Abstract
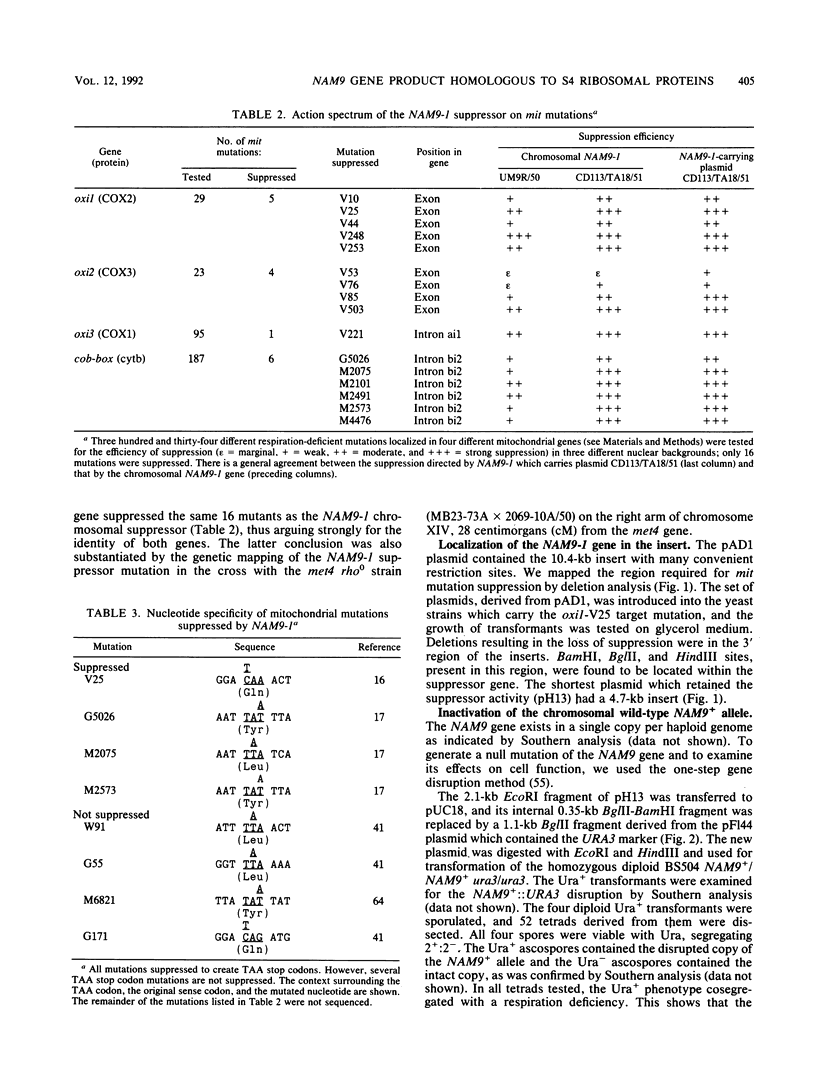
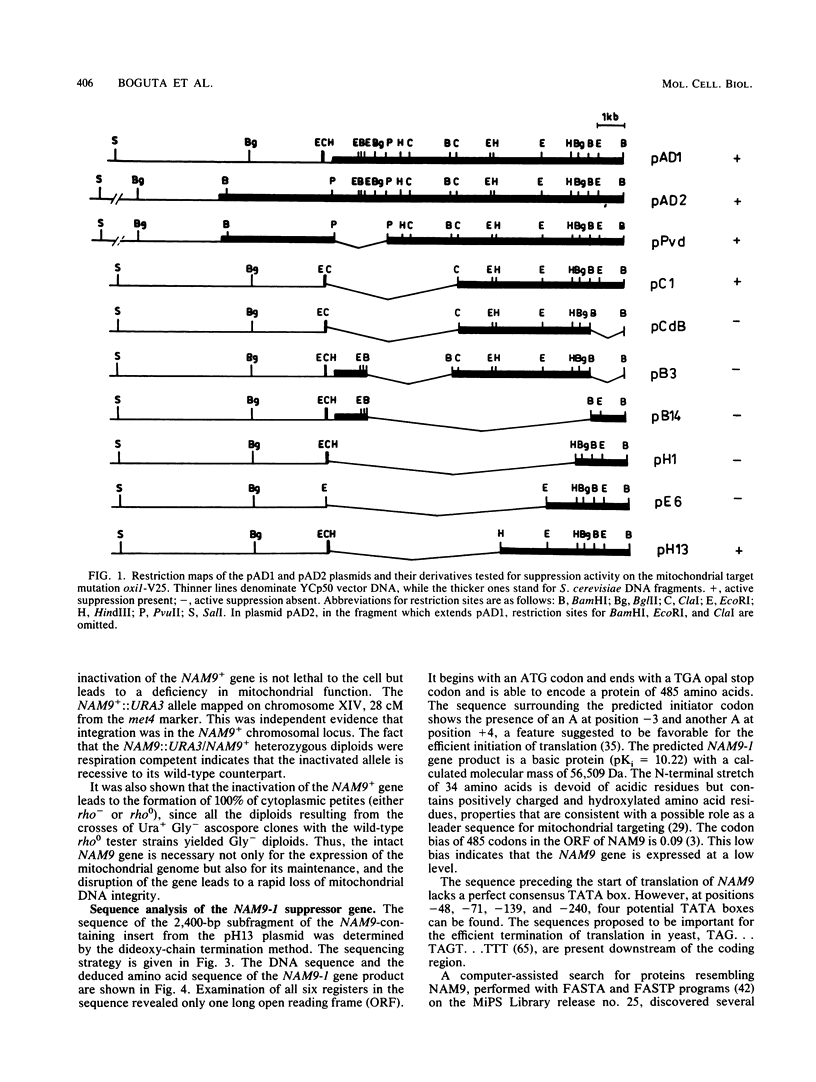
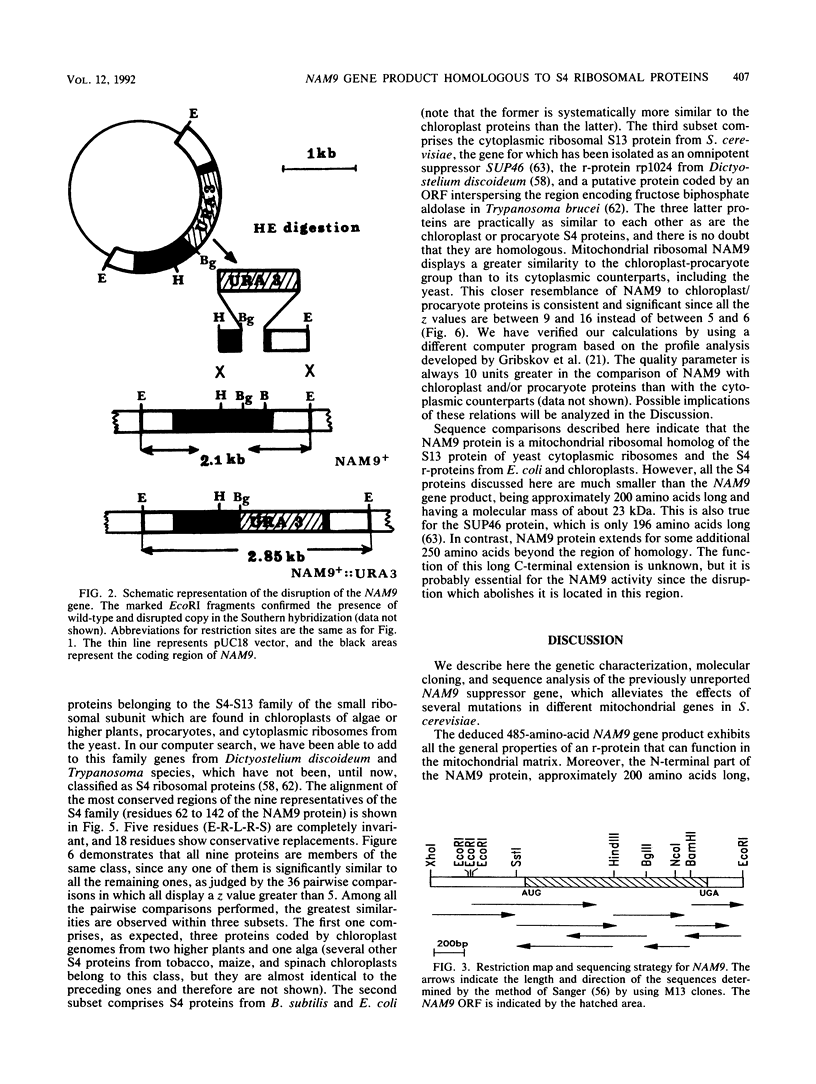
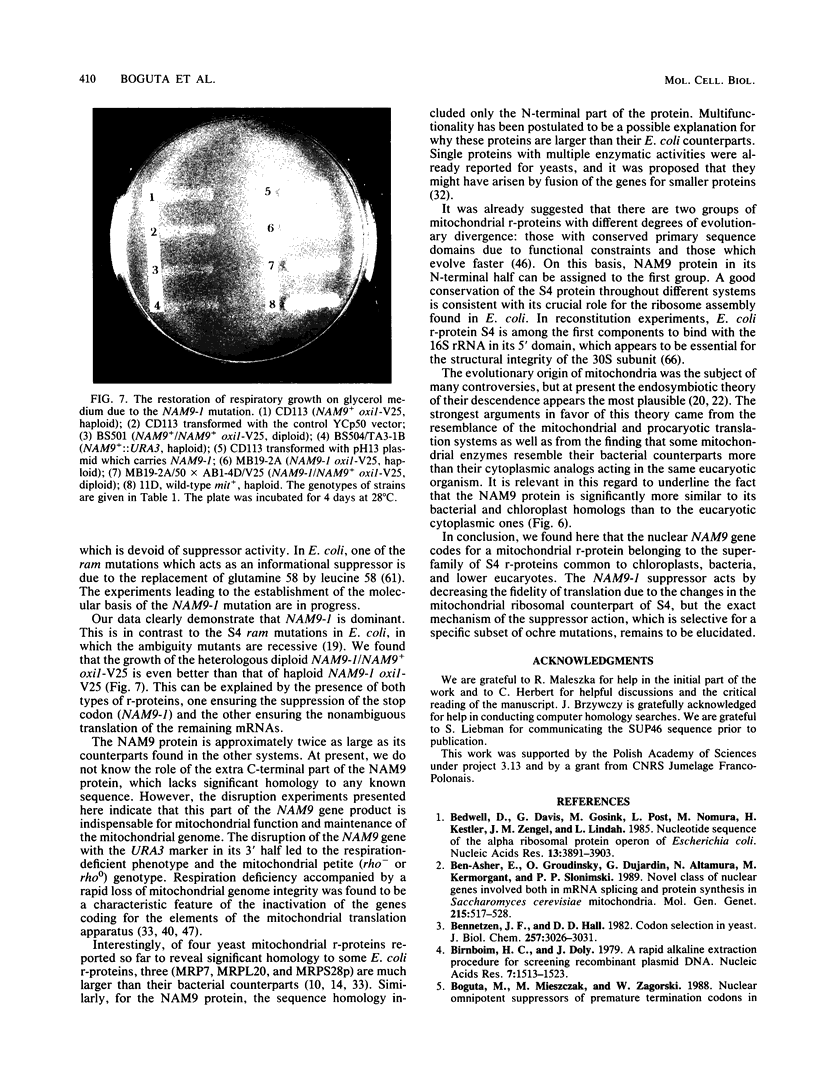
We report the genetic characterization, molecular cloning, and sequencing of a novel nuclear suppressor, the NAM9 gene from Saccharomyces cerevisiae, which acts on mutations of mitochondrial DNA. The strain NAM9-1 was isolated as a respiration-competent revertant of a mitochondrial mit mutant which carries the V25 ochre mutation in the oxi1 gene. Genetic characterization of the NAM9-1 mutation has shown that it is a nuclear dominant omnipotent suppressor alleviating several mutations in all four mitochondrial genes tested and has suggested its informational, and probably ribosomal, character. The NAM9 gene was cloned by transformation of the recipient oxi1-V25 mutant to respiration competence by using a gene bank from the NAM9-1 rho o strain. Orthogonal-field alternation gel electrophoresis analysis and genetic mapping localized the NAM9 gene on the right arm of chromosome XIV. Sequence analysis of the NAM9 gene showed that it encodes a basic protein of 485 amino acids with a presequence that could target the protein to the mitochondrial matrix. The N-terminal sequence of 200 amino acids of the deduced NAM9 product strongly resembles the S4 ribosomal proteins from chloroplasts and bacteria. Significant although less extensive similarity was found with ribosomal cytoplasmic proteins from lower eucaryotes, including S. cerevisiae. Chromosomal inactivation of the NAM9+ gene is not lethal to the cell but leads to respiration deficiency and loss of mitochondrial DNA integrity. We conclude that the NAM9 gene product is a mitochondrial ribosomal counterpart of S4 ribosomal proteins found in other systems and that the suppressor acts through decreasing the fidelity of translation.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Asher E. B., Groudinsky O., Dujardin G., Altamura N., Kermorgant M., Slonimski P. P. Novel class of nuclear genes involved in both mRNA splicing and protein synthesis in Saccharomyces cerevisiae mitochondria. Mol Gen Genet. 1989 Feb;215(3):517–528. doi: 10.1007/BF00427051. [DOI] [PubMed] [Google Scholar]
