Abstract
Nanos is a translational regulator required for the survival and maintenance of primordial germ cells during embryogenesis. Three nanos homologs are present in the genome of the sea urchin Strongylocentrotus purpuratus (Sp), and each nanos mRNA accumulates specifically in the small micromere (sMic) lineage. We found that a highly conserved element in the 3’ UTR of nanos2 is sufficient for reporter expression selectively in the sMic lineage: microinjection into a Sp fertilized egg of an RNA that contains the GFP open reading frame followed by Sp nanos2 3’UTR leads to selective reporter enrichment in the small micromeres in blastulae. The same result was seen with nanos2 from the sea urchin Hemicentrotus pulcherrimus (Hp). In both species, the 5’UTR alone is not sufficient for the sMic localization but it always increased the sMic reporter enrichment when present with the 3’UTR. We defined an element conserved between Hp and Sp in the nanos2 3’UTR which is necessary and sufficient for protein enrichment in the sMic, and refer to it as GNARLE (Global Nanos Associated RNA Lability Element). We also found that the nanos2 3’UTR is essential for the selective RNA retention in the small micromeres; GNARLE is required but not sufficient for this process. These results show that a combination of selective RNA retention and translational control mechanisms instills nanos accumulation uniquely in the sMic lineage.
Keywords: nanos, sea urchin, small micromeres, germ line, RNA retention, translational regulation
Introduction
Nanos is a RNA-binding protein containing two CCHC zinc-fingers, and was first described as a translational repressor in Drosophila (Cho et al., 2006; Irish et al., 1989). Although the nanos sequence is not highly conserved, nanos orthologs have been found in the germ line of all animals tested (e.g. C. elegans (Kraemer et al., 1999), Xenopus (Lai et al., 2011) and planarians (Wang et al., 2007)). Translational repression by nanos is mediated through interaction with pumilio, which binds RNAs containing a conserved motif in their 3’UTR, the Nanos Response Element (NRE; (Sonoda and Wharton, 1999; Wharton and Struhl, 1991). This function of nanos is involved in the regulation of various developmental processes; it was first characterized in Drosophila to regulate the differentiation of the anterior-posterior body axis through translational repression of the gap gene hunchback (Wang and Lehmann, 1991), and later shown to be required also for the continued production of egg chambers during oogenesis (Wang et al., 1994) and for primordial germ cell migration (Forbes and Lehmann, 1998). Nanos is required in both the male and female germ line of Drosophila; in the nanos mutant males, spermatogenesis is progressively affected and these males become sterile (Bhat, 1999). Similarly, nanos regulates primordial germ cell development and survival in C. elegans (Subramaniam and Seydoux, 1999), sea urchins (Juliano et al., 2010), zebrafish (Koprunner et al., 2001) and mice (Tsuda et al., 2003). In addition to these conserved functions in the germ line, nanos also functions in other multipotent cells. For example, the nanos related gene Cnnos1 in Hydra magnipapillata, is expressed in both multipotent stem cells and germ-line cells, but not in somatic cells (Mochizuki et al., 2000). In the polychaete annelid, Platynereis dumerilii, and the snail Ilyanassa obsoleta, multipotent cells of the embryos expressed nanos (Rabinowitz et al., 2008; Rebscher et al., 2007). Moreover, nanos has other functions in development e.g. in the Drosophila peripheral nervous system, in the dendritic arborization (da) neurons to maintain dendrite complexity (Brechbiel and Gavis, 2008), and at the larval neuro-muscular junction (Menon et al., 2009).
The expression of germ-line determination genes is highly regulated, and ectopic expression of these genes often induces cell cycle and developmental defects (Luo et al., 2011; Wu and Ruvkun, 2010). In Drosophila, C.elegans, zebrafish, and mouse, translation of nanos in the germ line requires its 3’ Untranslated Region (UTR) (D'Agostino et al., 2006; Gavis et al., 1996b; Koprunner et al., 2001; Saito et al., 2006; Suzuki et al., 2010). In Drosophila, the nanos 3’UTR mediates its RNA localization to the posterior pole of the syncytial embryo (Gavis et al., 1996a) and the protein rumpelstiltskin (rump), an heterogeneous nuclear ribonucleoprotein (hnRNP), binds nanos RNA directly to regulate its localization (Jain and Gavis, 2008). More recently, the argonaute family member, aubergine (aub) was also found to be a nanos RNA localization factor, independent of its function in RNA silencing. Aub interacts with nanos mRNA in vivo and co-purifies with rump in an RNA-dependent manner (Becalska et al., 2011). This nanos RNA localization element includes a 90 nucleotide translational control element (TCE) which mediates its translational repression (Gavis et al., 1996b) by forming two stem-loops which act independently of each other to repress translation at different times in development. Smaug and Glorund bind to each of the stem loop to control its translation, respectively, during oogenesis (Kalifa et al., 2006) and in embryogenesis (Smibert et al., 1996). Similarly, in C. elegans, nanos2 is translationally regulated by two independent stem loops in its 3’UTR (D'Agostino et al., 2006). Thus, nanos protein translation is highly safeguarded and may reflect its toxicity outside of the germ line, or multipotent cell environment.
Three nanos homologs are present in the genome of the sea urchin, Strongylocentrotus purpuratus (Sp), and each of them are expressed with differential timing in the small micromeres (Juliano et al., 2010), cells that contribute to the germ line (Yajima and Wessel, 2011). These cells are formed during embryogenesis at the 32-cell stage after two unequal cleavage divisions. In the blastula, the small micromeres reside at the vegetal plate where they divide once before being transported to the tip of the archenteron during gastrulation. The eight small micromere descendants are then partitioned into the left and right coelomic pouches, with the adult rudiment forming on the left side in the larva. Nanos1 and nanos2 are the first de novo mRNAs and proteins to accumulate in the sMics, and in both S.purpuratus and Hemicentrotus pulcherrimus (Hp), are required for adult rudiment formation (Fujii et al., 2009). In this study, we found that at least part of the mechanism for nanos2 RNA selective accumulation in the sMics stems from a post-transcriptional step of rapid mRNA turnover in all cells of the embryo, except the sMics, and that this essential information results from an RNA element in the 3’UTR of the mRNA that is highly conserved over the ~20 million years separating the last common ancestor of S. purpuratus and H.pulcherrimus. This element leads to mRNA turnover in all cells except the sMics, and is independent of miRNA – mediated decay.
Material and Methods
Animals
Strongylocentrotus purpuratus adults were housed in aquaria with artificial seawater (ASW) at 16 C (Coral Life Scientific Grade Marine Salt; Carson, CA). Gametes were acquired by either 0.5M KCl injection or by shaking. Eggs were collected in ASW or filtered seawater and sperm was collected dry. Embryos were cultured in filtered seawater at 16 C. Hemicentrotus pulcherrimus were harvested from Seto inland sea or from Tateyama Bay and their gametes were obtained by coelomic injection of 0.55 M KCl. Fertilized eggs were cultured in filtered sea water (FSW) containing 50 µg/ml of streptomycin sulfate and 100 µg/ml of penicillin G potassium at 16°C.
Plasmid constructions
For the GFP construct with Hp nanos2 UTRs (Full length), Hp nanos2 5’ and 3’UTRs were amplified using the primers described in Supplemental figure S1A (Fujii et al., 2006). These Hp nanos2 UTRs were subcloned into pGreenLantern2-derived plasmid containing the GFP open reading frame and the T7 promoter. For 3’deletions, Hp nanos2 3’UTR was amplified from the full length using the primers presented in Supplemental figure S1B. For 5’deletions, Hp nanos2 3’UTR was amplified using the primers described in Supplemental figure S1C. For constructs A–C, Hp nanos2 3’UTR A–C regions were amplified using the primer sets (primers F for Δ7 and R for Δ4 to amplify region A; primers F for Δ8 and R for Δ4 to amplify region B; primers F for Δ7 and R for Δ5 to amplify region C). These PCR products were digested with XbaI and SalI, and inserted into the corresponding sites. For internal deletions of Hp nanos2 3’UTR, inverse PCR was carried out with 5’3’UTR-GFP using the primer sets presented in Supplemental figure S1D.
