Abstract
Context
Clopidogrel therapy improves cardiovascular outcomes in patients with acute coronary syndromes and following percutaneous coronary intervention by inhibiting adenosine diphosphate (ADP)–dependent platelet activation. However, nonresponsiveness is widely recognized and is related to recurrent ischemic events.
Objective
To identify gene variants that influence clopidogrel response.
Design, Setting, and Participants
In the Pharmacogenomics of Antiplatelet Intervention (PAPI) Study (2006-2008), we administered clopidogrel for 7 days to 429 healthy Amish persons and measured response by ex vivo platelet aggregometry. A genome-wide association study was performed followed by genotyping the loss-of-function cytochrome P450 (CYP) 2C19*2 variant (rs4244285). Findings in the PAPI Study were extended by examining the relation of CYP2C19*2 genotype to platelet function and cardiovascular outcomes in an independent sample of 227 patients undergoing percutaneous coronary intervention.
Main Outcome Measure
ADP-stimulated platelet aggregation in response to clopidogrel treatment and cardiovascular events.
Results
Platelet response to clopidogrel was highly heritable (h2=0.73; P<.001). Thirteen single-nucleotide polymorphisms on chromosome 10q24 within the CYP2C18-CYP2C19-CYP2C9-CYP2C8 cluster were associated with diminished clopidogrel response, with a high degree of statistical significance (P=1.5 × 10−13 for rs12777823, additive model). The rs12777823 polymorphism was in strong linkage disequilibrium with the CYP2C19*2 variant, and was associated with diminished clopidogrel response, accounting for 12% of the variation in platelet aggregation to ADP (P=4.3 × 10−11). The relation between CYP2C19*2 genotype and platelet aggregation was replicated in clopidogrel-treated patients undergoing coronary intervention (P=.02). Furthermore, patients with the CYP2C19*2 variant were more likely (20.9% vs 10.0%) to have a cardiovascular ischemic event or death during 1 year of follow-up (hazard ratio, 2.42; 95% confidence interval, 1.18-4.99; P=.02).
Conclusion
CYP2C19*2 genotype was associated with diminished platelet response to clopidogrel treatment and poorer cardiovascular outcomes.
Dual Antiplatelet Therapy, including clopidogrel and aspirin, inhibits platelet function, preventing ischemic events and improving outcomes following acute coronary syndromes and percutaneous coronary intervention (PCI).1,2 To exert an antiplatelet effect, clopidogrel requires conversion to an active thiol metabolite (SR 26334) by hepatic cytochrome P450 (CYP) isoenzymes, which inhibit adenosine diphosphate (ADP)–stimulated platelet activation by irreversibly binding to platelet P2Y12 receptors.3-5
Variability in clopidogrel response is well established.6-8 Patients treated with clopidogrel who demonstrate higher ex vivo platelet reactivity are at increased risk of ischemic events.9-12 Variation in platelet function in response to clopidogrel has been associated with lipophilic statins, calcium channel blockers, proton pump inhibitors, St John's wort, and smoking.13-16 However, these factors account for only a small fraction of the variation in response. Recently, the loss-of-function CYP2C19 (GenBank 1557) *2 allele has been shown to be associated with a decreased activation of clopidogrel17,18 and antiplatelet effect19-23 and with increased cardiovascular events in patients receiving clopidogrel.18,24-26
To identify genes associated with variation in clopidogrel response, we performed a genome-wide association study of ADP-stimulated platelet aggregation in response to clopidogrel in the Old Order Amish, a relatively homogeneous founder population in which confounding factors, including medication usage and lifestyle variability, are minimized. Replication and extension of genetic findings and time-to-event analyses were performed in a population with high risk for cardiovascular disease recruited from a cardiac catheterization laboratory in Baltimore, Maryland.
Methods
Study Populations
Amish Pharmacogenomics of Antiplatelet Intervention Study
The Amish Pharmacogenomics of Antiplatelet Intervention (PAPI) Study (NCT0079936) recruited 429 generally healthy white participants 20 years or older between August 2006 and October 2008 (for additional details, see eMethods at http://www.jama.org). These individuals comprised a number of relative pairs informative for estimating heritabilities, including 105 parent-offspring pairs, 175 sibling pairs, 1 grandparent-grandchild pair, 48 avuncular pairs, and 12 first-cousin pairs. Medical and family histories, anthropometry, physical examinations, and blood samples after an overnight fast were obtained. Complete blood count with platelet number and levels of serum lipids (total cholesterol, high-density lipoprotein cholesterol, and triglycerides) were assayed by Quest Diagnostics (Horsham, Pennsylvania); levels of low-density lipoprotein cholesterol were calculated using the Friedewald equation.
After baseline platelet aggregation measurements were obtained, participants were given a 300-mg oral loading dose of clopidogrel followed by 75 mg per day for 6 days. Follow-up platelet aggregation studies were repeated 1 hour following the last dose of clopidogrel. A second follow-up platelet aggregation measurement was made later the same day, 1 hour after oral ingestion of 324 mg of chewable aspirin. Platelet function was assessed by optical aggregometry with a PAP8E Aggregometer (Bio/Data Corporation, Horsham, Pennsylvania) in platelet-rich plasma, stimulated with ADP (20 μmol/L) or arachidonic acid (1.6 mmol/L) (eMethods).
Sinai Hospital of Baltimore Study Patients
The Sinai Hospital of Baltimore Study (NCT00370045) enrolled 227 patients older than 18 years and undergoing nonemergent PCI between January 2004 and May 2007 (eMethods). Of these, 140 (61.7%) were white, 83 (36.6%) were African American, and 4 (1.8%) were other race/ethnicity. Information on race/ ethnicity was obtained by self-report. On the day of PCI, patients received a 600-mg (n=112) or 300-mg (n=25) clopidogrel loading dose; 90 were already receiving maintenance therapy with a 75-mg daily dose at the time of PCI and received no loading dose. There were no differences in baseline characteristics or in the long-term outcomes investigated in stratified analyses of acute clopidogrel dosing; thus, these groups were combined for further analyses.
Patients received bivalirudin or heparin therapy, either with (n=107) or without (n=120) eptifibatide.27,28 Anticoagulant therapy was discontinued at the completion of the procedure in all patients. All patients received 81 to 325 mg of aspirin daily for at least 1 week prior to PCI and 325 mg on the day of the procedure. To minimize the effects of acute anticoagulant therapy during PCI, platelet function was measured on the day of hospital discharge in patients not treated with eptifibatide or 5 days or more postdischarge in patients treated with eptifibatide.
In total, results of baseline platelet function studies were available for 143 patients not taking clopidogrel at the time of enrollment, and results of postclopidogrel platelet aggregation studies were available for 188 patients. Platelet aggregation was assessed in platelet-rich plasma after stimulation with 20 μmol/L of ADP or 2 mmol/L of arachidonic acid using a Chronolog Lumi-Aggregometer (Model 490-4D; Chronolog, Havertown, Pennsylvania), as described previously (eMethods).7
Aspirin (325 mg/d) and clopidogrel (75 mg/d) were prescribed for all patients at the time of hospital discharge, according to American College of Cardiology/American Heart Association guidelines.1 We assessed medication adherence by self-report and by review of source documents from hospitalizations for ischemic events.
