Abstract
In this review, we focus on the roles of long noncoding RNAs (lncRNAs), including cellular and viral lncRNAs, in virus replication in infected cells. We survey the interactions and functions of several cellular lncRNAs such as XIST, HOTAIR, NEAT1, BIC, and several virus-encoded lncRNAs.
Keywords: long noncoding RNAs, virus
1. Introduction
A small portion (less than 2%) of the human genome is used to encode about 25,000 protein-encoding genes, while based on the findings from genome tiling arrays and RNA sequencing, >70% of the human genome is transcribed into RNAs, with the vast majority of these RNAs being devoid of obvious protein-coding capacity [1], [2]. These numbers suggest that noncoding RNAs (ncRNAs) may not simply be effete materials, and they, like their protein counterparts, may play significant functional roles [3].
Operationally, ncRNAs can be grouped into small noncoding RNAs (sncRNAs) and long noncoding RNAs (lncRNAs) according to their length [4]. Within these two groups, there can be additional subclassifications of the moieties [5], [6].
Of these two RNA groups, sncRNAs are transcripts that are <200 nt in length. Housekeeping RNAs, such as transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs) (5S, 5.8S), fall within this noncoding category [7]. The first characterized ncRNAs, tRNAs, serve as adaptors between messenger RNAs (mRNAs) and proteins for the elongation of polypeptide; rRNAs are essential elements for protein translation [8], [9]. Other sncRNAs include small nucleolar RNAs and small nuclear (snRNAs) that play certain roles in rRNA modification and RNA splicing [10], [11]. Besides these entities, sncRNAs also include three major types of regulatory RNAs: piwi-interacting RNAs (piRNAs), microRNAs (miRNAs), and small interfering RNAs (siRNAs) [12], [13]. Of these regulatory RNAs, piRNAs are involved in the regulation of transposon activity and chromatin state in germline and somatic cells [14], [15]; miRNAs and cell endogenous siRNAs contribute to RNA interference (RNAi) or post-transcriptional gene silencing [16]. Approximately 2000 miRNAs are encoded by the human genome [17], [18]; miRNAs serve as guide RNAs in an RNA-induced silencing complex (RISC) to target mRNAs through imperfect complementarity leading to repression of translation or degradation of the mRNAs [19], [20], [21]. Similarly, cell endogenous siRNAs also participate with RISC proteins in silencing gene expression usually via perfect complementarity with mRNA targets [22]. The functional roles of sncRNAs have been reviewed extensively elsewhere [12], [18], [23], [24], [25], [26]. This current review focuses on the still less well-studied counterpart of sncRNAs and lncRNAs, and their interactions with viruses.
2. Long noncoding RNAs
The lncRNAs are transcripts that are >200 nt in length [5]. This class of RNAs includes intergenic ncRNAs, pseudogene transcripts, and many antisense RNAs [5], [6]. The majority of lncRNAs are transcribed by RNA polymerase II; they are 5′-capped, spliced, and polyadenylated, and are mRNA-like in many ways [27]. A small minority of lncRNAs are transcribed by RNA polymerase III; these include 7SK and 7SL [28], [29]. The 21A lncRNA is also an RNA Pol-III transcript, but it is not polyadenylated [30]. Additionally, several other features frequently define lncRNAs, such as epigenetic marks shared with protein-coding gene (H3K4me3 at the gene promoter and H3K36me3 throughout the gene body), splicing of multiple exons via canonical splice site motifs, regulation by transcription factors, and expression in a tissue-specific manner [6]. It is now increasingly understood that lncRNA sequences are abundant in the mammalian genome. To date, approximately 6700 lncRNAs have been identified in the human genome [31], [32], and it is estimated that 7000–23,000 lncRNAs putatively exist in the human genome [33], [34]. Whether many of these deduced lncRNAs are authentically expressed and serve functional roles remain to be demonstrated. As of November 2012, 194 lncRNAs have been recorded in the lncRNAdb, a database that archives lncRNAs reported in published literature [35].
3. Functions of lncRNAs
Accumulating data support the fact that lncRNAs contribute functions that affect many cellular processes [6], [27], [36], [37]. Extant findings indicate contributions of lncRNAs to both transcriptional and post-transcriptional regulations [6], [37]. Here, we discuss a few of the better characterized lncRNAs such as XIST, HOTAIR, H19, HMGA1-p, MALAT1, and NEAT1 among others. Many of these RNAs are expressed during tumorigenesis or disease pathogenesis, or in different stages of embryonic stem cell differentiation [36].
4. Transcriptional regulation
Several lncRNAs play roles in transcriptional regulation. XIST, which was first discovered by searching cDNA libraries [38], is perhaps the most well-known lncRNA. A double-hairpin RNA motif in the RepA domain in XIST binds polycomb repressive complex 2 (PRC2), the complex that has been shown to be recruited by many lncRNAs to target genes [39] and that propagates function leading to X chromosome inactivation, as observed in some breast cancers [40], [41], [42]. Hypomethylation of XIST in lymphoma and male testicular germ-cell tumors has also been described, although their functional significance needs more study [6], [43].
Another well-studied lncRNA is HOTAIR. The HOTAIR gene is located within the HoxC gene cluster on chromosome 12 [44]. The HOTAIR transcript represses the expression of genes in the HoxD gene cluster on chromosome 2. The 5′-domain of HOTAIR binds PRC2, while its 3′-domain interacts with the LSD1/CoRES/REST complex [44], [45], leading to methylation of histone H3 lysine 27 and demethylation of lysine 4, and gene repression by chromatin remodeling [45]. Mechanistically, HOTAIR serves as a modular scaffold for assembling a multiprotein complex [45].
Separately, it has been demonstrated that lncRNAs can regulate gene expression by increasing enhancer activity [4]. Evf-2, a 3.8-kilobase (kb) alternatively spliced form of Evf-1, is transcribed from the highly conserved region between the Dlx-5 and Dlx-6 genes, members of the Dlx/dll homeodomain-containing protein family [46]. Evf-2 specifically recruits DLX and MECP2 transcription factors to increase the transcriptional activity of the Dlx-5/6 enhancer [47].