- Bedwell D., Davis G., Gosink M., Post L., Nomura M., Kestler H., Zengel J. M., Lindahl L. Nucleotide sequence of the alpha ribosomal protein operon of Escherichia coli. Nucleic Acids Res. 1985 Jun 11;13(11):3891–3903. doi: 10.1093/nar/13.11.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguta M., Mieszczak M., Zagórski W. Nuclear omnipotent suppressors of premature termination codons in mitochondrial genes affect the 37S mitoribosomal subunit. Curr Genet. 1988 Feb;13(2):129–135. doi: 10.1007/BF00365647. [DOI] [PubMed] [Google Scholar]
- Claisse M. L., Pajot P. F. Presence of cytochrome c1 in cytoplasmic "petite" mutants of Saccharomyces cerevisiae. Eur J Biochem. 1974 Nov 1;49(1):49–59. doi: 10.1111/j.1432-1033.1974.tb03810.x. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde J., Fink G. R. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3651–3655. doi: 10.1073/pnas.73.10.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M. C., Fox T. D. Control of mitochondrial gene expression in Saccharomyces cerevisiae. Annu Rev Genet. 1990;24:91–113. doi: 10.1146/annurev.ge.24.120190.000515. [DOI] [PubMed] [Google Scholar]
- Dang H., Ellis S. R. Structural and functional analyses of a yeast mitochondrial ribosomal protein homologous to ribosomal protein S15 of Escherichia coli. Nucleic Acids Res. 1990 Dec 11;18(23):6895–6901. doi: 10.1093/nar/18.23.6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang H., Franklin G., Darlak K., Spatola A. F., Ellis S. R. Discoordinate expression of the yeast mitochondrial ribosomal protein MRP1. J Biol Chem. 1990 May 5;265(13):7449–7454. [PubMed] [Google Scholar]
- Dmochowska A., Dignard D., Henning D., Thomas D. Y., Bussey H. Yeast KEX1 gene encodes a putative protease with a carboxypeptidase B-like function involved in killer toxin and alpha-factor precursor processing. Cell. 1987 Aug 14;50(4):573–584. doi: 10.1016/0092-8674(87)90030-4. [DOI] [PubMed] [Google Scholar]
- Douglas S. E., Durnford D. G. Nucleotide sequence of the genes for ribosomal protein S4 and tRNA(Arg) from the chlorophyll c-containing alga Cryptomonas phi. Nucleic Acids Res. 1990 Apr 11;18(7):1903–1903. doi: 10.1093/nar/18.7.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujardin G., Pajot P., Groudinsky O., Slonimski P. P. Long range control circuits within mitochondria and between nucleus and mitochondria. I. Methodology and phenomenology of suppressors. Mol Gen Genet. 1980;179(3):469–482. doi: 10.1007/BF00271736. [DOI] [PubMed] [Google Scholar]