The 227 enrolled patients were contacted at the end of 1 and 12 months post-PCI to determine the occurrence of postdischarge cardiovascular ischemic events (eMethods). Of these patients, 95 were still taking clopidogrel after 1 year; 132 were not. A physician, blinded to the study results of the patient, adjudicated all end points through review of source documents obtained from medical records. Post-discharge ischemic events were defined as myocardial infarction (the occurrence of ischemic symptoms and a troponin I value greater than the upper limits of normal), ischemic stroke, stent thrombosis (definite stent thrombosis according to Academic Research Consortium criteria29), unplanned target vessel revascularization (revascularization of vessel treated at time of study enrollment), unplanned nontarget vessel revascularization (revascularization of a vessel different from that treated at time of study enrollment), hospitalization for coronary ischemia without revascularization (hospitalization for chest pain with evidence of ischemia on electrocardiogram and no evidence of myocardial infarction as measured by troponin I value), and death secondary to any cardiovascular cause.
The study protocols were approved by the respective institutional review boards at the University of Maryland and Sinai Hospital of Baltimore. Written informed consent was obtained from each participant; participants were compensated for their participation.
Genotype Analysis
Genotyping in the PAPI Study was performed with the Affymetrix GeneChip Human Mapping 500K or 1 M (version 6.0) arrays according to the manufacturer's instructions (Affymetrix Inc, Santa Clara, California). Genotype calls were performed using BRLMM (500K array) or Birdseed version 2 (1 M array). Single-nucleotide polymorphisms (SNPs) present on both arrays with a minor allele frequency greater than 1% were included in our analyses. The mean genotype call rate of these 400 230 SNPs was 98.7%. All SNPs within the region of interest on chromosome 10q24 were in Hardy-Weinberg equilibrium (P>.05), except rs12572351 (P = 1.89 × 10−5) and rs2860838 (P = 1.03 × 10−4). Follow-up genotyping of the known common loss-of-function CYP2C19*2 variant (rs4244285), as well as other functional variants of CYP2C19 (*3 [rs4986893], *5 [rs56337013], and *17 [rs12248560]) was performed using TaqMan SNP genotyping assays (Applied Biosystems, Foster City, California). The genotype concordance rate was greater than 98% in a subset of duplicate samples.
Statistical Analysis
Summary statistics (eg, means [SDs]) and frequencies for the Amish PAPI and Sinai Hospital of Baltimore populations were generated using SAS version 9.1 (SAS Institute Inc, Cary, North Carolina). For both studies, platelet aggregation was expressed as the maximum percentage change in light transmittance, using platelet-poor plasma as a referent.
Amish PAPI Study
We assessed the correlates (eg, age, sex, body mass index [BMI], lipid levels, and blood pressure) of clopidogrel response using a regression-based approach as implemented in the SOLAR version 4.07 (Southwest Foundation for Biomedical Research, San Antonio, Texas),30 in which we accounted for relatedness among study participants by including a polygenic component as a random effect. Triglyceride levels were logarithm-transformed for analysis and back-transformed for presentation. Distribution analyses were generated in SAS. All statistical tests were 2-sided.
Association analyses between SNPs and ADP-stimulated platelet aggregation following clopidogrel administration were performed under a variance component model that assesses the effect of genotype as an additive effect on the quantitative trait, while simultaneously estimating the effects of age, age2, sex, preclopidogrel platelet aggregation, and the aforementioned polygenic component.
The polygenic component was modeled using the relationship matrix derived from the complete Amish pedigree structure available through published genealogical records maintained by the church.31 The heritability of baseline platelet aggregation and clopidogrel response corresponds to the proportion of the trait variance accounted for by the polygenic component. The genomic control λ coefficient was 1.03; thus, the P values reported are unadjusted.
A power calculation indicated 80% power to detect SNPs with allele frequencies of 0.2 to 0.4 in the initial sample (n = 429), accounting for 8% to 9% of phenotypic variation at α =10−7. To determine whether the loss-of-function CYP2C19*2 variant could account for the chromosome 10q24 association signal, we estimated the independent effects of both rs12777823— the most highly associated SNP from the genome-wide association analysis— and the CYP2C19*2 variant on platelet aggregation by including both in the model simultaneously. Pairwise linkage disequilibrium correlation statistics (|D′| and r2) were computed using Haploview (http://www.broad.mit.edu).
Sinai Hospital of Baltimore Study
We estimated the effect of CYP2C19*2 genotype on preclopidogrel and postclopidogrel ADP-stimulated platelet aggregation under an additive genetic model by classifying participants according to whether they had 0, 1, or 2 risk alleles. The genotype effect was estimated using analysis of variance with adjustment for age, sex, race, study (Peri-Procedural Myocardial Infarction, Platelet Reactivity, Thrombin Generation, and Clot Strength: Differential Effects of Eptifibatide + Bivalirudin vs Bivalirudin study; Clopidogrel Loading With Eptifibatide to Arrest the Reactivity of Platelets [CLEAR PLATELETS−1] Study), and treatment group (clopidogrel dose and use of eptifibatide).
We similarly compared platelet aggregation values between participants who did and did not experience an event while taking clopidogrel. Lastly, we constructed survival curves to compare 1-year cardiovascular event-free survival between participants with (n = 67) and without (n=160) a CYP2C19*2 risk allele. We analyzed the effect of the allele on outcomes in relation to ongoing clopidogrel therapy at the time of the event. We used the proportional hazards model to estimate the relative hazard associated with having a risk allele on subsequent ischemic event rates, adjusting for age, sex, and race. This analysis was carried out both with and without ADP-stimulated aggregation included as a mediator variable.
Survival analysis stratified by CYP2C19*2 genotype was also performed in the subset of 95 patients still taking clopidogrel at the time of event or at 1 year of follow-up and the 132 patients who were not receiving clopidogrel at the time of event or at 1 year of follow-up.
Results
Amish PAPI Study
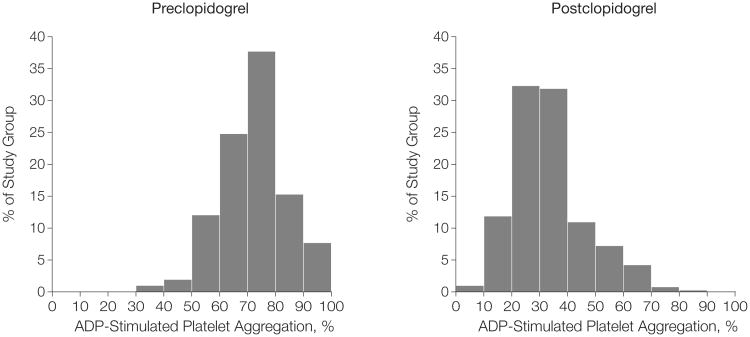
By design, Amish PAPI participants were generally healthy (Table 1). There was wide interindividual variability in ADP-stimulated platelet aggregation at baseline and after clopidogrel administration, with no clear cutoff to define resistance (Figure 1). Poorer clopidogrel response, as defined by ADP-stimulated aggregation after 7 days of clopidogrel administration, was associated with increasing age (3.8% of variance; 95% confidence interval [CI], 3.6%-4.1%; P<.001), greater BMI (2.3% of variance; 95% CI, 2.2%-2.4%; P = .005), higher triglyceride levels (1.3% of variance; 95% CI, 1.28%-1.35%; P = .01), and nominally lower levels of high-density lipoprotein cholesterol (1.0% of variance; 95% CI, 0.98%-1.03%; P = .04) (Table 2). The variation explained by these variables combined was less than 10%. The heritability of ADP-stimulated platelet aggregation at baseline and in response to clopidogrel was 0.33 (SE, 0.13; 95% CI, 0.08-0.58; P = .005) and 0.73 (SE, 0.12; 95% CI, 0.49-0.97; P < .001), respectively, suggesting a substantial genetic component.
