Abstract
Mammalian aging is associated with reduced tissue regeneration, increased degenerative disease, and cancer. Because stem cells regenerate many adult tissues and contribute to the development of cancer by accumulating mutations, age-related changes in stem cells likely contribute to age-related morbidity. Consistent with this, stem cell function declines with age in numerous tissues as a result of gate-keeping tumor suppressor expression, DNA damage, changes in cellular physiology, and environmental changes in tissues. It remains unknown whether declines in stem cell function during aging influence organismal longevity. However, mechanisms that influence longevity also modulate age-related morbidity, partly through effects on stem cells.
Introduction
Damage accumulates in biological macromolecules during aging, impairing cellular processes, tissue homeostasis, and organ function. This contributes to the onset of age-related diseases, including cognitive (Yankner et al., 2008), neoplastic (Hoeijmakers, 2009), immunologic (Dorshkind et al., 2009), and metabolic (Wallace, 2005) disorders. Age-related morbidity is determined partly by changes in nondividing differentiated cells, such as neurons (Lu et al., 2004), and partly by changes in mitotic cells, including stem cells, restricted progenitors, and differentiated cells (Sharpless and DePinho, 2007).
Stem cells persist throughout life in numerous mammalian tissues, replacing cells lost to homeostatic turnover, injury, and disease. However, stem cell function declines with age in a number of tissues, including the blood (Morrison et al., 1996b; de Haan et al., 1997; Chen et al., 2000), forebrain (Kuhn et al., 1996; Maslov et al., 2004; Molofsky et al., 2006), skeletal muscle (Conboy et al., 2003, 2005), and skin (Nishimura et al., 2005) (Table 1). These declines in stem cell function may contribute to degeneration and dysfunction in aging regenerative tissues (Sharpless and DePinho, 2007). Thus, age-related changes in the function of stem cells and other progenitors may contribute to some diseases of aging, particularly in regenerative tissues, even while other diseases of aging may not be influenced by stem cell aging at all.
Table 1.
Summary of Age-Related Changes in Various Mammalian Stem Cell Populations
| Stem Cell Population | Frequency | Proliferation | Differentiation | Other Defects |
|---|---|---|---|---|
| Hematopoietic | ↑ long-lived mouse strains, ↓ short-lived mouse strains | ↑ cycling, ↓ self-renewal | ↓ lymphoid, ↑ myeloid | ↓ homing, ↓ mobilization, ↓ engraftment, |
| Neural | ↓ lateral ventricle SVZ, ↓ dentate gyrus subgranular layer | ↓ cycling, ↓ self-renewal (SVZ) | ↓ neurogenesis, ↑ gliogenesis | |
| Muscle | ↓ satellite cells associated with muscle fibers | ↓ proliferation | ↓ myogenic, ↑ fibrogenic, ↑ adipogenic | |
| Melanonocyte stem cells in hair bulge | ↓ melanocyte stem cells | ↑ terminal differentiation of melanocytes |
It is unknown whether stem cell aging influences mammalian life span. However, in Drosophila genetic changes that improve homeostasis in the intestinal epithelium by blocking stem cell overproliferation and differentiation defects during aging do extend life span (Biteau et al., 2010). This raises the possibility that some age-related changes in mammalian stem cells promote homeostasis in aging tissues despite declines in stem cell function.
It is important to emphasize that stem cells are not the only mitotic cells that persist throughout life and whose aging might influence age-related diseases. Like stem cells, some restricted progenitors and differentiated cells are also perpetuated throughout life by intermittent self-renewing divisions. Such cells include pancreatic β cells and memory B and T cells. During aging, declines in the number or function of pancreatic β cells (Teta et al., 2005) and memory T cells (Liu et al., 2011) contribute to the development of type 2 diabetes (Butler et al., 2003) and reduced immune function (Dorshkind et al., 2009). There is at least some overlap in self-renewal mechanisms between these differentiated cells and stem cells (Luckey et al., 2006). This suggests that some of the mechanisms that regulate stem cell aging may also regulate the aging of mitotic differentiated cells, and both classes of progenitors may contribute to age-related morbidity.
Stem cells must change their properties throughout life to match the changing growth and regeneration demands of tissues. Stem cells divide rapidly during fetal development to support rapid growth. By young adulthood, growth has slowed or ceased in mammalian tissues and most stem cells are quiescent most of the time, intermittently dividing to maintain tissue homeostasis. In old adults, stem cells increase gate-keeping tumor suppressor expression. This may reduce the incidence of cancer in aging tissues, but also reduces regenerative capacity (Janzen et al., 2006; Krishnamurthy et al., 2006; Molofsky et al., 2006). These changes in stem cells likely reflect regulation by heterochronic genes—genes whose expression changes over time in a way that causes temporal changes in stem cell function (Nishino et al., 2008; Toledano et al., 2012). Heterochronic genes were originally identified as regulating the timing of developmental transitions in C. elegans (Ambros and Horvitz, 1984). This raises the question of whether the increase in tumor suppressor expression and the temporal changes in stem cell function in aging mammalian tissues are partly developmentally programmed.
Mitochondrial activity, tissue growth, and metabolic rates during development can also influence life span and the rates of cellular aging at later stages of life (Dillin et al., 2002). Thus, the aging of stem cells cannot be considered in isolation but rather in the context of temporal changes in stem cell and tissue properties that occur throughout life.
Like all cells, stem cell aging is determined partly by the accumulation of damage over time. Declines in stem cell function during aging can be precipitated by telomere shortening, DNA damage, and mitochondrial damage (Choudhury et al., 2007; Rossi et al., 2007; Sahin and Depinho, 2010) (Figure 1). Stem cell aging can be slowed by dietary restriction (Lee et al., 2000; Chen et al., 2003; Mair et al., 2010; Cerletti et al., 2012) and by exposure to humoral factors from a young parabiont (sharing circulation with an old mouse) (Conboy et al., 2005; Villeda et al., 2011). In this review we discuss all of these mechanisms that influence stem cell aging in the context of mechanisms that are known to influence general cellular aging and life span.
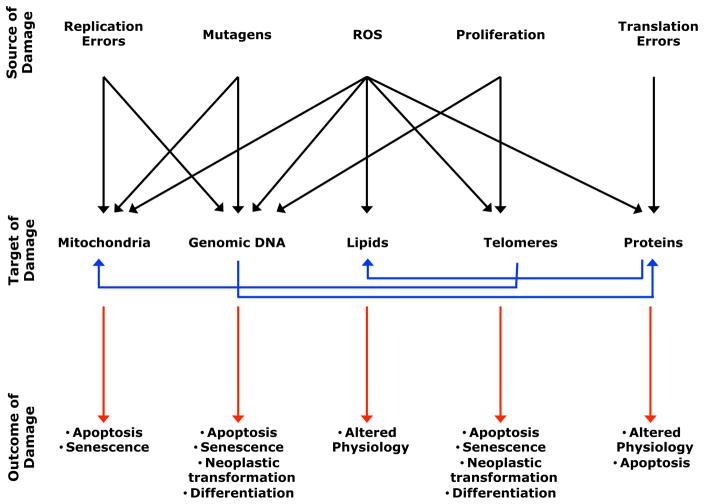
Figure 1. Multiple Sources of Damage to Biological Macromolecules Reduce Stem Cell Function during Aging.
Sources of damage (top row) including ROS, exogenous mutagens, proliferation, infidelity of DNA replication, and errors in protein translation can damage macromolecules or organelles within a cell (middle row). Damage accumulates in DNA, proteins, mitochondria, and lipids during aging and contributes to declines in stem cell function, tissue regeneration, and life span. The cellular consequences of this damage (bottom row) include cell death, cellular senescence, differentiation, altered cellular physiology, and cancer. All of these mechanisms are interrelated; damage to one component, such as telomeres, can influence the function of other components, such as mitochondria (Sahin and Depinho, 2010).
Gate-Keeping Tumor Suppressors
Gate-keeping tumor suppressors (such as p16Ink4a, p19Arf, and p53—see Figure 2) negatively regulate cellular proliferation and survival (Kinzler and Vogelstein, 1997). These gene products were first discovered by virtue of their role in cancer, but probably evolved to regulate homeostasis in normal tissues by regulating the proliferation and survival of normal cells. Their role in cancer reflects the ability of cancer cells to evade normal homeostatic controls by deleting these genes. Gatekeeping tumor suppressors tend to negatively regulate stem cell function (He et al., 2009) and regulate stem cell aging because their expression and/or function increase with age (Krishnamurthy et al., 2004, 2006; Janzen et al., 2006; Molofsky et al., 2006).
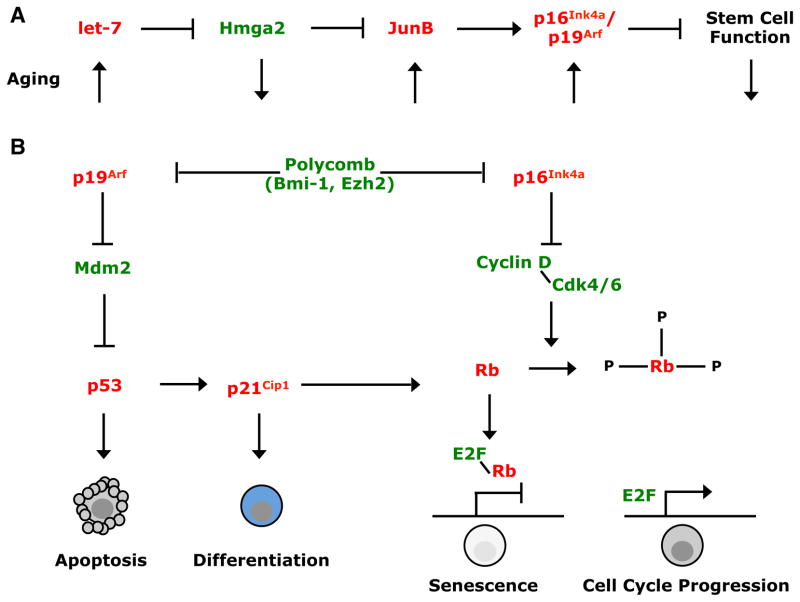
Figure 2. Heterochronic Genes Regulate Increases in the Expression of GateKeeping Tumor Suppressors and Declines in Aging Stem Cell Function.
Stem cell self-renewal and stem cell aging are regulated by networks of proto-oncogenes (green) and tumor suppressors (red).
(A) let-7 microRNA is an evolutionarily conserved heterochronic gene that regulates the timing of developmental events from C. elegans to mammals (Pasquinelli et al., 2000). let-7b expression increases with age in mammals, reducing the expression of the Hmga2 chromatin-associated factor, and increasing the expression of the JunB, p16Ink4a, and p19Arf tumor suppressors (Nishino et al., 2008). The increase in p16Ink4a expression during aging reduces stem cell function in multiple tissues (Janzen et al., 2006; Krishnamurthy et al., 2006; Molofsky et al., 2006). let-7 microRNA also increases with age in Drosophila, acting in the niche to non-cell-autonomously reduce spermatogonial stem cell function by impairing the secretion of Unpaired (Toledano et al., 2012).
(B) p16Ink4a and p19Arf expression are also repressed in mammalian stem cells by polycomb proteins, including Bmi-1 and Ezh2 (Jacobs et al., 1999; Lessard and Sauvageau, 2003; Molofsky et al., 2003; Park et al., 2003; Chen et al., 2009b). In the absence of Bmi-1, p16Ink4a and p19Arf expression are induced in postnatal stem cells from multiple tissues, inducing cell death, cellular senescence, or premature differentiation. These pathways illustrate how networks of proto-oncogenes and tumor suppressors regulate stem cell maintenance and homeostasis in adult tissues. The way in which proto-oncogenic and tumor suppressor signals are balanced within these networks changes with age in stem cells.
