “active chemotherapy” regimen 6 months prior to 3 months after index visit |
| Prior indication of Alzheimer’s disease |
| Blood dyscrasia (leukemia, myeloma, bone marrow failure, aplastic anemia, etc.) |
| “Active infection” in prior or subsequent 30 days |
| Other acute and chronic infections |
Technical information
Genotyping
Most subjects were genotyped on the Illumina Human660 W-Quadv1_A (660 W) genotyping platform. Samples from subjects who were self-reported (North-western) or observer-reported (Vanderbilt University) to have more recent African ancestry were genotyped on the Illumina Human1 M-Duo (1 M) genotyping platform. The 660 W array included 561,490 SNPs and 95,876 intensity-only probes, and the 1 M array consisted of a total of 1,199,187 loci. Genotyping calls for both platforms were made at CIDR and Broad using BeadStudio version 3.3.7 and Gentrain version 1.0. Vanderbilt University served as the eMERGE coordinating center, which conducted the QC/QA process for the genotypic data, with duplicate efforts at each site. Both samples and SNPs were assessed for quality and subsequently filtered from the production data if thresholds were not met. Cryptic relatedness was assessed for all sites and pairs at half-sibling level (τ = k1/4 = k2/2 = 1/8) or higher were randomly broken (by dropping one) before assessing whole-genome association. Subjects identified for filtering at each particular site through the quality control/quality assurance (QA/QC) process were subsequently filtered for the entire merged data set. The (QA/QC) process was performed using four general categories: (1) assessment of genotyping batch quality, (2) assessment of sample quality, (3) validating sample identity, and (4) assessment of SNP quality (Laurie et al. 2010; McCarty et al. 2011; Turner et al. 2011). Detailed QA/QC reports of all eMERGE projects are available for public access on the eMERGE web site.
The combined genotype data set was created at the Fred Hutchinson Cancer Research Center. All source projects were generated on TOP strand from Bead studio final report files, so (nominally) there was no need to flip strands before merging. The released SNPs in the Group Health Cooperative (s = 560,732) were used as the prototype and released SNPs from the other eMERGE projects were extracted. Merging these files on the SNPs that were called in every project yielded 532,566 variants in common. There were 1,564 strand ambiguous SNPs (A/T and C/G polymorphism) out of the 532,566 total intersecting SNPs. If the most frequent alleles in these generally agreed between source projects, then we concluded that the strands were concordant prior to merging.
Imputation
Imputation analyses of untyped SNPs were performed using BEAGLE version 3.3, a freely available software program written in Java (Browning and Browning 2009). We imputed the region surrounding the Duffy antigen/chemokine receptor gene (DARC) on chromosome 1, specifically to infer the rs2814778 genotypes for the 120 AA subjects not genotyped on the 1 M array. We used a cosmopolitan reference panel comprising phased HapMap3_r2 founder haplotypes from six Phase 3 populations: CEU, TSI, ASW, LWK, MKK, and YRI (Altshuler et al. 2010). Given the 19 Mb region of significance, we added a 5 Mb buffer to each end. Thus, the imputed result is a 29 Mb region (140.5–169.6 Mb, NCBI build 36), which encompasses 13,516 SNPs. For quality, we checked the concordance between the most likely imputed genotype and genotyped results for rs2814778 in the 1,373 overlapping individuals (typed on 1 M and AA subjects who we imputed), which was 99.1%. The estimated allele dosage (0 × P(AA) + 1 × P(AB) + 2 × P(BB) for rs2814778 was subsequently used in the association analysis for the 120 subjects without observed 1 M array genotypes.
Principal components analysis (PCA) for ancestry
We used principal components analysis (PCA), essentially as described by Patterson et al. (2006) (Patterson et al.2006), using independent, autosomal SNPs with missing call rates <5.0% and minor allele frequency >5.0% across the merged data set of 17,150 unique subjects (532,566 → 482,985 SNPs). To select independent SNPs, we utilized PLINK’s linkage disequilibrium (LD) pruning function (Purcell et al. 2007). For the first round of short-range LD pruning, we used the default settings of a 50 SNP window with a shift of five SNPs, and pair-wise genetic correlation with a threshold of r2 = 0.2. In a second round to remove long-range LD, we took the average number of SNPs over 5 Mb from the output of the first round as our window (s = 200), again with an iteration of 20 SNPs and a threshold of 0.2. The resulting 105,291 SNPs were used to generate the principal components. For PCA, we utilized an in-house program (SNPRelate), which has no limit to the number of SNPs used for the covariance matrix (http://cran.r-project.org/web/packages/SNPRelate/index.html).
Supplemental Figure S1 shows a plot of the first two eigenvectors from an analysis of 17,150 subjects from all sites, along with HapMap controls (CEU, YRI, CHB and JPT). The solid and dotted lines are the means and standard deviations (SD) for eigenvectors 1 and 2 for self-identified EA and AA subjects, respectively. The first eigenvector, accounting for 4.8% of the variance, separated the self-identified (all sites other than Vanderbilt) or observed-identified (Vanderbilt) EA subjects from the self-identified or observed-identified AA subjects. The second eigenvector, accounting for 0.3% of the variance, separated the self-identified Asian subjects from the other ancestry groups.
To define the EA sample, we identified all subjects with values less than three (+3) and greater than negative one (−1) SD from the mean of eigenvectors 1 and 2 of self-identified European ancestry, respectively (see Figure S1).
For the AA sample, we identified all subjects with values less than two (+2) and greater than negative one (−1) SD from the mean of eigenvector 1, and less than and greater than one (±1) SD for eigenvector 2 of self-identified African ancestry subjects, respectively. These boundaries were chosen as an attempt to select a homogenous set of ancestry groups based on visual inspection of the eigenvector plots. Respective numbers of self-identified and genetically determined ancestry are listed in Table 2 and discussed in the “Results” section.
Table 2.