- Fearon K., Mason T. L. Structure and regulation of a nuclear gene in Saccharomyces cerevisiae that specifies MRP7, a protein of the large subunit of the mitochondrial ribosome. Mol Cell Biol. 1988 Sep;8(9):3636–3646. doi: 10.1128/mcb.8.9.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fox T. D., Staempfli S. Suppressor of yeast mitochondrial ochre mutations that maps in or near the 15S ribosomal RNA gene of mtDNA. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1583–1587. doi: 10.1073/pnas.79.5.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorini L. The contrasting role of strA and ram gene products in ribosomal functioning. Cold Spring Harb Symp Quant Biol. 1969;34:101–109. doi: 10.1101/sqb.1969.034.01.016. [DOI] [PubMed] [Google Scholar]
- Gray M. W., Doolittle W. F. Has the endosymbiont hypothesis been proven? Microbiol Rev. 1982 Mar;46(1):1–42. doi: 10.1128/mr.46.1.1-42.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribskov M., McLachlan A. D., Eisenberg D. Profile analysis: detection of distantly related proteins. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4355–4358. doi: 10.1073/pnas.84.13.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohmann L., Graack H. R., Kitakawa M. Molecular cloning of the nuclear gene for mitochondrial ribosomal protein YmL31 from Saccharomyces cerevisiae. Eur J Biochem. 1989 Jul 15;183(1):155–160. doi: 10.1111/j.1432-1033.1989.tb14907.x. [DOI] [PubMed] [Google Scholar]
- Groudinsky O., Dujardin G., Slonimski P. P. Long range control circuits within mitochondria and between nucleus and mitochondria. II. Genetic and biochemical analyses of suppressors which selectively alleviate the mitochondrial intron mutations. Mol Gen Genet. 1981;184(3):493–503. doi: 10.1007/BF00352529. [DOI] [PubMed] [Google Scholar]