A regulatory pathway of heterochronic genes increases gatekeeping tumor suppressor expression in aging stem cells (Figure 2A). let-7 microRNA expression increases with age, probably in many types of stem cells, eliminating the expression of the high mobility group transcriptional regulator, Hmga2, in stem cells from old mice (Nishino et al., 2008). Hmga2 is a proto-oncogene and let-7 target. The loss of Hmga2 expression from neural stem cells reduces their frequency and self-renewal potential by increasing the expression of p16Ink4a and p19Arf (Nishino et al., 2008). p16Ink4a is a cyclin-dependent kinase inhibitor (Figure 2B) whose expression increases in aging mouse and human tissues (Krishnamurthy et al., 2004), reducing stem cell frequency and self-renewal potential in multiple tissues (Janzen et al., 2006; Krishnamurthy et al., 2006; Molofsky et al., 2006). Elevated p16Ink4a expression also depletes certain differentiated progenitors during aging, including pancreatic β cells (Krishnamurthy et al., 2006) and memory T cells (Liu et al., 2011).
p19Arf (p14Arf in humans) also increases with age in mouse tissues (Zindy et al., 1997; Krishnamurthy et al., 2004). p19Arf promotes p53 protein stability by inhibiting Mdm2-mediated degradation (Figure 2B). It has not yet been tested whether p19Arf negatively regulates stem cell function in aging tissues.
Beyond the mechanisms described above, there are likely to be a number of other mechanisms that regulate changes in p16Ink4a and p19Arf expression during aging, including yet undiscovered mechanisms. A decline in the expression of the poly-comb complex component, Ezh2, contributes to increased p16Ink4a and p19Arf expression in aging pancreatic β cells (Chen et al., 2009b). Whether a change in polycomb complex activity contributes generally to changes in p16Ink4a and p19Arf expression in aging stem cells remains untested.
The increase in gate-keeping tumor suppressor expression in aging tissues, and the onset of senescence in some aging cells, may oppose the increased incidence of cancer during aging (Campisi, 2005; Signer et al., 2008). Indeed, p16Ink4a and/or p19Arf deficiency increase the incidence of cancer in adult mice (Serrano et al., 1996; Kamijo et al., 1997), and humans with germline loss-of-function mutations in p16Ink4a/p14Arf have more adult-onset cancers (Ruas and Peters, 1998). However, it remains unclear whether the physiological increase in p16Ink4a and p19Arf expression in aging cells suppresses cancer or whether an even higher level of p16Ink4a and p19Arf expression, induced by oncogenic stimuli, is responsible for cancer suppression. Transgenic mice with an extra copy of the p16Ink4a/p19Arf/p15Ink4b and p53 loci have a lower cancer incidence, though this may reflect the inability of cancer cells to delete the extra copy of the locus rather than the modestly increased expression of these tumor suppressors under physiological conditions (García-Cao et al., 2002; Matheu et al., 2004, 2007).
While p53 expression promotes the maintenance of genomic integrity (Schoppy et al., 2010), the net effect of p53 in a wild-type background is to negatively regulate stem cell function, at least in hematopoietic stem cells (HSCs) from young adult mice (TeKippe et al., 2003), presumably by opposing cell cycle entry, blocking symmetric division, or inducing cell death (Cicalese et al., 2009; Liu et al., 2009b) (Figure 2 and Figure 3). Elevated p53 expression or constitutive p53 activation can deplete stem cells (Lee et al., 2010), cause premature aging, and shorten life span despite reducing cancer incidence (Tyner et al., 2002; Dumble et al., 2007; Gannon et al., 2011) (Figure 3). These effects in mice also appear to reflect similar functions in humans because a polymorphism in p53 that reduces p53 function increases cancer incidence and life span in humans (van Heemst et al., 2005). This suggests that increased p53 activity protects against cancer but can promote aging and shorten life span, at least when a certain threshold of activity is reached.
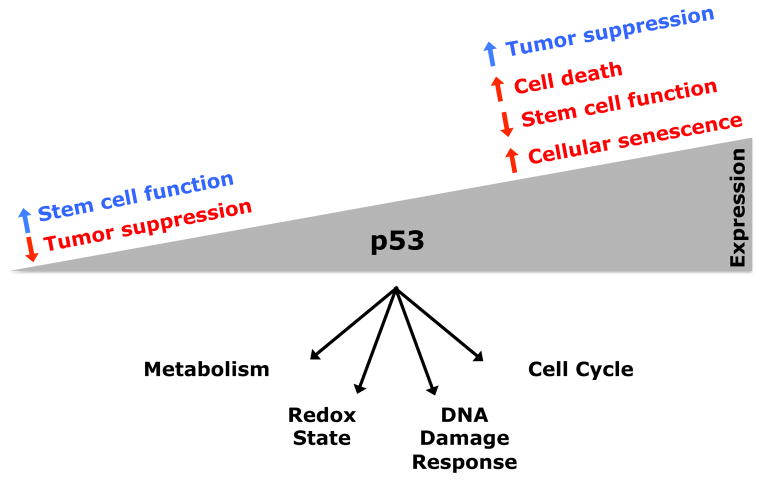
Figure 3. The Multifaceted and Context-Dependent Effects of p53 in Stem Cells.
The consequences of tumor suppressor expression in stem cells can be context dependent. p53 can have both positive (blue) and negative (red) effects on stem cell function, depending on context and expression level. When expressed at low levels, p53 can promote stem cell maintenance by promoting the maintenance of genomic integrity and by regulating metabolism. When expressed at high levels, p53 can promote stem cell depletion through cell death or cellular senescence. The aggregate effect of these functions influences longevity, cancer incidence, and tissue regeneration during aging (Tyner et al., 2002; TeKippe et al., 2003; van Heemst et al., 2005; Dumble et al., 2007; Schoppy et al., 2010; Gannon et al., 2011).
The functions of the p16Ink4a, p19Arf, and p53 tumor suppressors depend on expression level and context (Figure 3), promoting the maintenance of mitotically active cells in some contexts while promoting cell death or senescence in other contexts. For example, p53 promotes the maintenance of genomic integrity (Schoppy et al., 2010) and promotes tissue regeneration in Atr mutant mice by promoting DNA repair and/or by promoting the death of cells with DNA damage (Ruzankina et al., 2009); however, in response to oncogenic stimuli or telomere attrition, p53 depletes stem cells (Begus-Nahrmann et al., 2009; Lee et al., 2010). Increased p53 function in HSCs reduces proliferative potential but slows the expression of some molecular markers of aging (Chambers et al., 2007). Moreover, transgenic mice that constitutively express moderately increased levels of p15Ink4b, p16Ink4a, p19Arf, and/or p53 exhibit no signs of accelerated aging and may even show increased median life span that cannot be explained by reduced cancer incidence (García-Cao et al., 2002; Matheu et al., 2004, 2007). Not all normal cell proliferation in aging tissues is advantageous, as illustrated by atherosclerosis. Therefore, cancer suppression may not be the only function of gate-keeping tumor suppressors in aging stem/progenitor cells, as these tumor suppressors might also help sustain tissue homeostasis by suppressing pathological or dysplastic proliferation, or aberrant differentiation, in aging tissues.
p16Ink4a and p14Arf may also regulate human aging. A series of genome-wide association studies have found significant associations between polymorphisms in or near the p16Ink4a/p14Arf locus in humans and the risk of age-related diseases, including type 2 diabetes and heart disease (Jeck et al., 2012). However, it is not clear whether the polymorphisms increase or decrease p16Ink4a/p14Arf function. Overall, gate-keeping tumor suppressors have pleiotropic functions that promote stem cell function in some ways and negatively regulate stem cell function in other ways, with complex and context-dependent consequences for aging.
Care-Taking Tumor Suppressors and Genomic Integrity
Care-taking tumor suppressors, including DNA repair pathway components, promote stem cell function and tissue regeneration by maintaining genomic integrity (Kinzler and Vogelstein, 1997). Various forms of DNA damage accumulate throughout life as a result of DNA replication errors, exposure to endogenous mutagens such as reactive oxygen species (ROS), and exposure to exogenous mutagens such as UV light. To attenuate the accumulation of mutations, a DNA damage response network can sense DNA damage and activate a variety of repair mechanisms, including nucleotide excision repair, mismatch repair, nonhomologous end joining, and homologous recombination (Ciccia and Elledge, 2010). Activation of the DNA damage response network can transiently halt the cell cycle and repair damaged DNA through p53-dependent mechanisms. If the damage is too extensive to be repaired, the network can trigger the onset of senescence or cell death to eliminate the cells. Abundant cell death and senescence, however, can lead to tissue degeneration. Alternatively, unrepaired DNA damage can lead to the development of cancer, the incidence of which rises dramatically with age.
DNA repair pathway components thus delay cellular aging by maintaining genomic integrity. A number of single gene mutations that impair DNA repair cause segmental progeria syndromes. Segmental progeria syndromes are rare human diseases defined by reduced life span and premature aging phenotypes, including cataracts, osteoporosis, skin atrophy, hair graying, heart disease, cancer, cerebellum degeneration, and immunodeficiency. Segmental progeria syndromes caused by defects in DNA repair include Werner syndrome, a recessive trait caused by loss of function in the RecQ DNA helicase WRN, and Ataxia Telangiectasia, a recessive trait caused by loss of function in the DNA damage signaling protein ATM (Martin, 2005). Mice engineered to carry mutations in the genes associated with human progeroid syndromes similarly display phenotypes consistent with accelerated aging (Wong et al., 2003; Chang et al., 2004). The observation that progeria syndromes are frequently caused by defects in DNA repair suggests that DNA damage may be a fundamental underlying cause of aging.
Mice with loss-of-function mutations in DNA repair pathway components exhibit stem cell defects in multiple tissues. Loss of the DNA damage sensor ATM depletes HSCs (Ito et al., 2004), exacerbates the loss of melanocyte stem cells in response to low dose radiation (Inomata et al., 2009), and promotes the loss of undifferentiated spermatogonia (Takubo et al., 2008). Loss of a related DNA damage sensor, ATR, depletes HSCs and hair follicle stem cells (Ruzankina et al., 2007). Mice deficient in nucleotide excision repair (XpdTTD), mismatch repair (Msh2−/−), nonhomologous end joining (Lig4(Y288C) and Ku80−/−), or homologous recombination (Brca2−/−) all exhibit reduced HSC function (Reese et al., 2003; Navarro et al., 2006; Nijnik et al., 2007; Rossi et al., 2007). The mechanism by which deficiency for DNA repair genes depletes stem cells involves accumulation of DNA damage, induction of p53 and p21cip1 (Merritt et al., 1994; Choudhury et al., 2007; Takubo et al., 2008; Begus-Nahrmann et al., 2009), elevated ROS levels (Ito et al., 2004), and premature differentiation (Inomata et al., 2009; Wang et al., 2012).
DNA damage accumulates with age in HSCs and epidermal stem cells from mice (Rossi et al., 2007; Sotiropoulou et al., 2010). It remains to be determined whether the amount of DNA damage that accumulates with age in normal stem cells actually contributes to declines in stem cell function under physiological conditions. However, DNA damage in stem cells may nonetheless have profound consequences. Most somatic cells are post-mitotic and are therefore unlikely to be transformed into cancer cells by mutations or to pass their mutations onto progeny. Most dividing cells are short-lived and therefore produce a limited number of progeny or do not persist long enough to accumulate mutations over time. In contrast, stem cells remain mitotically active throughout life, generating large numbers of progeny in some tissues. Given that transformation of normal cells into cancer cells requires a series of mutations that accumulate over a period of years, the ability of stem cells to accumulate mutations and then expand the pool of mutated cells may be critical for the evolution of cancer in regenerative tissues (Rossi et al., 2008). Nonetheless, this does not mean that most cancers arise from stem cells. Even if most carcinogenic mutations accumulate in stem cells, the final mutation that causes frank transformation may occur in the numerically expanded restricted progenitors or differentiated cells that arise from stem cells.