Summary statistics of demographic data and phenotypes by eMERGE participating site and combined
| N | Group Health (N = 2,358) |
Marshfield Clinic (N = 3,883) |
Mayo Clinic (N = 2,809) |
Northwestern (N = 1,303) |
Vanderbilt (N = 3,570) |
Combined (N = 13,923) |
Test statistic | |
|---|---|---|---|---|---|---|---|---|
| Sex (M) | 13,923 | 42% (996) | 41% (1,595) | 62% (1,751) | 43% (556) | 38% (1,344) | 45% (6,242) | Chi-square = 453 df = 4 p < 0.001# |
| Median BMI | 13,923 | 23.6/26.4/29.6A 26.9 ± 5.0B |
25.6/28.6/32.4A 29.5 ± 5.9B |
25.2/28.1/3 1.6A 28.7 ± 5.3B |
24.9/28.9/34.5A 30.6 ± 8.2B |
25.9/28.9/33.0A 30.0 ± 7.2B |
25.0/28.5/31.9A 29.1 ± 6.4B |
F = 117 df = 4,13,918 p < 0.001† |
| Median age | 13,923 | 68.4/74.2/79.8A 74.1 ± 7.9B |
51.7/61.2/70.2A 60.8 ±12.2B |
57.7/63.3/70.2A 64.1 ± 9.1B |
43.3/53.3/61.4A 52.5 ±13.4B |
40.5/51.9/62.5A 51.2 ±16.2B |
51.6/61.9/71.1A 60.5 ±14.7B |
F = 1,444 df = 4,13,918 p < 0.001† |
| Self reported race | 13,923 | Chi-square = 4,167 df = 20 p < 0.001# |
||||||
| American Indian or Alaska Native |
0.2% (4) | 0.6% (22) | 0.2% (5) | 0.0% (0) | 0.0% (0) | 0.2% (31) | ||
| Asian | 2.7% (63) | 0.3% (11) | 0.1% (2) | 0.0% (0) | 0.1% (3) | 0.6% (79) | ||
| Black or African American |
3.2% (76) | 0.1% (3) | 0.3% (9) | 25.8% (336) | 33.9% (1212) | 11.8% (1,636) | ||
| Other | 1.1% (27) | 0.4% (14) | 0.4% (11) | 0.0% (0) | 0.1% (5) | 0.4% (57) | ||
| White | 92.7% (2,185) | 98.7% (3,833) | 94.5% (2,655) | 74.2% (967) | 55.5% (1,982) | 83.5% (11,622) | ||
| Unknown | 0.1% (3) | 0.0% (0) | 4.5% (127) | 0.0% (0) | 10.3% (368) | 3.6% (498) | ||
| GDA of African continent | 13,923 | 3% (62) | 0% (2) | 0% (6) | 22% (291) | 32% (1,126) | 11% (1,487) | Chi-square = 2,757 df = 4 p < 0.001# |
| GDA of European continent |
13,923 | 93% (2,189) | 99% (3,855) | 99% (2779) | 72% (937) | 64% (2,286) | 87% (12,046) | Chi-square = 2,780 df = 4 p < 0.001# |
| Self reported ethnicity | 13,923 | Chi-square = 3,433 df = 8 p < 0.001# |
||||||
| Hispanic or Latino | 0.5% (12) | 0.3% (10) | 0.2% (7) | 2.4% (31) | 0.1% (3) | 0.5% (63) | ||
| Not Hispanic or Latino | 91.7% (2,162) | 99.7% (3,873) | 55.4% (1,556) | 97.6% (1,272) | 89.6% (3,199) | 86.6% (12,062) | ||
| Unknown | 7.8% (184) | 0.0% (0) | 44.4% (1,246) | 0.0% (0) | 10.3% (368) | 12.9% (1,798) | ||
| Median leukocyte count [K/μl] |
13,695 | 5.7/6.6/7.6A 6.7 ±1.4B |
5.5/6.4/7.5A 6.6 ±1.5B |
5.4/6.3/7.5A 6.6 ±1.6B |
5.7/6.8/8.3A 7.1 ±2.0B |
5.8/6.9/8.4A 7.1 ±1.8B |
5.6/6.6/7.8A 6.8 ±1.7B |
F = 61 df = 4,13,690 p < 0.001# |
| Median neutrophils [%] | 10,978 | 58.1/64.0/69.0A 63.5 ± 8.1B |
55.0/60.0/65.0A 59.9 ± 7.2B |
55.2/60.2/65.2A 60.1 ± 7.6B |
55.0/61.0/67.0A 61.0 ± 9.4B |
53.2/60.2/66.6A 59.9 ±10.1B |
55.2/61.0/66.0A 60.7 ± 8.3B |
F = 73 df = 4,10973 p < 0.001† |
| Median lymphocytes [%] |
11,011 | 6.0/7.5/9.0A 7.6 ±2.4B |
24.0/28.0/33.0A 28.3 ± 6.7B |
22.7/27.4/32.3A 27.6 ± 7.0B |
22.2/28.0/33.2A 28.1 ± 8.6B |
23.5/29.2/35.7A 29.4 ± 9.2B |
23.0/27.9/33.0A 28.0 ± 7.6B |
F = 31 df = 4,11,006 p < 0.001† |
| Median monocytes [%] | 11,014 | 21.4/26.0/31.9A 26.6 ± 7.7B |
7.0/8.0/9.5A 8.4 ±1.9B |
7.3/8.5/9.7A 8.6 ±1.9B |
6.5/8.0/9.0A 7.9 ±2.2B |
5.5/6.7/8.4A 7.1 ±2.4B |
6.6/8.0/9.2A 8.0 ±2.2B |
F = 217 df = 4,11,009 p < 0.001† |
| Median eosinophils [%] | 10,895 | 0.0/1.0/2.5A 1.6 ±1.8B |
2.0/2.0/3.5A 2.7 ±1.6B |
1.8/2.6/3.7A 2.9 ±1.7B |
1.0/2.0/3.0A 2.4 ±1.7B |
1.3/2.0/3.1A 2.5 ±2.0B |
1.3/2.0/3.2A 2.5 ±1.8B |
F = 275 df = 4,10,890 p < 0.001† |
| Median basophils [%] | 10,847 | 0.00/0.00/0.73A 0.35 ±0.50B |
0.50/1.00/1.00A 0.81 ±0.49B |
0.40/0.55/0.70A 0.60 ±0.31B |
0.00/0.50/1,00A 0.55 ±0.48B |
0.30/0.50/0.70A 0.56 ±0.35B |
0.30/0.60/1.00A 0.61 ±0.46B |
F = 474 df = 4,10,842 p < 0.001† |
The number of records per site is listed under site name (N), and the total number of records was 13,923. If the number listed under the “N” header was different than the total, then the difference was the number of missing records for that variable
Numbers after percents are frequencies
Pearson test
Kruskal–Wallis test
a/b/c represent the lower quartile a, the median b, and the upper quartile c for continuous variables
x ± s represents X ± 1 SD
Statistics
To assess heterogeneity of potential phenotypes and covariates among sites, we used the summary function in the Hmisc package (http://cran.r-project.org/web/packages/Hmisc/index.html) (Harrell 2004) (R statistical computing) (Table 2). We used the Pearson Chi-square tests for categorical variables and the Kruskal–Wallis tests for continuous variables (>2 groups). Clinical characteristics deemed important for the association analyses were then analyzed for significant association to the phenotypes of interest using ordinary least squares. A majority of the subjects from all sites had multiple visits over many years for the duration of the EMR. While other within-subjects measures were considered, we assessed the median value of WBC and its differentials for a given subject, which provides a natural transformation of the data and is less sensitive to outlying extreme values than other summaries such as the mean. As a further check, we identified and excluded outliers whose median WBC value fell outside two standard deviations from the mean of the median values.
We analyzed total WBC and each of its differential components as quantitative phenotypes (ordinary least squares) adjusted for the following variables: (1) sex, (2) median BMI, (3) median age, (4) eigenvectors 1 and 2 derived from PCA, and for the joint analysis (5) study site. For the GDA-stratified analyses, eigenvectors 1 and 2 were dropped from the model. We analyzed each dependent variable with the given covariates and the genotype of one SNP coded as 0, 1 and 2 copies of the minor allele (additive genotypic model) in PLINK. We deemed genome-wide significance at a p value less than 5.0e–8, which approximated a Bonferroni correction for approximately 1,000,000 independent hypothesis tests (Schnabel et al. 2010).
Results
Descriptive statistics by eMERGE study site
Table 2 contains descriptive statistics of total WBC and its subtypes along with covariates by eMERGE study site and combined. Continuous variables are illustrated as box-percentile plots by study site in Supplemental Figure S2. NU (26%) and VU (34%) had the highest proportions of participants who self-identified or were observed-reported as having more recent African ancestry. Most (83%) participants self-identified or were observed-reported as having European ancestry. These numbers reflect the percentages of genetically determined ancestry groups (see “Methods”) for both the African and European continent. For AA subjects, NU and VU had the largest percentage at 22 and 32%, respectively. All other sites were at 3% or less. Only a small percentage of subjects from all sites combined (<1%) self-reported an ethnicity of Hispanic or Latino. The differences of WBC and the differentials by site are illustrated in Supplemental Figure S2, and listed in Table 2. There was a significant difference (p < 0.001) in median WBC, with NU and VU having the highest median values of 6.8 and 6.9 K/μl, respectively.