Table 1. Characteristics of Amish PAPI Study and Sinai Hospital of Baltimore Study Participants.
| Characteristic, Units | Amish PAPIa | Sinai Hospital of Baltimore | ||
|---|---|---|---|---|
| Men | Women | Men | Women | |
| No. (%) | 214 (49.9) | 215 (50.1) | 136 (59.9) | 91 (40.1) |
| Age, mean (SD), y | 43.9 (13.2) | 47.3 (14.4) | 62.5 (11.4) | 67.0 (11.0) |
| White, No. (%) | 214 (100) | 215 (100) | 93 (68.4) | 47 (51.6) |
| Body mass index, mean (SD)b | 26.0 (3.7) | 28.3 (5.6) | 30.1 (6.3) | 30.9 (7.1) |
| Blood pressure, mean (SD), mm Hg | ||||
| Systolic | 116.6 (12.0) | 117.4 (14.0) | 138.0 (19.9) | 144.6 (19.9) |
| Diastolic | 70.8 (7.4) | 69.7 (7.5) | 73.6 (13.8) | 70.3 (14.2) |
| Hypertension, No. (%)c | 11 (5.1) | 13 (6.0) | 102 (75.0) | 72 (79.1) |
| Lipids, mean (SD), mg/dL | ||||
| Total cholesterol | 206.0 (44.8) | 217.9 (52.2) | NA | NA |
| LDL-C | 137.7 (41.5) | 141.0 (48.9) | NA | NA |
| HDL-C | 54.7 (15.0) | 61.6 (15.2) | NA | NA |
| Triglyceridesd | 68.1 (40.4) | 76.0 (39.4) | NA | NA |
| Hypercholesterolemia, No. (%)e | 52 (24.3) | 59 (27.4) | 111 (81.6) | 72 (79.1) |
| Taking aspirin, No. (%) | 6 (2.8) | 3 (1.4) | 136 (100) | 91 (100) |
| Self-reported diabetes, No. (%) | 1 (0.5) | 2 (0.9) | 39 (28.7) | 44 (48.4) |
| Hematocrit, mean (SD), % | 41.3 (2.4) | 37.7 (2.3) | 41.8 (5.0) | 37.9 (4.5) |
| White blood cell count, median (IQR), × 1000/μL | 5.8 (5.1-6.7) | 5.8 (5.1-6.8) | 6.8 (5.7-8.2) | 7.5 (6.0-9.2) |
| Platelet count, mean (SD) × 100 000/μL | 240.9 (43.7) | 248.1 (50.3) | 223.6 (68.5) | 252.2 (71.4) |
| Current smoker, No. (%)f | 38 (17.8) | 0 | 40 (29.4) | 18 (19.8) |
Abbreviations: HDL-C, high-density lipoprotein cholesterol; IQR, interquartile range; LDL-C, low-density lipoprotein cholesterol; NA, not available; PAPI, Pharmacogenomics of Anti-Platelet Intervention.
SI conversion factors: To convert HDL-C, LDL-C, and total cholesterol values to mmol/L, multiply by 0.0259; triglyceride values to mmol/L, multiply by 0.0113.
For PAPI Study, all participants were withdrawn from prescription and nonprescription medications, vitamins, and supplements 7 days prior to and for the duration of the study. Participants taking prescription antihypertensive, lipid-lowering, and diabetes medications accounted for 0%, 1.6%, and 0% of participants, respectively.
Calculated as weight in kilograms divided by height in meters squared.
Defined as systolic blood pressure greater than 140 mm Hg or diastolic blood pressure greater than 90 mm Hg or taking prescription medication for previously diagnosed hypertension.
Logarithm-transformed for analysis and back-transformed for presentation.
Defined as LDL-C level greater than 160 mg/dL or taking prescription medication for previously diagnosed hyper-cholesterolemia.
Self-reported history of smoking cigarette, pipe, or cigar.
Figure 1. Distribution of Adenosine Diphosphate (ADP)–Stimulated (20 μmol/L) Platelet Aggregation Before and After 7 Days of Clopidogrel Administration in 429 Members of the Amish Pharmacogenomics of Anti-Platelet Intervention (PAPI) Study.
Class intervals include data greater than the lower limit and equal to the upper limit of each interval.
Table 2. Multivariate Analysis of Clopidogrel Response as Measured by Adenosine Diphosphate–Stimulated Platelet Aggregation in PAPI Study Participants (n = 429).
| Characteristic | β (SE) | P Valuea | Variance of Significant Predictors, % |
|---|---|---|---|
| Age | 0.19 (0.04) | <.001 | 3.8 |
| Sex | 1.73 (1.15)b | .13 | |
| Body mass index | 0.35 (0.12) | .005 | 2.3 |
| Lipids | |||
| Total cholesterol | 0.01 (0.01) | .27 | |
| HDL-C | −0.08 (0.04) | .04 | 1.0 |
| LDL-C | 0.02 (0.01) | .17 | |
| Log triglycerides | 0.03 (0.01) | .01 | 1.3 |
| Blood pressure | |||
| Systolic | 0.0 (0.05) | .21 | |
| Diastolic | 0.07 (0.08) | .42 |
Abbreviations: HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; PAPI, Pharmacogenomics of Anti-Platelet Intervention.
Adjusted for age, sex, and preclopidogrel adenosine diphosphate (20 μmol/L)–stimulated platelet aggregation; age adjusted for sex and preclopidogrel platelet aggregation; sex adjusted for age and preclopidogrel platelet aggregation.
Indicates that women tend to respond less well than men.
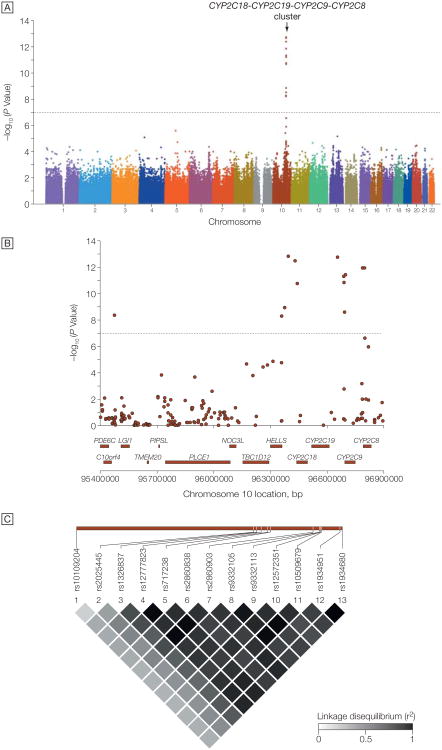
A genome-wide association analysis revealed a cluster of 13 SNPs spanning 1.5 megabases on chromosome 10q24, showing strong evidence for association with clopidogrel response, with P values <10−7 (Figure 2 and eTable 1; Q-Q plot shown in eFigure). These SNPs were in strong linkage disequilibrium with each other, eg, pairwise r2=0.35 to 0.99 with rs12777823, the most significantly associated SNP (P=1.5×10−13 for the additive model). The 9 participants homozygous for the minor allele of rs12777823 had the poorest response to clopidogrel, while the 300 homozygous for the major allele had the best response; the 117 heterozygous participants were intermediate between the 2 homozygous groups. None of the chromosome 10q24 SNPs associated with platelet aggregation after clopidogrel administration was associated with baseline platelet aggregation measures, consistent with this locus as a true determinant of clopidogrel response. No other genomic region revealed association signals that met or exceeded genome-wide significance (P<1.0×10−7) (Figure 2 and eTable 1). The full results of the genome-wide association study are available at http://medschool.umaryland.edu/endocrinology/supplementalInfo.asp.