Telomeres
Telomeres are specialized nucleoprotein caps that contain thousands of base pairs of repetitive DNA sequences that protect the ends of chromosomes from end-to-end fusions that induce DNA damage responses (Palm and de Lange, 2008; Sahin and Depinho, 2010). Because of the way DNA is replicated, telomeres shorten with each round of cell division such that the replicative potential of cells is limited by the length of their telomeres, unless the cells express telomerase, which can lengthen telomeres and increase replicative capacity. Telomeres shorten with age in many human cells, including HSCs (Vaziri et al., 1994, and references therein). When telomeres reach a critically short length, cells can exhibit genomic instability and undergo cell cycle arrest, senescence, or apoptosis (Chin et al., 1999; Choudhury et al., 2007; Begus-Nahrmann et al., 2009; Sperka et al., 2012). In addition to protecting against genomic instability, p53 activation following telomere dysfunction also impairs mitochondrial biogenesis, mitochondrial activity, and metabolic function (Sahin et al., 2011). It has been proposed that cellular aging is determined partly by telomere erosion, and partly by the DNA damage and loss of replicative potential that ensue (Harley et al., 1992).
Most human cells do not express telomerase, though telomerase is commonly expressed in cancer cells and other immortalized cells (Kim et al., 1994). Loss of telomerase function in mice reduces the regenerative capacity of proliferative organs (Lee et al., 1998), accelerates the development of aging phenotypes (like hair graying), reduces life span, and increases cancer incidence (particularly in the absence of p53) (Blasco et al., 1997; Rudolph et al., 1999; Artandi et al., 2000). Telomerase-deficient mice exhibit defects in stem cell function in the forebrain, epidermis, intestinal epithelium, and hematopoietic system through cell-autonomous (Lee et al., 1998; Allsopp et al., 2003; Choudhury et al., 2007; Ferrón et al., 2009; Jaskelioff et al., 2011) and non-cell-autonomous effects on stem cells (Ju et al., 2007). HSCs express telomerase (Morrison et al., 1996a), slowing, but not eliminating, the decline in telomere length with age (Vaziri et al., 1994). An important caveat is that profound defects are not apparent in germline telomerase-deficient mice for three generations after they are generated because inbred mice have relatively long telomeres (Kipling and Cooke, 1990). This suggests that telomerase is not required during a single generation in inbred mice under normal circumstances. In contrast, telomere length in young zebra finches is predictive of life span (Heidinger et al., 2012), suggesting that telomere length may limit life span in some species. This raises the question of whether telomere length limits proliferative potential during a normal human life span or whether telomere length only becomes limiting in the context of conditions that promote chronic tissue regeneration.
Defects in human telomerase function cause diseases with features of premature aging, including impaired regeneration of proliferative tissues (Lansdorp, 2009). Dyskeratosis congenita is a rare form of ectodermal dysplasia caused by very short telomeres that result from loss-of-function mutations in telomerase components or telomere binding proteins (reviewed by Walne and Dokal, 2009). Indeed, loss-of-function mutations in only a single copy of telomerase lead to accelerated telomere shortening and reduced tissue regeneration (Hao et al., 2005). Accelerated shortening of telomeres has also been observed in other conditions with premature aging phenotypes, including trisomy 21 (Vaziri et al., 1993). Telomere preservation is thus a key aspect of genomic integrity in which defects impair regeneration and accelerate aging.
Oxygen, Energy Metabolism, and ROS
Aging is proposed to result from cellular damage caused by free radicals, principally ROS generated as a consequence of oxidative phosphorylation in the mitochondrial electron transport chain (Wallace, 2005). ROS, such as superoxide and hydroxyl radical, are highly reactive and can damage mitochondrial and nuclear DNA, as well as proteins and lipids, by chemically modifying them. Oxidized macromolecules, such as 8-hydroxy-2-deoxyguanosine, accumulate with age in rats (Fraga et al., 1990). Increased expression of enzymes such as superoxide dismutases or catalase, which convert ROS into less reactive or nonreactive species, reduce the accumulation of oxidized macromolecules, increase maximum life span, and decrease the incidence of certain diseases of aging, including cancer (Wallace, 2005).
Stem cells appear to be particularly sensitive to elevated ROS levels. Under normal conditions, ROS can function as signaling molecules that regulate the differentiation of stem/progenitor cells, such as in Drosophila hematopoietic cells (Owusu-Ansah and Banerjee, 2009). However, ROS levels increase in HSCs with age, and prolonged treatment with the antioxidant N-acetyl-L-cysteine increases the replicative potential of HSCs upon serial transplantation in irradiated mice (Ito et al., 2006). Overexpression of superoxide dismutase in either stem cells or their supporting cells in the niche can prolong stem cell function during aging, as shown by work performed in the Drosophila ovary (Pan et al., 2007b).
Although the consequences of elevated ROS for stem cell function have been widely studied, we have only glimpses of how ROS levels are regulated in stem cells. FoxO transcription factors regulate metabolism and oxidative stress, partly by promoting the expression of antioxidant enzymes (Salih and Brunet, 2008). Conditional deletion of FoxO1, FoxO3, and FoxO4 in mice increases ROS levels and depletes HSCs and neural stem cells (Tothova et al., 2007; Paik et al., 2009). Treatment with N-acetyl-L-cysteine partially rescues the stem cell defects in these mice. FoxO3 appears to be particularly important, because deficiency for FoxO3 alone leads to oxidative stress and depletion of HSCs and neural stem cells (Miyamoto et al., 2007; Yalcin et al., 2008; Renault et al., 2009). Multiple other mechanisms promote stem cell maintenance at least partly by regulating oxidative stress, including the transcription factor Prdm16 (Chuikov et al., 2010), the polycomb family chromatin regulator, Bmi-1 (Liu et al., 2009a), and the DNA damage signaling molecule ATM (Ito et al., 2004; Maryanovich et al., 2012). There are likely to be many additional transcriptional and metabolic mechanisms that influence the generation and response to ROS.
Consistent with the sensitivity of stem cells to ROS, responses to oxygen levels and mitochondrial function are highly regulated in stem cells. The Hypoxia inducible factor 1α (Hif1α) transcription factor regulates stem cell function and aging. Under nor-moxic conditions, the E3 ubiquitin ligase von Hippel Lindau (VHL) targets Hif1α for degradation (Majmundar et al., 2010). However, Hif1α is stabilized in low oxygen conditions, activating the transcription of heat shock proteins, glucose transporters, and glycolytic enzymes that allow a cell to survive in a low oxygen environment. Some hematopoietic and neural stem cells are thought to reside in hypoxic microenvironments (Parmar et al., 2007), and Hif1α is stabilized within these cells to promote their maintenance. Deficiency for Hif1α depletes neurogenic progenitors in the dentate gyrus and HSCs during aging (Mazumdar et al., 2010; Takubo et al., 2010). However, increased stabilization of Hif1α by reduced VHL function also impairs HSC function, suggesting that Hif1α levels must be tightly controlled for stem cell maintenance (Takubo et al., 2010).
Mitochondrial function is regulated in concert with ROS levels. For example, the PGC-1 transcriptional coactivator is a potent activator of mitochondrial biogenesis and oxidative phosphorylation (Puigserver et al., 1998). To avoid inducing oxidative stress, PGC-1 also promotes the expression of ROS-detoxifying enzymes, including GPx1 and SOD2 (St-Pierre et al., 2006). Overexpression of PGC-1 in Drosophila intestinal stem cells is sufficient to increase life span in flies, delaying age-related changes in the intestine and improving tissue homeostasis during aging (Rera et al., 2011). The authors of this study speculated that PGC-1 function within stem cells may be an important determinant of aging and longevity. This idea has not yet been tested in mammals.
Defects in mitochondrial function, such as those caused by an error-prone mitochondrial DNA polymerase, can also accelerate aging phenotypes and reduce life span (Trifunovic et al., 2004). The progeroid phenotypes in these mice include defects in the function of hematopoietic and neural progenitors that can be partially rescued by N-acetyl-L-cysteine treatment (Norddahl et al., 2011; Ahlqvist et al., 2012). But while these studies demonstrate that mitochondrial defects can lead to phenotypes that are reminiscent of premature aging, they do not necessarily demonstrate that mutations to mitochondrial DNA are a mechanism underlying physiological aging because the rate of mitochondrial DNA mutations in aging wild-type mice is 500-fold lower than in the mitochondrial mutator mice (Vermulst et al., 2007).
Non-Cell-Autonomous Regulation of Cellular Aging
Extrinsic factors in the stem cell microenvironment regulate stem cell aging. Stem cells typically reside in specialized microenvironments that promote stem cell maintenance and regulate stem cell function (Morrison and Spradling, 2008). Aging of the niche cells can cause changes in stem cell function. In Drosophila, the number of germline stem cells, their mitotic activity, and the number of progeny all decline with age due to both cell autonomous and non-cell-autonomous changes (Wallenfang et al., 2006; Pan et al., 2007b). In the male testis, these changes are partially caused by changes within the niche, as hub cells from older animals express reduced levels of DE-cadherin and Unpaired, both of which are necessary for germline stem cell maintenance (Boyle et al., 2007). Reduced Unpaired expression is caused partly by an increase in mRNA degradation from let-7-targeting of IGF-II messenger RNA binding protein (IMP) expression in aging hub cells (Toledano et al., 2012). Overexpression of Unpaired in the hub cells of older males rescues the age-related decline in germline stem cell frequency (Boyle et al., 2007). Similarly, in the Drosophila ovary, E-cadherin and BMP expression within the niche decline with age, and genetically increasing expression can enhance the function of old stem cells (Pan et al., 2007b).
Work on muscle stem cells (a subpopulation of satellite cells) suggests that similar age-related changes in the microenvironment within mammalian tissues, as well as in circulation, reduce somatic stem cell function. Aging is associated with a reduced capacity for muscle regeneration after injury, partly as a result of reduced expression of Notch ligand by satellite muscle cells, which reduces satellite cell proliferation after injury (Conboy et al., 2003). Aging muscles also produce elevated levels of TGF-β, which impedes regeneration and satellite cell proliferation (Carlson et al., 2008). However, exposure of old mice to young systemic factors by making old and young mice parabiotic can rejuvenate stem cell function (Conboy et al., 2005). Exposure of satellite cells from old mice to serum from young mice increases Notch ligand expression and proliferation (Conboy et al., 2005). This demonstrates that age-related changes in stem cells are partially reversible and influenced by circulating factors that change with age.
A combination of cell-autonomous and non-cell-autonomous mechanisms regulate the aging of stem cells in other tissues as well. Within the nervous system astrocytes and neural stem cells in the dentate gyrus promote the self-renewal of stem cells and the expansion of neuroblasts by secreting Wnts (Song et al., 2002; Lie et al., 2005). Expression of the Wnt antagonist Dkk1 increases in the aging dentate gyrus, and conditional deletion of Dkk1 from neural stem and progenitor cells increases neural stem cell self-renewal, neurogenesis, and spatial learning and memory in old mice (Seib et al., 2013).