Effects of covariates on WBC
Median age, median BMI and sex were used as standard covariates for all multivariate models considered. The effects of BMI and sex on WBC are similar for the GDA-stratified and pooled analyses as illustrated in Supplemental Figure S3. Higher BMI correlated with higher WBC. On average, men had lower WBC than women. Age had suggestive effects in the stratified ancestry groups, although the confidence intervals for both effects cross β = 0.0. In the AA group, the higher ages inversely correlated with lower WBC. For the EA group, the higher the age correlated with higher WBC. There is virtually no effect of age on WBC in the pooled analysis, but the variance may have been captured in the significant site effects.
White blood cell count association
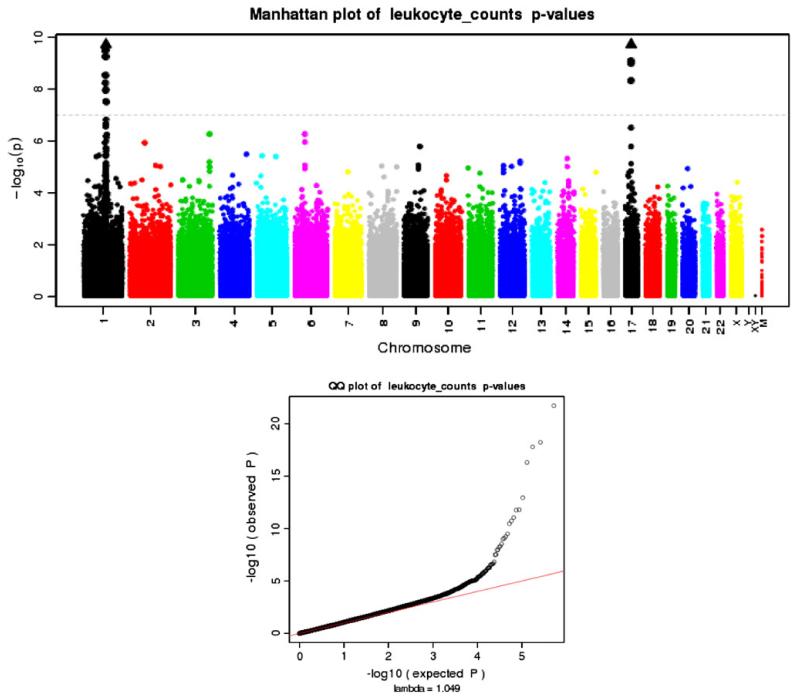
We performed a pooled and GDA-stratified association analyses as defined in the “Methods” section. The results of the major class of the WBCs, the neutrophils, strongly mimicked the WBC results as expected (data not shown). The results presented were classified into two categories, one for the AA subjects and one for the EA subjects. For the AA subjects, there was a strong genome-wide association centered on the 1q21–q22 region, specifically the Duffy antigen/chemokine receptor gene (DARC) detailed in the “Introduction” section. Figure 1 illustrates this peak that traverses the centromere on chromosome 1. The genome-wide significant association for the EA subjects was found at the 17q21 region, tagging several genes of interest. Both regions of significance were GDA-specific, but for completeness the SNP association results are presented in Table 3 for each stratum as well as the pooled analyses. We also performed association analysis by site as a sensitivity analysis and have presented the results in Supplemental Table 1. Effects sizes and p values by site are listed for the SNPs presented in Table 3 except for the Duffy SNP rs2814778. This variant was not genotyped for all sites and therefore is not provided. We also do not provide ancestry-stratified results for each site as PCA was performed across the entire consortium. In general, the site-specific results were suggestive of the joint results as expected. In general, it is more advantageous to perform a joint analysis over a replication-based analysis (Skol et al.2006). PCA across the entire consortium controlling for ancestry effects allowed for this joint analysis.
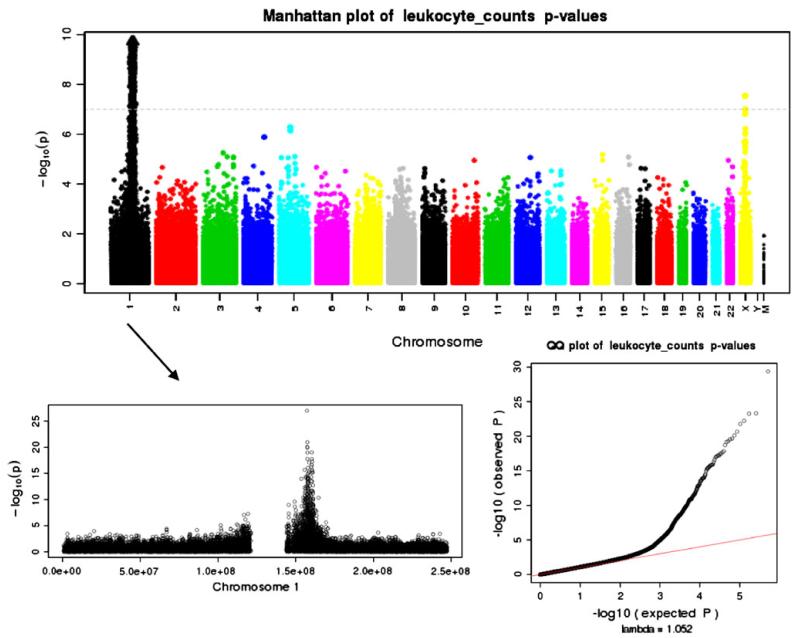
Fig. 1.
Manhattan and Q–Q plots of p values from the WBC association analyses of subjects with GDA of African continent
Table 3.