Figure 2. Genome-Wide Association Study of Adenosine Diphosphate–Stimulated Platelet Aggregation in Response to Clopidogrel.
A, Association (plotted as −log P value) of individual single-nucleotide polymorphisms distributed across the 22 autosomes. Horizontal dotted line indicates P=1.0 × 10−7. B, Enlargement of 1.5-megabase region on chromosome 10q24. Genes encoded in the region are shown below the plot. C, Linkage disequilibrium (r2) among the 13 single-nucleotide polymorphisms showing genome-wide significance with clopidogrel response. Increasing shades of gray represent increasing linkage disequilibrium, from white (r2=0) to black (r2=1).
The cluster of SNPs on chromosome 10q24 most significantly associated with clopidogrel response is located within and immediately flanking the CYP2C18–CYP2C19–CYP2C9– CYP2C8 gene cluster, which encodes a group of cytochrome P450 enzymes that play an important role in drug metabolism, including conversion of clopidogrel to its active metabolite.4,5,17,18,32-35 A guanine>adenine mutation in exon 5 of CYP2C19 (rs4244285, also known as CYP2C19*2) creates an aberrant splice site that leads to an altered reading frame at amino acid 215 and a premature stop codon 20 amino acids downstream, producing a nonfunctional truncated protein, lack of translation resulting from nonsense-mediated messenger RNA decay, or both.
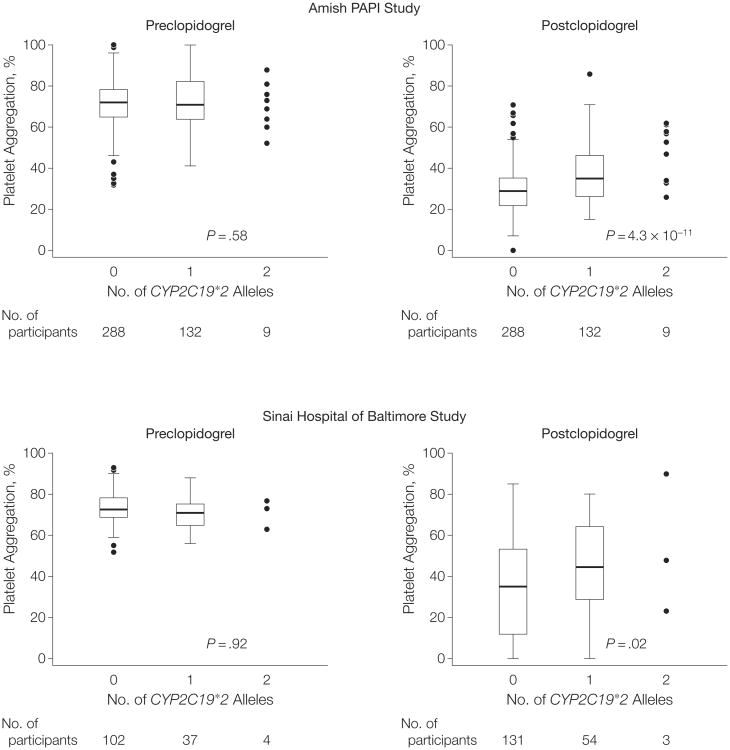
The frequency of CYP2C19*2 was 0.17 in the Amish PAPI population, similar to that found in other white populations. CYP2C19*2 was in high linkage disequilibrium with the cluster of SNPs on chromosome 10q24 identified in the genome-wide association study (eg, r2=0.87 with rs12777823). CYP2C19*2 was associated with ADP-stimulated platelet aggregation after clopidogrel administration, with a high degree of statistical significance (P=4.3 × 10−11 for the additive model) (Figure 3). ADP-stimulated platelet aggregation was reduced to 40.7%, 47.1%, and 65.4% of baseline in response to clopidogrel in participants with 0, 1, and 2 CYP2C19*2 alleles, respectively. The CYP2C19*2 genotype, present in its heterozygous and homozygous state in 30.8% and 2.1% of the PAPI study population, accounted for 12% of the variation in clopidogrel response. When we included CYP2C19*2 as a covariate, the association between chromosome 10q24 SNP rs12777823 and clopidogrel response was markedly attenuated (P=1.5 × 10−13 without adjustment for CYP2C19*2 genotype to P = 2.5 × 10−3 with adjustment for CYP2C19*2 genotype). These findings show that the loss-of-function CYP2C19*2 mutation accounts for most or all of the original 10q24 association signal.
Figure 3.
Association of CYP2C19*2 (rs4244285) Loss-of-Function Variant With Adenosine Diphosphate–Stimulated Platelet Aggregation Before and After Clopidogrel Administration in Participants in the Amish Pharmacogenomics of Antiplatelet Intervention (PAPI) Study and Sinai Hospital of Baltimore Study
The horizontal line in the middle of each box indicates the median; the top and bottom borders of each box indicate the interquartile range (IQR). The whiskers above and below the box indicate plus/minus 1.5 IQRs, respectively; the points beyond the whiskers indicate outliers beyond 1.5 IQRs, except for those carrying 2 alleles in which all data points are plotted.
Other previously described CYP2C19 loss-of-function variants, CYP2C19*3 and *5, were not polymorphic in the Amish population, but there may be additional variation in CYP2C19 or a nearby gene, further contributing to relatively small effects on response and accounting for the remainder of the association signal. The gain-of-function variant CYP2C19*17 had an allele frequency of 0.25 and was not associated with ADP-stimulated platelet aggregation, either at baseline or in response to clopidogrel (eTable 2).
Predictors (age, BMI, and levels of high-density lipoprotein cholesterol and triglycerides) and heritability (h2=0.83 [SE, 0.12], P<.001) of ADP-stimulated platelet aggregation during clopidogrel administration were very similar after a single 324-mg dose of aspirin was added. Addition of aspirin to clopidogrel resulted in potent inhibition of platelet aggregation in response to arachidonic acid, indicating effective inhibition of the cyclooxygenase pathway. CYP2C19*2 genotype had no effect on aspirin-induced inhibition of platelet aggregation of this pathway (eTable 3). However, the association observed between CYP2C19*2 genotype and ADP-stimulated platelet aggregation and clopidogrel persisted after administration of aspirin (P=7.9 × 10−13), suggesting that this drug combination, which is standard of care in patients with acute coronary syndromes and following PCI, does not overcome the effect of the CYP2C19*2 genotype on platelet function.
In addition to the CYP2C19*2 variant, Simon et al25 identified a variant in ABCB1 (GeneID 403879), which encodes a transporter that modulates clopidogrel absorption, to be associated with poorer clinical outcomes in patients receiving clopidogrel. In our study, this variant (rs1045642) was not associated with ADP-stimulated platelet aggregation at baseline or after clopidogrel treatment (P=.60).