Circulating blood-borne factors also regulate changes in stem cell function in the aging central nervous system where stem cell frequency, overall mitotic activity, and rates of neurogenesis decline profoundly with age in the mouse forebrain (Kuhn et al., 1996; Maslov et al., 2004; Molofsky et al., 2006). In heterochronic parabionts, rates of neurogenesis and other measures of neural function decline in the young parabiont and increase in the old parabiont (Villeda et al., 2011). These effects appear to be partially explained by an age-related increase in the level of CCL11 chemokine in the plasma, which is sufficient to reduce neurogenesis, learning, and memory when administered to young mice. Heterochronic parabiosis also enhances the rate of remyelination after experimentally induced demyelination in old mice, a process that normally declines with age (Ruckh et al., 2012). The enhanced remyelination is associated with the recruitment of blood monocytes from the young parabiont, suggesting that the young circulating agents that enhance the regeneration of aging tissues can be cellular as well as soluble factors.
The circulating hormones insulin and insulin-like growth factor 1 (Igf1) also regulate aging and stem cells. The insulin/Igf1 signaling pathway coordinates growth and development in response to nutrient availability by activating the phosphatidyli-nositol-3-kinase (PI3K) signaling pathway and inactivating FoxO transcription factors. In C. elegans, mutations in daf-2 (an IGFR ortholog) or other downstream signaling components extend life span in a manner that depends upon daf-16 (a FoxO ortholog) (Kenyon et al., 1993; Lin et al., 1997; Ogg et al., 1997). In Drosophila, reducing insulin signaling by ablating insulin receptors, insulin receptor substrates, insulin producing cells, or overexpression of dFOXO all extend life span (Clancy et al., 2001; Tatar et al., 2001; Broughton et al., 2005; Giannakou et al., 2007). Homozygosity for a polymorphism in FOXO3A is associated with longevity in humans (Willcox et al., 2008). Mice with reduced insulin or Igf1 signaling, systemically or only in certain tissues, all exhibit slowed aging and increased life span (Tatar et al., 2001). The ability of the insulin signaling pathway to regulate aging and life span is thus evolutionarily conserved (Tatar et al., 2003).
Insulin signaling is known to regulate stem cells, though stem cell aging in long-lived mutants has not yet been closely examined. In Drosophila, reduced insulin signaling leads to declines in germline stem cell proliferation and fecundity (LaFever and Drummond-Barbosa, 2005), though data on mammalian stem cells are surprisingly limited. It will be interesting to determine whether long-lived insulin pathway mutants have increased stem cell function and regenerative capacity in aging tissues.
The accumulation of senescent cells in aging tissues can also non-cell-autonomously affect the function of other cells. Senescence is a cellular state associated with an irreversible loss in the ability to divide. Senescent cells undergo a series of changes, including the secretion of inflammatory factors, growth regulators, proteases, and other signaling molecules (Coppé et al., 2010). These secreted factors affect other cells in the local environment, promoting senescence and inflammation and promoting or inhibiting tumor growth. Life-long clearance of senescent cells from adult mouse tissues resulting from genetic ablation of p16Ink4a-expressing cells delays the onset of pathologies in multiple aging tissues (Baker et al., 2011). Clearance of senescent cells only late in life did not improve age-related pathologies but did attenuate their progression. This raises the question of the extent to which p16Ink4a non-cell-autonomously or cell-autonomously influences stem cell function in aging tissues.
Dietary Restriction and TOR Signaling
Dietary restriction, defined as reducing food intake below ad libitum (free feeding) levels without causing malnutrition, extends life span in certain contexts while also delaying the onset of age-related pathologies (Mair and Dillin, 2008). Dietary restriction can also increase stem cell function or slow the decline in stem cell function during aging in multiple tissues (Lee et al., 2000; Mair et al., 2010). Short-term dietary restriction increases the frequency and function of satellite cells in skeletal muscle of both young and old mice, partly by increasing mitochondrial content and promoting oxidative metabolism (Cerletti et al., 2012). In at least one short-lived mouse strain, dietary restriction attenuates age-related declines in HSC frequency and reconstituting activity (Chen et al., 2003). However, these effects of dietary restriction may not be universal. Life span extension was not observed in certain mouse strains (Harrison and Archer, 1987) and was observed in monkeys in one study (Colman et al., 2009) but not in another (Mattison et al., 2012).
The effects of dietary restriction on aging and life span are thought to occur partly through modulation of target of rapamycin (TOR) signaling (Figure 4). TOR is a conserved serine/threonine kinase that promotes protein synthesis and cellular growth and is activated by signals that sense nutrient, growth factor, amino acid, and energy availability (Laplante and Sabatini, 2012). TOR is the kinase within at least two multiprotein complexes, TORC1 and TORC2, which contain the Raptor and Rictor binding partners, respectively (Laplante and Sabatini, 2012). Activated TORC1 promotes protein synthesis by phosphorylating ribosomal protein S6 kinase 1 (S6K1), which activates ribosome biogenesis partly by phosphorylating the ribosomal protein S6, and 4E-BP1, which frees eIF4E to bind 5′-capped mRNAs, recruiting them to the ribosomal initiation complex (Laplante and Sabatini, 2012) (Figure 4). In contrast, TORC2 promotes cell growth, proliferation, survival, and aspects of cellular metabolism by phosphorylating AKT (Sarbassov et al., 2005), SGK (García-Martínez and Alessi, 2008), and protein kinase C (Guertin et al., 2006) (Figure 4). TOR therefore has distinct functions in different signaling complexes.
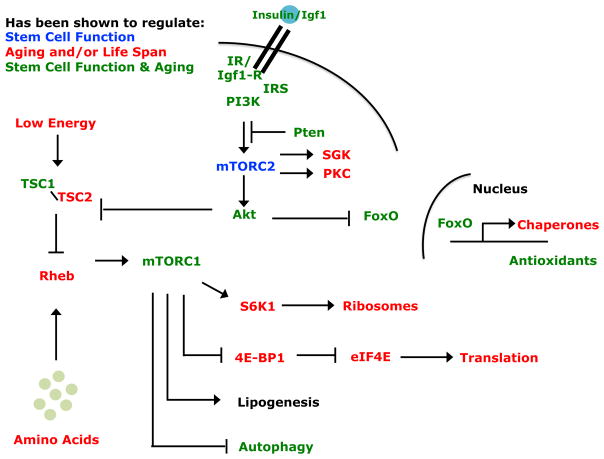
Figure 4. Many Components of the Insulin/PI3K Signaling Pathway Regulate Stem Cell Function and Aging.
A variety of tyrosine kinase receptors, including the insulin receptor, activate the PI3K pathway, which leads to the activation of both mTORC1 and mTOCR2 (Laplante and Sabatini, 2012). mTORC2 can phosphorylate and activate Akt, SGK, and protein kinase C (PKC). Activated Akt can phosphorylate FoxO transcription factors, restricting their localization to the cytosol. FoxOs that translocate to the nucleus can transcriptionally activate the expression of a variety of genes, including protein folding chaperones, antioxidant enzymes, and metabolic regulators (Salih and Brunet, 2008). Activated Akt can also activate mTORC1 by phosphorylating TSC2, which relieves the inhibitory effects of the TSC1/TSC2 complex on Rheb. mTORC1 activates mechanisms that promote protein translation and lipid and nucleic acid synthesis and inhibit autophagy. The components of these pathways that have not yet been studied in stem cells are likely to regulate stem cell function and perhaps even stem cell aging.
Reduced TOR signaling, and TORC1 signaling in particular, can slow organismal aging and extend life span (Figure 4). In C. elegans, heterozygosity for the Raptor ortholog daf-15 significantly increases maximum life span (Jia et al., 2004). Reducing the expression of various downstream targets of TORC1, such as rsks-1 (an S6K1 ortholog), ifg-1 (eIF4G ortholog), and other translation initiation complex factors, also extend C. elegans life span (Hansen et al., 2007; Pan et al., 2007a). In mammals, reduced mammalian TOR (mTOR) signaling also extends life span, such as is observed upon feeding mice the mTORC1 inhibitor rapamycin (Harrison et al., 2009). The beneficial effects of rapamycin on longevity were evident even when treatment was initiated at 270 days of age, suggesting that interventions late in life can influence health and longevity. Mice that are deficient for S6K1 also have increased life span and improved motor function, T cell abundance, bone volume, and insulin sensitivity in old age (Selman et al., 2009). TORC1 signaling is thus a key regulator of aging.
TOR also regulates stem cell function (Figure 4). Dietary restriction reduces mTOR signaling in Paneth cells (a component of the intestinal stem cell niche), which non-cell-autonomously increases the proliferation of intestinal stem cells (Yilmaz et al., 2012). Hyperactivation of mTORC1 and/or mTORC2 by deletion of Pten or TSC1 aberrantly increases the proliferation of neural stem/progenitor cells (Groszer et al., 2001) and HSCs (Yilmaz et al., 2006; Zhang et al., 2006; Gan et al., 2008). However, hyperactivation of mTOR in vivo leads to the depletion of some adult neural stem cells (Bonaguidi et al., 2011) as well as HSCs through mTORC1 and mTORC2-dependent mechanisms (Lee et al., 2010; Kalaitzidis et al., 2012; Magee et al., 2012).
The studies described above would predict that reduced mTOR signaling should also attenuate the decline in stem cell function during aging. However, there remain little data on this point. One study reported that mTORC1 signaling is increased in HSCs isolated from old mice, and that age-related declines in HSC reconstituting activity and lymphoid differentiation could be rescued by rapamycin treatment (Chen et al., 2009a). However, rapamycin treatment also increases HSC frequency in young mice, raising questions about the extent to which these effects reflected a rescue of aging phenotypes. Additional studies examining the consequences of reduced mTOR signaling in young and old stem cells are needed to address its role in stem cell aging.
Proteostasis
A major challenge for aging cells is homeostasis of the proteome (proteostasis) (Taylor and Dillin, 2011). Misfolded or damaged proteins can disrupt membranes, form toxic aggregates, and cause cell death (Bucciantini et al., 2002). Several age-related diseases are associated with protein misfolding, including Alzheimer’s disease, Parkinson’s disease, and Huntington’s disease (Ross and Poirier, 2004). Emerging evidence suggests that proteotoxic stress may be an underlying mechanism in metabolic disorders such as diabetes and a determinant of life span (Balch et al., 2008; Durieux et al., 2011; Vilchez et al., 2012b).
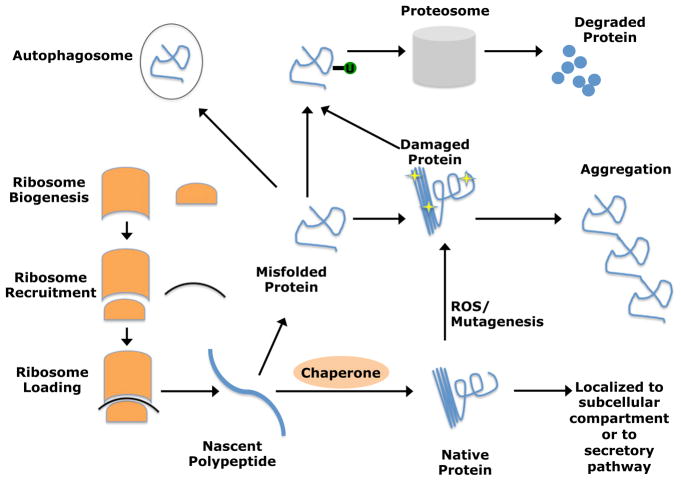
A complex network of cellular machinery regulates proteostasis by monitoring proteins throughout their life cycle (Figure 5). The rate at which proteins are produced is tightly controlled by stringent regulation of translation through control of ribosome biogenesis, ribosome recruitment, and ribosome loading (Gebauer and Hentze, 2004). Protein folding and localization are regulated by molecular chaperones, which trap nascent proteins in tight binding pockets to assist folding, to prevent unwanted aggregation, and to protect them from thermal or oxidative stress (Hartl et al., 2011). Unneeded, misfolded, damaged, and aggregated proteins can be eliminated by the ubiquitin proteasome system (Finley, 2009) or through autophagy (Rubinsztein et al., 2011). Each of these proteostasis mechanisms are capable of eliminating damaged proteins or triggering a more global response, such as cell cycle arrest or apoptosis in the context of severe proteotoxic stress (Ron and Walter, 2007; Boulon et al., 2010).
