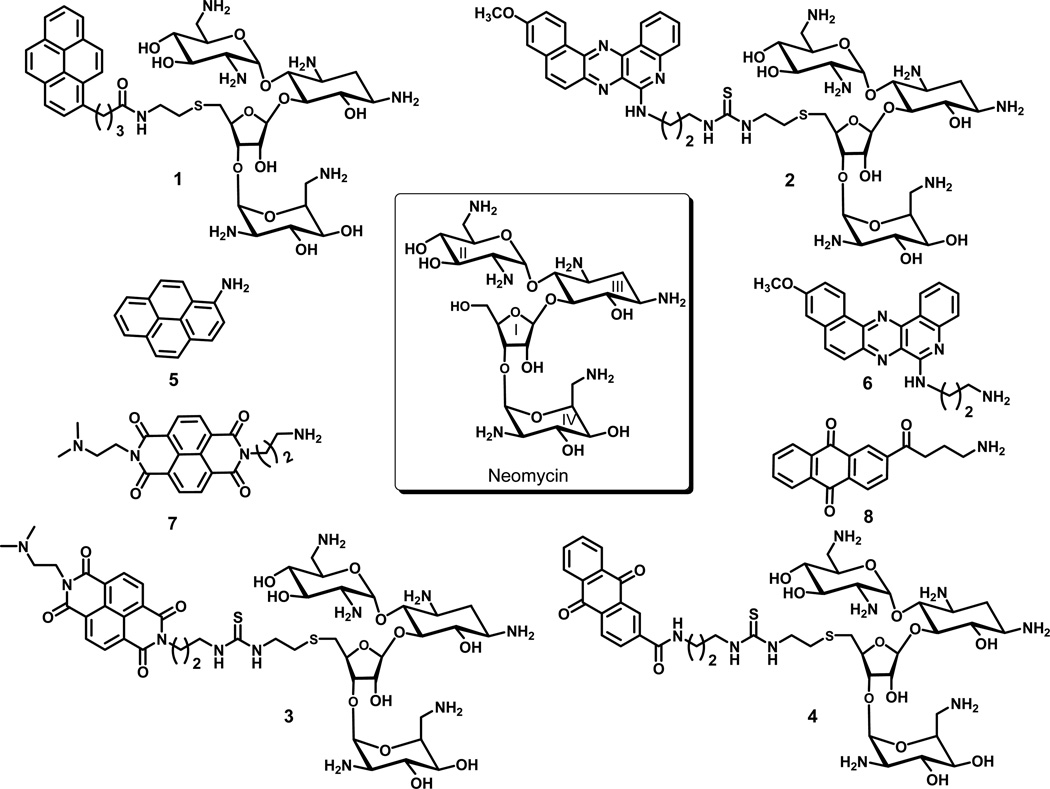
Abstract
Thermodynamic studies on the interactions between intercalator-neomycin conjugates and a DNA polynucleotide triplex [poly(dA)•2poly(dT)] were conducted. To draw a complete picture of such interactions, naphthalenedimide-neomycin (3) and anthraquinone-neomycin (4) were synthesized and used together with two other analogues, previously synthesized pyrene-neomycin (1) and BQQ-neomycin (2), in our investigations. A combination of experiments including UV denaturation, circular dichroism (CD) titration, differential scanning calorimetry (DSC), and isothermal titration calorimetry (ITC) revealed that all four conjugates (1–4) stabilized poly(dA)•2poly(dT) much greater than its parent compound, neomycin. UV melting experiments clearly showed that the temperature (Tm3→2) at which poly(dA)•2poly(dT) dissociated into poly(dA)•poly(dT) and poly(dT) increased dramatically (> 12 °C) in the presence of intercalator-neomycin (1–4) even at a very low concentration (2 µM). In contrast to intercalator-neomycin conjugates, the increment of Tm3→2 of poly(dA)•2poly(dT) induced by neomycin was negligible under the same conditions. The binding preference of intercalator-neomycin (1–4) to poly(dA)•2poly(dT) was also confirmed by competition dialysis and fluorescent intercalator displacement assay. Circular dichroism titration studies revealed that compound 1–4 had slightly larger binding site size (~7–7.5) with poly(dA)•2poly(dT) as compared to neomycin (~6.5). The thermodynamic parameters of these intercalator-neomycin conjugates with poly(dA)•2poly(dT) were derived from an integrated van’t Hoff equation using the Tm3→2 values, the binding site size numbers, and other parameters obtained from DSC and ITC. The binding affinity of all tested ligands with poly(dA)•2poly(dT) increased in the order neomycin < 1 < 3 < 4 < 2. Amongst them, the binding constant [(2.7 ± 0.3) × 108 M−1] of 2 with poly(dA)•2poly(dT) was the highest, almost 1000 fold more than that of neomycin. The binding of compounds 1–4 with poly(dA)•2poly(dT) was mostly enthalpy—driven and gave negative ΔCp values. The results described here suggest that the binding affinity of intercalator-neomycin conjugates to poly(dA)•2poly(dT) increases as a function of the surface area of the intercalator moiety.
Targeting the major groove of DNA duplex with a single-stranded DNA to result in a DNA triple helical structure has aroused much interest, especially in the past two decades, since its discovery in 1950’s(1). Such recognition obeys well-defined base pairing rules by making specific hydrogen bond contacts between nucleobases of the third strand and substituents on the exposed faces of the Watson-Crick base pairs, known as the purine and pyrimidine motifs, which have been examined by many investigators(2, 3, 4, 5, 6). DNA triple helical structures formed in vivo under physiological conditions have been observed and named as “H-DNA”, yet much remains to be learned about their biological functions(7, 8). A well-designed synthetic DNA oligonucleotide (also known as triplex-forming oligonucleotide, TFO) binds to DNA duplex in a sequence specific manner. Evidence suggesting that TFOs can effectively block the binding of a variety of proteins to DNA duplexes raises the level of importance of their potential therapeutical uses. Examples of triplex formation that inhibit cleavage of DNA mediated by topoiosomerase II and interfere with the unwinding function of DNA helicases are well documented(9, 10). Several groups have revealed inhibition of transcription by blockage of DNA or RNA polymerase using the triplex strategy in promoter (P1) of the c-myc oncogene(11, 12) and in the alpha subunit of interleukin-2 receptor (IL-2Rα)(13, 14).