- Grundy F. J., Henkin T. M. Cloning and analysis of the Bacillus subtilis rpsD gene, encoding ribosomal protein S4. J Bacteriol. 1990 Nov;172(11):6372–6379. doi: 10.1128/jb.172.11.6372-6379.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert C. J., Labouesse M., Dujardin G., Slonimski P. P. The NAM2 proteins from S. cerevisiae and S. douglasii are mitochondrial leucyl-tRNA synthetases, and are involved in mRNA splicing. EMBO J. 1988 Feb;7(2):473–483. doi: 10.1002/j.1460-2075.1988.tb02835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D. G., Sharp P. M. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988 Dec 15;73(1):237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- Hiratsuka J., Shimada H., Whittier R., Ishibashi T., Sakamoto M., Mori M., Kondo C., Honji Y., Sun C. R., Meng B. Y. The complete sequence of the rice (Oryza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol Gen Genet. 1989 Jun;217(2-3):185–194. doi: 10.1007/BF02464880. [DOI] [PubMed] [Google Scholar]
- Hurt E. C., Pesold-Hurt B., Suda K., Oppliger W., Schatz G. The first twelve amino acids (less than half of the pre-sequence) of an imported mitochondrial protein can direct mouse cytosolic dihydrofolate reductase into the yeast mitochondrial matrix. EMBO J. 1985 Aug;4(8):2061–2068. doi: 10.1002/j.1460-2075.1985.tb03892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro J., Ono B. I., Masurekar M., McLaughlin C. S., Sherman F. Altered ribosomal protein S11 from the SUP46 suppressor of yeast. J Mol Biol. 1981 Apr 15;147(3):391–397. doi: 10.1016/0022-2836(81)90491-5. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitakawa M., Grohmann L., Graack H. R., Isono K. Cloning and characterization of nuclear genes for two mitochondrial ribosomal proteins in Saccharomyces cerevisiae. Nucleic Acids Res. 1990 Mar 25;18(6):1521–1529. doi: 10.1093/nar/18.6.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Possible role of flanking nucleotides in recognition of the AUG initiator codon by eukaryotic ribosomes. Nucleic Acids Res. 1981 Oct 24;9(20):5233–5252. doi: 10.1093/nar/9.20.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruszewska A., Szcześniak B. Functional nuclear suppressor of mitochondrial oxi2 mutations in yeast. Curr Genet. 1985;10(2):87–93. doi: 10.1007/BF00636472. [DOI] [PubMed] [Google Scholar]
- Labouesse M., Dujardin G., Slonimski P. P. The yeast nuclear gene NAM2 is essential for mitochondrial DNA integrity and can cure a mitochondrial RNA-maturase deficiency. Cell. 1985 May;41(1):133–143. doi: 10.1016/0092-8674(85)90068-6. [DOI] [PubMed] [Google Scholar]