Sinai Hospital of Baltimore Study
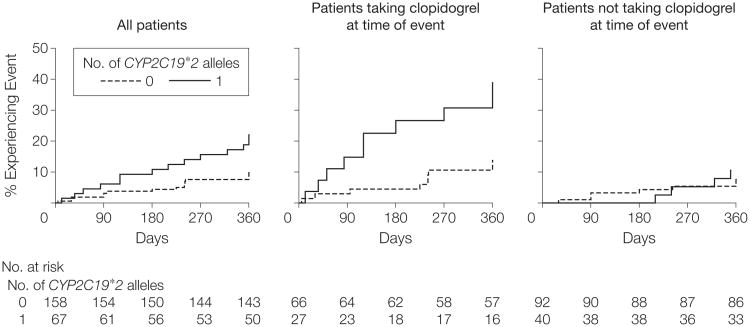
We next sought to replicate and extend our findings in a group of individuals from the general US population with a clinical indication for clopidogrel who underwent PCI and were recruited from a cardiac catheterization laboratory in Baltimore. Characteristics of these participants are shown in Table 1. Similar to the Amish PAPI Study group, those with the CYP2C19*2 genotype had no difference in baseline platelet aggregation (P=.58) but demonstrated greater residual platelet aggregation after clopidogrel therapy (P=.02) compared with those without the loss-of-function allele (Figure 3). After 1 year of follow-up, carriers of the CYP2C19*2 genotype had higher cardiovascular event rates compared with noncarriers (20.9% vs 10.0%; hazard ratio [HR], 2.42; 95% CI, 1.18-4.99; P=.02) (Figure 4). The increased event rate conferred by the CYP2C19*2 genotype was limited to the 95 participants still taking clopidogrel when the event occurred (HR, 3.40; 95% CI, 1.36-8.46; P=.004), with no increase in event rate in those not taking clopidogrel when the event occurred or at 1 year of follow-up (HR, 1.39; 95% CI, 0.39-4.88; P=.60). Inclusion of ADP-stimulated platelet aggregation as a covariate in the regression model markedly reduced the relation between the CYP2C19*2 genotype and cardiovascular outcome (HR, 1.58; 95% CI, 0.68-3.66; P=.29), suggesting that the genotype effect on clinical outcomes is mediated through decreased inhibition of platelet function.
Figure 4. Event-Free Survival Over 1 Year of Follow-up in Sinai Hospital of Baltimore Patients Treated With Clopidogrel Following Percutaneous Coronary Intervention, Stratified by CYP2C19*2 Genotype.
Postdischarge ischemic events were defined as myocardial infarction (the occurrence of ischemic symptoms and a troponin I value greater than upper limits of normal), ischemic stroke, stent thrombosis (definite stent thrombosis according to the Academic Research Consortium criteria29), unplanned target vessel revascularization (revascularization of vessel treated at time of study enrollment), unplanned nontarget vessel revascularization (revascularization of a vessel different from that treated at time of study enrollment), hospitalization for coronary ischemia without revascularization (hospitalization for chest pain with evidence of ischemia on electrocardiogram and no evidence of myocardial infarction as measured by troponin I value), and death secondary to any cardiovascular cause. Patients were further stratified into those who were taking clopidogrel when the event occurred or at 1 year of follow-up and those who were not. All analyses adjusted for age, sex, and race. For all patients, hazard ratio (HR)=2.42 (95% confidence interval [CI], 1.18-4.99; P=.02); for patients taking clopidogrel at time of event, HR=3.40 (95% CI, 1.36-8.46; P=.004); for patients not taking clopidogrel at time of event, HR=1.39 (95% CI, 0.39-4.88; P=.60).
Comment
Our study in a relatively large healthy drug-naive population indicates that clopidogrel response as defined by ADP-stimulated platelet aggregation is a normally distributed trait, with no clear cutoff to define resistance. Others have reported similar findings in patient populations with wide variation in atherosclerotic burden, medication usage, concurrent illnesses, and compliance—all potential confounding influences affecting clopidogrel response. Thus, clopidogrel response is likely caused by multiple factors, including potentially genetic factors. We found that increased age, BMI, and triglyceride levels and decreased levels of high-density lipoprotein cholesterol are predictors of poorer clopidogrel response; however, these factors combined account for less than 10% of the variation, suggesting other, undiscovered factors that contribute to variation in clopidogrel response.
Since all Amish persons are related, we were uniquely able to estimate heritability of clopidogrel response by determining the extent of variation in ADP-stimulated platelet aggregation that could be attributed to familial relatedness. Clopidogrel response was highly heritable. Indeed, in an agnostic genome-wide association analysis, we found compelling evidence for a major locus on chromosome 10q24 that influences clopidogrel response. This locus extends across the CYP2C18– CYP2C19–CYP2C9–CYP2C8 gene cluster. CYP2C19 was a strong biological candidate because this enzyme is a key activator of the antiplatelet function of clopidogrel. Indeed, follow-up genotyping indicated that the common loss-of-function CYP2C19*2 variant was associated with clopidogrel response and could account for most of the association signal detected in the initial genome-wide association study. The CYP2C19*2 genotype accounts for approximately 12% of the variation in clopidogrel response. With age, BMI, and lipid levels, approximately 22% of the variation in clopidogrel response can be explained.
Although substantial and highly significant, the majority of the variation in platelet response to clopidogrel remains unexplained. Presumably, other unmeasured factors that influence clopidogrel response (including potentially additional genetic variants, given the high heritability estimate) remain to be identified. CYP2C19*2 genotype was not associated with preclopidogrel platelet aggregation measures, and the postclopidogrel measures were associated with genotype even after adjusting for preclopidogrel measures, indicating a true association with clopidogrel response. Due to linkage disequilibrium across this region, we can not rule out that a variant other than CYP2C19*2 may be causative, but this is unlikely in light of the unequivocal effect of this mutation on enzyme production. Furthermore, it is possible that other variants in CYP2C19 or other CYP2C genes at this locus may also contribute to clopidogrel response.
Our study is unique in that we used an agnostic genome-wide approach. In our genome-wide association study, we detected no other region associated with clopidogrel response at or exceeding a genome-wide level of statistical significance, suggesting that common variants in other parts of the genome with similar or greater effect size are unlikely. However, we cannot rule out the possibility that common SNPs or other types of variants (eg, copy number variants, insertions/deletions), or rare variants with large effect size not tagged by those SNPs genotyped, may have been missed.
Genome-wide association studies are prone to false-positive results, owing to the large number of statistical tests performed. It is unlikely that our finding represents a false-positive result, because we replicated the association between CYP2C19*2 genotype and ADP-stimulated platelet aggregation in an independent sample. In addition to the replication of the initial association between CYP2C19*2 genotype and clopidogrel response, the Baltimore Sinai Hospital cohort demonstrates generalizability to an outbred population with significant atherosclerotic disease. Importantly, the association between CYP2C19*2 genotype and cardiovascular event–free survival in this high-risk population provides evidence for clinical relevance. A limitation is that the Sinai Hospital cohort was a mixed population that received differing regimens of antiplatelet agents in the acute setting (1-3 days). However, our stratified analyses did not detect any significant effect of acute management on the longer-term outcomes investigated. Furthermore, we cannot rule out the possibility that a subset of patients were nonadherent to clopidogrel. Our data show that there is no relationship between CYP2C19 genotype and platelet aggregation in the absence of clopidogrel. Thus, the differences in platelet aggregation we observed between genotypes once clopidogrel treatment was initiated strongly suggest that a large proportion of the population did indeed adhere to the clopidogrel regimen. Lastly, owing to limitations in sample size, we could not determine if CYP2C19 genotype was related to adverse bleeding events.