5. Post-transcriptional regulation
Recent studies have also provided insights into the post-transcriptional regulatory roles of lncRNAs. One view suggests that lncRNAs can be precursors of small RNAs, e.g., miRNAs [48]. H19, a 2.5-kb RNA polymerase II-dependent transcript, is an imprinting-associated lncRNA located on chromosome 11 [49]. The function of H19 has remained elusive since its discovery over 20 years ago. Recently, H19 has been reported to serve as the precursor of miR-675, which can act to moderate cell growth. The excision of miR-675 from H19 is under the control of the stress-response RNA-binding protein HuR; miR-675 is specifically expressed in the placenta from time of gestation and may function to limit placental growth [50].
Emerging reports suggest that pseudogenes can play important roles in regulating coding gene expression. For example, HMGA1-p, the pseudogene of HMGA1, encodes a transcript that competes with the HMGA1 3′-UTR for a critical RNA stability factor; this competition triggers a significant decline in the stability of HMGA1 mRNA [51]. In the case of PTEN, the transcribed pseudogene PTENP1 competes for miRNA-binding sites with the authentic PTEN RNA, thereby regulating the cellular abundance of PTEN mRNA [52].
Recently, Gong and Maquat [53] described a new functional mechanism of lncRNAs. The half-STAU1-binding site RNA 1 1/2-sbsRNA1 contains an Alu element that can base-pair with the Alu element in the 3′-UTR of SERPINE1 mRNA and FLJ21870 mRNA. This base-pairing between two Alu elements forms the binding site for Staufen 1 (STAU1) protein, which recognizes double-stranded RNA [54], resulting in STAU1-mediated mRNA decay [53].
The lncRNAs can also bind to cellular protein and modulate their localization and activity. One of the well-characterized examples is MALAT1, which regulates alternative splicing by modulating the phosphorylation of the serine/arginine splicing factors [55]. Another example is NRON (an ncRNA repressor of the nuclear factor of activated T cells), which is proposed to block specifically nuclear trafficking of transcription factor NFAT (the nuclear factor of activated T cells) as an RNA component of a protein complex that acts to repress NFAT activity [56].
6. Structural lncRNAs
There is also evidence that lncRNAs contribute structural/scaffolding functions. The lncRNA NEAT1, also known as MENɛ/β, was reported recently to be essential for the formation and maintenance of the nuclear substructure paraspeckles [48], [57], [58], [59]. Paraspeckles are found in the interchromatin space of a nucleus and serve as depots for RNA-binding proteins in the nucleus [60]. Similarly, the lncRNA Xlsirts is suggested to be necessary for maintaining the cytokeratin cytoskeleton in Xenopus oocytes [61], [62].
7. Cellular lncRNAs in virus-infected cells
Viruses are parasites that interact with their hosts. Since the functions of lncRNAs are highly pleiotropic, ranging from gene regulation to sncRNA precursors [4] and from cell development to cancer growth [6], [12], it is not surprising that lncRNAs may be involved in virus replication. Early studies of the relationship between ncRNAs and viruses mainly focused on sncRNAs, such as miRNAs [63], [64], while the roles of lncRNAs were not well studied. However, there is emerging evidence that cellular lncRNA expression can be regulated by virus infection. Thus, whole transcriptome analyses showed that during an infection by severe acute respiratory syndrome coronavirus, approximately 500 annotated lncRNAs and 1000 nonannotated genomic regions are differentially expressed in lung samples, and 40% of these changes are similarly observed during influenza virus infection and interferon treatment, indicating that many lncRNAs may be involved in regulating the host response to virus infection [65]. A concordant interpretation was separately proposed based on findings that the expression patterns of eight mRNA-like lncRNAs in immune tissues of chickens were changed after Marek's disease virus infection, similarly suggesting that they may play a role in host immune response [66]. Another example is PRINS (psoriasis susceptibility-related RNA gene induced by stress), which is increased by herpes simplex virus infection [67]. Separately, we recently profiled 83 disease-related lncRNAs in HIV-1-infected T cells and identified several lncRNAs that were changed in both Jurkat and MT4 cells, e.g., BIC, NEAT1, and PANDA [68].Our findings are also consistent with the overall notion that some lncRNAs serve in host responses to viral infections. Finally, studies have found a non-protein-coding infection-specific gene family called Pinci1, which is upregulated by Phytophthora infestans infection, in potatoes [69].
Although the lncRNAs discussed above have differential expression in virus-infected cells, their specific functions in virus replication remain incompletely characterized. In the following, we will discuss briefly some examples of lncRNAs whose roles in viral life cycle and viral pathogenesis are beginning to be better understood (Fig. 1 ).
Fig. 1.
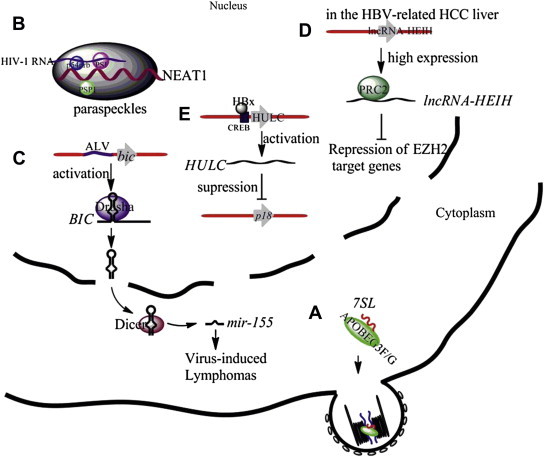
Illustrations of the functions of selected cellular lncRNAs in virus-infected cells. (A) The cytidinedeaminase APOBEC3G and APOBEC3F selectively interact with 7SL RNAs and are incorporated into virions. (B) The lncRNA NEAT1 serves as a structural scaffold for the nuclear substructure paraspeckles. Paraspeckle proteins PSF and p54nrb bind to HIV-1 RNA and retain the RNA in paraspeckles. (C) The integrated ALV activates bic gene expression by promoter insertion. BIC RNA, the precursor of miR-155, is suggested to be responsible for virus-induced lymphomas. (D) The lncRNA-HEIH, which is highly expressed in HBV-related HCC, recruits the PRC2 complex to repress EZH2 (an important subunit of the PRC2 complex) targeted genes. (E) HULC is upregulated by HBx protein through activation of the HULC promoter via CREB, leading to the suppression of the tumor suppressor gene p18. ALV = avian leukosis virus; CREB = cAMP responsive element binding protein; EZH2 = enhancer of zeste homolog 2; HBV = hepatitis B virus; HBx = hepatitis B virus X; HCC = hepatocellular carcinoma; lncRNAs = long noncoding RNAs; lncRNA-HEIH = lncRNA high expression in HCC; HULC = lncRNAs highly upregulated in liver cancer; PRC2 = polycomb repressive complex 2.