In spite of its promising sequence-specific recognition of DNA duplex, the triplex formation strategy, also known as Antigene, has some limitations(3, 15). DNA triplex is more difficult to form than DNA duplex due to the electrostatic repulsions between the negatively charged backbone(s) of TFO and DNA duplex. In addition, the formation of DNA triplex is energetically less favorable, and the resulting DNA triplex is less stable than its counterpart duplex.
Over the years, several strategies to overcome the instability of DNA triplex, such as modification of nucleobases and backbone groups of TFO(16, 17, 18) and utilization of small binding molecules(19, 20, 21, 22) have been investigated. Interestingly, many DNA duplex intercalators have been found to stabilize DNA triple helical structures although the selectivity of these molecules between DNA triplex and duplex is subtle(23). Helene and coworkers rationally designed and synthesized benzopyridoindole derivatives, which prefer to bind DNA triplex over duplex and increase the rate of triplex formation(24, 25). We have reported the discovery of first DNA triplex specific groove binder, neomycin (an antibiotic, Figure 1)(26, 27). Our results showed that amongst all the aminoglycosides, neomycin specifically stabilizes DNA triple helical structures such as poly(dA)•2poly(dT), AT-rich or mixed base of DNA oligonucleotide triplexes, and a 12 mer intramolecular DNA triplex while having less or no effect on DNA duplexes(26). Furthermore, modeling studies of a system including neomycin and a (dT)10•(dA)10•(dT)10 DNA triplex suggest that neomycin most likely binds into the Watson-Hoogsteen groove of DNA triplex with its ring I sitting in the center of the groove and ring II an IV bridging the two pyrimidine strands together(27). These reports expanded the number of nucleic acids that aminoglycosides have been shown to bind and suggested that aminoglycoside preference is for smaller A-form major grooves(28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43). Further expanding the family of DNA triplex binding ligands based on the structure of neomycin was achieved by conjugating intercalators such as pyrene and BQQ with neomycin to yield pyrene-neomycin (1) and BQQ-neomycin (2) (Figure 1). UV denaturation and circular dichroism studies of DNA triplexes with 1 and 2 reveal that these two conjugates enhance the stabilization of DNA triplexes much greater than their parent compound, neomycin(44, 45). In addition, covalent attachment of intercalators to neomycin seems not to alter the selectively binding of neomycin to DNA triplex over duplex. The observed enhancement of DNA triplex stabilization by 1 and 2 is attributed to the “dual recognition mode”, in which the neomycin moiety binds into the Watson-Hoogsteen groove when the intercalator inserts between the proximate base triplets to provide additional stabilization.
Figure 1.
The structures of neomycin and intercalator-neomycin conjugates.
Further understanding on interactions of intercalator-neomycin conjugates with DNA triplexes can be achieved by acquiring thermodynamic parameters of their binding events. For a complete study, we in this period have synthesized two additional intercalator-neomycin conjugates, naphthalenedimide-neomycin (3) and anthraquinone-neomycin (4). We probe the surface area of the DNA triplex using compound 1–4 with varying surface for stacking between the DNA triplets (Figure 1). Fluorescent intercalator displacement (FID) assays were first performed to estimate the relative affinities of these ligands to the DNA triplex. The binding enthalpies, entropies, and equilibrium constants of these intercalator-neomycin conjugates (1–4) with a DNA polynucleotide triplex, poly(dA)•2poly(dT) were derived from an integrated van’t Hoff equation(46) to exploit the effect of intercalator moiety and the important factors on the stabilization of DNA triplex.
Experimental Procedures
General Methods
Unless otherwise specified, chemicals were purchased from Aldrich (St. Louis, MO, USA) or Fisher Scientific (Pittsburgh, PA, USA) and used without further purification. Neomycin sulfate was purchased from ICN Biomedicals (Solon, OH, USA) and was used without further purification. MINI dialysis flotation devices were acquired from Pierce (Rockford, IL, USA). Polynucleotides were purchased from GE Healthcare (Piscataway, NJ, USA). The concentrations of the polynucleotide solutions were determined by UV spectroscopy, using the following molar extinction coefficients: ε264 = 8,520 M−1 cm−1 base−1 for poly(dT), ε260 = 6,000 M−1 cm−1 bp−1 for poly(dA)•poly(dT). Oligonucleotides were synthesized and purified by IDT (Coralville, IA, USA). 1H NMR spectra were collected on a JEOL ECA 500 MHz FT-NMR spectrometer (Tokyo, Japan). MS (MALDI-TOF) spectra were collected using a Kratos analytical KOMPACT SEQ mass spectrometer (Columbia, MD, USA). UV spectra were collected on a Varian Cary 1E UV-Vis spectrophotometer (Walnut Creek, CA, USA). FID assays were carried out using a Photon Technology International instrument (Lawrenceville, NJ, USA). Isothermal titration calorimetric measurements were performed on a MicroCal VP-ITC isothermal titration calorimeter (Piscataway, NJ, USA). Circular dichroism spectra were collected on a JASCO J-810 spectropolarimeter equipped with a thermoelectrically controlled cell holder (Easton, MD, USA). Differential scanning calorimetric measurements were carried out on a MicroCal VP-DSC differential scanning calorimeter (Piscataway, NJ, USA).
Preparation of 3a
To an anhydrous pyridine solution (2 mL) of compound 12 (18 mg, 0.014 mmol), were added compound 5 (7.6 mg, 0.015 mmol) and DMAP (catalytic amount). After stirring under N2 at room temperature overnight, the organic solvent was removed under vacuum. Flash chromatography of the residue (6% MeOH in CH2Cl2) yielded the desired product as white solid (15 mg, 86%). Rf = 0.33 (silica gel, 6% MeOH in CH2Cl2); 1H NMR (CD3OD) δ 8.74–8.77 (m, 4H), 5.40 (br, 1H), 5.37 (m, 1H), 5.11 (m, 1H), 4.90 (m, 1H), 4.23 (m, 1H), 4.09 (m, 2H), 3.82 (m, 4H), 3.76 (m, 2H), 3.67 (m, 2H), 3.44–3.56 (m, 6H), 3.00–3.30 (m, 9H), 2.84–2.86 (m, 4H), 2.70–2.78 (m, 6H), 2.69 (m, 4H), 2.35 (m, 6H), 2.03 (m, 2H), 1.94 (m, 1H), 1.40 (m, 54 H). MS (MALDI-TOF): calcd. for C77H119N11O28S2 1610.96, found 1611.56.