For the GFP constructs, Sp nanos2 5’ and 3’UTRs were amplified using the primers described in Supplemental figure S2A (Juliano et al., 2010). These Sp nanos2 UTRs were subcloned in a plasmid containing the GFP open reading frame, and the T7 promotor. Sp nanos2 3’UTR GNARLE region was amplified using the primers presented in Supplemental figure S2B, and inserted in the GFP containing plasmid. To make the Sp nanos2 3’UTR ΔGNARLE construct, two EcoRI restriction site were introduced in the UTR, at the beginning and at the end of GNARLE using the primers described in Supplemental figure S2C. Mutations were made using the QuickChange II Site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA). The plasmid was then digested by EcoRI to remove GNARLE, and then ligated. Sp nanos2 3’UTR NRRE was amplified using the primers presented in Supplemental figure S2D.
For the Renilla luciferase constructs, Sp nanos2 5’ and 3’UTRs were amplified (Supplemental figure S2E). These UTRs were then subcloned in a plasmid containing the Renilla luciferase open reading frame, and a SP6 promotor. The Sp nanos2 3’UTR GNARLE and ΔGNARLE were amplified from the corresponding GFP construct described above (Supplemental figure S2E).
In vitro RNA synthesis
Capped sense RNA were synthesized using the mMessage mMachine® T7 or Sp6 Kit (Ambion, Austin, TX) yielding RNA concentrations between 0.5 and 2µg/µl. Each RNA was co-injected with mCherry flanked with β-globin UTRs. Injection solutions contain: 20% glycerol, 1×1012 copies of a GFP RNA, 1.1012 copies of the mCherry RNA. Approximately 2 pl of each RNA mixture was injected into each fertilized egg.
Morpholino approach
The morpholino against dicer 5′ GGACTCGATGGTGGCTCATCCATTC 3′ was previously described (Song et al., 2011). Each embryo received approximately 24nM of the dicer morpholino.
Microinjections
Microinjections of zygotes were performed as previously described (Cheers and Ettensohn, 2004). In brief, eggs were de-jellied with acidic sea water (pH 5.0) for 10 min, washed with filtered sea water three times, rowed with a mouth pipette onto protamine sulfate-coated 60 × 15 mm petri dishes, fertilized in the presence of 1 mM 3-AT, and injected using the Femto Jet ® injection system (Eppendorf; Hamburg, Germany). 1 × 90 mm glass capillaries with filaments (Narishige; Tokyo, Japan) were pulled on a vertical needle puller for injections (Narishige; Tokyo, Japan). Injected embryos were cultured in sea water at 16 °C.
Whole mount RNA in situ hybridization (WMISH)
Antisense DIG-labeled RNA probes against GFP and mCherry were constructed using a DIG RNA labeling kit (Roche; Indianapolis, IN). WMISH experiments were performed as previously described (Minokawa et al., 2004) and the alkaline phosphatase reaction was carried out for 1h. All steps were carried out in 96-well round-bottom PVC plates (ThermoFisher Scientific; Rockford, IL). Samples were imaged on a Zeiss Axiovert 200M microscope equipped with a Zeiss color AxioCam MRc5 camera (Carl Zeiss, Inc.; Thornwood, NY).
Real-time quantitative PCR (QPCR)
RNA was extracted from 100 mock-injected and 100 Sp-nanos morpholino-injected embryos, collected at 24 hpf, using the RNeasy Micro Kit (Qiagen; Valencia, CA). cDNA was prepared using the TaqMan ® Reverse Transcription Reagents kit (Applied Biosystems; Foster City, CA). QPCR was performed on the 7300 Real-Time PCR system (Applied Biosystems; Foster City, CA) with the SYBR Green PCR Master Mix Kit (Applied Biosystems; Foster City, CA). Sp-nanos 2 primer set is described in (Juliano et al., 2006): F (5’-GCAAGAACAACGGAGAGAGC-3’) and R (5’- CCGCATAATGGACAGGTGTA-3’). 4 embryo equivalents were used as template. Experiments were run in triplicate and the data were normalized to ubiquitin RNA levels.
Reporter Fluorescence
Injected embryos were cultured as described above and samples were collected at indicated stages of development. Embryos were imaged on an LSM 510 laser scanning confocal microscope (Carl Zeiss, Inc.; Thornwood, NY).
Dual luciferase assay
Strongylocentrotus purpuratus fertilized eggs were injected, as described above, with a solution containing 1×1012 copies of a Renilla luciferase RNA, 1×1012 copies of a Firefly luciferase RNA, 20% glycerol, and 1mM Alexafluor 488-dextran to allow visualization of injected eggs. For each measurement, 100 injected embryos were collected at the blastula stage. Renilla and Firefly luminescence were measured using the Dual luciferase assay kit (Promega) in a Lumat LB 9501 luminometer (Berthold Technologies, Germany).
Results
The nanos2 3’UTR directs selective protein accumulation in the small micromeres
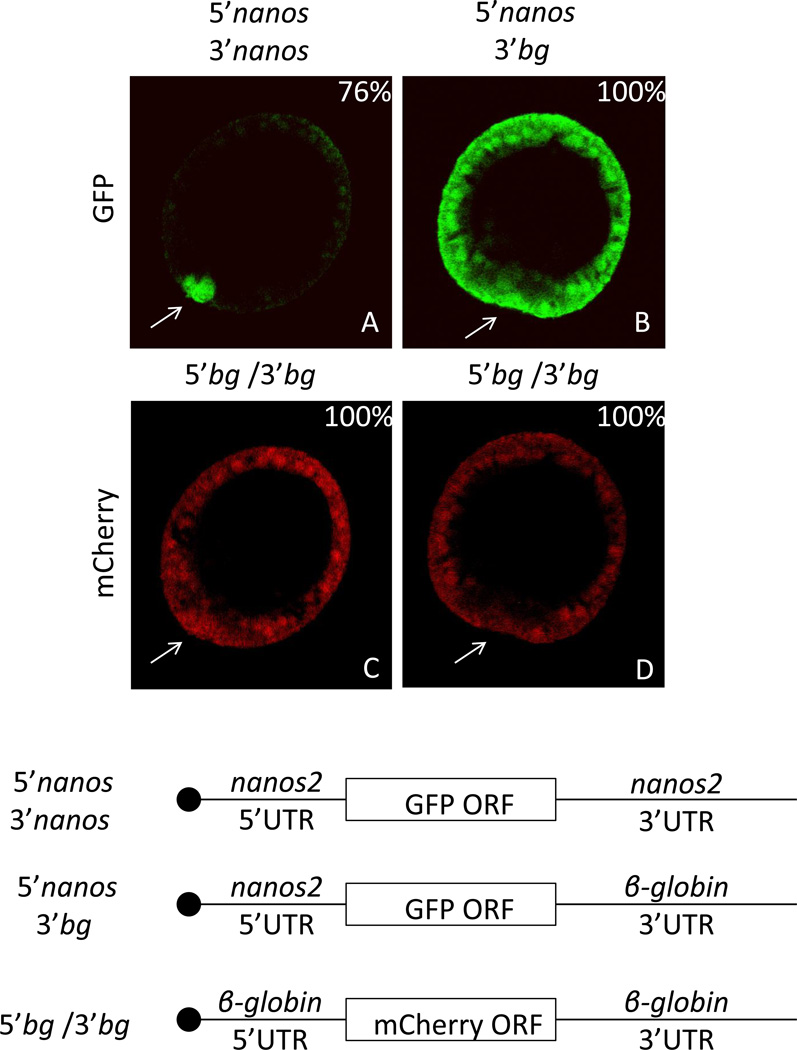
Transcripts and protein of nanos2 accumulate selectively in the small micromere lineage in two sea urchin species, H.pulcherrimus (Hp) and S.purpuratus (Sp) (Fujii et al., 2009; Juliano et al., 2010). In analyzing these mRNAs we found a region of the 3’UTR that was highly conserved between these two species. To test if the nanos2 3’UTRs were involved in this selective accumulation, two constructs were developed, both contained the Sp nanos2 5’UTR and GFP open reading frame (ORF). The Sp nanos2 3’UTR was fused to the first one, and the Xenopus β-globin 3’ UTR was fused to the second one (Figure 1). The corresponding RNAs were in vitro transcribed, and injected into Sp fertilized eggs. An RNA comprising mCherry ORF flanked by Xenopus β-globin UTRs was co-injected as a control reporter. At the mesenchyme blastula stage (approximately 24 hours post fertilization) the mCherry protein was found in all cells. In contrast, the co-injected GFP-encoding fluorescence was enriched in the small micromeres when it contained the Sp nanos2 5’ and 3’ UTRs. The GFP signal resulting from the Xenopus β-globin 3’UTR RNA, however, accumulated uniformly in all cells. The same observations were made by injecting Hp constructs in Hemicentrotus pulcherimus fertilized eggs (data not shown). These results indicate that the nanos2 UTRs are required for the selective translation of the encoded protein in the small micromeres.