8. 7SL
The 7SL, a 300-nt RNA transcribed by RNA Pol-III, is the architectural RNA component of the signal recognition particle (SRP) ribonucleoprotein complex [70], [71], which is a universally conserved ribonucleoprotein that directs the traffic of proteins within the cell and allows them to be secreted [72]. In mammals, six SRP proteins, named SRP9, SRP14, SRP19, SRP54, SRP68, and SRP72, assemble on 7SL and form SRPs [73], [74].
The 7SL was first detected in avian and murine oncogenic RNA virus particles and then was found to be packaged by a broad range of retroviruses [75], [76]. Tian et al [77] showed that 7SL RNA is more selectively packaged into HIV-1 virions than other abundant Pol-III-transcribed RNAs, such as Y RNAs, 7SK RNA, U6 snRNA, and cellular mRNAs. Interestingly, 7SL has been suggested to participate as a cofactor in the innate antiviral function of host cytidine deaminases such as cytidine deaminases APOBEC3G (A3G) and APOBEC3F (A3F) [78], [79]. Wang et al [78] demonstrated that A3G selectively interacts with 7SL RNAs and both are incorporated into virions, while A3G mutants that reduce 7SL RNA binding but maintain wild-type levels of mRNA and tRNA binding are packaged poorly and have impaired antiviral activity. Reducing 7SL RNA packaging by overexpression of SRP19 proteins inhibits 7SL RNA, and A3G and A3F virion packaging, and impairs their antiviral functions [78], [79]. Moreover, virion packaging of both A3G and cellular 7SL RNA was mapped to the same regions in the HIV-1 nucleocapsid (NC) domain [77].
9. NEAT1
NEAT1 serves as a structural scaffold for nuclear paraspeckles [80]. It has two isoforms: NEAT1_1 (3.7 kb in human) and NEAT1_2 (23 kb in human); the isoforms are also named ΜΕΝɛ and ΜΕΝβ [60]. Besides NEAT1 RNA, paraspeckles contain more than 30 nuclear proteins including p54nrb, PSF, and PSPC1, which are all RNA-binding proteins [60]. Despite much progress, the function of paraspeckles is still not well defined, but they are suggested to be involved in regulation of gene expression through nuclear RNA retention [81].
NEAT1 expression was reported to be increased in the central nervous system of mice during their infection with Japanese encephalitis virus (JEV) or Rabies virus [82]. More interestingly, several cellular proteins that play roles in HIV-1 replication are found in paraspeckles (e.g., PSF, p54nrb, and Matrin 3) [83], [84]. Recently, we identified NEAT1 as one of several lncRNAs whose expression is changed by HIV-1 infection, and we reported that the knockdown of NEAT1 enhances virus production through increased nuclear to cytoplasmic export of Rev-dependent INS-containing HIV-1 mRNAs [68].
10. BIC
BIC was first identified as an ncRNA upregulated by avian leukosis virus infection [85]. The integrated provirus activates bic gene expression by promoter insertion, resulting in high levels of expression of BIC RNA, which was suggested to be responsible for virus-induced lymphomas [85]. More recently, BIC was found to be the precursor of oncogenic miR-155 [86], which is induced by several oncogenic viruses, e.g., Epstein–Barr virus, hepatitis C virus, and reticuloendotheliosis virus strain T [87], [88], [89]. Interestingly, Kaposi's sarcoma-associated herpesvirus, a gammaherpesvirus, and Marek's disease virus, an avian alphaherpesvirus, encode viral miRNAs, miR-K11, and miR-M4, as functional orthologs of miR-155 [90], [91]. Of interest, it has been shown that Epstein–Barr virus attenuates NF-κB signaling and stabilizes latent virus persistence by inducing miR-155 [92]. In hepatitis C virus infection, upregulated miR-155 has been demonstrated to promote hepatocarcinogenesis by activating Wnt signaling [88]. Reticuloendotheliosis virus strain T induces miR-155 to target JARID2, a cell-cycle regulator that is a part of a histone methyltransferase complex, in order to promote cell survival [89]. Induction of miR-155 by virus infection may not only be due to the induction of BIC RNA, but also arise from enhanced RNA processing [93].
11. Long ncRNA high expression in HCC and lncRNAs highly upregulated in liver cancer
Hepatitis B virus (HBV) infection is a major cause of hepatocellular carcinoma (HCC) [94]. Recent research has shown that some lncRNAs are aberrantly expressed in HBV-related HCCs. Two of these lncRNAs, lncRNA-HEIH (lncRNA high expression in HCC) and HULC (lncRNA highly upregulated in liver cancer), are reported to play key roles in HBV-related hepatocarcinogenesis [95], [96]. Of these two lncRNAs, lncRNA-HEIH was reported to play a key role in G0/G1 arrest and be associated with enhancer of zeste homolog 2 (EZH2, an important subunit of the PRC2 complex), resulting in the repression of EZH2 target genes [95]. The other lncRNA HULC, a ∼500 nt mRNA-like ncRNA, was reported to be upregulated by the hepatitis B virus X (HBx) protein through activation of the HULC promoter via cAMP responsive element binding protein. Upregulated HULC promotes proliferation of hepatoma cells through suppressing a tumor suppressor gene p18 [96].