Preparation of 3
In a 10 mL round-bottomed flask, 3a (15 mg, 0.090 mmol) was dissolved in a 1: 1 mixture of TFA/CH2Cl2 (2 mL) and stirred at room temperature for 3 h. The solvent was removed under vacuum and the residue was dissolved in DI water (20 mL). The aqueous layer was washed with ether (20 mL × 3). Lyophilization yielded 3 as yellow solid (9 mg, 90%). 1H NMR (CD3OD) δ 8.74–8.77 (m, 4H), 5.40 (br, 1H), 5.37 (m, 1H), 5.11 (m, 1H), 4.90 (m, 1H), 4.90 (q, 2H) 4.23 (m, 2H), 4.09 (m, 2H), 3.82 (m, 6H), 3.76 (m, 6H), 3.67 (m, 2H), 3.44–3.56 (m, 6H), 3.00–3.30 (m, 12H), 2.84–2.86 (m, 6H), 2.70–2.78 (m, 8H), 2.69 (m, 4H), 2.45 (m, 2H), 2.03 (m, 2H), 1.94 (m, 1H), 1.45 (m, 2H). MS (MALDI-TOF): calcd. for C47H71N11O16S2Na+ (M + Na)+ 1133.3, found 1134.4.
Preparation of 4a
To an anhydrous pyridine solution (5 mL) of compound 12 (10.7 mg, 0.008 mmol), were added compound 6 (9.3 mg, 0.027 mmol) and DMAP (catalytic amount). After stirring at room temperature overnight, the organic solvent was removed under vacuum. Flash chromatography of the residue (5% MeOH in CH2Cl2) yielded the desired product as white solid (7.1 mg, 86%). Rf = 0.60 (silica gel, 10% MeOH in CH2Cl2); 1H NMR (CD3OD) δ 8.60 (s, 1H), 8.20–8.40 (br, 1H), 5.37 (m, 1H), 5.11 (m, 1H), 4.90 (m, 1H), 4.23 (m, 1H), 4.09 (m, 2H), 3.82 (m, 4H), 3.76 (m, 2H), 3.67 (m, 2H), 3.44–3.56 (m, 6H), 3.00–3.30 (m, 9H), 2.84–2.86 (m, 4H), 2.70–2.78 (m, 6H), 2.69 (m, 4H), 2.35 (m, 6H), 2.03 (m, 2H), 1.94 (m, 1H), 1.40 (m, 54 H). MS (MALDI-TOF): calcd. for C74H113N9O27S2Na+ (M + Na)+ 1646.86, found 1647.19.
Preparation of 4
In a 10 mL round-bottomed flask, 4a (7.1 mg, 0.090 mmol) was dissolved in a 1: 1 mixture of TFA/CH2Cl2 (2 mL) and stirred at room temperature for 3 h. The solvent was removed under vacuum and the residue was dissolved in DI water (20 mL). The aqueous layer was washed with ether (20 mL × 3). Lyophilization yielded 3 as yellow solid (9 mg, 90%). 1H NMR (CD3OD) δ 8.60 (s, 1H), 8.20–8.40 (m, 4H), 7.8–7.84 (m, 2H), 5.40 (br, 1H), 5.37 (m, 1H), 5.11 (m, 1H), 4.90 (m, 1H), 4.50 (m, 2H) 4.23 (m, 1H), 4.09 (m, 2H), 3.82 (m, 4H), 3.76 (m, 2H), 3.67 (m, 2H), 3.44–3.56 (m, 8H), 3.00–3.30 (m, 10H), 2.84–2.86 (m, 4H), 2.70–2.78 (m, 6H), 2.69 (m, 4H), 2.35 (m, 6H), 2.03 (m, 2H), 1.94 (m, 1H). MS (MALDI-TOF): calcd. For C44H65N9O15S2Na+ (M + Na)+ 1047.2, found 1048.3.
Preparation of poly(dA)•2poly(dT)
The poly(dA)•poly(dT) duplex (100 µM/triplet) and poly(dT) (100 µM/triplet) were dissolved in a mixture (1.5 mL) of sodium cacodylate (SC, 10 mM, pH 6.8), KCl (150 mM), and EDTA (0.5 mM). The mixture was heated at 90°C for 10 min, slowly cooling down to room temperature, and incubated at 4°C for 12 h to maximize the DNA triplex formation.
Isothermal Titration Calorimetry (ITC)
To a mixture (1.42 mL) of poly(dA)•2poly(dT) DNA solution (100 µM), sodium cacodylate (10 mM, pH 6.8), KCl (150 mM), and EDTA (0.5 mM) in a sample cell at 20°C, was constantly injected an aliquot of mixture (10 µL) of the ligand of interest (1–200µM) , sodium cacodylate (10 mM, pH 6.8), KCl (150 mM), and EDTA (0.5 mM) through a rotary syringe. The interval time between each injection was 300 s and the duration time of each injection was 20 s. The syringe rotated at 260 rpm. The calorimetric spectrum was recorded after each injection and was processed using Origin (V 5.0). The spectra obtained from injection of the ligand solution at the same concentration into a buffer solution (10 mM sodium cacodylate, 150 mM KCl, and 0.5 mM EDTA, pH 6.8) at 20°C was used as blanks. Integration of the area under the each heat burst curve yielded the heat given off upon each injection.
To calculate the actual heat produced from binding of ligand to DNA triplex, the following equation was used.
ΔHactual: The actual heat produced from binding of ligand to DNA triplex
ΔHmeasured: The measured heat from the titration curve
ΔHblank: The heat produced from injection of ligand into the buffer solution (blank)
UV denaturation
The UV denaturation samples (1 mL) were prepared by mixing the poly(dA)•2poly(dT) DNA (15 µM/triplet), sodium cacodylate (10 mM, pH 6.8), KCl (150 mM), and EDTA (0.5 mM) and one of the intercalator-neomycin conjugates (1–4) at various concentrations (0, 1, 2, 4, 10, 15, and 25 µM). The UV melting spectra of these samples in 1 cm path length quartz cuvettes were recorded at 260 nm and 280 nm as a function of temperature (5–95°C, heating rate: 0.2 °C/min). The melting temperature was determined as the one which has the peak value in the first derivative of the melting curve.