Using a candidate gene approach, Hulot et al22 were the first to report an association between CYP2C19*2 genotype and ADP-stimulated platelet aggregation in response to clopidogrel. More recently, CYP2C19*2 genotype was associated with poorer clinical outcomes in patients with coronary artery disease treated with clopidogrel and aspirin.18,24-26 In patients taking clopidogrel and aspirin following myocardial infarction, those carrying the CYP2C19*2 genotype were more likely to experience a second cardiovascular event, with an HR (3.69) similar to that found in our study.24 In the Therapeutic Outcomes by Optimizing Platelet Inhibition With Prasugrel–Thrombolysis in Myocardial Infarction (TRITON-TIMI) 38 trial, patients carrying the CYP2C19*2 genotype had lower plasma levels of the active metabolite of clopidogrel, reduced maximal platelet aggregation in response to clopidogrel, and a 53% higher composite primary efficacy outcome of the risk of cardiovascular events and death.18 In a nationwide French registry of patients with acute myocardial infarction treated with clopidogrel, poorer outcomes were observed in patients who carried 2 CYP2C19 loss-of-function alleles (HR, 1.98).25 Unlike our study and the other studies, heterozygous persons from this French registry did not appear to have a significantly increased event rate, suggesting a more modest effect of the genotype on clopidogrel response in these patients.
The loss-of-function CYP2C19*2 genotype is common in diverse populations. In white populations, approximately 24% have at least 1 CYP2C19*2 allele. The frequency of this allele is somewhat lower in Mexican Americans (≈ 18% with at least 1 CYP2C19*2 allele), higher in African Americans (≈33% with at least 1 copy), and markedly higher in Asian populations (≈51% with at least 1 copy).36-38 Thus, clopidogrel resistance due to this variant may be particularly important in Asian and African American populations. However, the strength of effect of CYP2C19*2 genotype on clopidogrel response may depend on other factors such as genetic background or environmental exposures, which may differ among ethnic groups. Unfortunately, our sample size was not sufficient to examine ethnicity-specific differences in CYP2C19 genotype effects on clopidogrel response. Additional studies in diverse populations will be necessary.
The commonly prescribed proton pump inhibitors omeprazole and esomeprazole, and some other medications commonly coprescribed with clopidogrel, such as cimetidine and fluoxitine, are potent inhibitors of CYP2C19, introducing the possibility that they may attenuate the antiplatelet activity of clopidogrel, especially in individuals heterozygous or homozygous for the CYP2C19*2 genotype.14,39-41 We could not test this hypothesis directly, because all participants in the Amish PAPI Study were drug-naive, and the number of participants taking proton pump inhibitors in the Sinai Hospital of Baltimore sample was too small.
CYP2C19 genotype may prove useful in helping clinicians choose the most effective antiplatelet therapy and dose for a given individual. Those with the CYP2C19*2 genotype may benefit more from an antiplatelet regimen that does not include clopidogrel, such as the third-generation thienopyridine prasugrel, or ticagrelor and cangrelor. Like clopidogrel, these agents inhibit ADP-stimulated platelet aggregation but are not as dependent on CYP2C19 for activation. Genotype-directed decisions regarding which antiplatelet agent to use in a specific patient may also have an important economic impact if costs of equally efficacious medications differ greatly. Whether CYP2C19*2 carriers may benefit from increased dosing of clopidogrel is not yet known.
Conclusions
We report the first genome-wide association study of clopidogrel response and show that the common loss-of-function CYP2C19*2 variant is a major determinant of ADP-stimulated platelet aggregation. Individuals with this genotype have reduced protection from clopidogrel in preventing cardiovascular disease–related events following PCI. Prospective randomized clinical trials will be necessary to determine the efficacy of CYP2C19 genotype–directed therapy in evidence-based clinical decision making.
Acknowledgments
Funding/Support: This study was supported by National Institutes of Health grants NIH U01 GM074518 and U01 HL084756, the Clinical Nutrition Research Unit of Maryland (P30 DK072488), the University of Maryland General Clinical Research Center (M01 RR 16500), the Baltimore Veterans Administration Geriatric Research and Education Clinical Center, and Sinai Hospital of Baltimore.
Role of the Sponsors: The funding organizations had no role in the design and conduct of the study; the collection, analysis, and interpretation of the data; or the preparation, review, or approval of the manuscript.
Footnotes
Author Contributions: Drs Shuldiner and Gurbel had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Shuldiner, Post, Faraday, Herzog, Gurbel.
Acquisition of data: Shuldiner, Bliden, Gandhi, Horenstein, Damcott, Pakyz, Tantry, Herzog, Gurbel.
Analysis and interpretation of data: Shuldiner, O'Connell, Bliden, Ryan, Damcott, Tantry, Gibson, Pollin, Post, Parsa, Mitchell, Faraday, Herzog, Gurbel.
Drafting of the manuscript: Shuldiner, Bliden, Tantry, Herzog, Gurbel.
Critical revision of the manuscript for important intellectual content: Shuldiner, O'Connell, Gandhi, Ryan, Horenstein, Damcott, Pakyz, Tantry, Gibson, Pollin, Post, Parsa, Mitchell, Faraday, Herzog, Gurbel.
Statistical analysis: O'Connell, Bliden, Mitchell, Gurbel.
Obtained funding: Shuldiner, Gurbel.
Administrative, technical, or material support: Shuldiner, Bliden, Ryan, Damcott, Pakyz, Tantry, Gibson, Herzog, Gurbel.
Study supervision: Shuldiner, Bliden, Faraday, Gurbel.
Financial Disclosures: Dr Faraday reported that he is a coinventor of a patent application on novel anti-thrombotic agents and their methods of use. Dr Gurbel reported receiving grant support from Schering-Plough, AstraZeneca, Bayer Healthcare, Sanofi-Aventis, Portola Pharmaceuticals, Daiichi-Sankyo, and Lilly; and receiving honoraria/consulting income from Schering-Plough, AstraZeneca, Bayer Healthcare, Sanofi-Aventis, Portola Pharmaceuticals, Daiichi-Sankyo, Lilly, and Pozen. No other authors reported disclosures.
Additional Information: eMethods, eTables 1 through 3, and eFigures 1 and 2 are available at http://www.jama.com.
Additional Contributions: We gratefully acknowledge our Amish liaisons and field workers and the extraordinary cooperation and support of the Amish community, without which these studies would not have been possible.
Contributor Information
Dr Alan R. Shuldiner, Division of Endocrinology, Diabetes and Nutrition, Department of Medicine, University of Maryland School of Medicine, Baltimore; Geriatric Research and Education Clinical Center, Veterans Administration Medical Center, Baltimore.
Dr Jeffrey R. O'Connell, Division of Endocrinology, Diabetes and Nutrition, Department of Medicine, University of Maryland School of Medicine, Baltimore.
Mr Kevin P. Bliden, Sinai Center for Thrombosis Research, Sinai Hospital of Baltimore, Baltimore.
Dr Amish Gandhi, Division of Endocrinology, Diabetes and Nutrition, Department of Medicine, University of Maryland School of Medicine, Baltimore.
Ms Kathleen Ryan, Division of Endocrinology, Diabetes and Nutrition, Department of Medicine, University of Maryland School of Medicine, Baltimore.
Dr Richard B. Horenstein, Division of Endocrinology, Diabetes and Nutrition, Department of Medicine, University of Maryland School of Medicine, Baltimore.
Dr Coleen M. Damcott, Division of Endocrinology, Diabetes and Nutrition, Department of Medicine, University of Maryland School of Medicine, Baltimore.
Ms Ruth Pakyz, Division of Endocrinology, Diabetes and Nutrition, Department of Medicine, University of Maryland School of Medicine, Baltimore.