Figure 5. Proteostasis Is Required for Cellular Homeostasis during Aging.
Proteostasis is regulated by protein translation rates, which are controlled by ribosome biogenesis, recruitment, and loading. Chaperones promote folding of nascent polypeptides or re-folding of misfolded proteins to prevent protein aggregation. Misfolded or damaged proteins can be ubiquitylated and targeted for proteosomal degradation or engulfed and degraded by auto-phagosomes. Interventions that promote proteostasis can slow aging, reduce the incidence of age-related diseases, and increase life span (Cohen et al., 2009; Durieux et al., 2011; Taylor and Dillin, 2011). These mechanisms are likely to influence tissue regeneration and stem cell function during aging, but this remains largely unstudied.
The accumulation of damaged proteins during aging suggests that the capacity to regulate proteostasis declines with age. Protein damage can occur by misfolding, aggregation, glycation, carbonylation, or oxidation, or from translation errors, genetic mutations (Chiti et al., 2003), and reactive metabolites (Berlett and Stadtman, 1997). Mutations and damage from reactive metabolites accumulate with age. In addition, some proteostasis mechanisms are known to decline during aging, including the endoplasmic reticulum stress response (Ben-Zvi et al., 2009) and autophagy (Rubinsztein et al., 2011). Furthermore, interventions that promote proteostasis can slow aging, reduce the incidence of age-related diseases, and increase life span (Cohen et al., 2009; Durieux et al., 2011; Taylor and Dillin, 2011). Decreasing translation by reducing ribosomal protein levels in yeast increases replicative life span (Chiocchetti et al., 2007), and reduced expression of a variety of translation initiation factors increases life span in C. elegans (Pan et al., 2007a) and mammals (Selman et al., 2009).
Many mechanisms that regulate aging and proteostasis also regulate stem cells (Buckley et al., 2012; Vilchez et al., 2012a). Autophagy is likely to be important for HSC maintenance, because deletion of either Atg7 (Mortensen et al., 2011) or Fip200 (Liu et al., 2010), both of which are necessary for autophagy, increases ROS levels and depletes HSCs. FoxO, which promotes longevity and stem cell function, transcriptionally activates the expression of multiple protein-folding chaperones (Murphy et al., 2003; Oh et al., 2006; Demontis and Perrimon, 2010), some of which regulate aging (Tatar et al., 1997; Walker and Lithgow, 2003; Morley and Morimoto, 2004); however, it is unclear to what extent the chaperones regulate stem cell function or stem cell aging. FOXO4 promotes proteasome activity in human embryonic stem cells, promoting proteostasis and maintenance of pluripotency (Vilchez et al., 2012a). In addition, mTOR is a potent activator of protein translation and inhibitor of autophagy (Laplante and Sabatini, 2012), but it is not clear to what extent these proteostasis pathways mediate the effects of mTOR on stem cell function or aging. It will be important to characterize the mechanisms that regulate proteostasis in stem cells to determine whether they differ from other cells and whether they influence changes in stem cell function during aging.
Conclusions
Many aspects of cellular physiology are regulated differently in stem cells as compared to other kinds of cells (He et al., 2009). Some regulators of stem cell self-renewal are broadly required by many types of dividing cells while other key self-renewal regulators do not regulate the proliferation of restricted progenitors in the same tissues. This suggests that some mechanisms that regulate stem cell aging may broadly regulate the aging of many cells, while other mechanisms will preferentially regulate stem cell aging. So far, the data suggest that the mechanisms that promote the onset of aging phenotypes in other cells (DNA damage, ROS, proteotoxicity, circulating factors from old mice, and telomere erosion) also reduce stem cell function in a manner that is consistent with premature aging, at least in certain tissues (Figure 1). Nonetheless, the details of how these mechanisms influence stem cells may differ from how they influence restricted progenitors and postmitotic cells. For example, gate-keeping tumor suppressors may regulate different cells in different ways, potentially slowing the onset of aging phenotypes in some cells while accelerating aging phenotypes in others. It is not surprising that such mechanisms would influence dividing and nondividing cells in different ways; however, there are also likely to be less obvious differences in how these mechanisms influence stem cells versus restricted progenitors.
There are also likely to be important undiscovered mechanisms, including those that preferentially regulate stem cell aging. For example, there is a centrosome orientation checkpoint in Drosophila that prevents spermatogonial stem cells from dividing unless the centrosomes align in a way that facilitates asymmetric division (Cheng et al., 2008). The frequency of stem cells with misaligned centrosomes increases with age, reducing stem cell activity and spermatogenesis. While this is a major cause of the physiological decline in spermatogenesis that occurs with age in flies it remains uncertain whether a similar checkpoint contributes to mammalian stem cell aging. This work illustrates the existence of previously unsuspected mechanisms that regulate declines in stem cell function with age, and that do not fit neatly into the themes emphasized by common theories of aging. Much work remains to be done to understand the mechanisms that regulate stem cell aging. Elucidating these mechanisms will be critical to understanding how regenerative capacity is preserved in certain tissues throughout adult life, and why that capacity declines with age.
Acknowledgments
This work was supported by the Howard Hughes Medical Institute, the Cancer Prevention and Research Institute of Texas, and the National Institute on Aging (R37 AG024945). R.A.J.S. is supported by fellowships from the Leukemia & Lymphoma Society (5541-11) and the Canadian Institutes of Health Research (MFE-106993). We apologize to authors whose papers we could not cite due to space limitations.
References
- Ahlqvist KJ, Hämaäläinen RH, Yatsuga S, Uutela M, Terzioglu M, Götz A, Forsström S, Salven P, Angers-Loustau A, Kopra OH, et al. Somatic progenitor cell vulnerability to mitochondrial DNA mutagenesis underlies progeroid phenotypes in Polg mutator mice. Cell Metab. 2012;15:100–109. doi: 10.1016/j.cmet.2011.11.012. [DOI] [PubMed] [Google Scholar]
- Allsopp RC, Morin GB, DePinho R, Harley CB, Weissman IL. Telomerase is required to slow telomere shortening and extend replicative lifespan of HSCs during serial transplantation. Blood. 2003;102:517–520. doi: 10.1182/blood-2002-07-2334. [DOI] [PubMed] [Google Scholar]
- Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- Artandi SE, Chang S, Lee SL, Alson S, Gottlieb GJ, Chin L, DePinho RA. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–645. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- Baker DJ, Wijshake T, Tchkonia T, LeBrasseur NK, Childs BG, van de Sluis B, Kirkland JL, van Deursen JM. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479:232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Begus-Nahrmann Y, Lechel A, Obenauf AC, Nalapareddy K, Peit E, Hoffmann E, Schlaudraff F, Liss B, Schirmacher P, Kestler H, et al. p53 deletion impairs clearance of chromosomal-instable stem cells in aging telomere-dysfunctional mice. Nat Genet. 2009;41:1138–1143. doi: 10.1038/ng.426. [DOI] [PubMed] [Google Scholar]
- Ben-Zvi A, Miller EA, Morimoto RI. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc Natl Acad Sci USA. 2009;106:14914–14919. doi: 10.1073/pnas.0902882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlett BS, Stadtman ER. Protein oxidation in aging, disease, and oxidative stress. J Biol Chem. 1997;272:20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- Biteau B, Karpac J, Supoyo S, Degennaro M, Lehmann R, Jasper H. Lifespan extension by preserving proliferative homeostasis in Drosophila. PLoS Genet. 2010;6:e1001159. doi: 10.1371/journal.pgen.1001159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- Bonaguidi MA, Wheeler MA, Shapiro JS, Stadel RP, Sun GJ, Ming GL, Song H. In vivo clonal analysis reveals self-renewing and multipotent adult neural stem cell characteristics. Cell. 2011;145:1142–1155. doi: 10.1016/j.cell.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. The nucleolus under stress. Mol Cell. 2010;40:216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle M, Wong C, Rocha M, Jones DL. Decline in self-renewal factors contributes to aging of the stem cell niche. Cell Stem Cell. 2007;1:470–478. doi: 10.1016/j.stem.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Broughton SJ, Piper MD, Ikeya T, Bass TM, Jacobson J, Driege Y, Martinez P, Hafen E, Withers DJ, Leevers SJ, Partridge L. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc Natl Acad Sci USA. 2005;102:3105–3110. doi: 10.1073/pnas.0405775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson CM, Stefani M. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- Buckley SM, Aranda-Orgilles B, Strikoudis A, Apostolou E, Loizou E, Moran-Crusio K, Farnsworth CL, Koller AA, Dasgupta R, Silva JC, et al. Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell Stem Cell. 2012;11:783–798. doi: 10.1016/j.stem.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler AE, Janson J, Bonner-Weir S, Ritzel R, Rizza RA, Butler PC. Beta-cell deficit and increased beta-cell apoptosis in humans with type 2 diabetes. Diabetes. 2003;52:102–110. doi: 10.2337/diabetes.52.1.102. [DOI] [PubMed] [Google Scholar]
- Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Carlson ME, Hsu M, Conboy IM. Imbalance between pSmad3 and Notch induces CDK inhibitors in old muscle stem cells. Nature. 2008;454:528–532. doi: 10.1038/nature07034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerletti M, Jang YC, Finley LW, Haigis MC, Wagers AJ. Short-term calorie restriction enhances skeletal muscle stem cell function. Cell Stem Cell. 2012;10:515–519. doi: 10.1016/j.stem.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Shaw CA, Gatza C, Fisk CJ, Donehower LA, Goodell MA. Aging hematopoietic stem cells decline in function and exhibit epigenetic dysregulation. PLoS Biol. 2007;5:e201. doi: 10.1371/journal.pbio.0050201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Multani AS, Cabrera NG, Naylor ML, Laud P, Lombard D, Pathak S, Guarente L, DePinho RA. Essential role of limiting telomeres in the pathogenesis of Werner syndrome. Nat Genet. 2004;36:877–882. doi: 10.1038/ng1389. [DOI] [PubMed] [Google Scholar]
- Chen J, Astle CM, Harrison DE. Genetic regulation of primitive hematopoietic stem cell senescence. Exp Hematol. 2000;28:442–450. doi: 10.1016/s0301-472x(99)00157-5. [DOI] [PubMed] [Google Scholar]
- Chen J, Astle CM, Harrison DE. Hematopoietic senescence is postponed and hematopoietic stem cell function is enhanced by dietary restriction. Exp Hematol. 2003;31:1097–1103. doi: 10.1016/s0301-472x(03)00238-8. [DOI] [PubMed] [Google Scholar]