- Lazowska J., Jacq C., Slonimski P. P. Sequence of introns and flanking exons in wild-type and box3 mutants of cytochrome b reveals an interlaced splicing protein coded by an intron. Cell. 1980 Nov;22(2 Pt 2):333–348. doi: 10.1016/0092-8674(80)90344-x. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Masurekar M., Palmer E., Ono B. I., Wilhelm J. M., Sherman F. Misreading of the ribosomal suppressor SUP46 due to an altered 40 S subunit in yeast. J Mol Biol. 1981 Apr 15;147(3):381–390. doi: 10.1016/0022-2836(81)90490-3. [DOI] [PubMed] [Google Scholar]
- Matsushita Y., Kitakawa M., Isono K. Cloning and analysis of the nuclear genes for two mitochondrial ribosomal proteins in yeast. Mol Gen Genet. 1989 Oct;219(1-2):119–124. doi: 10.1007/BF00261166. [DOI] [PubMed] [Google Scholar]
- McMullin T. W., Haffter P., Fox T. D. A novel small-subunit ribosomal protein of yeast mitochondria that interacts functionally with an mRNA-specific translational activator. Mol Cell Biol. 1990 Sep;10(9):4590–4595. doi: 10.1128/mcb.10.9.4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers A. M., Crivellone M. D., Tzagoloff A. Assembly of the mitochondrial membrane system. MRP1 and MRP2, two yeast nuclear genes coding for mitochondrial ribosomal proteins. J Biol Chem. 1987 Mar 5;262(7):3388–3397. [PubMed] [Google Scholar]
- Myers A. M., Pape L. K., Tzagoloff A. Mitochondrial protein synthesis is required for maintenance of intact mitochondrial genomes in Saccharomyces cerevisiae. EMBO J. 1985 Aug;4(8):2087–2092. doi: 10.1002/j.1460-2075.1985.tb03896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. A., Reed S. I. Isolation of genes by complementation in yeast: molecular cloning of a cell-cycle gene. Proc Natl Acad Sci U S A. 1980 Apr;77(4):2119–2123. doi: 10.1073/pnas.77.4.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Partaledis J. A., Mason T. L. Structure and regulation of a nuclear gene in Saccharomyces cerevisiae that specifies MRP13, a protein of the small subunit of the mitochondrial ribosome. Mol Cell Biol. 1988 Sep;8(9):3647–3660. doi: 10.1128/mcb.8.9.