Summary of effects of loci that reached genome-wide significance for the AA, EA and pooled analyses
| AA | EA | Pooled (eig.1&2) | |
|---|---|---|---|
| 1q21, DARC, adjusted for site, median age, sex and median BMI (rs2814778-rs12075: r2 = 0.394) rs12075, missense; (154G → A), G [Gly] → D [Asp] | |||
| β Estimate (S.E.) | 1.27 (0.12) | −0.04 (0.02) | 7.95e–04 (0.02) |
| p value | 4.92e–24 | 3.55e–02 | 0.97 |
| Sample MAF (G); A is the ancestral allele | 0.08 | 0.42 | 0.38 |
| Includes dosages of 120 imputed genotypes rs2814778, promoter UTR-5 | |||
| β Estimate (S.E.) | 1.35 (0.08) | NA | NA |
| p value | 6.71e–55 | NA | NA |
| Sample MAF (A); A is the ancestral allele | 0.18 | NA | NA |
| Includes dosages of 120 imputed genotypes, both Duffy SNPs and interaction term rs2814778, promoter UTR-5 | |||
| β Estimate (S.E.) | 1.37 (0.11) | NA | NA |
| p value | 7.19e–33 | NA | NA |
| rs12075, missense | |||
| β Estimate (S.E.) | 0.82 (0.34) | NA | NA |
| p value | 1.42e–02 | NA | NA |
| rs2814778, promoter UTR-5 * rs12075, missense | |||
| β Estimate (S.E.) | −0.61 (0.24) | NA | NA |
| p value | 1.21e–02 | NA | NA |
| 17q21.1, GSDMA, adjusted for site, median age, sex and median BMI (rs3894194-rs3859192: r2 = 0.492) rs3894194, missense; (171G → A), R [Arg] → Q [Gln] | |||
| β Estimate (S.E.) | 0.03 (0.08) | 0.11 (0.02) | 0.10 (0.02) |
| p value | 0.68 | 2.01e–07 | 2.29e–07 |
| Sample MAF (A); C is the ancestral allele | 0.29 | 0.46 | 0.44 |
| rs3859192, intronic | |||
| β Estimate (S.E.) | 0.03 (0.08) | 0.14 (0.02) | 0.14 (0.02) |
| p value | 0.68 | 1.75e–12 | 1.75e–12 |
| Sample MAF (A); A is the ancestral allele | 0.33 | 0.47 | 0.45 |
| 17q21.1, PSMD3, adjusted for median age, sex, site and median bmi (rs3859192GSDMA-rs4065321: r2 = 0.458) rs4065321, intronic | |||
| β Estimate (S.E.) | 0.10 (0.07) | 0.14 (0.02) | 0.14 (0.02) |
| p value | 0.16 | 3.47e–11 | 1.43e–12 |
| Sample MAF (G); C is the ancestral allele | 0.36 | 0.45 | 0.44 |
| 17q21.1, MED24, adjusted for median age, sex, site and median bmi (rs3859192GSDMA-rs4065321: r2 = 0.410) rs9916158, intronic | |||
| β Estimate (S.E.) | −0.01 (0.08) | −0.13 (0.02) | −0.13 (0.02) |
| p value | 0.89 | 4.92e–10 | 8.86e–10 |
| Sample MAF (A); T is the ancestral allele | 0.23 | 0.38 | 0.36 |
Duffy antigen/chemokine receptor gene (DARC)
Figures 1, 2 and 3 illustrate the Manhattan and Q–Q plots of p values from the AA, EA and pooled analyses, respectively. We also provide in Fig. 1 a zoomed-in plot of chromosome 1 that shows chromosomal width of the extensive association peak. As illustrated in the Q–Q plot, there was marginal genomic inflation, lambda = 1.052, most likely driven by the extensive chromosome 1 region of significance, which is likely attributable to a selective sweep for Duffy as noted. While many genes fell within the peak (19 MB), the most significant result (p value ≈ e–24) was a missense mutation (rs12075) found in the DARC (“Duffy”) gene. Table 3 outlines the SNP association results for rs12075 on WBC. The pooled analysis model included eigenvectors 1 and 2 derived from the PCA. The AA subjects with the minor allele for rs12075 on average had a higher median WBC (β = 1.28, S.E. = 0.12). The EA association result were suggestive (p value = 0.04), but the effect was small and in the opposite direction (β = −0.04, S.E. = 0.02). As rs12075 is the SNP that is responsible for the FYA and FYB Duffy phenotypes, we also assessed the corresponding Duffy variant rs2814778 producing the null phenotype from subjects genotyped on the 1 M genotyping platform.
Fig. 2.
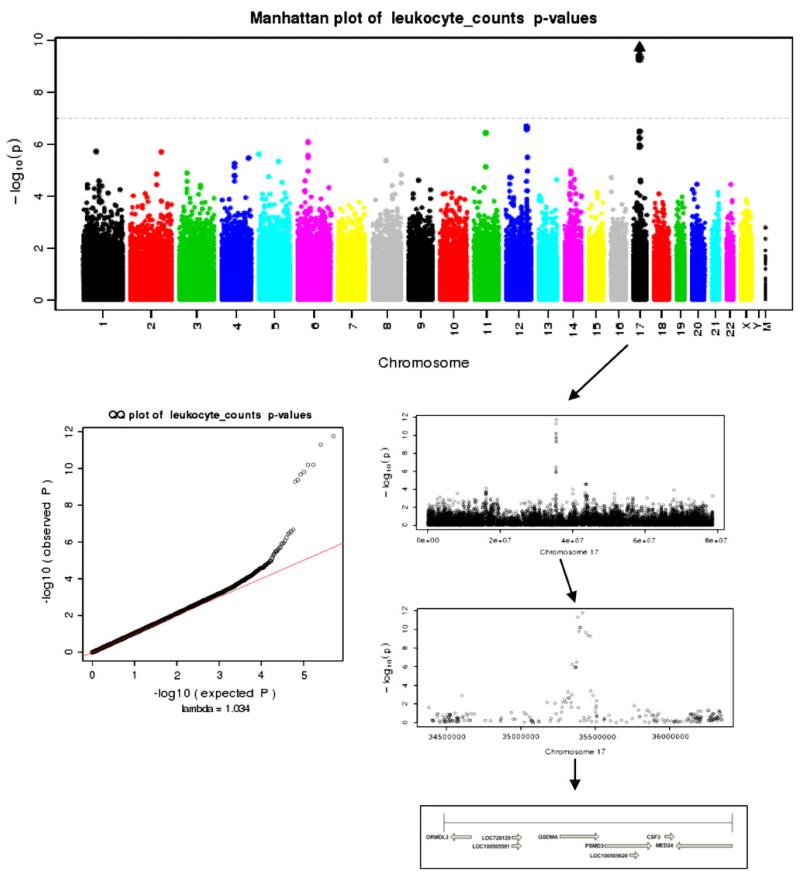
Manhattan and Q–Q plots of p values from the WBC association analyses of subjects with GDA of European continent
Fig. 3.
Manhattan and Q–Q plots of p values from the pooled WBC association analyses of subjects from all eMERGE sites
The majority of these samples were self-reported or observed-reported as African ancestry at NU and VU, respectively. For the AA subjects not genotyped on the 1 M (n = 120), we included the imputed dosages in the analyses (see “Methods”). The AA association for rs2814778 (the null variant) was more significant (p value = 6.71e–55) than the Duffy missense mutation rs12075 with a similar effect size and direction (β = 1.35, S.E. = 0.08). As expected, this allele was very rare in subjects of European ancestry.
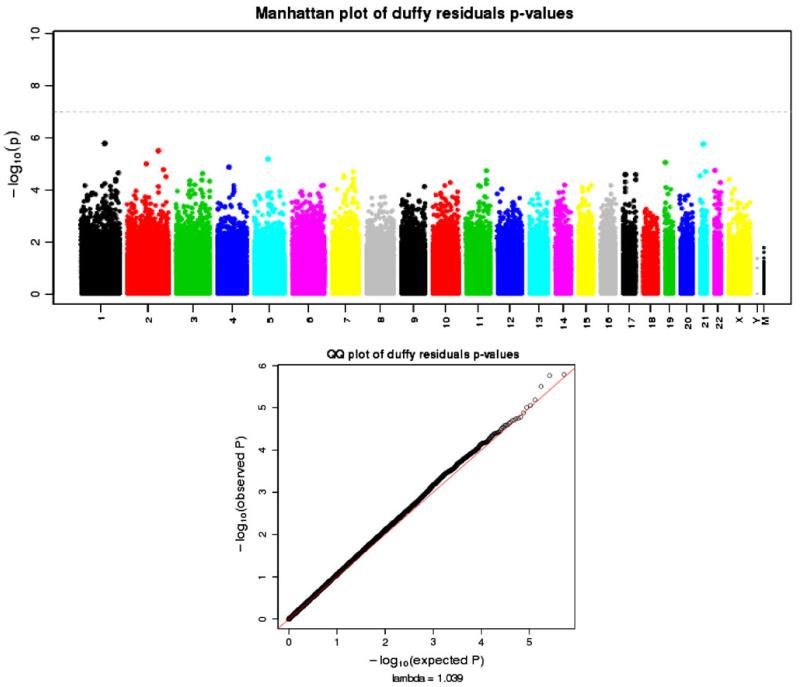
We were able to explore a novel multi-SNP Duffy model with an interaction term because we had genotypes for both rs2814778 and rs12075 in the AA subjects (see Table 3). The LD for these two SNPs in the genotyped 1 M subjects was small (r2 = 0.394). When both SNPs were added to the model, the rs12075 effect was no longer significant (p value = 0.63), while the rs2814778 effect remained significant at a genome-wide level (p value = 1.62e–32). When the interaction term was added (see Table 3), the rs2814778 effect (β = 1.37, S.E. = 0.11) remained significant (p value = 7.19e–33), while both rs12075 and the interaction between the SNPs were moderately significant, although not at a genome-wide level (p value ≈ 0.001). This model appears to account for a proportion of the WBC variance on our AA sample (multiple r2 = 0.203). Residuals from this two-SNP, interaction model were extracted and analyzed as a phenotype. Figure 4 illustrates the Manhattan and Q–Q plot of the p values generated from the analyses. There were no significant genome-wide associations with moderate inflation (lambda = 1.039), including the previous evidence of X-linked associations illustrated in Fig. 1. Two untranslated 3′SNPs (rs2209549, rs7052314) tagged two genes KIAA2022, ZDHHC15, respectively. Both hypothetical protein LOC340533 (KIAA2022) and zinc finger, DHHC-type containing 15 isoform 1 (ZDHHC15) have been linked to X-linked mental retardation. A relationship if any between these genes and WBC is no longer supported. Also tagged was UPRT by SNP rs12832571 with unknown function. This gene encodes uracil phosphoribosyl-transferase, which plays an important part of nucleotide metabolism, specifically the pyrimidine salvage pathway (Kent et al. 2002).