Figure 1.
Sp nanos2 3’UTR is sufficient for the selective protein accumulation in the small micromeres. Synthetic mRNA containing the GFP open reading frame flanked by (A) Sp nanos2 5’ and 3’UTRs or (B) Sp nanos2 5’UTR and Xenopus β-globin 3’UTR were co-injected with mCherry mRNA (C and D) containing Xenopus β-globin 5’ and 3’UTRs in Sp fertilized eggs. GFP (green) and mCherry (red) fluorescence were assayed in the same embryos 24 hours post-fertilization at mesenchyme blastula: A and C represent the same embryo, B and D represent another one. For GFP images, A and B were obtained using the same settings (laser intensity, pin-hole opening). For mCherry images, C and D were also taken using the same settings. bg indicates β-globin UTRs. The arrows are pointing toward the small micromeres. The blastula are presented in the same orientation in the subsequent figures. Approximately one hundred blastulas were visualized after injection of each construct, the corresponding percentages of representative embryos are indicated in the right corner. Cartoons of the injected RNAs are presented at the bottom (the black circle representing the m7GTP cap).
A 357 nucleotide element located in the nanos2 3’UTR is sufficient for selective reporter accumulation in the small micromeres
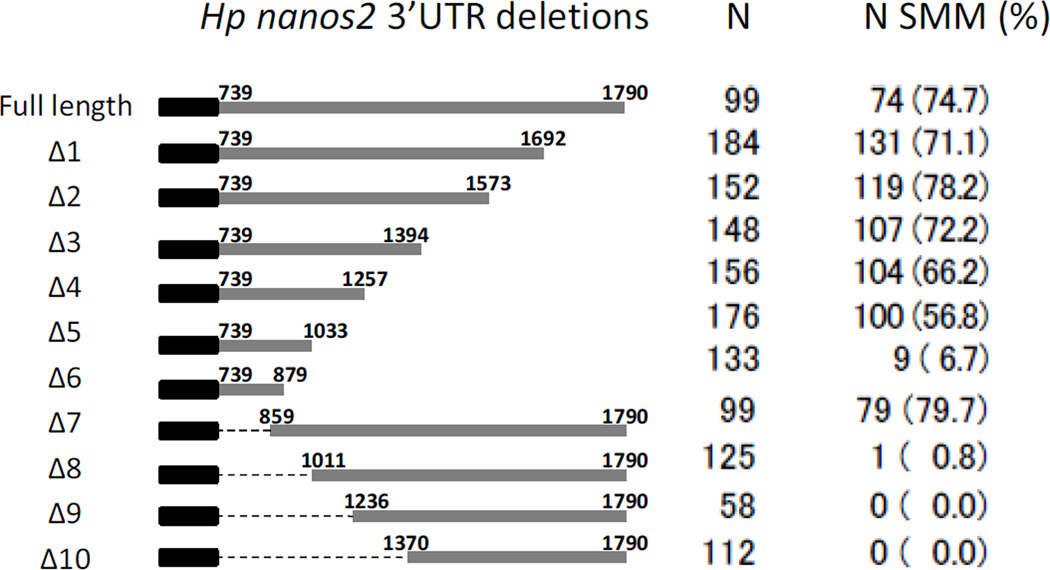
To test which part of the nanos2 3’UTR is important for the selective protein expression, ten deletion mutants were made using the Hp nanos2. All constructs contained the Hp nanos2 5’UTR, followed by GFP, and then by different regions of Hp nanos2 3’UTR. RNAs were injected in Hp fertilized eggs, and the GFP signal was monitored at the blastula stage (Figure 2). In Hp, the full length nanos2 3’UTR is 1051 nucleotide long, from nucleotide position 739 of the nanos2 transcript to 1790. Of the embryos injected with the RNA comprising the full length Hp nanos2 3’UTR, 74.7% had a selective GFP signal in the small micromeres at the blastula stage (Figure 2 Full length). Similar percentages were obtained with three 3’deletions (Δ1/2/3), and with one 5’deletion (Δ7), demonstrating that the last 396 nucleotides and the first 120 (859-739) nucleotides of Hp nanos2 3’UTRs were expendable for the selective protein expression. This percentage progressively decreased after injection of Δ4, Δ5, Δ6 and reached 0% in the Δ9 mutant. These results show that the sequence located between nucleotides 859 and 1236 in Hp nanos2 3’UTR is essential for the selective protein expression in the small micromeres. Moreover, the RNA with a 3’UTR containing only the nucleotides located between 859 and 1257, was sufficient to give a selective GFP expression in most of the injected embryos (Figure 3A).
Figure 2.
A proximal element in the Hp nanos2 3’UTR is required for selective protein accumulation in the small micromeres. Synthetic mRNAs were made using Hp nanos2 3’UTR and GFP followed by different deletions of Hp nanos2 3’UTR. These RNAs were injected into Hp fertilized eggs, and the numbers of injected embryos having a GFP signal enriched in the small micromeres at the blastula stage was monitored under the fluorescence scope. N indicates the number of injected embryos used for each RNA. N sMic indicates the number of injected embryos having a protein enrichment in the small micromeres, the corresponding percentages are represented in the parentheses (%).
Figure 3.
The nucleotides localized between the position 880 and 1236 of Hp nanos2 3’UTR are essential for protein enrichment in the small micromeres. Synthetic mRNAs were made using Hp nanos2 3’UTR and the GFP ORF followed by different deletions of Hp nanos2 3’UTR (A and B). These RNAs were injected in Hp fertilized eggs, and the numbers of injected embryos having a GFP signal enriched in the small micromeres at the blastula stage was monitored under the fluorescence microscope. N indicates the number of injected embryos used for each RNA construct. N sMic indicates the number of injected embryos having protein enrichment in the small micromeres, the corresponding percentages are represented in parentheses (%). The graphs represent the percentage of injected embryos showing protein enrichment within the small micromeres for each RNA.
These results are complemented by experiments using additional Hp nanos2 3’UTR deletion constructs (Figure 3B). Deletion of the nucleotides 880 to 1236 (ΔA) completely abolished the selective expression of GFP in the small micromeres. Similarly, partial deletions in this element at the 3’end (ΔB) or at the 5’end (ΔC) did not give a selective GFP expression in any of the injected embryos. Altogether, these results indicate that nucleotides 880 to 1236 represent the element required for selective protein expression in the small micromeres. This element will be noted GNARLE in the following experiments: Global Nanos Associated RNA Lability Element.