Circular Dichroism (CD) titration measurements
To a mixture (1.8 mL) of poly(dA)•2poly(dT) DNA solution (15 µM/triplet), sodium cacodylate (10 mM, pH 6.8), KCl (150 mM), and EDTA (0.5 mM) in a 1-cm path length quartz cuvette at 20 °C, were injected aliquots (0.6–40 µL) of intercalator-neomycin stock aqueous solution. The solution was then mixed by gentle inversion of the close capped cuvette several times. The interval time between each injection was 10 minutes. The circular dichroism spectra were recorded as a function of wavelength (200–350 nm).
Competition dialysis measurements
Nucleic acid solutions were prepared in the phosphate buffer (pH 7.0) and NaCl (185 mM) to make a final concentration of 75 µM (per monomeirc unit of each polymer). The nucleic acid solution (200 µL) was placed into a MINI dialysis units and then allowed to equilibrate against a phosphate buffer (400 mL) of ligand (1 µM) at room temperature for 72 h. At the end of the experiment, nucleic acids samples (180 µL) were carefully removed and were taken to a final concentration of sodium dodecyl sulfate [SDS, 1% (w/v)]. After equilibration for 2 h, the concentration of ligand was determined spectroscopically. An appropriate correction was made due to volume changes.
The nucleic acids(47) used in the experiments are as follows: 16s A-site rRNA, i-motif, poly(dA)●poly(rU), poly(rA)●poly(dT), poly(dG)●poly(rC), poly(dA-dT), C.Perifigens DNA, M. Lysodeikticus DNA, Calf Thymus DNA, poly(dA)●2poly(dT), poly(dA)●poly(dT), poly(dA), poly(dT), poly(rA)●2poly(rU), poly(rA)●poly(rU), poly(rA), and poly(rU).
Fluorescence Intercalator Displacement Assay (FID)
Thiazole orange (700 nM) was dissolved in a buffer solution (10 mM sodium cacodylate, 150 mM KCl, 0.5 mM EDTA, pH 6.8 or 5.5) and its fluorescence was recorded (Ex: 504 nm, Em: 520–600 nm). The triplex deoxynucleotide hairpin 5’-dA12-x-dT12-x-dT12-3’ (where x= hexaethyleneglycol) was added into the triazole solution to make a final DNA concentration of 100 nM (per strand), and the fluorescence was measured again and normalized to 100% relative fluorescence. A concentrated solution of compound, aminoglycoside, (100 µM to 10 mM) was added, and the fluorescence was measured after incubation at 10 °C for 5 min. The addition of compound was continued until the fluorescence reached saturation. For all titrations, final concentrations were corrected for dilution (less than 5% of the total volume).
For the FID assay using the plate reader, the procedures are described as follows. The solution of poly(dA)•2poly(dT) was incubated with thiazole orange for 30 min prior to use for FID titration. Each well of 96-well plate was loaded with poly(dA)•2poly(dT) solution (200 µL each). A concentrated solution of ligand (1–8), (9.35 µM to 935 µM) was added, and the fluorescence was measured after incubation for 5 min. The 96-well plate was read in triplicate on Carry eclipse plate reader fluorometer with advanced reads software (Ex. 504 nm, Em. 532 nm, cutoff filter at 430–1100 nm). (No ligand = 100% fluorescence, no DNA= 0% fluorescence). Fluorescence readings are reported as % fluorescence relative to control wells.
Results and Discussion
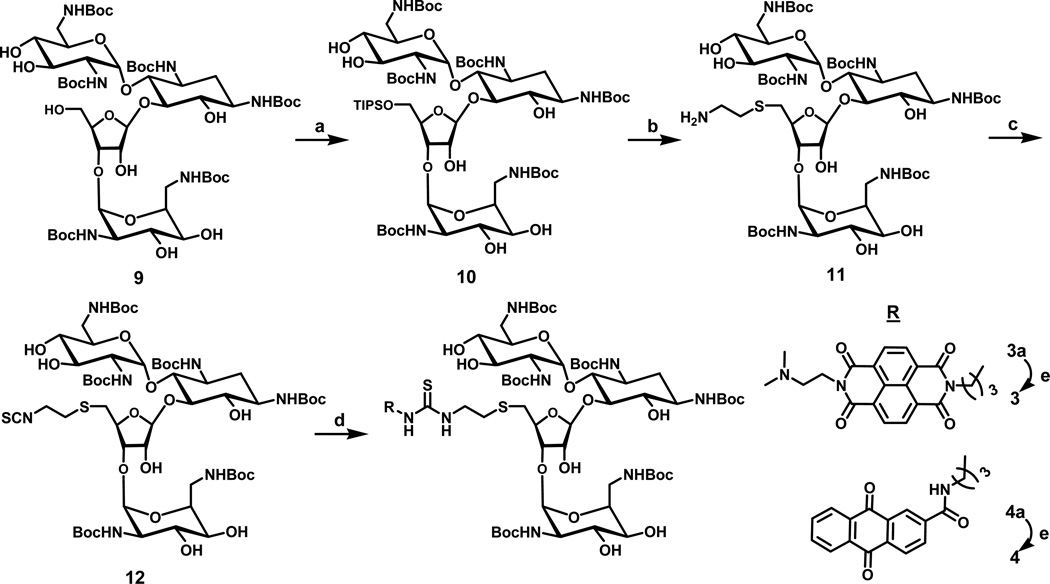
Synthesis of naphthalenedimide-neomycin and anthraquinone-neomycin conjugate
We recently reported the preparation of two intercalator-neomycin conjugates (1 and 2) by tethering the intercalator moiety at the 5’-OH position of neomycin(44, 45). The synthesis of 3 and 4 in the present study adopted the same synthetic strategy, in which the 5’-OH position of neomycin was selected to tether the intercalators because of the ease of modification at this position(48)(45)(49). Typically, reaction of t-Boc protected neomycin with triisopropylbenzenesulfonyl chloride followed by treatment of 1,2-aminothiolethane to yield compound 11 (Scheme 1). The amino group of 11 was further converted into an isothiocyanate group by reacting with 1,1’-thiocarbonyldi-2-(1H)-pyridone (TCDP) to yield 12(37). Subsequently reacting 12 with intercalators (7 or 8) yielded 3a or 4a with a thiourea linkage in decent yields. Treatment of 3a and 4a with trifluoroacetic acid (TFA) removed the acid labile Boc-protecting groups to produce 3 and 4 in quantitative yields. It is noteworthy that compound 12 has broad applications in terms of conjugation of neomycin with other moieties containing amino groups (Scheme 1)(37, 51).