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Novick P., Thomas J. H., Botstein D., Fink G. R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60(2-3):237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Rosset R., Gorini L. A ribosomal ambiguity mutation. J Mol Biol. 1969 Jan 14;39(1):95–112. doi: 10.1016/0022-2836(69)90336-2. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolińska U. Mitochondrial mutagenesis in yeast: mutagenic specificity of EMS and the effects of RAD9 and REV3 gene products. Mutat Res. 1987 Aug;179(2):167–174. doi: 10.1016/0027-5107(87)90307-1. [DOI] [PubMed] [Google Scholar]
- Steel L. F., Smyth A., Jacobson A. Nucleotide sequence and characterization of the transcript of a Dictyostelium ribosomal protein gene. Nucleic Acids Res. 1987 Dec 23;15(24):10285–10298. doi: 10.1093/nar/15.24.10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A., Dieckmann C. L. PET genes of Saccharomyces cerevisiae. Microbiol Rev. 1990 Sep;54(3):211–225. doi: 10.1128/mr.54.3.211-225.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A., Myers A. M. Genetics of mitochondrial biogenesis. Annu Rev Biochem. 1986;55:249–285. doi: 10.1146/annurev.bi.55.070186.001341. [DOI] [PubMed] [Google Scholar]
- Ullrich A., Shine J., Chirgwin J., Pictet R., Tischer E., Rutter W. J., Goodman H. M. Rat insulin genes: construction of plasmids containing the coding sequences. Science. 1977 Jun 17;196(4296):1313–1319. doi: 10.1126/science.325648. [DOI] [PubMed] [Google Scholar]
- Vijayasarathy S., Ernest I., Itzhaki J. E., Sherman D., Mowatt M. R., Michels P. A., Clayton C. E. The genes encoding fructose bisphosphate aldolase in Trypanosoma brucei are interspersed with unrelated genes. Nucleic Acids Res. 1990 May 25;18(10):2967–2975. doi: 10.1093/nar/18.10.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss-Brummer B., Hüttenhofer A., Kaudewitz F. Leakiness of termination codons in mitochondrial mutants of the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1984;198(2):62–68. doi: 10.1007/BF00328702. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]
- van Acken U. Proteinchemical studies on ribosomal proteins S4 and S12 from ram (ribosomal ambiguity) mutants of Escherichia coli. Mol Gen Genet. 1975 Sep 15;140(1):61–68. doi: 10.1007/BF00268989. [DOI] [PubMed] [Google Scholar]