12. Theiler's murine encephalomyelitis virus persistence candidate gene 1
Previously, the Tmevp3 locus, located on the telomeric region of chromosome 10, was shown to control the persistence of Theiler's virus in the central nervous system of mice [97]. More recently, an ncRNA Tmevpg1 (Theiler's murine encephalomyelitis virus persistence candidate gene 1) was identified, at that locus through a positional cloning approach, as a candidate gene for controlling the persistence of Theiler's virus [98]. The promoter of Tmevpg1 contains binding sites for E2A and the Ets family of transcription factors, indicating that it is regulated by transcription factors involved in the immune system [99]. Since Tmevpg1 and its human ortholog, TMEVPG1, are located in a cluster of cytokine genes that includes the genes for gamma interferon and homologs of interleukin-10, Tmevpg1 is suggested to be involved in the control of Interferon-gammagene (Ifng) expression [98]. However, its precise role requires further characterization.
13. Virus-encoded lncRNA
To date, over 200 miRNAs encoded by several virus families have been identified [18]. Similarly, several lncRNAs encoded by viruses have also been discovered [100], [101] (Table 1 ) [102], [103], [104], [105], [106], [107], [108], [109], [110]. Although each is less than 200 nt in size, Epstein–Barr virus-encoded RNAs (EBERs) [102], herpesvirus saimiri U-rich RNAs (HSURs) [103], [104], and virus-associated RNA I and II (VA I and II) encoded by adenovirus [105], [106], [107], [108] are sometimes also referred to as viral lncRNAs, because they are significantly longer than viral miRNAs [111]. Here, we focus an illustrative discussion on two viral lncRNAs, β2.7 and sfRNA, which are longer than 200 nt in size.
Table 1.
Virus-encoded lncRNAs.
| Length | Name | Virus | Characteristics | References |
|---|---|---|---|---|
| <200 nt | EBERs | Epstein–Barr virus | ∼170 nt, play roles in oncogenesis and modulate innate immune signaling | [102] |
| (EBER1, EBER2) | ||||
| HSURs | Herpesvirus saimiri | HSUR1 (143 nt), HSUR2 (115 nt);HSUR1 directs degradation of miR-27 to manipulate host T-cell gene expression | [103], [104] | |
| (HUSR1, HUSR2) | ||||
| VA I and II | Human adenovirus | ∼160 nt, block PKR activity, avoiding phosphorylation of eIF-2α and inhibition of viral mRNA translation; can be processed by Dicer into small RNAs that are incorporated into RISC | [105], [106], [107], [108] | |
| >200 nt | β2.7 | Human cytomegalovirus | 2.7 kb, binds to the mitochondrial enzyme complex I, protecting virus-infected cells from apoptosis, resulting in continued ATP production | [100], [109] |
| sfRNA | Flaviviruses | 0.3–0.5 kb, produced from the incomplete degradation of the viral genome by the host exonuclease XRN1 and required for virus-induced cytopathicity and pathogenicity | [110] |
ATP = adenosine triphosphate; EBERs = Epstein–Barr virus-encoded RNAs; HSURs = Herpesvirus saimiri U-rich RNAs; lncRNAs = long noncoding RNAs; mRNA = messenger RNA; PKR = double-stranded RNA-activated protein kinase; RISC = RNA-induced silencing complex; VA I and II = virus-associated RNA I and II.
14. Viral lncRNA β2.7
β2.7 RNA, a highly conserved 2.7-kb transcript of human cytomegalovirus, accounts for more than 20% of total viral gene transcription during the early phase of infection [100]. Since the replication rate of a β2.7 deletion mutant virus is similar to that of a wild-type virus, the β2.7 gene was considered not to be essential for virus replication in vitro [112]. However, recently, by Northwestern screening of a human cDNA library with a β2.7 probe, Reeves et al [109] found that β2.7 binds directly to the mitochondrial enzyme complex I (reduced nicotinamide adenine dinucleotide–ubiquinone oxidoreductase). This binding protects virus-infected cells from apoptosis and results in continued adenosine triphosphate (ATP) production, which is critical for the successful completion of the viral life cycle. The β2.7 RNA can also protect rat aortic endothelial cells from ischemia/reperfusion injury-induced apoptosis by reducing the formation of reactive oxygen species [113].
15. Subgenomic flavivirus RNA
The subgenomic flavivirus RNA (sfRNA), 0.3–0.5 kb, is derived from the 3′ untranslated region of the RNA genome of flaviviruses, a large group of single-stranded, positive-sense RNA viruses that includes several human pathogenic viruses, such as yellow fever virus, JEV, and West Nile virus (WNV) [101], [114], [115]. The sfRNA has been demonstrated to be produced from the incomplete degradation of the viral genome by the host 5′ to 3′ exonuclease XRN1. The rigid secondary structure stem-loop II located at the beginning of the 3′-UTR of the above viral genomes is resistant to nuclease XRN1 degradation and results in the production of sfRNA [116]. Production of sfRNA has been shown to increase the replication efficiency of WNVs and is important for virus-induced cytopathicity in cell culture and also for viral pathogenicity in mice [110], [117]. However, the exact mechanisms explaining how sfRNA leads to increased virus replication and cell death remain elusive. sfRNA is also found in JEV infection. In this setting, sfRNA becomes apparent at the time during which minus-strand RNA (antigenome) reaches a plateau, suggesting a role for sfRNA in the regulation of antigenome synthesis. The presence of sfRNA may inhibit antigenome synthesis and may exert a negative effect on JEV translation [118].
16. Perspective
In recent years, technological advances have made it possible to investigate the expression of whole transcriptomes in an unbiased manner. This new capability has driven the discovery of an increasing number of lncRNAs. Nonetheless, our knowledge regarding the functions of these lncRNA transcripts remains quite limited [119]. Because lncRNAs have diverse functions, they likely represent important bioentities that merit further investigation. Here, we have focused on a few examples of lncRNAs, including cellular and viral lncRNAs. Our brief survey shows that we are at the initial stages of uncovering their functions and their relationships with viruses. This review is meant to serve as a brief illustrative introduction to lncRNAs, which we hope may spur interest by readers for conducting further studies on these interesting and important biomolecules.