Dr Udaya S. Tantry, Sinai Center for Thrombosis Research, Sinai Hospital of Baltimore, Baltimore.
Mr Quince Gibson, Division of Endocrinology, Diabetes and Nutrition, Department of Medicine, University of Maryland School of Medicine, Baltimore.
Dr Toni I. Pollin, Division of Endocrinology, Diabetes and Nutrition, Department of Medicine, University of Maryland School of Medicine, Baltimore.
Dr Wendy Post, Division of Cardiology, Department of Medicine Johns Hopkins University School of Medicine, Baltimore.
Dr Afshin Parsa, Division of Nephrology, Department of Medicine, University of Maryland School of Medicine, Baltimore.
Dr Braxton D. Mitchell, Division of Endocrinology, Diabetes and Nutrition, Department of Medicine, University of Maryland School of Medicine, Baltimore.
Dr Nauder Faraday, Department of Anesthesiology and Critical Care Medicine, Johns Hopkins University School of Medicine, Baltimore.
Dr William Herzog, Sinai Center for Thrombosis Research, Sinai Hospital of Baltimore, Baltimore; Division of Cardiology, Department of Medicine Johns Hopkins University School of Medicine, Baltimore.
Dr Paul A. Gurbel, Sinai Center for Thrombosis Research, Sinai Hospital of Baltimore, Baltimore.
References
- 1.Antman EM, Hand M, Armstrong PW, et al. Canadian Cardiovascular Society; American Academy of Family Physicians; American College of Cardiology; American Heart Association. 2007 focused update of the ACC/AHA 2004 guidelines for the management of patients with ST-elevation myocardial infarction: a report of the American College of Cardiology/ American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol. 2008;51(2):210–247. doi: 10.1016/j.jacc.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 2.King SB, III, Smith SC, Jr, Hirshfeld JW, Jr, et al. 2005 WRITING COMMITTEE MEMBERS. 2007 Focused Update of the ACC/AHA/SCAI 2005 Guideline Update for Percutaneous Coronary Intervention: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines: 2007 Writing Group to Review New Evidence and Update the ACC/AHA/SCAI 2005 Guideline Update for Percutaneous Coronary Intervention, Writing on Behalf of the 2005 Writing Committee. Circulation. 2008;117(2):261–295. doi: 10.1161/CIRCULATIONAHA.107.188208. [DOI] [PubMed] [Google Scholar]
- 3.Gurbel PA, Tantry US. Clopidogrel resistance? Thromb Res. 2007;120(3):311–321. doi: 10.1016/j.thromres.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 4.Hollopeter G, Jantzen HM, Vincent D, et al. Identification of the platelet ADP receptor targeted by antithrombotic drugs. Nature. 2001;409(6817):202–207. doi: 10.1038/35051599. [DOI] [PubMed] [Google Scholar]
- 5.Savi P, Pereillo JM, Uzabiaga MF, et al. Identification and biological activity of the active metabolite of clopidogrel. Thromb Haemost. 2000;84(5):891–896. [PubMed] [Google Scholar]
- 6.Angiolillo DJ, Fernandez-Ortiz A, Bernardo E, et al. Variability in individual responsiveness to clopidogrel: clinical implications, management, and future perspectives. J Am Coll Cardiol. 2007;49(14):1505–1516. doi: 10.1016/j.jacc.2006.11.044. [DOI] [PubMed] [Google Scholar]
- 7.Gurbel PA, Bliden KP, Hiatt BL, O'Connor CM. Clopidogrel for coronary stenting: response variability, drug resistance, and the effect of pretreatment platelet reactivity. Circulation. 2003;107(23):2908–2913. doi: 10.1161/01.CIR.0000072771.11429.83. [DOI] [PubMed] [Google Scholar]
- 8.Gurbel PA, Tantry US. Drug insight: clopidogrel nonresponsiveness. Nat Clin Pract Cardiovasc Med. 2006;3(7):387–395. doi: 10.1038/ncpcardio0602. [DOI] [PubMed] [Google Scholar]
- 9.Angiolillo DJ, Alfonso F. Platelet function testing and cardiovascular outcomes: steps forward in identifying the best predictive measure. Thromb Haemost. 2007;98(4):707–709. [PubMed] [Google Scholar]
- 10.Bliden KP, Dichiara J, Tantry US, Bassi AK, Chaganti SK, Gurbel PA. Increased risk in patients with high platelet aggregation receiving chronic clopidogrel therapy undergoing percutaneous coronary intervention: is the current antiplatelet therapy adequate? J Am Coll Cardiol. 2007;49(6):657–666. doi: 10.1016/j.jacc.2006.10.050. [DOI] [PubMed] [Google Scholar]
- 11.Buonamici P, Marcucci R, Migliorini A, et al. Impact of platelet reactivity after clopidogrel administration on drug-eluting stent thrombosis. J Am Coll Cardiol. 2007;49(24):2312–2317. doi: 10.1016/j.jacc.2007.01.094. [DOI] [PubMed] [Google Scholar]
- 12.Gurbel PA, Bliden KP, Guyer K, et al. Platelet reactivity in patients and recurrent events post-stenting: results of the PREPARE POST-STENTING Study. J Am Coll Cardiol. 2005;46(10):1820–1826. doi: 10.1016/j.jacc.2005.07.041. [DOI] [PubMed] [Google Scholar]
- 13.Farid NA, Small DS, Payne CD, et al. Effect of a torvastatin on the pharmacokinetics and pharmacodynamics of prasugrel and clopidogrel in healthy subjects. Pharmacotherapy. 2008;28(12):1483–1494. doi: 10.1592/phco.28.12.1483. [DOI] [PubMed] [Google Scholar]
- 14.Gilard M, Arnaud B, Le Gal G, Abgrall JF, Boschat J. Influence of omeprazol on the antiplatelet action of clopidogrel associated to aspirin. J Thromb Haemost. 2006;4(11):2508–2509. doi: 10.1111/j.1538-7836.2006.02162.x. [DOI] [PubMed] [Google Scholar]
- 15.Lau WC, Gurbel PA, Watkins PB, et al. Contribution of hepatic cytochrome P450 3A4 metabolic activity to the phenomenon of clopidogrel resistance. Circulation. 2004;109(2):166–171. doi: 10.1161/01.CIR.0000112378.09325.F9. [DOI] [PubMed] [Google Scholar]
- 16.Siller-Matula JM, Lang I, Christ G, Jilma B. Calcium-channel blockers reduce the antiplatelet effect of clopidogrel. J Am Coll Cardiol. 2008;52(19):1557–1563. doi: 10.1016/j.jacc.2008.07.055. [DOI] [PubMed] [Google Scholar]
- 17.Kim KA, Park PW, Hong SJ, Park JY. The effect of CYP2C19 polymorphism on the pharmacokinetics and pharmacodynamics of clopidogrel: a possible mechanism for clopidogrel resistance. Clin Pharmacol Ther. 2008;84(2):236–242. doi: 10.1038/clpt.2008.20. [DOI] [PubMed] [Google Scholar]
- 18.Mega JL, Close SL, Wiviott SD, et al. Cytochrome P-450 polymorphisms and response to clopidogrel. N Engl J Med. 2009;360(4):354–362. doi: 10.1056/NEJMoa0809171. [DOI] [PubMed] [Google Scholar]