Scheme 1.
a) Triisopropylbenzenesulfonyl chloride, pyridine, r.t. b) 1,2-aminothiolethane, Na, and EtOH c) TCDP, DCM, r.t. d) 7 or 8, CH2Cl2. e) TFA/CH2CH2.
FID assay as a rapid probe for conjugate affinities
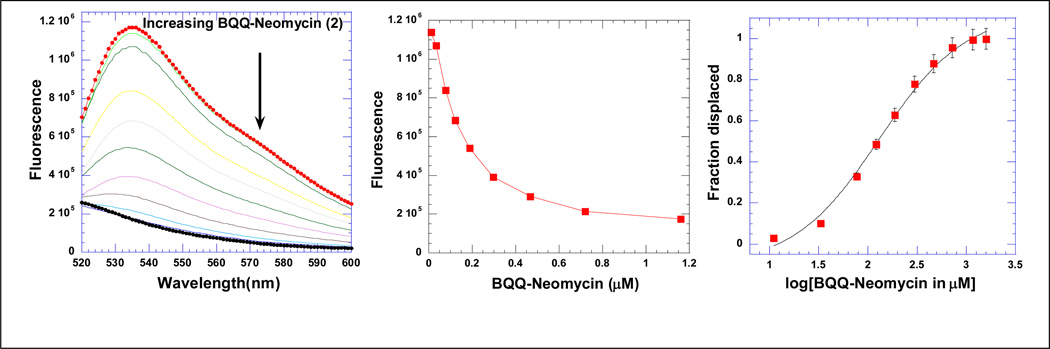
Fluorescence intercalator displacement (FID) method, a technique complementing the limitations of ITC in calculating the intrinsic affinities, has been introduced by Boger(52). In this method, an intercalator such as ethidium bromide (EB) or thiazole orange (TO) intercalates into a DNA triplex to form a complex solution. The bound intercalator fluoresces more significantly than the free intercalator. Titration of a competitive ligand that has a high binding affinity to DNA triplex into the solution will in principle displace the bound intercalator. The amount of the displaced bound intercalator can be measured as a function of decrease in fluorescence, Ethidium bromide, which intercalates into nucleic acids with a moderate affinity(52), is known to be displaced by a competitive ligand. Thiazole orange was used in our assay because it gave much higher fluorescence enhancement (3000-fold increment) upon intercalation into DNA than ethidium bromide does (20-fold increment) (53).
The displacement assays were first performed as complete titrations on a fluorometer (Figure 1a) and were then run (as triplicates) on a 96 well plate reader at a salt concentration of 150 mM KCl (Figure 1a–c). The polynucleotide triplex poly(dA)•2poly(dT) and a smaller intramolecular triplex hairpin 5’-dA12-x-dT12-x-dT12-3’ were used to assess the affinities of the conjugates. The AC50 values reported in Table 1 represent the ligand concentrations required to displace 50% of thiazole orange from the triplex, as measured by a 50% reduction of the initial bound thizaole orange fluorescence. The AC50 values for different intercalators and intercalator-neomycin conjugates are tabulated in Table 1. BQQ-neomycin 2 has the lowest AC50 value amongst all the intercalator-neomycin conjugates, suggesting that it binds to the DNA triplex more tightly than other conjugates. The rank order for binding to poly(dA)•2poly(dT) given by the AC50 values is neomycin (3 µM) < 3 (1.56 µM)< 1 (366 nM) < 4 (138 nM) < 2 (124 nM). A similar trend of binding affinity was observed in the experiments of the 5'-dA12-x-dT12-x-dT12-3' triplex. The AC50 of neomycin (47.4 µM) is approximately 100-fold higher than BQQ-neomcyin 2 (379 nM) and approximately 50 fold higher than anthraquinone-neomycin 4 (AC50 = 713 nM). Surprisingly, the AC50 value of pyrene-neomycin 1 is smaller than that of naphthalenedimide-neomcyin 3 even though 3 has a larger surface area to interact with DNA than 1. It is noteworthy that the trend of AC50 values for intercalators follows the same pattern as that for intercalator-neomcyin conjugates. In particular, the rationally designed triplex specific ligand BQQ has the lowest AC50 value, indicating the highest affinity towards the DNA triplex amongst the tested intercalators. When the FID experiments were carried out with poly(dA)•poly(dT) duplex, thiazole orange could not be displaced by 10 mM neomycin, suggesting that the affinity of the aminoglycoside is 2–3 orders of magnitude lower for the AT rich DNA duplex as compared to its affinity for the AT rich DNA triplex.
Table 1.
AC50 values of various neomycin conjugates to DNA triplex. Buffer conditions: 150 mM KCl, 10 mM SC, 0.5 mM EDTA. [5’-dA12-x-dT12-x-dT12-3’] = 100 nM/strand, [TO] = 700 nM. [poly(dA)•2poly(dT)] = 0.88 µM/bt, [TO] = 1.25 µm.