- Chen C, Liu Y, Liu Y, Zheng P. mTOR regulation and therapeutic rejuvenation of aging hematopoietic stem cells. Sci Signal. 2009a;2:ra75. doi: 10.1126/scisignal.2000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Gu X, Su IH, Bottino R, Contreras JL, Tarakhovsky A, Kim SK. Polycomb protein Ezh2 regulates pancreatic beta-cell Ink4a/Arf expression and regeneration in diabetes mellitus. Genes Dev. 2009b;23:975–985. doi: 10.1101/gad.1742509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Türkel N, Hemati N, Fuller MT, Hunt AJ, Yamashita YM. Centrosome misorientation reduces stem cell division during ageing. Nature. 2008;456:599–604. doi: 10.1038/nature07386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L, Artandi SE, Shen Q, Tam A, Lee SL, Gottlieb GJ, Greider CW, DePinho RA. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97:527–538. doi: 10.1016/s0092-8674(00)80762-x. [DOI] [PubMed] [Google Scholar]
- Chiocchetti A, Zhou J, Zhu H, Karl T, Haubenreisser O, Rinnerthaler M, Heeren G, Oender K, Bauer J, Hintner H, et al. Ribosomal proteins Rpl10 and Rps6 are potent regulators of yeast replicative life span. Exp Gerontol. 2007;42:275–286. doi: 10.1016/j.exger.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Chiti F, Stefani M, Taddei N, Ramponi G, Dobson CM. Rationalization of the effects of mutations on peptide and protein aggregation rates. Nature. 2003;424:805–808. doi: 10.1038/nature01891. [DOI] [PubMed] [Google Scholar]
- Choudhury AR, Ju Z, Djojosubroto MW, Schienke A, Lechel A, Schaetzlein S, Jiang H, Stepczynska A, Wang C, Buer J, et al. Cdkn1a deletion improves stem cell function and lifespan of mice with dysfunctional telomeres without accelerating cancer formation. Nat Genet. 2007;39:99–105. doi: 10.1038/ng1937. [DOI] [PubMed] [Google Scholar]
- Chuikov S, Levi BP, Smith ML, Morrison SJ. Prdm16 promotes stem cell maintenance in multiple tissues, partly by regulating oxidative stress. Nat Cell Biol. 2010;12:999–1006. doi: 10.1038/ncb2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicalese A, Bonizzi G, Pasi CE, Faretta M, Ronzoni S, Giulini B, Brisken C, Minucci S, Di Fiore PP, Pelicci PG. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell. 2009;138:1083–1095. doi: 10.1016/j.cell.2009.06.048. [DOI] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy DJ, Gems D, Harshman LG, Oldham S, Stocker H, Hafen E, Leevers SJ, Partridge L. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292:104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- Cohen E, Paulsson JF, Blinder P, Burstyn-Cohen T, Du D, Estepa G, Adame A, Pham HM, Holzenberger M, Kelly JW, et al. Reduced IGF-1 signaling delays age-associated proteotoxicity in mice. Cell. 2009;139:1157–1169. doi: 10.1016/j.cell.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman RJ, Anderson RM, Johnson SC, Kastman EK, Kosmatka KJ, Beasley TM, Allison DB, Cruzen C, Simmons HA, Kemnitz JW, Weindruch R. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325:201–204. doi: 10.1126/science.1173635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy IM, Conboy MJ, Smythe GM, Rando TA. Notch-mediated restoration of regenerative potential to aged muscle. Science. 2003;302:1575–1577. doi: 10.1126/science.1087573. [DOI] [PubMed] [Google Scholar]
- Conboy IM, Conboy MJ, Wagers AJ, Girma ER, Weissman IL, Rando TA. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature. 2005;433:760–764. doi: 10.1038/nature03260. [DOI] [PubMed] [Google Scholar]
- Coppé JP, Desprez PY, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan G, Nijhof W, Van Zant G. Mouse strain-dependent changes in frequency and proliferation of hematopoietic stem cells during aging: correlation between lifespan and cycling activity. Blood. 1997;89:1543–1550. [PubMed] [Google Scholar]
- Demontis F, Perrimon N. FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell. 2010;143:813–825. doi: 10.1016/j.cell.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- Dorshkind K, Montecino-Rodriguez E, Signer RA. The ageing immune system: is it ever too old to become young again? Nat. Rev Immunol. 2009;9:57–62. doi: 10.1038/nri2471. [DOI] [PubMed] [Google Scholar]
- Dumble M, Moore L, Chambers SM, Geiger H, Van Zant G, Goodell MA, Donehower LA. The impact of altered p53 dosage on hematopoietic stem cell dynamics during aging. Blood. 2007;109:1736–1742. doi: 10.1182/blood-2006-03-010413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrón SR, Marqués-Torrejón MA, Mira H, Flores I, Taylor K, Blasco MA, Fariñas I. Telomere shortening in neural stem cells disrupts neuronal differentiation and neuritogenesis. J Neurosci. 2009;29:14394–14407. doi: 10.1523/JNEUROSCI.3836-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga CG, Shigenaga MK, Park JW, Degan P, Ames BN. Oxidative damage to DNA during aging: 8-hydroxy-2′-deoxyguanosine in rat organ DNA and urine. Proc Natl Acad Sci USA. 1990;87:4533–4537. doi: 10.1073/pnas.87.12.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan B, Sahin E, Jiang S, Sanchez-Aguilera A, Scott KL, Chin L, Williams DA, Kwiatkowski DJ, DePinho RA. mTORC1-dependent and -independent regulation of stem cell renewal, differentiation, and mobilization. Proc Natl Acad Sci USA. 2008;105:19384–19389. doi: 10.1073/pnas.0810584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon HS, Donehower LA, Lyle S, Jones SN. Mdm2-p53 signaling regulates epidermal stem cell senescence and premature aging phenotypes in mouse skin. Dev Biol. 2011;353:1–9. doi: 10.1016/j.ydbio.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Cao I, García-Cao M, Martín-Caballero J, Criado LM, Klatt P, Flores JM, Weill JC, Blasco MA, Serrano M. “Super p53” mice exhibit enhanced DNA damage response, are tumor resistant and age normally. EMBO J. 2002;21:6225–6235. doi: 10.1093/emboj/cdf595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Martínez JM, Alessi DR. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1) Biochem J. 2008;416:375–385. doi: 10.1042/BJ20081668. [DOI] [PubMed] [Google Scholar]
- Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakou ME, Goss M, Jacobson J, Vinti G, Leevers SJ, Partridge L. Dynamics of the action of dFOXO on adult mortality in Drosophila. Aging Cell. 2007;6:429–438. doi: 10.1111/j.1474-9726.2007.00290.x. [DOI] [PubMed] [Google Scholar]
- Groszer M, Erickson R, Scripture-Adams DD, Lesche R, Trumpp A, Zack JA, Kornblum HI, Liu X, Wu H. Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science. 2001;294:2186–2189. doi: 10.1126/science.1065518. [DOI] [PubMed] [Google Scholar]
- Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- Hao LY, Armanios M, Strong MA, Karim B, Feldser DM, Huso D, Greider CW. Short telomeres, even in the presence of telomerase, limit tissue renewal capacity. Cell. 2005;123:1121–1131. doi: 10.1016/j.cell.2005.11.020. [DOI] [PubMed] [Google Scholar]
- Harley CB, Vaziri H, Counter CM, Allsopp RC. The telomere hypothesis of cellular aging. Exp Gerontol. 1992;27:375–382. doi: 10.1016/0531-5565(92)90068-b. [DOI] [PubMed] [Google Scholar]
- Harrison DE, Archer JR. Genetic differences in effects of food restriction on aging in mice. J Nutr. 1987;117:376–382. doi: 10.1093/jn/117.2.376. [DOI] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- He S, Nakada D, Morrison SJ. Mechanisms of stem cell self-renewal. Annu Rev Cell Dev Biol. 2009;25:377–406. doi: 10.1146/annurev.cellbio.042308.113248. [DOI] [PubMed] [Google Scholar]
- Heidinger BJ, Blount JD, Boner W, Griffiths K, Metcalfe NB, Monaghan P. Telomere length in early life predicts lifespan. Proc Natl Acad Sci USA. 2012;109:1743–1748. doi: 10.1073/pnas.1113306109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- Inomata K, Aoto T, Binh NT, Okamoto N, Tanimura S, Wakayama T, Iseki S, Hara E, Masunaga T, Shimizu H, Nishimura EK. Genotoxic stress abrogates renewal of melanocyte stem cells by triggering their differentiation. Cell. 2009;137:1088–1099. doi: 10.1016/j.cell.2009.03.037. [DOI] [PubMed] [Google Scholar]
- Ito K, Hirao A, Arai F, Matsuoka S, Takubo K, Hamaguchi I, Nomiyama K, Hosokawa K, Sakurada K, Nakagata N, et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 2004;431:997–1002. doi: 10.1038/nature02989. [DOI] [PubMed] [Google Scholar]
- Ito K, Hirao A, Arai F, Takubo K, Matsuoka S, Miyamoto K, Ohmura M, Naka K, Hosokawa K, Ikeda Y, Suda T. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat Med. 2006;12:446–451. doi: 10.1038/nm1388. [DOI] [PubMed] [Google Scholar]
- Jacobs JJL, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- Janzen V, Forkert R, Fleming HE, Saito Y, Waring MT, Dombkowski DM, Cheng T, DePinho RA, Sharpless NE, Scadden DT. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;443:421–426. doi: 10.1038/nature05159. [DOI] [PubMed] [Google Scholar]
- Jaskelioff M, Muller FL, Paik JH, Thomas E, Jiang S, Adams AC, Sahin E, Kost-Alimova M, Protopopov A, Cadiñanos J, et al. Telomerase reactivation reverses tissue degeneration in aged telomerase-deficient mice. Nature. 2011;469:102–106. doi: 10.1038/nature09603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck WR, Siebold AP, Sharpless NE. Review: a meta-analysis of GWAS and age-associated diseases. Aging Cell. 2012;11:727–731. doi: 10.1111/j.1474-9726.2012.00871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia K, Chen D, Riddle DL. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development. 2004;131:3897–3906. doi: 10.1242/dev.01255. [DOI] [PubMed] [Google Scholar]
- Ju Z, Jiang H, Jaworski M, Rathinam C, Gompf A, Klein C, Trumpp A, Rudolph KL. Telomere dysfunction induces environmental alterations limiting hematopoietic stem cell function and engraftment. Nat Med. 2007;13:742–747. doi: 10.1038/nm1578. [DOI] [PubMed] [Google Scholar]
- Kalaitzidis D, Sykes SM, Wang Z, Punt N, Tang Y, Ragu C, Sinha AU, Lane SW, Souza AL, Clish CB, et al. mTOR complex 1 plays critical roles in hematopoiesis and Pten-loss-evoked leukemogenesis. Cell Stem Cell. 2012;11:429–439. doi: 10.1016/j.stem.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, Grosveld G, Sherr CJ. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kim NW, Piatzszek MA, Prowse KR, Harley CB, West MD, Ho PLC, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- Kinzler KW, Vogelstein B. Cancer-susceptibility genes. Gatekeepers and caretakers. Nature. 1997;386:761–763. doi: 10.1038/386761a0. [DOI] [PubMed] [Google Scholar]
- Kipling D, Cooke HJ. Hypervariable ultra-long telomeres in mice. Nature. 1990;347:400–402. doi: 10.1038/347400a0. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy J, Torrice C, Ramsey MR, Kovalev GI, Al-Regaiey K, Su L, Sharpless NE. Ink4a/Arf expression is a biomarker of aging. J Clin Invest. 2004;114:1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy J, Ramsey MR, Ligon KL, Torrice C, Koh A, Bonner-Weir S, Sharpless NE. p16INK4a induces an age-dependent decline in islet regenerative potential. Nature. 2006;443:453–457. doi: 10.1038/nature05092. [DOI] [PubMed] [Google Scholar]