Fig. 4.
Manhattan and Q–Q plots of p values from the analyses residuals derived from the Duffy multivariate model described in the “Results” section. Subjects of African ancestry were analyzed
Supplemental Figure 4 illustrates the difference of WBC by Duffy phenotypes (Fya + b+, Fya+b−, Fya−b+, Fya−b−) in the subjects of African ancestry. As expected, the neutrophils showed the same associations with Duffy antigens as the total leukocyte count with Duffy null (Fya−b−) having a reduced count. Notably, the lymphocytes, eosinophils, monocytes, and basophils demonstrated the opposite trend with Duffy null being associated with higher counts.
17q21.1 region
There was a genome-wide significant association with median WBC for EA subjects centered on the 17q21.1 region. There were several genes tagged by the genomewide significant SNPs presented in Table 3, all located in this region. Figure 2 illustrates the Manhattan and Q–Q plot of the p values generated from the analyses (lambda = 1.034). Figure 2 also provides a zoomed-in plot of chromosome 17, and further of the 17q21.1 region illustrating genes and directions of transcription. Increasing in genomic coordinates, GSDMA is one of the genes tagged with the most significant association for both the EA (p value ≈ e–12) and pooled (p value ≈ e–11) analyses. The pooled sample minor allele for this intronic variant (rs3859192) was also the ancestral allele (A), but the frequency was 0.45. In the EA sample, higher numbers of copies of this allele were associated with higher WBC levels (β = 0.14, S.E. = 0.02), and this association was present in the overall pooled sample (β = 0.14, S.E. = 0.02). This SNP was in moderate LD (r2 = 0.492) with a missense variant (rs3894194) that was suggestive in our EA (2.01e–07) and pooled (2.29e–07) samples (see Table 3). Higher numbers of copies of the minor allele were associated with higher values of WBC. For the EA sample β = 0.11 (S.E. = 0.02), and for the pooled sample β = 0.10 (S.E. = 0.02). Moving towards 3′ as illustrated in Fig. 2, the next significant association was found in the PSMD3 gene, intronic SNP rs4065321. As listed in Table 3, this variant is in moderate LD with the GSDMA intronic SNP rs3859192 (r2 = 0.458). The MAFs (0.45 and 0.44), effect sizes (both β = 0.14) and significance (p value = 3.47e–11 and 1.43e–12) were similar to rs3859192 in the EA and pooled analyses. On the anti-sense strand, there was a genome-wide significant intronic SNP (rs9916158) found in the MED24 gene. As outlined in Table 3, this SNP was significant in the EA (p value = 4.92e–10) and pooled analyses (p value = 8.86e–10). The sample minor allele (A) was associated with lower median WBC. For the EA sample β = −0.13 (S.E. = 0.02), and for the pooled sample β = −0.13 (S.E. = 0.02). This variant is in moderate LD with the GSDMA intronic SNP rs3859192 (r2 = 0.410). None of the 17q21.1 SNPs associated with WBC in EA analyses were found to be significantly associated in analyses of AA (all p value > 0.05).
Discussion
Through the eMERGE Network, we were able to successfully mine electronic medical records linked to five U.S. cohorts encompassing 13,923 subjects for WBC. This EMR algorithm, which attempted to remove acute and chronic influences on WBC levels, was developed at one site and confirmed at the other network sites. Our findings extend previous reports of two regions of interest unique to subjects of genetically determined ancestry to the African or European continents and additionally identify a novel interaction for the former. Of interest, we were able to detect these ancestry-specific associations in pooled analyses. Further, these results may inform a previously reported asthma association on chromosome 17q21.1 and its impact on the expression of ORMDL3 or GSDMB genes (Halapi et al. 2010; Kabesch 2010; Moffatt et al. 2007,2010; Verlaan et al. 2009).
Duffy antigen/chemokine receptor gene (DARC)
Our chromosome 1 association with WBC in the AA sample provided unified support of previous findings through admixture mapping (Nalls et al. 2008) and SNP associations (Reich et al. 2009; Schnabel et al. 2010). These included the Duffy null polymorphism (rs2814778) with its association with the major WBC component, neutrophils (Reich et al. 2009), and the Duffy missense mutation (rs12075) responsible for the two principal antigens, Fya and Fyb, and its association with monocyte chemoattractant protein-1 (MCP-1) (Schnabel et al. 2010). The direction of effect in our results support the hypothesis that DARC on red blood cells acts as a chemokine sink, limiting the stimulation of leukocytes by IL-8 in the blood (Pruenster and Rot 2006). Chemokines play a key role in trafficking leukocytes in the blood. The two Duffy variants interact to account for WBC variance observed in our AA sample, as evidenced by the lack of association from the genome-wide analyses of the residuals derived from the novel two-SNP, interaction model described in the “Results” section (Fig. 4). Still unexplained is the X chromosome peak in the AA subjects. This signal was also eliminated when “Duffy” was controlled for in the residual model derived from the two-SNP, interaction model. While we continue to study these effects, we suspect they may represent an artifact of different patterns of admixture in males versus females of recent African origin. The atypically broad region of association on chromosome 1 demonstrated in Fig. 1 may be due to selective sweep in the genetic region surrounding Duffy in geographic areas with high prevalence of P. vivax and P. knowlesi. This selective sweep also leads to spurious associations in regional genes not relevant to WBC. Such associations could have been misleading had Duffy not been typed. This suggests the need for caution in interpreting associations in atypically broad regions of association.