GNARLE is conserved, necessary, and sufficient for selective protein expression in the small micromeres
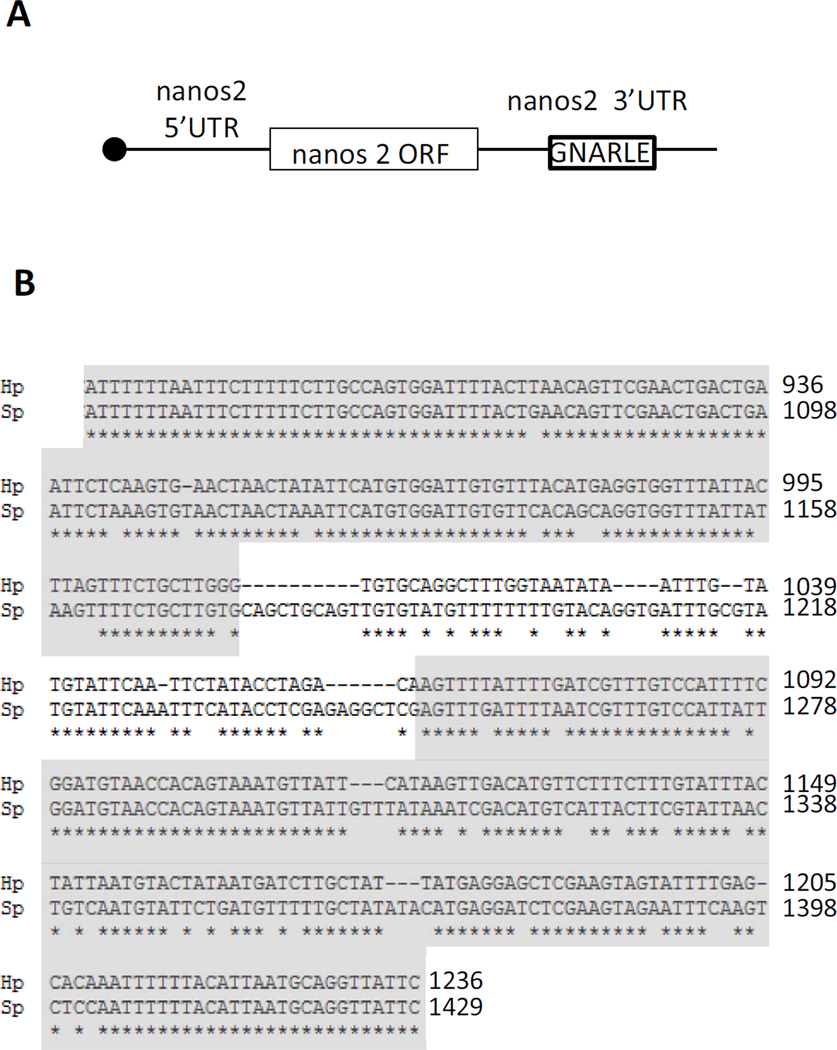
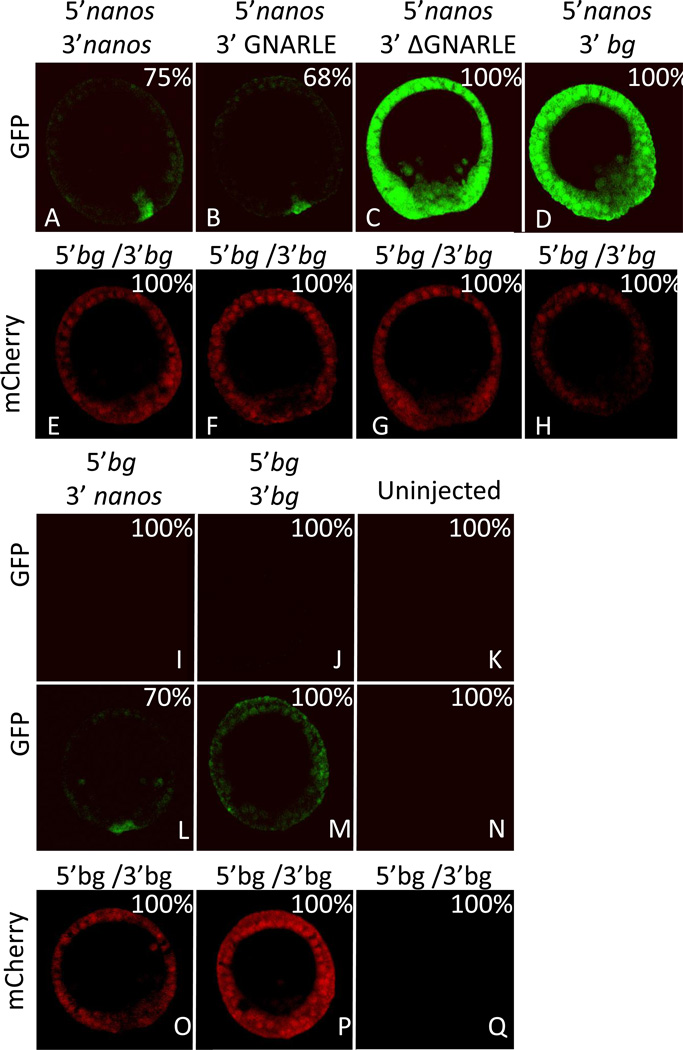
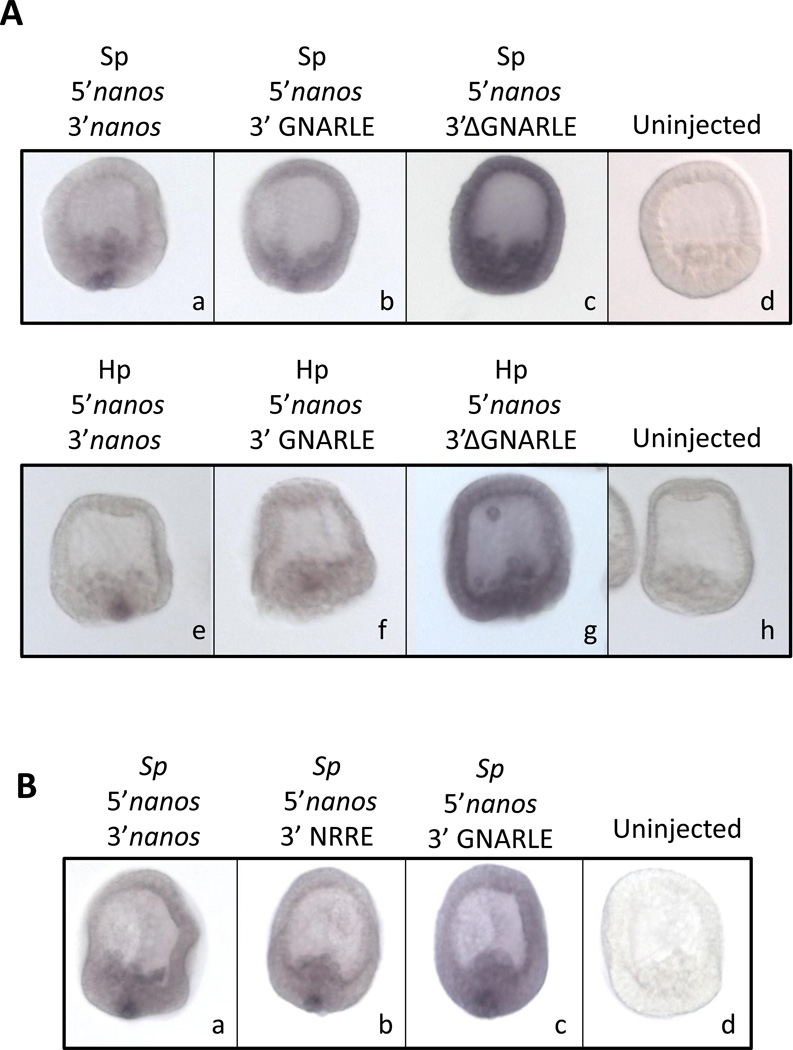
A sequence of 388 nucleotides in the 3’UTR of the nanos2 transcript from Sp aligned closely with the Hp GNARLE (80% identity; Figure 4). GNARLE contains two highly conserved regions, highlighted in grey, which shares 92 and 82% nucleotide identity between Hp and Sp, greater even than the coding region of nanos between these species, whereas regions of the 3’ UTR flanking GNARLE are not conserved between these species. To test if Sp nanos2 GNARLE is involved in the protein accumulation in the small micromeres, we injected RNAs containing Sp nanos2 5’UTR, the GFP ORF and Sp nanos2 3’UTR either full length, GNARLE only, or deleted in GNARLE (ΔGNARLE) (Figure 5). The GFP protein produced after injection of the full length Sp nanos2 3’UTR RNA, was strongly enriched in the small micromeres (Figure 5 A). Interestingly, the Sp nanos2 GNARLE RNA gave a similar GFP enrichment in the small micromeres (Figure 5 B), in contrast to the Sp nanos2 ΔGNARLE RNA in which GFP fluorescence was detected at a strong level in all cells (Figure 5 C). Similar results were obtained after injection of the Hp-derived RNA in Sp (Figure S3) although the selectivity of with or without the GNARLE was not as great as seen in Sp. Perhaps this minor enrichment difference reflects an importance in the regions of sequence for which differences between the species occurs. Overall however, these results suggest that the GNARLE is necessary and sufficient for selective protein enrichment in the small micromeres in the sea urchin Sp.
Figure 4.
(A) Schematic representation of nanos2 RNA, indicating the location of GNARLE in the 3’UTR. (B) Alignment of Hp and Sp nanos2 GNARLE using clustalW2. The stars below the sequences represent the identical nucleotides between Hp and Sp. The highly conserved regions are highlighted in grey.
Figure 5.
Sp GNARLE is necessary and sufficient for selective protein accumulation in the small micromeres. Synthetic mRNAs containing GFP open reading frame were injected in Sp fertilized eggs. The GFP ORF was preceded by either (A, B, C, D) Sp nanos2 5’UTR or (I, J) Xenopus β-globin 5’UTR, and followed by either (A, I, L) Sp nanos2 full length 3’UTR, (B) Sp nanos2 GNARLE 3’UTR, (C) Sp nanos2 ΔGNARLE 3’UTR, or (D, J, M) Xenopus β-globin 3’UTR. Uninjected embryos (K, N, Q) were used as a control. Each GFP mRNA was co-injected with a mRNA containing mCherry ORF (E, F, G, H, O, P) flanked by Xenopus β-globin 5’ and 3’UTRs. GFP (green) and mCherry (red) fluorescence were assayed in the same embryos 24 hrs post-fertilization at mesenchyme blastula following microinjection of synthetic mRNAs. For GFP images, A, B, C, D, I, J, and K were obtained using the same settings (laser intensity, pin-hole opening) at the microscope. L and M were obtained by increasing the laser intensity on the embryos shown in I and J respectively. For mCherry images, all the pictures were also taken using the same settings (E, F, G, H, O, P, Q). bg indicates β-globin UTRs. Approximately one hundred blastulas were visualized after injection of each construct, the corresponding percentages of representative embryos are indicated in the right corner.