Acknowledgments
The work in KTJ's laboratory was supported by intramural funding from NIAID, NIH; the IATAP program from the Office of the Director, NIH; and funding from the Bill and Melinda Gates Foundation. We thank members of the Jeang Laboratory for their critical reading of this writing.
References
- 1.Birney E., Stamatoyannopoulos J.A., Dutta A., Guigo R., Gingeras T.R., Margulies E.H. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stein L.D. Human genome: end of the beginning. Nature. 2004;431:915–916. doi: 10.1038/431915a. [DOI] [PubMed] [Google Scholar]
- 3.Costa F.F. Non-coding RNAs: meet thy masters. Bioessays. 2010;32:599–608. doi: 10.1002/bies.200900112. [DOI] [PubMed] [Google Scholar]
- 4.Huang Y., Liu N., Wang J.P., Wang Y.Q., Yu X.L., Wang Z.B. Regulatory long non-coding RNA and its functions. J Physiol Biochem. 2012;68(4):611–618. doi: 10.1007/s13105-012-0166-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibb E.A., Brown C.J., Lam W.L. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prensner J.R., Chinnaiyan A.M. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spadaro P.A., Bredy T.W. Emerging role of non-coding RNA in neural plasticity, cognitive function, and neuropsychiatric disorders. Front Genet. 2012;3:132. doi: 10.3389/fgene.2012.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phizicky E.M., Hopper A.K. tRNA biology charges to the front. Genes Dev. 2010;24:1832–1860. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stults D.M., Killen M.W., Pierce H.H., Pierce A.J. Genomic architecture and inheritance of human ribosomal RNA gene clusters. Genome Res. 2008;18:13–18. doi: 10.1101/gr.6858507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kishore S., Stamm S. Regulation of alternative splicing by snoRNAs. Cold Spring Harb Symp Quant Biol. 2006;71:329–334. doi: 10.1101/sqb.2006.71.024. [DOI] [PubMed] [Google Scholar]
- 11.Hernandez N. Small nuclear RNA genes: a model system to study fundamental mechanisms of transcription. J Biol Chem. 2001;276:26733–26736. doi: 10.1074/jbc.R100032200. [DOI] [PubMed] [Google Scholar]
- 12.Taft R.J., Pang K.C., Mercer T.R., Dinger M., Mattick J.S. Non-coding RNAs: regulators of disease. J Pathol. 2010;220:126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 13.Jeang K.T. RNAi in the regulation of mammalian viral infections. BMC Biol. 2012;10:58. doi: 10.1186/1741-7007-10-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malone C.D., Hannon G.J. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghildiyal M., Zamore P.D. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carthew R.W., Sontheimer E.J. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bentwich I., Avniel A., Karov Y., Aharonov R., Gilad S., Barad O. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 18.Klase Z., Houzet L., Jeang K.T. MicroRNAs and HIV-1: complex interactions. J Biol Chem. 2012;287:40884–40890. doi: 10.1074/jbc.R112.415448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krol J., Loedige I., Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 21.Houzet L., Klase Z., Yeung M.L., Wu A., Le S.Y., Quinones M. The extent of sequence complementarity correlates with the potency of cellular miRNA-mediated restriction of HIV-1. Nucleic Acids Res. 2012;40:11684–11696. doi: 10.1093/nar/gks912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 23.Berkhout B., Jeang K.T. MicroRNAs in viral gene regulation. Biochim Biophys Acta. 2011;1809:587. doi: 10.1016/j.bbagrm.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matera A.G., Terns R.M., Terns M.P. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 25.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 26.Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 28.Raha D., Wang Z., Moqtaderi Z., Wu L., Zhong G., Gerstein M. Close association of RNA polymerase II and many transcription factors with Pol III genes. Proc Natl Acad Sci U S A. 2010;107:3639–3644. doi: 10.1073/pnas.0911315106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bredow S., Kleinert H., Benecke B.J. Sequence and factor requirements for faithful in vitro transcription of human 7SL DNA. Gene. 1990;86:217–225. doi: 10.1016/0378-1119(90)90282-v. [DOI] [PubMed] [Google Scholar]
- 30.Pagano A., Castelnuovo M., Tortelli F., Ferrari R., Dieci G., Cancedda R. New small nuclear RNA gene-like transcriptional units as sources of regulatory transcripts. PLoS Genet. 2007;3:e1. doi: 10.1371/journal.pgen.0030001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gutschner T., Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jia H., Osak M., Bogu G.K., Stanton L.W., Johnson R., Lipovich L. Genome-wide computational identification and manual annotation of human long noncoding RNA genes. RNA. 2010;16:1478–1487. doi: 10.1261/rna.1951310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guttman M., Amit I., Garber M., French C., Lin M.F., Feldser D. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lipovich L., Johnson R., Lin C.Y. MacroRNA underdogs in a microRNA world: evolutionary, regulatory, and biomedical significance of mammalian long non-protein-coding RNA. Biochim Biophys Acta. 2010;1799:597–615. doi: 10.1016/j.bbagrm.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 35.Amaral P.P., Clark M.B., Gascoigne D.K., Dinger M.E., Mattick J.S. lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res. 2011;39:D146–D151. doi: 10.1093/nar/gkq1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X., Wu Z., Fu X., Han W. Long noncoding RNAs: insights from biological features and functions to diseases. Med Res Rev. 2012 doi: 10.1002/med.21254. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 37.Nie L., Wu H.J., Hsu J.M., Chang S.S., Labaff A.M., Li C.W. Long non-coding RNAs: versatile master regulators of gene expression and crucial players in cancer. Am J Transl Res. 2012;4:127–150. [PMC free article] [PubMed] [Google Scholar]