| Aminoglycoside conjugate | AC50 [5’-dA12-x-dT12-x-dT12-3’] |
AC50 [poly(dA)•2poly(dT)] |
|---|---|---|
| 6 | 1.59±0.04 µM | |
| 8 | 4.4±0.7 µM | |
| 7 | 7.2±0.9 µM | |
| 2 | 379±86 nM | 124±18 nM |
| 4 | 713±123 nM | 138±20 nM |
| 1 | 1.78±0.32 µM | 366±13 nM |
| 3 | 5.37±1.46 µM | 1.56±0.29 µM |
| Neomycin | 47.4±2.1 µM | 3.0±0.6 µM |
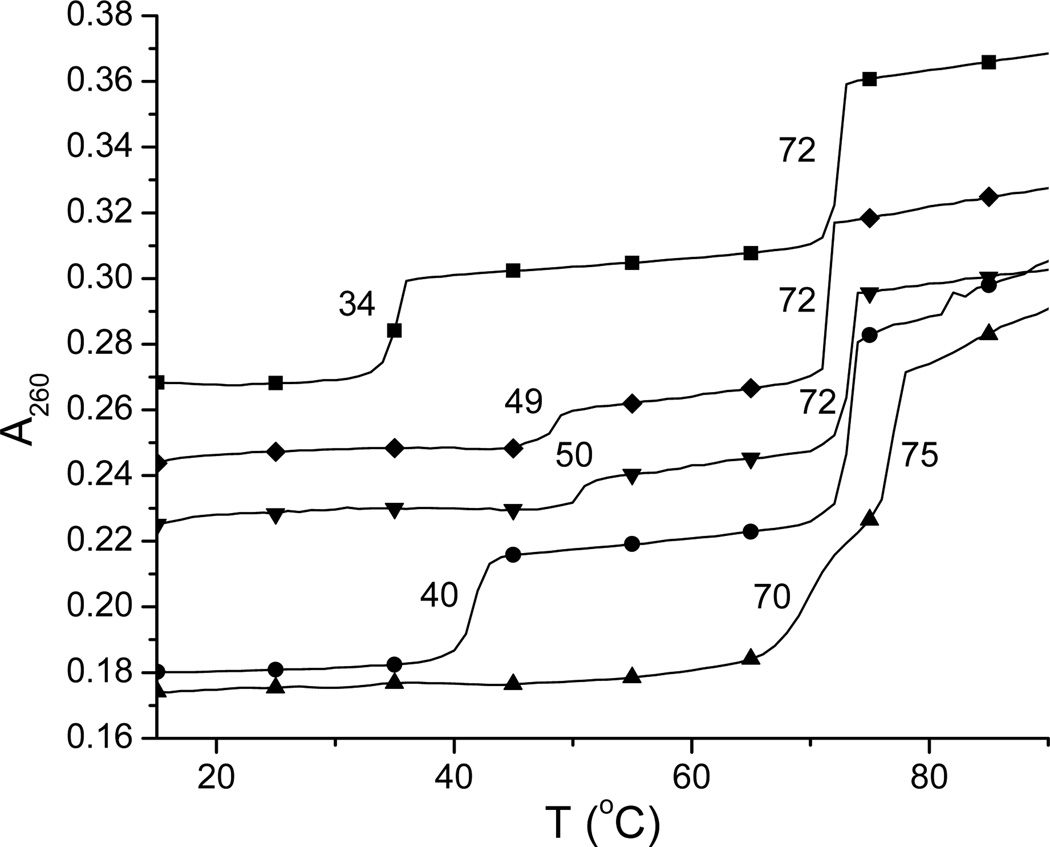
UV denaturation of poly(dA)•2poly(dT) with intercalator-neomycin conjugates
The thermal stability of DNA triple-helical structures can be examined using UV denaturation. Biphasic transitions are commonly observed when the UV spectrum of DNA triplex is monitored as a function of temperature, which represent dissociation of DNA triplex into its counterpart duplex at lower temperature and further dissociation of the resulting DNA duplex into single stranded structures at higher temperature. Measuring the melting temperatures of DNA triplex in the presence of binding ligands is a quick and unambiguous way to simultaneously determine the relative strength of ligands on stabilization of DNA triplex and the selectivity of ligands between DNA triplex and duplex. DNA polynucleotide triplex such as poly(dA)•2poly(dT) was used as a model DNA triplex in UV denaturation because of its clear transitions of triplex into duplex and duplex into random coil structures. Under our experiemental conditions, in the absence of ligands, the melting temperatures of poly(dA)•2poly(dT) dissociating into poly(dA)• poly(dT) and poly(dT) and poly(dA)•poly(dT) dissociating into poly(dA) and poly(dT) were 34 and 72 °C, respectively. Our UV denaturation results suggested that all four intercalator-neomycin conjugates dramatically stabilized poly(dA)•2poly(dT) and had little effect on its duplex, poly(dA)•poly(dT) (Table 2). Amongst the four, compound 2 showed the greatest stabilization on poly(dA)•2poly(dT). In the presence of 2 (4 µM), only one melting transition at 86 °C was observed (Figure 1 of the supporting information), indicating the merging of triplex to duplex and duplex to single strand transitions(54). In contrast, the UV melting temperatures representing the triplex to duplex transition in the presence of neomycin, 1, 3, or 4 at the same concentration (4 µM) were 40, 58, 70, and 74 °C (Table 2). In all cases, the changes in DNA duplex melting temperature were subtle. The stabilization effect on poly(dA)•2poly(dT) seems to result from the covalent attachement of intercalator and neomycin since a mixture of intercalator and neomycin does not show the equivalent stabilization. For instance, the melting temperature of poly(dA)•2poly(dT) in the presence of 7 (4 µM) and neomycin (4 µM) was 50 °C, about 20 °C lower than that in the presence of 3 at the same concentration (4 µM) (Figure 3). All of the described results here indicate that the strength of stabilizing poly(dA)•2poly(dT) increases in the order neomycin < 1 < 3 < 4 < 2. Interestingly, the strength of stabilizing poly(dA)•2poly(dT) by intercalators 5, 6, 7, and 8 at 4 µM showed the same order as that by intercalator-neomycin conjugates (Table 1, 2 of the supporting information), suggesting that the increasing surface area of the intercalator moiety is clearly an important factor in recognition of the DNA triplex.
Table 2.
The UV melting temperatures recorded when poly(dA)•2poly(dT) dissociates in the presence of intercalator-neomycin conjugates (1–4) at various concentrations. Tm3→2: The melting temperatures representing the transition when poly(dA)•2poly(dT) dissociates into poly(dA)•poly(dT) and poly(dT). Tm2→1: The melting temperatures representing the transition when poly(dA)•poly(dT) dissociates into poly(dA) and poly(dT).
| Concentration of 1 (µM) | Tm3→2 | Tm2→1 |
|---|---|---|
| 0 | 34 | 72 |
| 1 | 43 | 72 |
| 2 | 56 | 72 |
| 4 | 59 | 73 |
| Concentration of 2 (µM) | Tm3→2 | Tm2→1 |
| 0 | 34 | 72 |
| 2 | 80 (Tm3→1) | - |
| 4 | 86 (Tm3→1) | - |
| Concentration of 3 (µM) | Tm3→2 | Tm2→1 |
| 0 | 34 | 72 |
| 1 | 54 | 72 |
| 2 | 65 | 73 |
| 4 | 70 | 75 |
| Concentration of 4 (µM) | Tm3→2 | Tm2→1 |
| 0 | 34 | 72 |
| 1 | 60 | 74 |
| 2 | 68 | 74 |
| 4 | 74 | 80 |
Figure 3.
Representative UV melting profiles of poly(dA)•2poly(dT) at 260 nm in the absence (■) and presence of neomycin (4 µM, ♦), neomycin + 7 (4 µM each, ▼), 7 (4 µM, ●), and 3 (4 µM, ▲). Experimental conditions: sodium cacodylate buffer (10 mM, pH 6.8), KCl (150 mM), and EDTA (0.5 mM). [DNA] = 15 µM per base triplet. The y-axis has been artificially offset to differentiate the melting curves.