Moreover, the RNA containing Sp nanos2 5’UTR and Xenopus β-globin 3’UTR demonstrates that the 5’UTR by itself is not sufficient to give a protein enrichment in the small micromeres (Figure 5 D). Of note here is that an RNA containing Xenopus β-globin 5’UTR and Sp nanos2 3’UTR did not give any detectable fluorescence using the same detection parameters on the microscope as used for adjacent experiments (Figure 5 I). Nevertheless, by increasing the detection sensitivity, a GFP signal was still found enriched in the small micromeres (Figure 5 L), in contrast to the GFP signal obtained everywhere after injection of the GFP RNA containing the Xenopus β-globin 5’ and 3’ UTRs (Figure 5 J,M). These results indicate that Sp nanos2 5’UTR plays a substantial role in stimulation of protein synthesis.
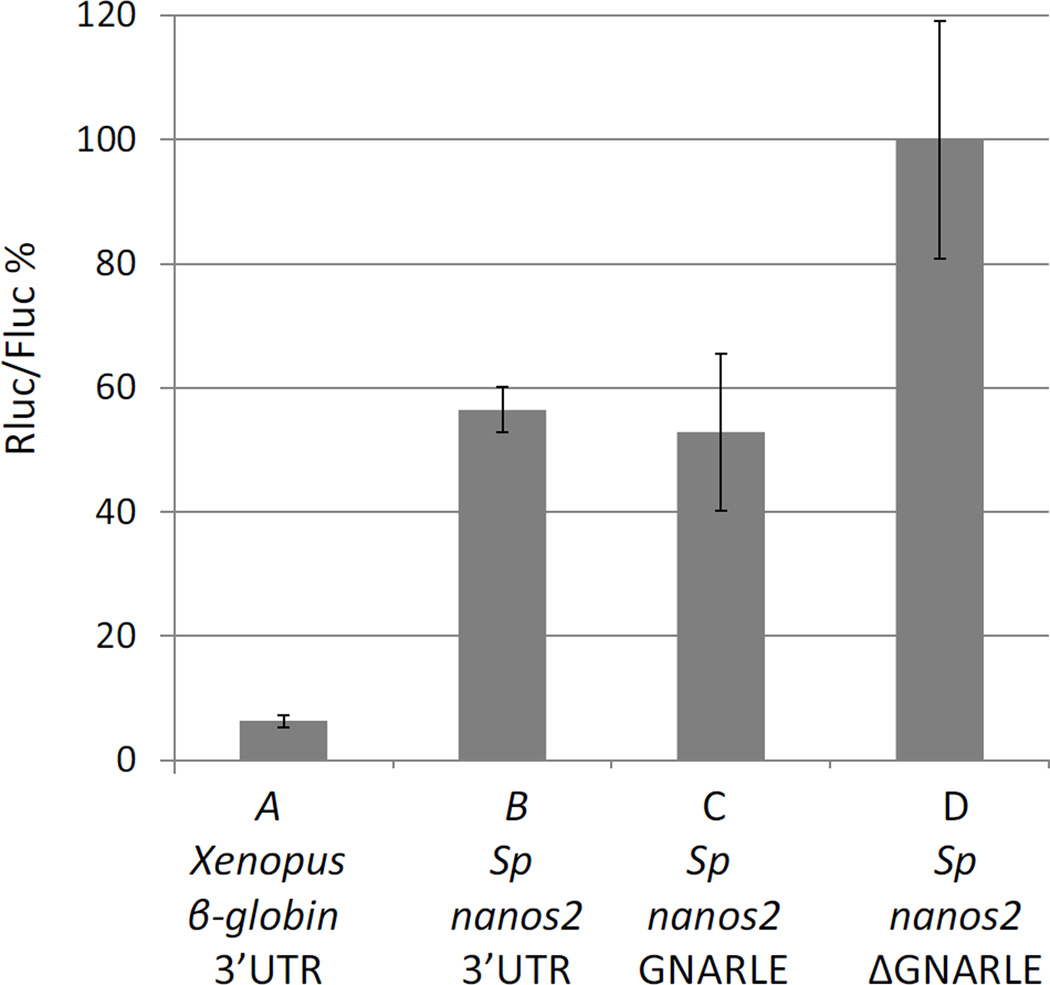
Protein synthesis obtained with Sp nanos2 UTRs was tested at the same time using a luciferase assay (Figure 6). Sp fertilized eggs were injected with RNAs containing Sp nanos2 5’UTR followed by the Renilla luciferase (Rluc) ORF. Three 3’UTRs were tested: Sp nanos2 3’UTR full length, the GNARLE alone, or the 3’UTR ΔGNARLE. At the mesenchyme blastula stage, a high level of Rluc was measured after injection of the ΔGNARLE RNA, and this ratio was set to 100%. In contrast, RNAs containing Sp nanos2 3’UTR full length or the GNARLE only, gave respectively an activity of 56% and 52%. Moreover, the control Rluc RNA, containing the Xenopus β-globin 5’ and 3’UTRs, only gave an activity of 6%. First, these results indicate that there is more Rluc produced when the injected RNA does not contain the GNARLE. Secondly, the protein synthesis obtained with Xenopus β-globin UTRs is lower than the one obtained with nanos2 UTRs. These results quantitatively supported the conclusions made by observations of fluorescence in vivo (Figure 5).
Figure 6.
Deletion of the GNARLE increases protein synthesis. Synthetic mRNAs, containing Renilla luciferase (Rluc) ORF preceded by (A) Xenopus β-globin 5’UTR or (B,C,D) Sp nanos2 5’UTR and followed by (A) Xenopus β-globin 3’UTR, (B) Sp nanos2 full length 3’UTR, (C) Sp nanos2 GNARLE 3’UTR, or (D) Sp nanos2 ΔGNARLE 3’UTR, were injected in Sp fertilized eggs. An mRNA containing the Firefly luciferase (Fluc) ORF flanked by Xenopus β-globin 5’ and 3’UTRs was co-injected. Luminescence was measured at mesenchyme blastula. The ratio Rluc/Fluc was determined, and the results are shown in percentages considering the ratio obtained for Sp ΔGNARLE (D) as 100%. Error bars indicate the standard deviation from three technical replicates.