- 19.Brandt JT, Close SL, Iturria SJ, et al. Common polymorphisms of CYP2C19 and CYP2C9 affect the pharmacokinetic and pharmacodynamic response to clopidogrel but not prasugrel. J Thromb Haemost. 2007;5(12):2429–2436. doi: 10.1111/j.1538-7836.2007.02775.x. [DOI] [PubMed] [Google Scholar]
- 20.Fontana P, Hulot JS, De Moerloose P, Gaussem P. Influence of CYP2C19 and CYP3A4 gene polymorphisms on clopidogrel responsiveness in healthy subjects. J Thromb Haemost. 2007;5(10):2153–2155. doi: 10.1111/j.1538-7836.2007.02722.x. [DOI] [PubMed] [Google Scholar]
- 21.Frere C, Cuisset T, Morange PE, et al. Effect of cytochrome p450 polymorphisms on platelet reactivity after treatment with clopidogrel in acute coronary syndrome. Am J Cardiol. 2008;101(8):1088–1093. doi: 10.1016/j.amjcard.2007.11.065. [DOI] [PubMed] [Google Scholar]
- 22.Hulot JS, Bura A, Villard E, et al. Cytochrome P450 2C19 loss-of-function polymorphism is a major determinant of clopidogrel responsiveness in healthy subjects. Blood. 2006;108(7):2244–2247. doi: 10.1182/blood-2006-04-013052. [DOI] [PubMed] [Google Scholar]
- 23.Malek LA, Kisiel B, Spiewak M, et al. Coexisting polymorphisms of P2Y12 and CYP2C19 genes as a risk factor for persistent platelet activation with clopidogrel. Circ J. 2008;72(7):1165–1169. doi: 10.1253/circj.72.1165. [DOI] [PubMed] [Google Scholar]
- 24.Collet JP, Hulot JS, Pena A, et al. Cytochrome P450 2C19 polymorphism in young patients treated with clopidogrel after myocardial infarction: a cohort study. Lancet. 2009;373(9660):309–317. doi: 10.1016/S0140-6736(08)61845-0. [DOI] [PubMed] [Google Scholar]
- 25.Simon T, Verstuyft C, Mary-Krause M, et al. French Registry of Acute ST-Elevation and Non-ST-Elevation Myocardial Infarction (FAST-MI) Investigators. Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med. 2009;360(4):363–375. doi: 10.1056/NEJMoa0808227. [DOI] [PubMed] [Google Scholar]
- 26.Trenk D, Hochholzer W, Fromm MF, et al. Cytochrome P450 2C19 681G>A polymorphism and high on-clopidogrel platelet reactivity associated with adverse 1-year clinical outcome of elective percutaneous coronary intervention with drug-eluting or baremetal stents. J Am Coll Cardiol. 2008;51(20):1925–1934. doi: 10.1016/j.jacc.2007.12.056. [DOI] [PubMed] [Google Scholar]
- 27.ESPRIT Investigators. Novel dosing regimen of eptifibatide in planned coronary stent implantation (ESPRIT): a randomised, placebo-controlled trial. Lancet. 2000;356(9247):2037–2044. doi: 10.1016/S0140-6736(00)03400-0. [DOI] [PubMed] [Google Scholar]
- 28.Lincoff AM, Bittl JA, Harrington RA, et al. REPLACE-2 Investigators. Bivalirudin and provisional glycoprotein IIb/IIIa blockade compared with heparin and planned glycoprotein IIb/IIIa blockade during percutaneous coronary intervention: REPLACE-2 randomized trial. JAMA. 2003;289(7):853–863. doi: 10.1001/jama.289.7.853. [DOI] [PubMed] [Google Scholar]
- 29.Applegate RJ, Sacrinty MT, Little WC, Santos RM, Gandhi SK, Kutcher MA. Incidence of coronary stent thrombosis based on academic research consortium definitions. Am J Cardiol. 2008;102(6):683–688. doi: 10.1016/j.amjcard.2008.04.050. [DOI] [PubMed] [Google Scholar]
- 30.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62(5):1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agarwala R, Biesecker LG, Hopkins KA, Francomano CA, Schäffer AA. Software for constructing and verifying pedigrees within large genealogies and an application to the Old Order Amish of Lancaster County. Genome Res. 1998;8(3):211–221. doi: 10.1101/gr.8.3.211. [DOI] [PubMed] [Google Scholar]
- 32.Brandt JT, Payne CD, Wiviott SD, et al. A comparison of prasugrel and clopidogrel loading doses on platelet function: magnitude of platelet inhibition is related to active metabolite formation. Am Heart J. 2007;153(1):66.e9–66.e16. doi: 10.1016/j.ahj.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 33.Takahashi M, Pang H, Kawabata K, Farid NA, Kurihara A. Quantitative determination of clopidogrel active metabolite in human plasma by LC-MS/MS. J Pharm Biomed Anal. 2008;48(4):1219–1224. doi: 10.1016/j.jpba.2008.08.020. [DOI] [PubMed] [Google Scholar]
- 34.Umemura K, Furuta T, Kondo K. The common gene variants of CYP2C19 affect pharmacokinetics and pharmacodynamics in an active metabolite of clopidogrel in healthy subjects. J Thromb Haemost. 2008;6(8):1439–1441. doi: 10.1111/j.1538-7836.2008.03050.x. [DOI] [PubMed] [Google Scholar]
- 35.Wallentin L, Varenhorst C, James S, et al. Prasugrel achieves greater and faster P2Y12 receptor-mediated platelet inhibition than clopidogrel due to more efficient generation of its active metabolite in aspirin-treated patients with coronary artery disease. Eur Heart J. 2008;29(1):21–30. doi: 10.1093/eurheartj/ehm545. [DOI] [PubMed] [Google Scholar]
- 36.Luo HR, Poland RE, Lin KM, Wan YJ. Genetic polymorphism of cytochrome P450 2C19 in Mexican Americans: a cross-ethnic comparative study. Clin Pharmacol Ther. 2006;80(1):33–40. doi: 10.1016/j.clpt.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 37.Xiao ZS, Goldstein JA, Xie HG, et al. Differences in the incidence of the CYP2C19 polymorphism affecting the S-mephenytoin phenotype in Chinese Han and Bai populations and identification of a new rare CYP2C19 mutant allele. J Pharmacol Exp Ther. 1997;281(1):604–609. [PubMed] [Google Scholar]
- 38.Takakubo F, Kuwano A, Kondo I. Evidence that poor metabolizers of (S)-mephenytoin could be identified by haplotypes of CYP2C19 in Japanese. Pharmacogenetics. 1996;6(3):265–267. doi: 10.1097/00008571-199606000-00011. [DOI] [PubMed] [Google Scholar]
- 39.Gurbel PA, Lau WC, Tantry US. Omeprazole: a possible new candidate influencing the antiplatelet effect of clopidogrel. J Am Coll Cardiol. 2008;51(3):261–263. doi: 10.1016/j.jacc.2007.07.090. [DOI] [PubMed] [Google Scholar]
- 40.Ho PM, Maddox TM, Wang L, et al. Risk of adverse outcomes associated with concomitant use of clopidogrel and proton pump inhibitors following acute coronary syndrome. JAMA. 2009;301(9):937–944. doi: 10.1001/jama.2009.261. [DOI] [PubMed] [Google Scholar]
- 41.Small DS, Farid NA, Li YG, et al. Effect of ranitidine on the pharmacokinetics and pharmacodynamics of prasugrel and clopidogrel. Curr Med Res Opin. 2008;24(8):2251–2257. doi: 10.1185/03007990802205985. [DOI] [PubMed] [Google Scholar]