17q21.1 region
Our 17q21.1 association results in the EA subjects provided further evidence that this region may play a role in regulating inflammatory processes and disorders. In our EA subjects, we identified a genome-wide significant region that tagged three separate genes: (1) GSDMA, (2) MED24, and (3) PSMD3. The HaemGen consortium used meta-analysis to identify one variant (rs17609240) with an association with WBC of genome-wide significance (Soranzo et al. 2009); this variant was on 17q21.1 near ORMDL3, a known susceptibility locus for childhood asthma (childhood asthma [MIM 610075]) (Moffatt et al. 2007; Soranzo et al. 2009). A study of WBC in Japanese individuals identified another SNP (rs4065321) in the same region (Kamatani et al. 2010). Others have linked this region to neutrophil count levels (Okada et al. 2010), and to the inflammatory disorders asthma (Halapi et al. 2010; Moffatt et al. 2007, 2010; Verlaan et al. 2009), Crohn’s disease (IBD22 [MIM 612380]) (Barrett et al. 2008) and type 1 diabetes (IDDM [MIM 222100]) (Barrett et al.2009). Other genes in the same region include GSDMA, MED24, and PSMD3. PSMD3 encodes one of the non-ATPase subunits of the 19S regulator lid for the multicatalytic proteinase complex that is involved in many cellular functions including inflammatory responses, and apoptosis (Kent et al. 2002). Another gene in close proximity is CSF3, which encodes colony stimulating factor 3, a cytokine controlling the production, differentiation and function of granulocytes (Hollard et al. 1975; Soranzo et al.2009). ORMDL3 has been identified as a potential risk factor for asthma (Breslow et al. 2010; Halapi et al. 2010; Kabesch 2010; Moffatt et al. 2010; Verlaan et al. 2009). Through a combination of global and target studies, Breslow et al. (2010) identified Orm proteins as homeostatic regulators of sphingolipid biosynthesis. Moffatt et al. identified genetic 17q21.1 variants regulating ORMDL3 expression and its contribution to the risk of childhood asthma (Moffatt et al. 2007). Two of these variants associated with asthma (rs3894194 and rs3859192) are missense and intronic SNPs found in GSDMA that were associated to WBC in our EA sample. With associations to multiple inflammatory diseases in addition to WBC and its differential, this 17q21.1 region is an excellent candidate for pleiotropy analyses (Kabesch 2010; Verlaan et al.2009).
Our own and the other reported associations in the 17q21.1 region are located near the European-enriched 17q21.31 MAPT inversion polymorphism region (Zody et al. 2008). Allergic and autoimmune diseases, including asthma, type 1 diabetes and Crohn’s disease, are not evenly distributed among continents, countries, or ethnic groups (Bach 2002) and this inversion could play a role. The incidence of disease decreases from north to south in the Northern Hemisphere (Bach 2002). Further investigation of whether this inversion and the lack of recombination surrounding this region may play a role in any 17q21 association is warranted to determine if these associations are an artifact of admixture.
Conclusion
Our results provided further evidence that variation in WBC levels does have a genetic component. While Duffy is likely the source of association in the AA subjects, further elucidation of the biological mechanism driving the EA association in the 17q21.1 region is needed. These regions are excellent candidates for exploring pathway enrichment as well as gene-gene interaction for genes with SNPs meeting genome-wide significance. In particular, we find that two SNPs previously associated with asthma (rs3894194 and rs3859192) were also associated with WBC. Joint study of these SNP effects on asthma and WBC is warranted to determine if these are independent effects. Such studies will inform the reported relationship of WBC to multiple chronic diseases. Finally, we demonstrated that phenotypes such as WBC can be mined from existing EMRs and translated between sites with different systems and coding schemes, which allowed efficient study of multiple phenotypes considering the same genotyped subjects.
Supplementary Material
Acknowledgments
The authors are grateful to all the participants in the eMERGE study. They also acknowledge Xiuwen Zheng and the fast PCA program to make principal component analysis on this many subjects achievable. This study was supported by the following U01 grants from the National Human Genome Research Institute (NHGRI), a component of the National Institutes of Health (NIH), Bethesda, MD, USA: (1) HG004610, AG06781 (Group Health Cooperative), (2) HG04599 (Mayo Clinic), (3) HG004608 (Marshfield Clinic), (4) HG004609 (Northwestern University), (5) HG004438 (CIDR), (6) HG004424 (BROAD), and (7) HG004603 (Vanderbilt University). Additional support was provided by the University of Washington’s Northwest Institute of Genetic Medicine from Washington State Life Sciences Discovery funds (Grant 265508).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00439-011-1103-9) contains supplementary material, which is available to authorized users.
Conflict of interest None of the authors have a financial interest related to this work.
Contributor Information
David R. Crosslin, Department of Medicine, Division of Medical Genetics, University of Washington, Seattle, WA 98195, USA; Department of Biostatistics, University of Washington, Floor 15, UW Tower, Campus Mail Box 359461, Seattle, WA 98195, USA
Andrew McDavid, Public Health Sciences Division, Fred Hutchinson Cancer Research Center, Seattle, WA 98109, USA.
Noah Weston, Group Health Research Institute, Seattle, WA 98124, USA.
Sarah C. Nelson, Department of Biostatistics, University of Washington, Floor 15, UW Tower, Campus Mail Box 359461, Seattle, WA 98195, USA
Xiuwen Zheng, Department of Biostatistics, University of Washington, Floor 15, UW Tower, Campus Mail Box 359461, Seattle, WA 98195, USA.
Eugene Hart, Group Health Research Institute, Seattle, WA 98124, USA.
Mariza de Andrade, Division of Biomedical Statistics and Informatics, Mayo Clinic, Rochester, MN 55905, USA.
Iftikhar J. Kullo, Division of Cardiovascular Diseases, Mayo Clinic, Rochester, MN 55905, USA
Catherine A. McCarty, Essentia Institute of Rural Health, Duluth, MN 55805, USA
Kimberly F. Doheny, Center for Inherited Disease Research, Johns Hopkins University, Baltimore, MD 21224, USA
Elizabeth Pugh, Center for Inherited Disease Research, Johns Hopkins University, Baltimore, MD 21224, USA.
Abel Kho, Divisions of General Internal Medicine and Health and Biomedical Informatics, Northwestern University, Chicago, IL 60611, USA.
M. Geoffrey Hayes, Division of Endocrinology, Metabolism, and Molecular Medicine, Feinberg School of Medicine, Northwestern University, Chicago, IL 60611, USA.
Stephanie Pretel, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD 20894, USA.
Alexander Saip, Vanderbilt Institute for Clinical and Translational Research, Vanderbilt University, Nashville, TN 37232, USA.
Marylyn D. Ritchie, Department of Biochemistry and Molecular Biology, The Pennsylvania State University, University Park, PA 16802, USA
Dana C. Crawford, Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, TN 37232, USA; Center for Human Genetics Research, Vanderbilt University, Nashville, TN 37232, USA
Paul K. Crane, Department of Medicine, Division of General Internal Medicine, University of Washington, Seattle, WA 98195, USA
Katherine Newton, Group Health Research Institute, Seattle, WA 98124, USA.
Rongling Li, Office of Population Genomics, National Human Genome Research Institute, National Institutes of Health, Bethesda, MD 20892, USA.
Daniel B. Mirel, Program in Medical and Population Genetics, Broad Institute of Harvard and MIT, Cambridge, MA 02142, USA
Andrew Crenshaw, Program in Medical and Population Genetics, Broad Institute of Harvard and MIT, Cambridge, MA 02142, USA.