The 3’UTR of nanos2 also directs selective RNA retention in the small micromeres
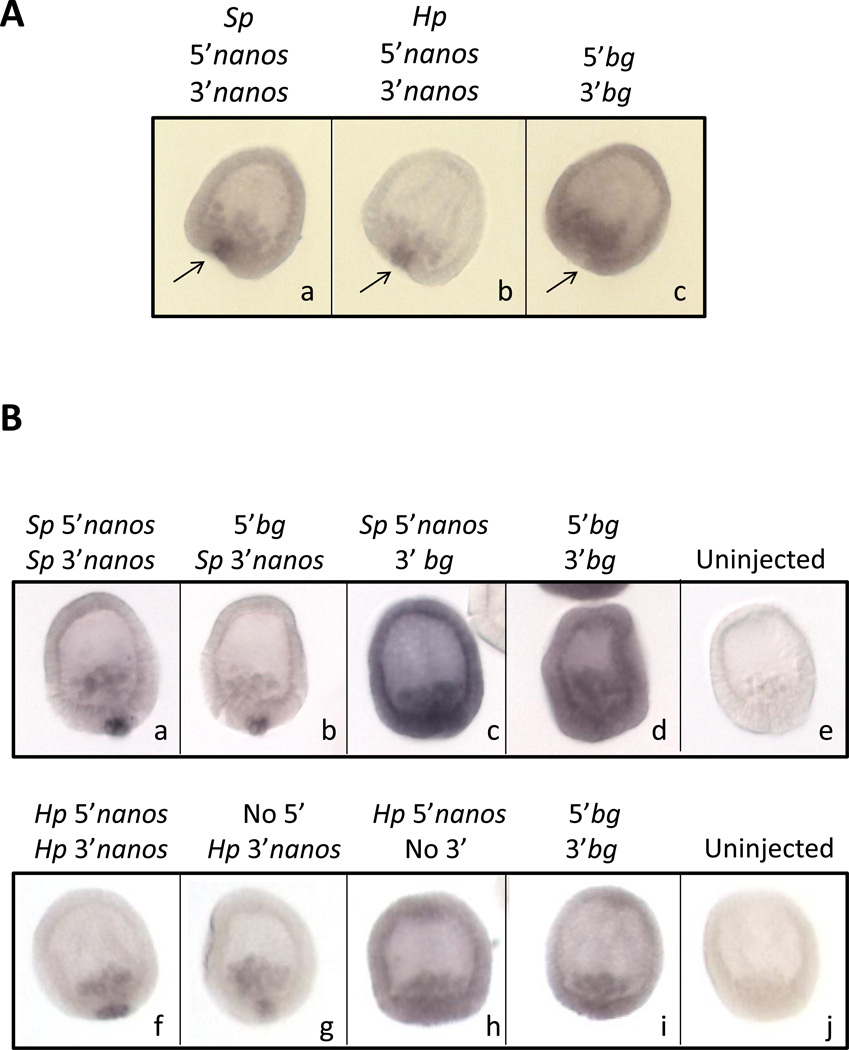
Endogenous nanos2 transcripts are localized in the small micromeres in these 2 sea urchin species, H.pulcherrimus and S.purpuratus (Fujii et al., 2009; Juliano et al., 2010). Preliminary results of transcriptional regulation of the nanos2 gene suggest that nanos2 is more broadly transcribed in the embryo (unpublished observations, M. Yajima), suggesting that a post transcriptional regulation is required to prevent nanos2 expression outside of the small micromeres. To test if nanos2 UTRs are required for the localization of nanos2 transcript in the small micromeres, we injected two RNAs containing the GFP ORF flanked by either Sp or Hp nanos2 5’ and 3’UTRs (Figure 7A). In contrast to the β -globin control RNA which is detected everywhere, the RNAs containing nanos2 UTRs from Sp or Hp were both found selectively in the small micromeres at the MB stage. Analysis of earlier stages of development using the Sp nanos2 UTR sequence indicated that the injected RNA is present in every cell during the first few hours after fertilization (Figure S4), and then is degraded in the non-small micromere cells; i.e. the RNA is protected from degradation in the small micromeres. These results suggest that the nanos2 UTRs are regulated in the small micromeres differently than in other cells of the embryo.
Figure 7.
Sp and Hp nanos2 3’UTRs are sufficient for the selective retention of RNA in the small micromeres. In situ hybridization on Sp mesenchyme blastula embryos using a probe against GFP, after injection of synthetic mRNAs in Sp fertilized eggs. (A) Synthetic mRNAs were made using the GFP ORF flanked by (a) Sp nanos2 5’ and 3’UTRs, (b) Hp nanos2 5’ and 3’UTRs, or (c) Xenopus β-globin UTRs. The arrows are pointing toward the small micromeres. (B) One set of synthetic mRNAs was made using the GFP ORF flanked by (a) Sp nanos2 5’ and 3’UTRs, (b) Xenopus β-globin 5’UTR and Sp nanos2 3’UTR, (c) Sp nanos2 5’UTR and Xenopus β-globin 3’UTR, or (d) Xenopus β-globin 5’ and 3’UTRs. A second set of synthetic mRNAs was made using the GFP ORF surrounded by (f) Hp nanos2 5’and 3’UTRs, (g) Hp nanos2 3’UTR, (h) Hp nanos2 5’UTR, or by (i) Xenopus β-globin UTRs. Uninjected embryos are shown in (e) and (j). Approximately one hundred blastulas were visualized after injection of each construct, the representative embryos are presented.
We next tested if the 5’ and 3’UTR of nanos2 are required for its RNA retention in the small micromeres, or if only one of them was sufficient. To test separately nanos2 5’UTR and 3’UTRs, we made two constructs for each sea urchin species: the first one containing nanos2 5’UTR followed by GFP ORF and Xenopus β-globin 3’UTR, and the second one containing the Xenopus β-globin 5’UTR followed by the GFP ORF and nanos2 3’UTR (Figure 7B). Only the RNAs containing nanos2 3’UTR are retained selectively in the small micromere at the MB stage. RNAs containing only nanos2 5’UTR were found everywhere, similar to the control and the Sp-derived and Hp-derived constructs gave similar results. These results show that in sea urchin, nanos2 3’UTR is sufficient for the early and selective retention of the RNA in the small micromeres.
The GNARLE is required but not sufficient for the selective RNA retention in small micromeres
To test if the GNARLE involved in the selective protein expression is also essential for RNA retention in the small micromeres, we injected Sp fertilized eggs with RNAs containing nanos2 5’UTR, GFP ORF followed by the nanos2 3’UTR full length, the GNARLE only or the 3’UTR ΔGNARLE. RNA retention was monitored by WMISH (Figure 8A) and similar results were obtained using Sp nanos2 or Hp nanos2. The RNA which contains only the GNARLE did not give a strong RNA retention in the small micromeres but is still selectively retained in the small micromere cells. In contrast, the RNA ΔGNARLE is more stable than the RNA containing the full length 3’UTR or the GNARLE, and is present at a high level in the entire blastula. These results suggest that the GNARLE is required for the retention of nanos2 RNA in the small micromeres, but is not as effective as the full length 3’UTR in this phenotype.
Figure 8.
The GNARLE is not sufficient for RNA retention in small micromeres. In situ hybridization on Sp mesenchyme blastula, using a RNA in situ probe against GFP, after injection of synthetic mRNAs in Sp fertilized eggs. (A) Synthetic mRNAs were made using the GFP ORF surrounded by (a,e) nanos2 5’ and 3’UTRs, (b,f) nanos2 5’UTR and GNARLE 3’UTR, (c,g) nanos2 5’UTR and ΔGNARLE 3’UTR. UTRs from Sp nanos2 (a,b,c) were used in one set of synthetic mRNAs, and UTRs from Hp nanos2 (e,f,g) were used in a second set. Uninjected embryos are shown in (d) and (h). (B) Synthetic mRNAs were made using the GFP ORF surrounded by (a.b.c) Sp nanos2 5’UTR, and either (a) Sp nanos2 full length 3’UTR, (b) Sp NRRE 3’UTR, (c) Sp GNARLE 3’UTR. Uninjected embryos are shown in (d). Approximately one hundred embryos were visualized after injection of each construct, and representative embryos are presented.
To better define the element sufficient for the selective RNA retention in the small micromeres, we arbitrarily made several constructs containing GNARLE and its surrounding nucleotides. After RNA injection followed by WMISH, we identified a minimal sequence: the NRRE for Nanos2 RNA Retention Element. This sequence of Sp nanos2 3’UTR includes 55 nucleotides before and 69 nucleotides after the GNARLE (Figure S5). The RNA retention obtained after injection of RNAs containing the Sp nanos2 5’UTR, followed by the GFP ORF and different parts of Sp nanos2 3’UTR: the full length, the NRRE or the GNARLE is presented in Figure 8B. As previously described in Figure 8A, the RNA containing the full length 3’UTR gave a strong selective RNA retention in the small micromeres, in contrast to the GNARLE which only gave a partial RNA retention. Interestingly, the NRRE RNA gave a pattern very similar to the one obtained with the full length 3’UTR. These results indicate that the NRRE is the element important for RNA retention in small micromeres.
Discussion
Nanos has a dominant function in cells to repress translation of mRNAs containing a Nanos Response Element (NRE). The best example of NRE in cells is in the cyclin B mRNA, and in this way, primordial germ cells are slow to divide during embryogenesis (Asaoka-Taguchi et al., 1999). Only after reaching the gonad and depleting the nanos protein do the germ cells begin to divide more rapidly. Ectopic nanos expression has a detrimental effect in embryos (e.g. Luo et al., 2011) and it is not surprising then that the nanos gene is carefully regulated at every level. Here we show a dramatic regulatory step at retention of the mRNA. We found that nanos2 3’UTR contains a Global Nanos Associated RNA Lability Element (GNARLE), which is required for selective RNA retention and protein enrichment in the small micromeres. We find this functionality conserved in two sea urchin species, Strongylocentrotus purpuratus and Hemicentrotus pulcherimus, which diverged less than 20 million years ago (Lee, 2003).