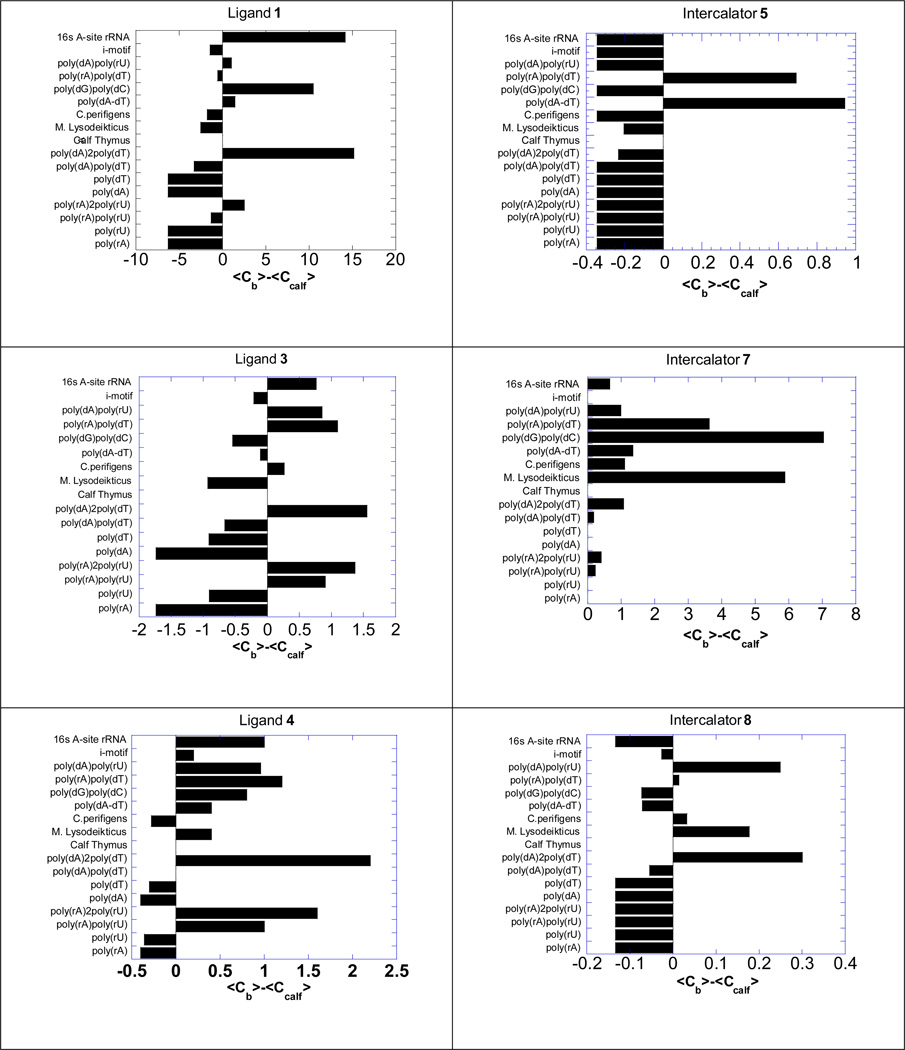
Competition dialysis of intercalator-neomycin conjugates with different structures of nucleic acids
Competition dialysis is a quick and convenient method developed by Chaires(47, 55) for simultaneously determining the binding preference of ligands to different structures of nucleic acids. The nucleic acid solutions (75 µM per monomeirc unit of each polymer) in mini-dialysis units are allowed to equilibrate against large excess of desired ligand (1 µM). After dissociating the bound ligand off nucleic acids using a surfactant (SDS), the concentration of the ligand is measured spectroscopically. The amount of the detected ligand in each nucleic acid solution corresponds to its binding affinity toward this nucleic acid. Our previous competition dialysis study revealed that 2 binds preferentially toward DNA or RNA triple helical structures while having minimum binding to the corresponding single-stranded and duplex nucleic acids. In the present study, competition dialysis experiments of 1, 3 and 4 as well as their corresponding intercalators (5, 7, and 8) against 20 different types of nucleic acids including G-quadruplex and triplex are carried out.
The concentration of the ligand in each individual nucleic acid solution is presented by subtracting the ligand concentration in the calf thymus DNA solution, a typical B-form nucleic acid structure (Figure 4). Calf thymus DNA is chosen because according to the UV denaturation studies, these intercalator-neomycin conjugates have minimum binding affinity to it. Our results show that in general, compound 1, 3, and 4 do not bind to single-stranded nucleic acids. Just like 2, all three intercalator-neomycin conjugates tested here bind preferentially toward poly(dA)●2poly(dT) while having little binding affinity to poly(dA)●poly(dT). This observation unambiguously supports our previous conclusions that intercalator-neomycin conjugates favors DNA triple helical structures. Compound 1 and 4 bind to poly(dG)●poly(dC) while 3 does not bind to it at all. All three intercalator-neomycin conjugates to some extent have binding affinity toward DNA/RNA hybrid sequences, such as poly(dA)●poly(rU), which are know to be A-form nucleic acid structures(49).The competition dialysis results from the intercalators (5, 7, and 8) show different patterns from those of 1, 3, and 4, suggesting that the selectivity in binding of 1, 3, and 4 toward different nucleic acids results from the combination of groove binding and stacking interactions, not simply from the base stacking interactions of the intercalator.
Figure 4.
Competition dialysis results of 1, 3 and 4 as well as their corresponding intercalators 5, 7, and 8 (1 µM) with various types of nucleic acids (75 µM). Buffer: Na2HPO4 (6 mM), NaH2PO4 (2 mM), Na2EDTA (1 mM), NaCl (185 mM), and pH 7.0.