- Kuhn HG, Dickinson-Anson H, Gage FH. Neurogenesis in the dentate gyrus of the adult rat: age-related decrease of neuronal progenitor proliferation. J Neurosci. 1996;16:2027–2033. doi: 10.1523/JNEUROSCI.16-06-02027.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFever L, Drummond-Barbosa D. Direct control of germline stem cell division and cyst growth by neural insulin in Drosophila. Science. 2005;309:1071–1073. doi: 10.1126/science.1111410. [DOI] [PubMed] [Google Scholar]
- Lansdorp PM. Telomeres and disease. EMBO J. 2009;28:2532–2540. doi: 10.1038/emboj.2009.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Blasco MA, Gottlieb GJ, Horner JW, 2nd, Greider CW, DePinho RA. Essential role of mouse telomerase in highly proliferative organs. Nature. 1998;392:569–574. doi: 10.1038/33345. [DOI] [PubMed] [Google Scholar]
- Lee J, Duan W, Long JM, Ingram DK, Mattson MP. Dietary restriction increases the number of newly generated neural cells, and induces BDNF expression, in the dentate gyrus of rats. J Mol Neurosci. 2000;15:99–108. doi: 10.1385/JMN:15:2:99. [DOI] [PubMed] [Google Scholar]
- Lee JY, Nakada D, Yilmaz OH, Tothova Z, Joseph NM, Lim MS, Gilliland DG, Morrison SJ. mTOR activation induces tumor suppressors that inhibit leukemogenesis and deplete hematopoietic stem cells after Pten deletion. Cell Stem Cell. 2010;7:593–605. doi: 10.1016/j.stem.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- Lie DC, Colamarino SA, Song HJ, Desire L, Mira H, Consiglio A, Lein ES, Jessberger S, Lansford H, Dearie AR, Gage FH. Wnt signalling regulates adult hippocampal neurogenesis. Nature. 2005;437:1370–1375. doi: 10.1038/nature04108. [DOI] [PubMed] [Google Scholar]
- Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- Liu J, Cao L, Chen J, Song S, Lee IH, Quijano C, Liu H, Keyvanfar K, Chen H, Cao LY, et al. Bmi1 regulates mitochondrial function and the DNA damage response pathway. Nature. 2009a;459:387–392. doi: 10.1038/nature08040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Elf SE, Miyata Y, Sashida G, Liu Y, Huang G, Di Giandomenico S, Lee JM, Deblasio A, Menendez S, et al. p53 regulates hematopoietic stem cell quiescence. Cell Stem Cell. 2009b;4:37–48. doi: 10.1016/j.stem.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Lee JY, Wei H, Tanabe O, Engel JD, Morrison SJ, Guan JL. FIP200 is required for the cell-autonomous maintenance of fetal hematopoietic stem cells. Blood. 2010;116:4806–4814. doi: 10.1182/blood-2010-06-288589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Johnson SM, Fedoriw Y, Rogers AB, Yuan H, Krishnamurthy J, Sharpless NE. Expression of p16(INK4a) prevents cancer and promotes aging in lymphocytes. Blood. 2011;117:3257–3267. doi: 10.1182/blood-2010-09-304402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, Yankner BA. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- Luckey CJ, Bhattacharya D, Goldrath AW, Weissman IL, Benoist C, Mathis D. Memory T and memory B cells share a transcriptional program of self-renewal with long-term hematopoietic stem cells. Proc Natl Acad Sci USA. 2006;103:3304–3309. doi: 10.1073/pnas.0511137103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee JA, Ikenoue T, Nakada D, Lee JY, Guan KL, Morrison SJ. Temporal changes in PTEN and mTORC2 regulation of hematopoietic stem cell self-renewal and leukemia suppression. Cell Stem Cell. 2012;11:415–428. doi: 10.1016/j.stem.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair W, Dillin A. Aging and survival: the genetics of life span extension by dietary restriction. Annu Rev Biochem. 2008;77:727–754. doi: 10.1146/annurev.biochem.77.061206.171059. [DOI] [PubMed] [Google Scholar]
- Mair W, McLeod CJ, Wang L, Jones DL. Dietary restriction enhances germline stem cell maintenance. Aging Cell. 2010;9:916–918. doi: 10.1111/j.1474-9726.2010.00602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GM. Genetic modulation of senescent phenotypes in Homo sapiens. Cell. 2005;120:523–532. doi: 10.1016/j.cell.2005.01.031. [DOI] [PubMed] [Google Scholar]
- Maryanovich M, Oberkovitz G, Niv H, Vorobiyov L, Zaltsman Y, Brenner O, Lapidot T, Jung S, Gross A. The ATM-BID pathway regulates quiescence and survival of haematopoietic stem cells. Nat Cell Biol. 2012;14:535–541. doi: 10.1038/ncb2468. [DOI] [PubMed] [Google Scholar]
- Maslov AY, Barone TA, Plunkett RJ, Pruitt SC. Neural stem cell detection, characterization, and age-related changes in the subventricular zone of mice. J Neurosci. 2004;24:1726–1733. doi: 10.1523/JNEUROSCI.4608-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheu A, Pantoja C, Efeyan A, Criado LM, Martín-Caballero J, Flores JM, Klatt P, Serrano M. Increased gene dosage of Ink4a/Arf results in cancer resistance and normal aging. Genes Dev. 2004;18:2736–2746. doi: 10.1101/gad.310304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheu A, Maraver A, Klatt P, Flores I, Garcia-Cao I, Borras C, Flores JM, Viña J, Blasco MA, Serrano M. Delayed ageing through damage protection by the Arf/p53 pathway. Nature. 2007;448:375–379. doi: 10.1038/nature05949. [DOI] [PubMed] [Google Scholar]
- Mattison JA, Roth GS, Beasley TM, Tilmont EM, Handy AM, Herbert RL, Longo DL, Allison DB, Young JE, Bryant M, et al. Impact of caloric restriction on health and survival in rhesus monkeys from the NIA study. Nature. 2012;489:318–321. doi: 10.1038/nature11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumdar J, O’Brien WT, Johnson RS, LaManna JC, Chavez JC, Klein PS, Simon MC. O2 regulates stem cells through Wnt/β-catenin signalling. Nat Cell Biol. 2010;12:1007–1013. doi: 10.1038/ncb2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt AJ, Potten CS, Kemp CJ, Hickman JA, Balmain A, Lane DP, Hall PA. The role of p53 in spontaneous and radiation-induced apoptosis in the gastrointestinal tract of normal and p53-deficient mice. Cancer Res. 1994;54:614–617. [PubMed] [Google Scholar]
- Miyamoto K, Araki KY, Naka K, Arai F, Takubo K, Yamazaki S, Matsuoka S, Miyamoto T, Ito K, Ohmura M, et al. Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell. 2007;1:101–112. doi: 10.1016/j.stem.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, Slutsky SG, Joseph NM, He S, Pardal R, Krishnamurthy J, Sharpless NE, Morrison SJ. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006;443:448–452. doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley JF, Morimoto RI. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell. 2004;15:657–664. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Spradling AC. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132:598–611. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Prowse KR, Ho P, Weissman IL. Telomerase activity in hematopoietic cells is associated with self-renewal potential. Immunity. 1996a;5:207–216. doi: 10.1016/s1074-7613(00)80316-7. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Wandycz AM, Akashi K, Globerson A, Weissman IL. The aging of hematopoietic stem cells. Nat Med. 1996b;2:1011–1016. doi: 10.1038/nm0996-1011. [DOI] [PubMed] [Google Scholar]
- Mortensen M, Soilleux EJ, Djordjevic G, Tripp R, Lutteropp M, Sadighi-Akha E, Stranks AJ, Glanville J, Knight S, Jacobsen SE, et al. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. J Exp Med. 2011;208:455–467. doi: 10.1084/jem.20101145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Navarro S, Meza NW, Quintana-Bustamante O, Casado JA, Jacome A, McAllister K, Puerto S, Surrallés J, Segovia JC, Bueren JA. Hematopoietic dysfunction in a mouse model for Fanconi anemia group D1. Mol Ther. 2006;14:525–535. doi: 10.1016/j.ymthe.2006.05.018. [DOI] [PubMed] [Google Scholar]
- Nijnik A, Woodbine L, Marchetti C, Dawson S, Lambe T, Liu C, Rodrigues NP, Crockford TL, Cabuy E, Vindigni A, et al. DNA repair is limiting for haematopoietic stem cells during ageing. Nature. 2007;447:686–690. doi: 10.1038/nature05875. [DOI] [PubMed] [Google Scholar]
- Nishimura EK, Granter SR, Fisher DE. Mechanisms of hair graying: incomplete melanocyte stem cell maintenance in the niche. Science. 2005;307:720–724. doi: 10.1126/science.1099593. [DOI] [PubMed] [Google Scholar]
- Nishino J, Kim I, Chada K, Morrison SJ. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell. 2008;135:227–239. doi: 10.1016/j.cell.2008.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norddahl GL, Pronk CJ, Wahlestedt M, Sten G, Nygren JM, Ugale A, Sigvardsson M, Bryder D. Accumulating mitochondrial DNA mutations drive premature hematopoietic aging phenotypes distinct from physiological stem cell aging. Cell Stem Cell. 2011;8:499–510. doi: 10.1016/j.stem.2011.03.009. [DOI] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- Oh SW, Mukhopadhyay A, Dixit BL, Raha T, Green MR, Tissenbaum HA. Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat Genet. 2006;38:251–257. doi: 10.1038/ng1723. [DOI] [PubMed] [Google Scholar]
- Owusu-Ansah E, Banerjee U. Reactive oxygen species prime Drosophila haematopoietic progenitors for differentiation. Nature. 2009;461:537–541. doi: 10.1038/nature08313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik JH, Ding Z, Narurkar R, Ramkissoon S, Muller F, Kamoun WS, Chae SS, Zheng H, Ying H, Mahoney J, et al. FoxOs cooperatively regulate diverse pathways governing neural stem cell homeostasis. Cell Stem Cell. 2009;5:540–553. doi: 10.1016/j.stem.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- Pan KZ, Palter JE, Rogers AN, Olsen A, Chen D, Lithgow GJ, Kapahi P. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell. 2007a;6:111–119. doi: 10.1111/j.1474-9726.2006.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L, Chen S, Weng C, Call G, Zhu D, Tang H, Zhang N, Xie T. Stem cell aging is controlled both intrinsically and extrinsically in the Drosophila ovary. Cell Stem Cell. 2007b;1:458–469. doi: 10.1016/j.stem.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- Parmar K, Mauch P, Vergilio JA, Sackstein R, Down JD. Distribution of hematopoietic stem cells in the bone marrow according to regional hypoxia. Proc Natl Acad Sci USA. 2007;104:5431–5436. doi: 10.1073/pnas.0701152104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Müller P, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Reese JS, Liu L, Gerson SL. Repopulating defect of mismatch repair-deficient hematopoietic stem cells. Blood. 2003;102:1626–1633. doi: 10.1182/blood-2002-10-3035. [DOI] [PubMed] [Google Scholar]
- Renault VM, Rafalski VA, Morgan AA, Salih DA, Brett JO, Webb AE, Villeda SA, Thekkat PU, Guillerey C, Denko NC, et al. FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell. 2009;5:527–539. doi: 10.1016/j.stem.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rera M, Bahadorani S, Cho J, Koehler CL, Ulgherait M, Hur JH, Ansari WS, Lo T, Jr, Jones DL, Walker DW. Modulation of longevity and tissue homeostasis by the Drosophila PGC-1 homolog. Cell Metab. 2011;14:623–634. doi: 10.1016/j.cmet.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med Suppl. 2004;10:S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- Rossi DJ, Bryder D, Seita J, Nussenzweig A, Hoeijmakers J, Weissman IL. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature. 2007;447:725–729. doi: 10.1038/nature05862. [DOI] [PubMed] [Google Scholar]
- Rossi DJ, Jamieson CH, Weissman IL. Stems cells and the pathways to aging and cancer. Cell. 2008;132:681–696. doi: 10.1016/j.cell.2008.01.036. [DOI] [PubMed] [Google Scholar]