The GNARLE is required for RNA retention in the small micromeres and its deletion leads to a high stabilization of the injected RNA in the entire embryo, meaning that this element is required for the degradation of the RNA in the non-small micromere cells. In terms of nanos regulation, mRNA retention and translation globally would be detrimental since the cyclin mRNAs needed for rapid embryo cell division would be repressed by nanos and its ubiquitous pumillio partner protein (in preparation). On the other hand, the GNARLE by itself is not sufficient for optimal RNA retention in the small micromeres, suggesting that the surrounding nucleotides defined in the NRRE are important to protect the RNA from degradation in the small micromeres. A combination of degradation in the non-small micromeres, and protection in the small micromeres, seems to be required for this strong selective RNA retention. Moreover, even if the GNARLE doesn’t give a strong selective RNA retention, this element is sufficient for high protein enrichment in the small micromeres, suggesting a role of the GNARLE in inhibiting the translation in the non-small micromeres, and/or stimulating the translation in the small micromeres. Therefore, the GNARLE plays a role in both promoting selective RNA retention and translational activity of nanos in the small micromeres.
Several overlapping mechanisms may contribute to selective nanos protein accumulation. The GNARLE may interact specifically with one or several proteins involved in RNA stability and/or protein translation. This element could be binding, directly or indirectly, proteins such as nucleases, or deadenylases to destabilize the transcripts in the non-small micromere cells, and proteins which could inhibit translation of nanos in the non-small micromere cells. Moreover, in various organisms, the nanos 3’UTR is also known to be regulated by the binding of miRNAs and piRNAs in its 3’UTR. During zebrafish embryogenesis, nanos expression is required for the germ-line development, and is restricted to the primordial germ cells. MicroRNA-430 targets the 3’UTR of nanos1 RNA to reduce its polyA tail length, its stability and its translation in the somatic cells (Mishima et al., 2006; Takeda et al., 2009). In Drosophila, nanos is expressed as a gradient that emanates from the posterior pole of the embryos. Recently, the piRNAs pathway was shown to be required for nanos mRNA deadenylation and decay as well as translational repression in Drosophila embryos (Rouget et al., 2010). Aubergine, one of the argonaute proteins in the piRNA pathway, is present in a complex with the RNA binding protein smaug, the deadenylase CCR4, nanos mRNA and piRNAs that target nanos 3’UTR. Small RNAs from different developmental stages of the sea urchin S. purpuratus embryos were recently identified (Song et al., 2011). However, perturbation of the miRNA formation by injection of a Dicer morpholino in Sp embryos did not affect the level of nanos2 transcript significantly, indicating that miRNAs are not responsible for the decay of nanos2 RNA outside of the small micromeres (Figure S6).
Our results show that Sp nanos2 5’UTR is not sufficient for selective RNA retention or protein enrichment but it strongly increases the level of protein synthesis compared to the Xenopus β-globin 5’UTR. This strong expression is independent of the 3’UTR used. The sea urchin translational machinery might be recognizing and scanning more efficiently the Sp nanos2 5’UTR than Xenopus β-globin 5’UTR which is used as a control in RNA injection experiments. Sp nanos2 5’UTR seems to be important to increase protein translation and could also be a useful tool to over-express protein after RNA injection in sea urchin embryos.
In many cases, translation is controlled by cis-regulatory sequences within the 5’ and 3’UTRs of the transcripts (Chatterjee and Pal, 2009), but the open reading frames could also be important to regulate gene expression. For example, in Xenopus, nanos (Xcat2) is transcribed during early oogenesis and becomes localized to the germ plasm, a subcellular compartment bearing the germ cell determinants. nanos RNA localization depends on a cis-acting element within its 3’UTR (Kloc et al., 2000). Recently, a new mechanism was discovered to be regulating nanos translation in Xenopus oocytes. nanos contains an RNA secondary structural element immediately downstream of the AUG start site which is both necessary and sufficient to prevent ribosome scanning during oogenesis (Luo et al., 2011). This inhibition could be relieved after fertilization by a developmentally regulated activator to allow the ribosome loading. The nanos2 open reading frame was not included in the injected RNAs used in this study, but its function in nanos2 expression is currently under investigations.
We show here that in the sea urchin, nanos2 3’UTR is sufficient for its selective protein enrichment. This study provides new insights on how gene expression is regulated in the sea urchin small micromeres. Another well-known protein selectively expressed in the sea urchin small micromeres is vasa, a DEAD box RNA helicase. In early development, vasa mRNA is present uniformly throughout all cells of the embryos, but vasa protein accumulates selectively in the small micromeres (Voronina et al., 2008). It was shown recently that vasa coding sequence is sufficient for its selective enrichment. The E3 ubiquitin ligase, gustavus, binds vasa to induce its proteolysis in all cells except the small micromeres (Gustafson et al., 2011). Thus, regulation of the various germ line components necessary to direct this cell type to contribute to the germ line already has multiple different pre- and post-transcriptional regulatory mechanisms essential for the process. Indeed, use of these many regulatory steps may in itself be a selection mechanism for culling cells deficient in any one of a diverse set of determination pathways.
Supplementary Material
A highly conserved element in the untranslated region of the nanos mRNA was found.
This element functions in protein expression and selective mRNA retention.
This mechanism, added to others reported, makes nanos one of the most highly controlled germ line-determinants.
Acknowledgments
The authors are grateful to the NIH and the NSF for supporting this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Asaoka-Taguchi M, Yamada M, Nakamura A, Hanyu K, Kobayashi S. Maternal Pumilio acts together with Nanos in germline development in Drosophila embryos. Nature cell biology. 1999;1:431–437. doi: 10.1038/15666. [DOI] [PubMed] [Google Scholar]
- Becalska AN, Kim YR, Belletier NG, Lerit DA, Sinsimer KS, Gavis ER. Aubergine is a component of a nanos mRNA localization complex. Dev Biol. 2011;349:46–52. doi: 10.1016/j.ydbio.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat KM. The posterior determinant gene nanos is required for the maintenance of the adult germline stem cells during Drosophila oogenesis. Genetics. 1999;151:1479–1492. doi: 10.1093/genetics/151.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brechbiel JL, Gavis ER. Spatial regulation of nanos is required for its function in dendrite morphogenesis. Curr Biol. 2008;18:745–750. doi: 10.1016/j.cub.2008.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Pal JK. Role of 5'- and 3'-untranslated regions of mRNAs in human diseases. Biol Cell. 2009;101:251–262. doi: 10.1042/BC20080104. [DOI] [PubMed] [Google Scholar]
- Cheers MS, Ettensohn CA. Rapid microinjection of fertilized eggs. Methods Cell Biol. 2004;74:287–310. doi: 10.1016/s0091-679x(04)74013-3. [DOI] [PubMed] [Google Scholar]
- Cho PF, Gamberi C, Cho-Park YA, Cho-Park IB, Lasko P, Sonenberg N. Cap-dependent translational inhibition establishes two opposing morphogen gradients in Drosophila embryos. Curr Biol. 2006;16:2035–2041. doi: 10.1016/j.cub.2006.08.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Agostino I, Merritt C, Chen PL, Seydoux G, Subramaniam K. Translational repression restricts expression of the C. elegans Nanos homolog NOS-2 to the embryonic germline. Dev Biol. 2006;292:244–252. doi: 10.1016/j.ydbio.2005.11.046. [DOI] [PubMed] [Google Scholar]
- Forbes A, Lehmann R. Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development. 1998;125:679–690. doi: 10.1242/dev.125.4.679. [DOI] [PubMed] [Google Scholar]
- Fujii T, Mitsunaga-Nakatsubo K, Saito I, Iida H, Sakamoto N, Akasaka K, Yamamoto T. Developmental expression of HpNanos, the Hemicentrotus pulcherrimus homologue of nanos. Gene Expr Patterns. 2006;6:572–577. doi: 10.1016/j.modgep.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Fujii T, Sakamoto N, Ochiai H, Fujita K, Okamitsu Y, Sumiyoshi N, Minokawa T, Yamamoto T. Role of the nanos homolog during sea urchin development. Dev Dyn. 2009;238:2511–2521. doi: 10.1002/dvdy.22074. [DOI] [PubMed] [Google Scholar]