- 38.Brown C.J., Ballabio A., Rupert J.L., Lafreniere R.G., Grompe M., Tonlorenzi R. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 39.Khalil A.M., Guttman M., Huarte M., Garber M., Raj A., Rivea Morales D. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao J., Sun B.K., Erwin J.A., Song J.J., Lee J.T. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sirchia S.M., Tabano S., Monti L., Recalcati M.P., Gariboldi M., Grati F.R. Misbehaviour of XIST RNA in breast cancer cells. PLoS One. 2009;4:e5559. doi: 10.1371/journal.pone.0005559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ganesan S., Silver D.P., Greenberg R.A., Avni D., Drapkin R., Miron A. BRCA1 supports XIST RNA concentration on the inactive X chromosome. Cell. 2002;111:393–405. doi: 10.1016/s0092-8674(02)01052-8. [DOI] [PubMed] [Google Scholar]
- 43.Kawakami T., Okamoto K., Ogawa O., Okada Y. XIST unmethylated DNA fragments in male-derived plasma as a tumour marker for testicular cancer. Lancet. 2004;363:40–42. doi: 10.1016/S0140-6736(03)15170-7. [DOI] [PubMed] [Google Scholar]
- 44.Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng J., Bi C., Clark B.S., Mady R., Shah P., Kohtz J.D. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bond A.M., Vangompel M.J., Sametsky E.A., Clark M.F., Savage J.C., Disterhoft J.F. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci. 2009;12:1020–1027. doi: 10.1038/nn.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilusz J.E., Sunwoo H., Spector D.L. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gabory A., Jammes H., Dandolo L. The H19 locus: role of an imprinted non-coding RNA in growth and development. Bioessays. 2010;32:473–480. doi: 10.1002/bies.200900170. [DOI] [PubMed] [Google Scholar]
- 50.Keniry A., Oxley D., Monnier P., Kyba M., Dandolo L., Smits G. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. 2012;14:659–665. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chiefari E., Iiritano S., Paonessa F., Le Pera I., Arcidiacono B., Filocamo M. Pseudogene-mediated posttranscriptional silencing of HMGA1 can result in insulin resistance and type 2 diabetes. Nat Commun. 2010;1:40. doi: 10.1038/ncomms1040. [DOI] [PubMed] [Google Scholar]
- 52.Poliseno L., Salmena L., Zhang J., Carver B., Haveman W.J., Pandolfi P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gong C., Maquat L.E. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3' UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim Y.K., Furic L., Parisien M., Major F., DesGroseillers L., Maquat L.E. Staufen1 regulates diverse classes of mammalian transcripts. EMBO J. 2007;26:2670–2681. doi: 10.1038/sj.emboj.7601712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tripathi V., Ellis J.D., Shen Z., Song D.Y., Pan Q., Watt A.T. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Willingham A.T., Orth A.P., Batalov S., Peters E.C., Wen B.G., Aza-Blanc P. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 57.Clemson C.M., Hutchinson J.N., Sara S.A., Ensminger A.W., Fox A.H., Chess A. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sunwoo H., Dinger M.E., Wilusz J.E., Amaral P.P., Mattick J.S., Spector D.L. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sasaki Y.T., Ideue T., Sano M., Mituyama T., Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci U S A. 2009;106:2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakagawa S., Hirose T. Paraspeckle nuclear bodies—useful uselessness? Cell Mol Life Sci. 2012;69:3027–3036. doi: 10.1007/s00018-012-0973-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kloc M., Wilk K., Vargas D., Shirato Y., Bilinski S., Etkin L.D. Potential structural role of non-coding and coding RNAs in the organization of the cytoskeleton at the vegetal cortex of Xenopus oocytes. Development. 2005;132:3445–3457. doi: 10.1242/dev.01919. [DOI] [PubMed] [Google Scholar]
- 62.Kloc M., Bilinski S., Dougherty M.T. Organization of cytokeratin cytoskeleton and germ plasm in the vegetal cortex of Xenopus laevis oocytes depends on coding and non-coding RNAs: three-dimensional and ultrastructural analysis. Exp Cell Res. 2007;313:1639–1651. doi: 10.1016/j.yexcr.2007.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ouellet D.L., Provost P. Current knowledge of microRNAs and noncoding RNAs in virus-infected cells. Methods Mol Biol. 2010;623:35–65. doi: 10.1007/978-1-60761-588-0_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grassmann R., Jeang K.T. The roles of microRNAs in mammalian virus infection. Biochim Biophys Acta. 2008;1779:706–711. doi: 10.1016/j.bbagrm.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peng X., Gralinski L., Armour C.D., Ferris M.T., Thomas M.J., Proll S. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. MBio. 2010;1:e00206–e00210. doi: 10.1128/mBio.00206-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ahanda M.L., Ruby T., Wittzell H., Bed'Hom B., Chausse A.M., Morin V. Non-coding RNAs revealed during identification of genes involved in chicken immune responses. Immunogenetics. 2009;61:55–70. doi: 10.1007/s00251-008-0337-8. [DOI] [PubMed] [Google Scholar]
- 67.Sonkoly E., Bata-Csorgo Z., Pivarcsi A., Polyanka H., Kenderessy-Szabo A., Molnar G. Identification and characterization of a novel, psoriasis susceptibility-related noncoding RNA gene, PRINS. J Biol Chem. 2005;280:24159–24167. doi: 10.1074/jbc.M501704200. [DOI] [PubMed] [Google Scholar]
- 68.Zhang Q., Chen C.-Y., Yedavalli V.S., Jeang K.-T. NEAT1 long non-coding RNA and paraspeckle bodies modulate HIV-1 post-transcriptional expression. MBio. 2013;4(1):e00596–e00612. doi: 10.1128/mBio.00596-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Avrova A.O., Whisson S.C., Pritchard L., Venter E., De Luca S., Hein I. A novel non-protein-coding infection-specific gene family is clustered throughout the genome of Phytophthora infestans. Microbiology. 2007;153(Pt 3):747–759. doi: 10.1099/mic.0.2006/002220-0. [DOI] [PubMed] [Google Scholar]