Eric B. Larson, Group Health Research Institute, Seattle, WA 98124, USA
Chris S. Carlson, Public Health Sciences Division, Fred Hutchinson Cancer Research Center, Seattle, WA 98109, USA
Gail P. Jarvik, Department of Medicine, Division of Medical Genetics, University of Washington, Seattle, WA 98195, USA
References
- Altshuler D, Gibbs R, Peltonen L, Dermitzakis E, Schaffner S, Yu F, Bonnen P, de Bakker P, Deloukas P, Gabriel S, Gwilliam R, Hunt S, Inouye M, Jia X, Palotie A, Parkin M, Whittaker P, Chang K, Hawes A, Lewis L, Ren Y, Wheeler D, Muzny D, Barnes C, Darvishi K, Hurles M, Korn J, Kristiansson K, Lee C, McCarrol S, Nemesh J, Keinan A, Montgomery S, Pollack S, Price A, Soranzo N, Gonzaga-Jauregui C, Anttila V, Brodeur W, Daly M, Leslie S, McVean G, Moutsianas L, Nguyen H, Zhang Q, Ghori M, McGinnis R, McLaren W, Takeuchi F, Grossman S, Shlyakhter I, Hostetter E, Sabeti P, Adebamowo C, Foster M, Gordon D, Licinio J, Manca M, Marshall P, Matsuda I, Ngare D, Wang V, Reddy D, Rotimi C, Royal C, Sharp R, Zeng C, Brooks L, McEwen J, Consortium IH. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. doi:10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med. 2002;347:911–920. doi: 10.1056/NEJMra020100. doi:10.1056/NEJMra020100. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM, Bitton A, Dassopoulos T, Datta LW, Green T, Griffiths AM, Kistner EO, Murtha MT, Regueiro MD, Rotter JI, Schumm LP, Steinhart AH, Targan SR, Xavier RJ, Libioulle C, Sandor C, Lathrop M, Belaiche J, Dewit O, Gut I, Heath S, Laukens D, Mni M, Rutgeerts P, Van Gossum A, Zelenika D, Franchimont D, Hugot JP, de Vos M, Vermeire S, Louis E, Cardon LR, Anderson CA, Drummond H, Nimmo E, Ahmad T, Prescott NJ, Onnie CM, Fisher SA, Marchini J, Ghori J, Bumpstead S, Gwilliam R, Tremelling M, Deloukas P, Mansfield J, Jewell D, Satsangi J, Mathew CG, Parkes M, Georges M, Daly MJ. Genomewide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. doi:10.1038/ng.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, Julier C, Morahan G, Nerup J, Nierras C, Plagnol V, Pociot F, Schuilenburg H, Smyth DJ, Stevens H, Todd JA, Walker NM, Rich SS. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41:703–707. doi: 10.1038/ng.381. doi:10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow DK, Collins SR, Bodenmiller B, Aebersold R, Simons K, Shevchenko A, Ejsing CS, Weissman JS. Orm family proteins mediate sphingolipid homeostasis. Nature. 2010;463:1048–1053. doi: 10.1038/nature08787. doi:10.1038/nature08787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning B, Browning S. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. doi:10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean L. Blood groups and red cell antigens. 2005. [Google Scholar]
- Gudbjartsson DF, Bjornsdottir US, Halapi E, Helgadottir A, Sulem P, Jonsdottir GM, Thorleifsson G, Helgadottir H, Steinthorsdottir V, Stefansson H, Williams C, Hui J, Beilby J, Warrington NM, James A, Palmer LJ, Koppelman GH, Heinzmann A, Krueger M, Boezen HM, Wheatley A, Altmuller J, Shin HD, Uh ST, Cheong HS, Jonsdottir B, Gislason D, Park CS, Rasmussen LM, Porsbjerg C, Hansen JW, Backer V, Werge T, Janson C, Jonsson UB, Ng MC, Chan J, So WY, Ma R, Shah SH, Granger CB, Quyyumi AA, Levey AI, Vaccarino V, Reilly MP, Rader DJ, Williams MJ, van Rij AM, Jones GT, Trabetti E, Malerba G, Pignatti PF, Boner A, Pescollderungg L, Girelli D, Olivieri O, Martinelli N, Ludviksson BR, Ludviksdottir D, Eyjolfsson GI, Arnar D, Thorgeirsson G, Deichmann K, Thompson PJ, Wjst M, Hall IP, Postma DS, Gislason T, Gulcher J, Kong A, Jonsdottir I, Thorsteinsdottir U, Stefansson K. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet. 2009;41:342–347. doi: 10.1038/ng.323. doi:10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- Halapi E, Gudbjartsson DF, Jonsdottir GM, Bjornsdottir US, Thorleifsson G, Helgadottir H, Williams C, Koppelman GH, Heinzmann A, Boezen HM, Jonasdottir A, Blondal T, Gudjonsson SA, Thorlacius T, Henry AP, Altmueller J, Krueger M, Shin HD, Uh ST, Cheong HS, Jonsdottir B, Ludviksson BR, Ludviksdottir D, Gislason D, Park CS, Deichmann K, Thompson PJ, Wjst M, Hall IP, Postma DS, Gislason T, Kong A, Jonsdottir I, Thorsteinsdottir U, Stefansson K. A sequence variant on 17q21 is associated with age at onset and severity of asthma. Eur J Hum Genet. 2010;18:902–908. doi: 10.1038/ejhg.2010.38. doi:10.1038/ejhg.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrell FE. Statistical tables and plots using S and LaTeX. 2004. [Google Scholar]
- Hollard D, Berthier R, Douady F. Granulopoiesis and its regulation. Sem Hop. 1975;51:643–651. [PubMed] [Google Scholar]
- Kabesch M. Novel asthma-associated genes from genome-wide association studies: what is their significance? Chest. 2010;137:909–915. doi: 10.1378/chest.09-1554. doi:10.1378/chest.09-1554. [DOI] [PubMed] [Google Scholar]
- Kamatani Y, Matsuda K, Okada Y, Kubo M, Hosono N, Daigo Y, Nakamura Y, Kamatani N. Genome-wide association study of hematological and biochemical traits in a Japanese population. Nat Genet. 2010;42:210–215. doi: 10.1038/ng.531. doi:10.1038/ng.531. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. doi:10.1101/gr.229102 (Article published online before print in May 2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie CC, Doheny KF, Mirel DB, Pugh EW, Bierut LJ, Bhangale T, Boehm F, Caporaso NE, Cornelis MC, Edenberg HJ, Gabriel SB, Harris EL, Hu FB, Jacobs KB, Kraft P, Landi MT, Lumley T, Manolio TA, McHugh C, Painter I, Paschall J, Rice JP, Rice KM, Zheng X, Weir BS. Quality control and quality assurance in genotypic data for genome-wide association studies. Genet Epidemiol. 2010;34:591–602. doi: 10.1002/gepi.20516. doi:10.1002/gepi.20516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madjid M, Awan I, Willerson JT, Casscells SW. Leukocyte count and coronary heart disease: implications for risk assessment. J Am Coll Cardiol. 2004;44:1945–1956. doi: 10.1016/j.jacc.2004.07.056. doi:10.1016/j.jacc.2004.07.056. [DOI] [PubMed] [Google Scholar]
- McCarty CA, Chisholm RL, Chute CG, Kullo IJ, Jarvik GP, Larson EB, Li R, Masys DR, Ritchie MD, Roden DM, Struewing JP, Wolf WA. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. doi:10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, Depner M, von Berg A, Bufe A, Rietschel E, Heinzmann A, Simma B, Frischer T, Willis-Owen SA, Wong KC, Illig T, Vogelberg C, Weiland SK, von Mutius E, Abecasis GR, Farrall M, Gut IG, Lathrop GM, Cookson WO. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. doi:10.1038/nature 06014. [DOI] [PubMed] [Google Scholar]
- Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, von Mutius E, Farrall M, Lathrop M, Cookson WO. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–1221. doi: 10.1056/NEJMoa0906312. doi:10.1056/NEJMoa 0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalls MA, Wilson JG, Patterson NJ, Tandon A, Zmuda JM, Huntsman S, Garcia M, Hu D, Li R, Beamer BA, Patel KV, Akylbekova EL, Files JC, Hardy CL, Buxbaum SG, Taylor HA, Reich D, Harris TB, Ziv E. Admixture mapping of white cell count: genetic locus responsible for lower white blood cell count in the Health ABC and Jackson Heart studies. Am J Hum Genet. 2008;82:81–87. doi: 10.1016/j.ajhg.2007.09.003. doi:10.1016/j.ajhg.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Kamatani Y, Takahashi A, Matsuda K, Hosono N, Ohmiya H, Daigo Y, Yamamoto K, Kubo M, Nakamura Y, Kamatani N. Common variations in PSMD3-CSF3 and PLCB4 are associated with neutrophil count. Hum Mol Genet. 2010;19:2079–2085. doi: 10.1093/hmg/ddq080. doi:10.1093/hmg/ddq080. [DOI] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. Population structure and eigenanalysis. Plos Genetics. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. doi:10.1371/journal.pgen. 0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips C, Salas A, Sanchez JJ, Fondevila M, Gomez-Tato A, Alvarez-Dios J, Calaza M, de Cal MC, Ballard D, Lareu MV, Carracedo A. Inferring ancestral origin using a single multiplex assay of ancestry-informative marker SNPs. Forensic Sci Int Genet. 2007;1:273–280. doi: 10.1016/j.fsigen.2007.06.008. doi:10.1016/j.fsigen.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Pruenster M, Rot A. Throwing light on DARC. Biochem Soc Trans. 2006;34:1005–1008. doi: 10.1042/BST0341005. doi:10.1042/BST0341005. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. doi:10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Nalls MA, Kao WH, Akylbekova EL, Tandon A, Patterson N, Mullikin J, Hsueh WC, Cheng CY, Coresh J, Boerwinkle E, Li M, Waliszewska A, Neubauer J, Li R, Leak TS, Ekunwe L, Files JC, Hardy CL, Zmuda JM, Taylor HA, Ziv E, Harris TB, Wilson JG. Reduced neutrophil count in people of African descent is due to a regulatory variant in the Duffy antigen receptor for chemokines gene. Plos Genetics. 2009;5:e1000360. doi: 10.1371/journal.pgen.1000360. doi:10.1371/journal.pgen.1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden DM, Pulley JM, Basford MA, Bernard GR, Clayton EW, Balser JR, Masys DR. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008;84:362–369. doi: 10.1038/clpt.2008.89. doi:10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel RB, Baumert J, Barbalic M, Dupuis J, Ellinor PT, Durda P, Dehghan A, Bis JC, Illig T, Morrison AC, Jenny NS, Keaney JF, Jr, Gieger C, Tilley C, Yamamoto JF, Khuseyinova N, Heiss G, Doyle M, Blankenberg S, Herder C, Walston JD, Zhu Y, Vasan RS, Klopp N, Boerwinkle E, Larson MG, Psaty BM, Peters A, Ballantyne CM, Witteman JC, Hoogeveen RC, Benjamin EJ, Koenig W, Tracy RP. Duffy antigen receptor for chemokines (Darc) polymorphism regulates circulating concentrations of monocyte chemoattractant protein-1 and other inflammatory mediators. Blood. 2010;115:5289–5299. doi: 10.1182/blood-2009-05-221382. doi:10.1182/blood-2009-05-221382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar A, Mitchell P, Rochtchina E, Tan J, Wang JJ. Association between circulating white blood cell count and long-term incidence of age-related macular degeneration: the Blue Mountains Eye Study. Am J Epidemiol. 2007;165:375–382. doi: 10.1093/aje/kwk022. doi:10.1093/aje/kwk022. [DOI] [PubMed] [Google Scholar]
- Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38:209–213. doi: 10.1038/ng1706. doi:10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- Soranzo N, Spector TD, Mangino M, Kuhnel B, Rendon A, Teumer A, Willenborg C, Wright B, Chen L, Li M, Salo P, Voight BF, Burns P, Laskowski RA, Xue Y, Menzel S, Altshuler D, Bradley JR, Bumpstead S, Burnett MS, Devaney J, Doring A, Elosua R, Epstein SE, Erber W, Falchi M, Garner SF, Ghori MJ, Goodall AH, Gwilliam R, Hakonarson HH, Hall AS, Hammond N, Hengstenberg C, Illig T, Konig IR, Knouff CW, McPherson R, Melander O, Mooser V, Nauck M, Nieminen MS, O’Donnell CJ, Peltonen L, Potter SC, Prokisch H, Rader DJ, Rice CM, Roberts R, Salomaa V, Sambrook J, Schreiber S, Schunkert H, Schwartz SM, Serbanovic-Canic J, Sinisalo J, Siscovick DS, Stark K, Surakka I, Stephens J, Thompson JR, Volker U, Volzke H, Watkins NA, Wells GA, Wichmann HE, Van Heel DA, Tyler-Smith C, Thein SL, Kathiresan S, Perola M, Reilly MP, Stewart AF, Erdmann J, Samani NJ, Meisinger C, Greinacher A, Deloukas P, Ouwehand WH, Gieger C. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat Genet. 2009;41:1182–1190. doi: 10.1038/ng.467. doi:10.1038/ng.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner S, Armstrong LL, Bradford Y, Carlson CS, Crawford DC, Crenshaw AT, de Andrade M, Doheny KF, Haines JL, Hayes G, Jarvik G, Jiang L, Kullo IJ, Li R, Ling H, Manolio TA, Matsumoto M, McCarty CA, McDavid AN, Mirel DB, Paschall JE, Pugh EW, Rasmussen LV, Wilke RA, Zuvich RL, Ritchie MD. Quality control procedures for genome-wide association studies. Curr Protoc Hum Genet Chapter 1: Unit1 19. 2011 doi: 10.1002/0471142905.hg0119s68. doi:10.1002/0471142905.hg0119s68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verlaan DJ, Berlivet S, Hunninghake GM, Madore AM, Lariviere M, Moussette S, Grundberg E, Kwan T, Ouimet M, Ge B, Hoberman R, Swiatek M, Dias J, Lam KC, Koka V, Harmsen E, Soto-Quiros M, Avila L, Celedon JC, Weiss ST, Dewar K, Sinnett D, Laprise C, Raby BA, Pastinen T, Naumova AK. Allele-specific chromatin remodeling in the ZPBP2/GSDMB/ORMDL3 locus associated with the risk of asthma and autoimmune disease. Am J Hum Genet. 2009;85:377–393. doi: 10.1016/j.ajhg.2009.08.007. doi:10.1016/j.ajhg.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijenberg MP, Feskens EJ, Kromhout D. White blood cell count and the risk of coronary heart disease and all-cause mortality in elderly men. Arterioscler Thromb Vasc Biol. 1996;16:499–503. doi: 10.1161/01.atv.16.4.499. [DOI] [PubMed] [Google Scholar]
- Zody MC, Jiang Z, Fung HC, Antonacci F, Hillier LW, Cardone MF, Graves TA, Kidd JM, Cheng Z, Abouelleil A, Chen L, Wallis J, Glasscock J, Wilson RK, Reily AD, Duckworth J, Ventura M, Hardy J, Warren WC, Eichler EE. Evolutionary toggling of the MAPT 17q21.31 inversion region. Nat Genet. 2008;40(9):1076–1083. doi: 10.1038/ng.193. doi:10.1038/ng.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.