- Ruas M, Peters G. The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochim Biophys Acta. 1998;1378:F115–F177. doi: 10.1016/s0304-419x(98)00017-1. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Mariño G, Kroemer G. Autophagy and aging. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- Ruckh JM, Zhao JW, Shadrach JL, van Wijngaarden P, Rao TN, Wagers AJ, Franklin RJ. Rejuvenation of regeneration in the aging central nervous system. Cell Stem Cell. 2012;10:96–103. doi: 10.1016/j.stem.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph KL, Chang S, Lee HW, Blasco M, Gottlieb GJ, Greider C, DePinho RA. Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell. 1999;96:701–712. doi: 10.1016/s0092-8674(00)80580-2. [DOI] [PubMed] [Google Scholar]
- Ruzankina Y, Pinzon-Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, Zediak VP, Velez M, Bhandoola A, Brown EJ. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell. 2007;1:113–126. doi: 10.1016/j.stem.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzankina Y, Schoppy DW, Asare A, Clark CE, Vonderheide RH, Brown EJ. Tissue regenerative delays and synthetic lethality in adult mice after combined deletion of Atr and Trp53. Nat Genet. 2009;41:1144–1149. doi: 10.1038/ng.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin E, Depinho RA. Linking functional decline of telomeres, mitochondria and stem cells during ageing. Nature. 2010;464:520–528. doi: 10.1038/nature08982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin E, Colla S, Liesa M, Moslehi J, Müller FL, Guo M, Cooper M, Kotton D, Fabian AJ, Walkey C, et al. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 2011;470:359–365. doi: 10.1038/nature09787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salih DA, Brunet A. FoxO transcription factors in the maintenance of cellular homeostasis during aging. Curr Opin Cell Biol. 2008;20:126–136. doi: 10.1016/j.ceb.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Schoppy DW, Ruzankina Y, Brown EJ. Removing all obstacles: a critical role for p53 in promoting tissue renewal. Cell Cycle. 2010;9:1313–1319. doi: 10.4161/cc.9.7.11194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seib D, Corsini NS, Ellwanger K, Plaas C, Mateos A, Pitzer C, Niehrs C, Celikel T, Martin-Villalba A. Loss of Dickkopf-1 restores neurogenesis in old age and counteracts cognitive decline. Cell Stem Cell. 2013;12:204–214. doi: 10.1016/j.stem.2012.11.010. this issue. [DOI] [PubMed] [Google Scholar]
- Selman C, Tullet JM, Wieser D, Irvine E, Lingard SJ, Choudhury AI, Claret M, Al-Qassab H, Carmignac D, Ramadani F, et al. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science. 2009;326:140–144. doi: 10.1126/science.1177221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- Sharpless NE, DePinho RA. How stem cells age and why this makes us grow old. Nat Rev Mol Cell Biol. 2007;8:703–713. doi: 10.1038/nrm2241. [DOI] [PubMed] [Google Scholar]
- Signer RA, Montecino-Rodriguez E, Witte ON, Dorshkind K. Aging and cancer resistance in lymphoid progenitors are linked processes conferred by p16Ink4a and Arf. Genes Dev. 2008;22:3115–3120. doi: 10.1101/gad.1715808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Stevens CF, Gage FH. Astroglia induce neurogenesis from adult neural stem cells. Nature. 2002;417:39–44. doi: 10.1038/417039a. [DOI] [PubMed] [Google Scholar]
- Sotiropoulou PA, Candi A, Mascré G, De Clercq S, Youssef KK, Lapouge G, Dahl E, Semeraro C, Denecker G, Marine JC, Blanpain C. Bcl-2 and accelerated DNA repair mediates resistance of hair follicle bulge stem cells to DNA-damage-induced cell death. Nat Cell Biol. 2010;12:572–582. doi: 10.1038/ncb2059. [DOI] [PubMed] [Google Scholar]
- Sperka T, Song Z, Morita Y, Nalapareddy K, Guachalla LM, Lechel A, Begus-Nahrmann Y, Burkhalter MD, Mach M, Schlaudraff F, et al. Puma and p21 represent cooperating checkpoints limiting self-renewal and chromosomal instability of somatic stem cells in response to telomere dysfunction. Nat Cell Biol. 2012;14:73–79. doi: 10.1038/ncb2388. [DOI] [PubMed] [Google Scholar]
- St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jäger S, Handschin C, Zheng K, Lin J, Yang W, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- Takubo K, Ohmura M, Azuma M, Nagamatsu G, Yamada W, Arai F, Hirao A, Suda T. Stem cell defects in ATM-deficient undifferentiated spermatogonia through DNA damage-induced cell-cycle arrest. Cell Stem Cell. 2008;2:170–182. doi: 10.1016/j.stem.2007.10.023. [DOI] [PubMed] [Google Scholar]
- Takubo K, Goda N, Yamada W, Iriuchishima H, Ikeda E, Kubota Y, Shima H, Johnson RS, Hirao A, Suematsu M, Suda T. Regulation of the HIF-1alpha level is essential for hematopoietic stem cells. Cell Stem Cell. 2010;7:391–402. doi: 10.1016/j.stem.2010.06.020. [DOI] [PubMed] [Google Scholar]
- Tatar M, Khazaeli AA, Curtsinger JW. Chaperoning extended life. Nature. 1997;390:30. doi: 10.1038/36237. [DOI] [PubMed] [Google Scholar]
- Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, Garofalo RS. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science. 2001;292:107–110. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- Tatar M, Bartke A, Antebi A. The endocrine regulation of aging by insulin-like signals. Science. 2003;299:1346–1351. doi: 10.1126/science.1081447. [DOI] [PubMed] [Google Scholar]
- Taylor RC, Dillin A. Aging as an event of proteostasis collapse. Cold Spring Harb Perspect Biol. 2011;3:3. doi: 10.1101/cshperspect.a004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TeKippe M, Harrison DE, Chen J. Expansion of hematopoietic stem cell phenotype and activity in Trp53-null mice. Exp Hematol. 2003;31:521–527. doi: 10.1016/s0301-472x(03)00072-9. [DOI] [PubMed] [Google Scholar]
- Teta M, Long SY, Wartschow LM, Rankin MM, Kushner JA. Very slow turnover of beta-cells in aged adult mice. Diabetes. 2005;54:2557–2567. doi: 10.2337/diabetes.54.9.2557. [DOI] [PubMed] [Google Scholar]
- Toledano H, D’Alterio C, Czech B, Levine E, Jones DL. The let-7-Imp axis regulates ageing of the Drosophila testis stem-cell niche. Nature. 2012;485:605–610. doi: 10.1038/nature11061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, Cullen DE, McDowell EP, Lazo-Kallanian S, Williams IR, Sears C, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly-Y M, Gidlöf S, Oldfors A, Wibom R, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- Tyner SD, Venkatachalam S, Choi J, Jones S, Ghebranious N, Igelmann H, Lu X, Soron G, Cooper B, Brayton C, et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- van Heemst D, Mooijaart SP, Beekman M, Schreuder J, de Craen AJ, Brandt BW, Slagboom PE, Westendorp RG Long Life study group. Variation in the human TP53 gene affects old age survival and cancer mortality. Exp Gerontol. 2005;40:11–15. doi: 10.1016/j.exger.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Vaziri H, Schächter F, Uchida I, Wei L, Zhu X, Effros R, Cohen D, Harley CB. Loss of telomeric DNA during aging of normal and trisomy 21 human lymphocytes. Am J Hum Genet. 1993;52:661–667. [PMC free article] [PubMed] [Google Scholar]
- Vaziri H, Dragowska W, Allsopp RC, Thomas TE, Harley CB, Lansdorp PM. Evidence for a mitotic clock in human hematopoietic stem cells: loss of telomeric DNA with age. Proc Natl Acad Sci USA. 1994;91:9857–9860. doi: 10.1073/pnas.91.21.9857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermulst M, Bielas JH, Kujoth GC, Ladiges WC, Rabinovitch PS, Prolla TA, Loeb LA. Mitochondrial point mutations do not limit the natural lifespan of mice. Nat Genet. 2007;39:540–543. doi: 10.1038/ng1988. [DOI] [PubMed] [Google Scholar]
- Vilchez D, Boyer L, Morantte I, Lutz M, Merkwirth C, Joyce D, Spencer B, Page L, Masliah E, Berggren WT, et al. Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature. 2012a;489:304–308. doi: 10.1038/nature11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilchez D, Morantte I, Liu Z, Douglas PM, Merkwirth C, Rodrigues AP, Manning G, Dillin A. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature. 2012b;489:263–268. doi: 10.1038/nature11315. [DOI] [PubMed] [Google Scholar]
- Villeda SA, Luo J, Mosher KI, Zou B, Britschgi M, Bieri G, Stan TM, Fainberg N, Ding Z, Eggel A, et al. The ageing systemic milieu negatively regulates neurogenesis and cognitive function. Nature. 2011;477:90–94. doi: 10.1038/nature10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker GA, Lithgow GJ. Lifespan extension in C. elegans by a molecular chaperone dependent upon insulin-like signals. Aging Cell. 2003;2:131–139. doi: 10.1046/j.1474-9728.2003.00045.x. [DOI] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallenfang MR, Nayak R, DiNardo S. Dynamics of the male germline stem cell population during aging of Drosophila melanogaster. Aging Cell. 2006;5:297–304. doi: 10.1111/j.1474-9726.2006.00221.x. [DOI] [PubMed] [Google Scholar]
- Walne AJ, Dokal I. Advances in the understanding of dyskeratosis congenita. Br J Haematol. 2009;145:164–172. doi: 10.1111/j.1365-2141.2009.07598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Sun Q, Morita Y, Jiang H, Gross A, Lechel A, Hildner K, Guachalla LM, Gompf A, Hartmann D, et al. A differentiation checkpoint limits hematopoietic stem cell self-renewal in response to DNA damage. Cell. 2012;148:1001–1014. doi: 10.1016/j.cell.2012.01.040. [DOI] [PubMed] [Google Scholar]
- Willcox BJ, Donlon TA, He Q, Chen R, Grove JS, Yano K, Masaki KH, Willcox DC, Rodriguez B, Curb JD. FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci USA. 2008;105:13987–13992. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KK, Maser RS, Bachoo RM, Menon J, Carrasco DR, Gu Y, Alt FW, DePinho RA. Telomere dysfunction and Atm deficiency compromises organ homeostasis and accelerates ageing. Nature. 2003;421:643–648. doi: 10.1038/nature01385. [DOI] [PubMed] [Google Scholar]
- Yalcin S, Zhang X, Luciano JP, Mungamuri SK, Marinkovic D, Vercherat C, Sarkar A, Grisotto M, Taneja R, Ghaffari S. Foxo3 is essential for the regulation of ataxia telangiectasia mutated and oxidative stress-mediated homeostasis of hematopoietic stem cells. J Biol Chem. 2008;283:25692–25705. doi: 10.1074/jbc.M800517200. [DOI] [PubMed] [Google Scholar]
- Yankner BA, Lu T, Loerch P. The aging brain. Annu Rev Pathol. 2008;3:41–66. doi: 10.1146/annurev.pathmechdis.2.010506.092044. [DOI] [PubMed] [Google Scholar]
- Yilmaz OH, Valdez R, Theisen BK, Guo W, Ferguson DO, Wu H, Morrison SJ. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441:475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- Yilmaz OH, Katajisto P, Lamming DW, Gültekin Y, Bauer-Rowe KE, Sengupta S, Birsoy K, Dursun A, Yilmaz VO, Selig M, et al. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature. 2012;486:490–495. doi: 10.1038/nature11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Grindley JC, Yin T, Jayasinghe S, He XC, Ross JT, Haug JS, Rupp D, Porter-Westpfahl KS, Wiedemann LM, et al. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature. 2006;441:518–522. doi: 10.1038/nature04747. [DOI] [PubMed] [Google Scholar]
- Zindy F, Quelle DE, Roussel MF, Sherr CJ. Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene. 1997;15:203–211. doi: 10.1038/sj.onc.1201178. [DOI] [PubMed] [Google Scholar]