- 70.Walter P., Blobel G. Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum. Nature. 1982;299:691–698. doi: 10.1038/299691a0. [DOI] [PubMed] [Google Scholar]
- 71.Zwieb C., van Nues R.W., Rosenblad M.A., Brown J.D., Samuelsson T. A nomenclature for all signal recognition particle RNAs. RNA. 2005;11:7–13. doi: 10.1261/rna.7203605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lutcke H. Signal recognition particle (SRP), a ubiquitous initiator of protein translocation. Eur J Biochem. 1995;228:531–550. doi: 10.1111/j.1432-1033.1995.tb20293.x. [DOI] [PubMed] [Google Scholar]
- 73.Kuglstatter A., Oubridge C., Nagai K. Induced structural changes of 7SL RNA during the assembly of human signal recognition particle. Nat Struct Biol. 2002;9:740–744. doi: 10.1038/nsb843. [DOI] [PubMed] [Google Scholar]
- 74.Walter P., Blobel G. Subcellular distribution of signal recognition particle and 7SL-RNA determined with polypeptide-specific antibodies and complementary DNA probe. J Cell Biol. 1983;97:1693–1699. doi: 10.1083/jcb.97.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Keene S.E., King S.R., Telesnitsky A. 7SL RNA is retained in HIV-1 minimal virus-like particles as an S-domain fragment. J Virol. 2010;84:9070–9077. doi: 10.1128/JVI.00714-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bishop J.M., Levinson W.E., Sullivan D., Fanshier L., Quintrell N., Jackson J. The low molecular weight RNAs of Rous sarcoma virus. II. The 7 S RNA. Virology. 1970;42:927–937. doi: 10.1016/0042-6822(70)90341-7. [DOI] [PubMed] [Google Scholar]
- 77.Tian C., Wang T., Zhang W., Yu X.F. Virion packaging determinants and reverse transcription of SRP RNA in HIV-1 particles. Nucleic Acids Res. 2007;35:7288–7302. doi: 10.1093/nar/gkm816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang T., Tian C., Zhang W., Luo K., Sarkis P.T., Yu L. 7SL RNA mediates virion packaging of the antiviral cytidinedeaminase APOBEC3G. J Virol. 2007;81:13112–13124. doi: 10.1128/JVI.00892-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang T., Tian C., Zhang W., Sarkis P.T., Yu X.F. Interaction with 7SL RNA but not with HIV-1 genomic RNA or P bodies is required for APOBEC3F virion packaging. J Mol Biol. 2008;375:1098–1112. doi: 10.1016/j.jmb.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 80.Bond C.S., Fox A.H. Paraspeckles: nuclear bodies built on long noncoding RNA. J Cell Biol. 2009;186:637–644. doi: 10.1083/jcb.200906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen L.L., Carmichael G.G. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Saha S., Murthy S., Rangarajan P.N. Identification and characterization of a virus-inducible non-coding RNA in mouse brain. J Gen Virol. 2006;87(Pt 7):1991–1995. doi: 10.1099/vir.0.81768-0. [DOI] [PubMed] [Google Scholar]
- 83.Yedavalli V.S., Jeang K.T. Matrin 3 is a co-factor for HIV-1 Rev in regulating post-transcriptional viral gene expression. Retrovirology. 2011;8:61. doi: 10.1186/1742-4690-8-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zolotukhin A.S., Michalowski D., Bear J., Smulevitch S.V., Traish A.M., Peng R. PSF acts through the human immunodeficiency virus type 1 mRNA instability elements to regulate virus expression. Mol Cell Biol. 2003;23:6618–6630. doi: 10.1128/MCB.23.18.6618-6630.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tam W., Ben-Yehuda D., Hayward W.S. bic, a novel gene activated by proviral insertions in avian leukosis virus-induced lymphomas, is likely to function through its noncoding RNA. Mol Cell Biol. 1997;17:1490–1502. doi: 10.1128/mcb.17.3.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chung K.H., Hart C.C., Al-Bassam S., Avery A., Taylor J., Patel P.D. Polycistronic RNA polymerase II expression vectors for RNA interference based on BIC/miR-155. Nucleic Acids Res. 2006;34:e53. doi: 10.1093/nar/gkl143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Linnstaedt S.D., Gottwein E., Skalsky R.L., Luftig M.A., Cullen B.R. Virally induced cellular microRNA miR-155 plays a key role in B-cell immortalization by Epstein–Barr virus. J Virol. 2010;84:11670–11678. doi: 10.1128/JVI.01248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang Y., Wei W., Cheng N., Wang K., Li B., Jiang X. Hepatitis C virus-induced up-regulation of microRNA-155 promotes hepatocarcinogenesis by activating Wnt signaling. Hepatology. 2012;56:1631–1640. doi: 10.1002/hep.25849. [DOI] [PubMed] [Google Scholar]
- 89.Bolisetty M.T., Dy G., Tam W., Beemon K.L. Reticuloendotheliosis virus strain T induces miR-155, which targets JARID2 and promotes cell survival. J Virol. 2009;83:12009–12017. doi: 10.1128/JVI.01182-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gottwein E., Mukherjee N., Sachse C., Frenzel C., Majoros W.H., Chi J.T. A viral microRNA functions as an orthologue of cellular miR-155. Nature. 2007;450:1096–1099. doi: 10.1038/nature05992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhao Y., Yao Y., Xu H., Lambeth L., Smith L.P., Kgosana L. A functional microRNA-155 ortholog encoded by the oncogenic Marek's disease virus. J Virol. 2009;83:489–492. doi: 10.1128/JVI.01166-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lu F., Weidmer A., Liu C.G., Volinia S., Croce C.M., Lieberman P.M. Epstein–Barr virus-induced miR-155 attenuates NF-kappaB signaling and stabilizes latent virus persistence. J Virol. 2008;82:10436–10443. doi: 10.1128/JVI.00752-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Eis P.S., Tam W., Sun L., Chadburn A., Li Z., Gomez M.F. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci U S A. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.But D.Y., Lai C.L., Yuen M.F. Natural history of hepatitis-related hepatocellular carcinoma. World J Gastroenterol. 2008;14:1652–1656. doi: 10.3748/wjg.14.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yang F., Zhang L., Huo X.S., Yuan J.H., Xu D., Yuan S.X. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679–1689. doi: 10.1002/hep.24563. [DOI] [PubMed] [Google Scholar]