- Gavis ER, Curtis D, Lehmann R. Identification of cis-acting sequences that control nanos RNA localization. Dev Biol. 1996a;176:36–50. doi: 10.1006/dbio.1996.9996. [DOI] [PubMed] [Google Scholar]
- Gavis ER, Lunsford L, Bergsten SE, Lehmann R. A conserved 90 nucleotide element mediates translational repression of nanos RNA. Development. 1996b;122:2791–2800. doi: 10.1242/dev.122.9.2791. [DOI] [PubMed] [Google Scholar]
- Gustafson EA, Yajima M, Juliano CE, Wessel GM. Post-translational regulation by gustavus contributes to selective Vasa protein accumulation in multipotent cells during embryogenesis. Dev Biol. 2011;349:440–450. doi: 10.1016/j.ydbio.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish V, Lehmann R, Akam M. The Drosophila posterior-group gene nanos functions by repressing hunchback activity. Nature. 1989;338:646–648. doi: 10.1038/338646a0. [DOI] [PubMed] [Google Scholar]
- Jain RA, Gavis ER. The Drosophila hnRNP M homolog Rumpelstiltskin regulates nanos mRNA localization. Development. 2008;135:973–982. doi: 10.1242/dev.015438. [DOI] [PubMed] [Google Scholar]
- Juliano CE, Voronina E, Stack C, Aldrich M, Cameron AR, Wessel GM. Germ line determinants are not localized early in sea urchin development, but do accumulate in the small micromere lineage. Developmental biology. 2006;300:406–415. doi: 10.1016/j.ydbio.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Juliano CE, Yajima M, Wessel GM. Nanos functions to maintain the fate of the small micromere lineage in the sea urchin embryo. Dev Biol. 2010;337:220–232. doi: 10.1016/j.ydbio.2009.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalifa Y, Huang T, Rosen LN, Chatterjee S, Gavis ER. Glorund, a Drosophila hnRNP F/H homolog, is an ovarian repressor of nanos translation. Dev Cell. 2006;10:291–301. doi: 10.1016/j.devcel.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Kloc M, Bilinski S, Pui-Yee Chan A, Etkin LD. The targeting of Xcat2 mRNA to the germinal granules depends on a cis-acting germinal granule localization element within the 3'UTR. Dev Biol. 2000;217:221–229. doi: 10.1006/dbio.1999.9554. [DOI] [PubMed] [Google Scholar]
- Koprunner M, Thisse C, Thisse B, Raz E. A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev. 2001;15:2877–2885. doi: 10.1101/gad.212401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer B, Crittenden S, Gallegos M, Moulder G, Barstead R, Kimble J, Wickens M. NANOS-3 and FBF proteins physically interact to control the sperm-oocyte switch in Caenorhabditis elegans. Curr Biol. 1999;9:1009–1018. doi: 10.1016/s0960-9822(99)80449-7. [DOI] [PubMed] [Google Scholar]
- Lai F, Zhou Y, Luo X, Fox J, King ML. Nanos1 functions as a translational repressor in the Xenopus germline. Mech Dev. 2011;128:153–163. doi: 10.1016/j.mod.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH. Molecular phylogenies and divergence times of sea urchin species of Strongylocentrotidae, Echinoida. Mol Biol Evol. 2003;20:1211–1221. doi: 10.1093/molbev/msg125. [DOI] [PubMed] [Google Scholar]
- Luo X, Nerlick S, An W, King ML. Xenopus germline nanos1 is translationally repressed by a novel structure-based mechanism. Development. 2011;138:589–598. doi: 10.1242/dev.056705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon KP, Andrews S, Murthy M, Gavis ER, Zinn K. The translational repressors Nanos and Pumilio have divergent effects on presynaptic terminal growth and postsynaptic glutamate receptor subunit composition. J Neurosci. 2009;29:5558–5572. doi: 10.1523/JNEUROSCI.0520-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minokawa T, Rast JP, Arenas-Mena C, Franco CB, Davidson EH. Expression patterns of four different regulatory genes that function during sea urchin development. Gene Expr Patterns. 2004;4:449–456. doi: 10.1016/j.modgep.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Mishima Y, Giraldez AJ, Takeda Y, Fujiwara T, Sakamoto H, Schier AF, Inoue K. Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr Biol. 2006;16:2135–2142. doi: 10.1016/j.cub.2006.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Sano H, Kobayashi S, Nishimiya-Fujisawa C, Fujisawa T. Expression and evolutionary conservation of nanos-related genes in Hydra. Dev Genes Evol. 2000;210:591–602. doi: 10.1007/s004270000105. [DOI] [PubMed] [Google Scholar]
- Rabinowitz JS, Chan XY, Kingsley EP, Duan Y, Lambert JD. Nanos is required in somatic blast cell lineages in the posterior of a mollusk embryo. Curr Biol. 2008;18:331–336. doi: 10.1016/j.cub.2008.01.055. [DOI] [PubMed] [Google Scholar]
- Rebscher N, Zelada-Gonzalez F, Banisch TU, Raible F, Arendt D. Vasa unveils a common origin of germ cells and of somatic stem cells from the posterior growth zone in the polychaete Platynereis dumerilii. Dev Biol. 2007;306:599–611. doi: 10.1016/j.ydbio.2007.03.521. [DOI] [PubMed] [Google Scholar]
- Rouget C, Papin C, Boureux A, Meunier AC, Franco B, Robine N, Lai EC, Pelisson A, Simonelig M. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature. 2010;467:1128–1132. doi: 10.1038/nature09465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Fujimoto T, Maegawa S, Inoue K, Tanaka M, Arai K, Yamaha E. Visualization of primordial germ cells in vivo using GFP-nos1 3'UTR mRNA. Int J Dev Biol. 2006;50:691–699. doi: 10.1387/ijdb.062143ts. [DOI] [PubMed] [Google Scholar]
- Smibert CA, Wilson JE, Kerr K, Macdonald PM. smaug protein represses translation of unlocalized nanos mRNA in the Drosophila embryo. Genes Dev. 1996;10:2600–2609. doi: 10.1101/gad.10.20.2600. [DOI] [PubMed] [Google Scholar]
- Song JL, Stoeckius M, Maaskola J, Friedlander M, Stepicheva N, Juliano C, Lebedeva S, Thompson W, Rajewsky N, Wessel GM. Select microRNAs are essential for early development in the sea urchin. Dev Biol. 2011 doi: 10.1016/j.ydbio.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J, Wharton RP. Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev. 1999;13:2704–2712. doi: 10.1101/gad.13.20.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam K, Seydoux G. nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development. 1999;126:4861–4871. doi: 10.1242/dev.126.21.4861. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Saba R, Sada A, Saga Y. The Nanos3-3'UTR is required for germ cell specific NANOS3 expression in mouse embryos. PLoS One. 2010;5:e9300. doi: 10.1371/journal.pone.0009300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda Y, Mishima Y, Fujiwara T, Sakamoto H, Inoue K. DAZL relieves miRNA-mediated repression of germline mRNAs by controlling poly(A) tail length in zebrafish. PLoS One. 2009;4:e7513. doi: 10.1371/journal.pone.0007513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda M, Sasaoka Y, Kiso M, Abe K, Haraguchi S, Kobayashi S, Saga Y. Conserved role of nanos proteins in germ cell development. Science. 2003;301:1239–1241. doi: 10.1126/science.1085222. [DOI] [PubMed] [Google Scholar]
- Voronina E, Lopez M, Juliano CE, Gustafson E, Song JL, Extavour C, George S, Oliveri P, McClay D, Wessel G. Vasa protein expression is restricted to the small micromeres of the sea urchin, but is inducible in other lineages early in development. Dev Biol. 2008;314:276–286. doi: 10.1016/j.ydbio.2007.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Dickinson LK, Lehmann R. Genetics of nanos localization in Drosophila. Dev Dyn. 1994;199:103–115. doi: 10.1002/aja.1001990204. [DOI] [PubMed] [Google Scholar]
- Wang C, Lehmann R. Nanos is the localized posterior determinant in Drosophila. Cell. 1991;66:637–647. doi: 10.1016/0092-8674(91)90110-k. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zayas RM, Guo T, Newmark PA. nanos function is essential for development and regeneration of planarian germ cells. Proc Natl Acad Sci U S A. 2007;104:5901–5906. doi: 10.1073/pnas.0609708104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton RP, Struhl G. RNA regulatory elements mediate control of Drosophila body pattern by the posterior morphogen nanos. Cell. 1991;67:955–967. doi: 10.1016/0092-8674(91)90368-9. [DOI] [PubMed] [Google Scholar]
- Wu X, Ruvkun G. Cancer. Germ cell genes and cancer. Science. 2010;330:1761–1762. doi: 10.1126/science.1200772. [DOI] [PubMed] [Google Scholar]
- Yajima M, Wessel GM. Small micromeres contribute to the germline in the sea urchin. Development. 2011;138:237–243. doi: 10.1242/dev.054940. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.