- 96.Du Y., Kong G., You X., Zhang S., Zhang T., Gao Y. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J Biol Chem. 2012;287:26302–26311. doi: 10.1074/jbc.M112.342113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bihl F., Brahic M., Bureau J.F. Two loci, Tmevp2 and Tmevp3, located on the telomeric region of chromosome 10, control the persistence of Theiler's virus in the central nervous system of mice. Genetics. 1999;152:385–392. doi: 10.1093/genetics/152.1.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vigneau S., Rohrlich P.S., Brahic M., Bureau J.F. Tmevpg1, a candidate gene for the control of Theiler's virus persistence, could be implicated in the regulation of gamma interferon. J Virol. 2003;77:5632–5638. doi: 10.1128/JVI.77.10.5632-5638.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vigneau S., Levillayer F., Crespeau H., Cattolico L., Caudron B., Bihl F. Homology between a 173-kb region from mouse chromosome 10, telomeric to the Ifng locus, and human chromosome 12q15. Genomics. 2001;78:206–213. doi: 10.1006/geno.2001.6656. [DOI] [PubMed] [Google Scholar]
- 100.Greenaway P.J., Wilkinson G.W. Nucleotide sequence of the most abundantly transcribed early gene of human cytomegalovirus strain AD169. Virus Res. 1987;7:17–31. doi: 10.1016/0168-1702(87)90055-4. [DOI] [PubMed] [Google Scholar]
- 101.Urosevic N., van Maanen M., Mansfield J.P., Mackenzie J.S., Shellam G.R. Molecular characterization of virus-specific RNA produced in the brains of flavivirus-susceptible and -resistant mice after challenge with Murray Valley encephalitis virus. J Gen Virol. 1997;78(Pt 1):23–29. doi: 10.1099/0022-1317-78-1-23. [DOI] [PubMed] [Google Scholar]
- 102.Iwakiri D., Takada K. Role of EBERs in the pathogenesis of EBV infection. Adv Cancer Res. 2010;107:119–136. doi: 10.1016/S0065-230X(10)07004-1. [DOI] [PubMed] [Google Scholar]
- 103.Lee S.I., Murthy S.C., Trimble J.J., Desrosiers R.C., Steitz J.A. Four novel U RNAs are encoded by a herpesvirus. Cell. 1988;54:599–607. doi: 10.1016/s0092-8674(88)80004-7. [DOI] [PubMed] [Google Scholar]
- 104.Buck A.H., Perot J., Chisholm M.A., Kumar D.S., Tuddenham L., Cognat V. Post-transcriptional regulation of miR-27 in murine cytomegalovirus infection. RNA. 2010;16:307–315. doi: 10.1261/rna.1819210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mathews M.B., Shenk T. Adenovirus virus-associated RNA and translation control. J Virol. 1991;65:5657–5662. doi: 10.1128/jvi.65.11.5657-5662.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wahid A.M., Coventry V.K., Conn G.L. The PKR-binding domain of adenovirus VA RNAI exists as a mixture of two functionally non-equivalent structures. Nucleic Acids Res. 2009;37:5830–5837. doi: 10.1093/nar/gkp595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xu N., Segerman B., Zhou X., Akusjarvi G. Adenovirus virus-associated RNAII-derived small RNAs are efficiently incorporated into the RNA-induced silencing complex and associate with polyribosomes. J Virol. 2007;81:10540–10549. doi: 10.1128/JVI.00885-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Andersson M.G., Haasnoot P.C., Xu N., Berenjian S., Berkhout B., Akusjarvi G. Suppression of RNA interference by adenovirus virus-associated RNA. J Virol. 2005;79:9556–9565. doi: 10.1128/JVI.79.15.9556-9565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Reeves M.B., Davies A.A., McSharry B.P., Wilkinson G.W., Sinclair J.H. Complex I binding by a virally encoded RNA regulates mitochondria-induced cell death. Science. 2007;316:1345–1348. doi: 10.1126/science.1142984. [DOI] [PubMed] [Google Scholar]
- 110.Pijlman G.P., Funk A., Kondratieva N., Leung J., Torres S., van der Aa L. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe. 2008;4:579–591. doi: 10.1016/j.chom.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 111.Sullivan C.S. New roles for large and small viral RNAs in evading host defences. Nat Rev Genet. 2008;9:503–507. doi: 10.1038/nrg2349. [DOI] [PubMed] [Google Scholar]
- 112.McSharry B.P., Tomasec P., Neale M.L., Wilkinson G.W. The most abundantly transcribed human cytomegalovirus gene (beta 2.7) is non-essential for growth in vitro. J Gen Virol. 2003;84(Pt 9):2511–2516. doi: 10.1099/vir.0.19298-0. [DOI] [PubMed] [Google Scholar]
- 113.Zhao J., Sinclair J., Houghton J., Bolton E., Bradley A., Lever A. Cytomegalovirus beta2.7 RNA transcript protects endothelial cells against apoptosis during ischemia/reperfusion injury. J Heart Lung Transplant. 2010;29:342–345. doi: 10.1016/j.healun.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 114.Lin K.C., Chang H.L., Chang R.Y. Accumulation of a 3′-terminal genome fragment in Japanese encephalitis virus-infected mammalian and mosquito cells. J Virol. 2004;78:5133–5138. doi: 10.1128/JVI.78.10.5133-5138.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Scherbik S.V., Paranjape J.M., Stockman B.M., Silverman R.H., Brinton M.A. RNase L plays a role in the antiviral response to West Nile virus. J Virol. 2006;80:2987–2999. doi: 10.1128/JVI.80.6.2987-2999.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Funk A., Truong K., Nagasaki T., Torres S., Floden N., Balmori Melian E. RNA structures required for production of subgenomic flavivirus RNA. J Virol. 2010;84:11407–11417. doi: 10.1128/JVI.01159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Silva P.A., Pereira C.F., Dalebout T.J., Spaan W.J., Bredenbeek P.J. An RNA pseudoknot is required for production of yellow fever virus subgenomic RNA by the host nuclease XRN1. J Virol. 2010;84:11395–11406. doi: 10.1128/JVI.01047-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fan Y.H., Nadar M., Chen C.C., Weng C.C., Lin Y.T., Chang R.Y. Small noncoding RNA modulates Japanese encephalitis virus replication and translation in trans. Virol J. 2011;8:492. doi: 10.1186/1743-422X-8-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Guttman M., Rinn J.L. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]