Determining the binding site size of intercalator-neomycin conjugates with poly(dA)•2poly(dT) using UV thermal denaturation, CD and fluorescence spectroscopy
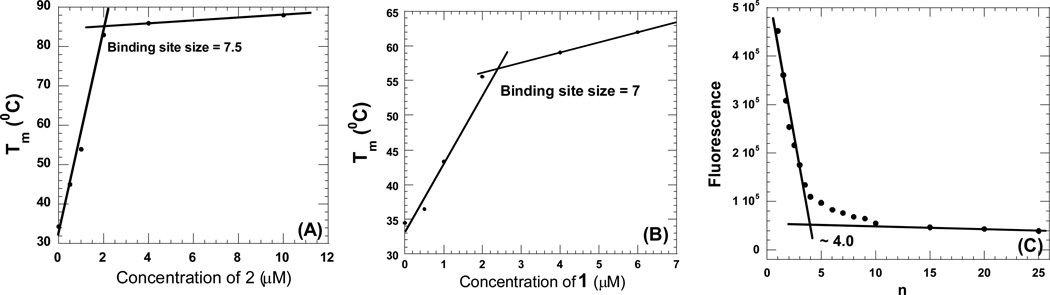
In order to understand the binding of above described intercalator-neomycin conjugates with poly(dA)•2poly(dT), we sought to investigate the binding site sizes between 1, 2, 3, and 4 with poly(dA)•2poly(dT) using UV thermal denaturation and circular dichroism (CD) spectroscopy. Binding of the intercalator-neomycin conjugates (1–4) with poly(dA)•2poly(dT) slightly altered the conformation of DNA triplex secondary structure, which was consequently detected by circular dichroism spectroscopy. A much larger change in Tm was observed using thermal denaturation studies (Table 2). Tm changes varied linearly with an increase in concentration of the intercalator-neomycin conjugates and a plot of Tm vs. ligand concentration was made to determine the binding site size. A transition point resulting from the slope change was clearly observed in all of the plots, when the primary available sites in poly(dA)•2poly(dT) are saturated by the intercalator-neomycin conjugate. Our results suggest that the binding site size between 1, 2, 3, and 4 with poly(dA)•2poly(dT) is 7, 7.5, 7.5, and 7.5, respectively (Figure 5). The values of binding site size described here are reasonable compared to that of neomycin and conjugates (~6–6.5) reported previously using CD spectroscopy (44, 45)(27). The observed similarity in binding site sizes implies a similarity in binding mode of compound 1–4 with poly(dA)•2poly(dT).
Figure 5.
(A–B) Representative plots for binding site size determination of intercalator-neomycin conjugates with poly(dA)•2poly(dT) as determined by UV thermal denaturation. (C) Plot for determination of binding site determination of Poly(dA)•2poly(dT) and naphthalene diimide. Graph shows the change in fluorescence of naphthalenedimide when titrated with Poly(dA)•2poly(dT). Naphthalenedimide was excited at 356 nm and the emission scans were recorded from 370–500 nm. [Poly(dA)•2poly(dT)] = 15 µM/bt. [naphthalene diimide] = 0.5 µm to 15µm. [T] = 10 °C. n = base triplet/drug. Buffer conditions: sodium cacodylate (10 mM), EDTA (0.5 mM), KCl (150 mM), pH (5.5).
Extrapolating thermodynamic parameters from the interactions between intercalator-neomycin conjugates and poly(dA)●2poly(dT)
Ultimate understanding of interactions between intercalator-neomycin conjugates and poly(dA)•2poly(dT) is achieved by acquiring a complete set of thermodynamic parameters. A method for extrapolating thermodynamic parameters from experimental data was developed and optimized by McGhee and others(46, 56), in which the following equation is used.
where Tm0 is the melting temperature representing the dissociation of DNA triplex into duplex in the absence of ligand, Tm the melting temperature representing the dissociation of DNA triplex into duplex in the presence of intercalator-neomycin conjugate, R is the gas constant, ΔHHS is the enthalpy change corresponding to the dissociation of DNA triplex into duplex in the absence of ligand, L is the free ligand concentration at Tm which is estimated as one-half of the total drug concentration, and n the binding site size. The melting temperatures were determined using UV spectroscopy as described above. The enthalpy change (ΔHHS) was measured using differential scanning calorimetry (DSC), in which the heat produced from the dissociation of DNA triplex was plotted as a function of temperature (20–90 °C). The binding site size was obtained from UV thermal denaturation and CD titration experiments as described above. After solving this equation, the association constant at Tm (KTm) in the presence of intercalator-neomycin conjugate at a desired concentration was obtained. The more meaningful and useful association constants (KT) at different temperatures for analysis of interactions between intercalator-neomycin conjugate and poly(dA)•2poly(dT) were further derived from the following integrated van’t Hoff equation using the calculated KTm values.
where ΔHT is the observed enthalpy change associated with the binding of intercalator-neomycin conjugate to poly(dA)•2poly(dT) and ΔCp is the heat capacity change in the binding. ΔHT was determined using isothermal titration calorimetry (ITC), in which small aliquots of buffered intercalator-neomycin conjugate were injected into a solution containing large excess of poly(dA)•2poly(dT) in terms of possible numbers of binding sites. ΔCp was derived from the slope of the linear relationship in which the binding enthalpy (ΔHT) was plotted as a function of temperature (T).
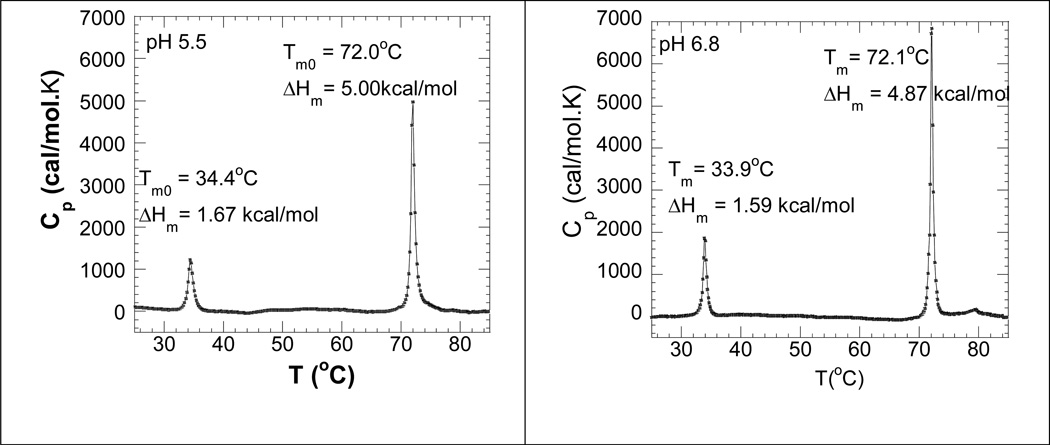
All of the experiments used for calculations of thermodynamic parameters were conducted at two different pH values (6.8 and 5.5). The ΔHHS values measured by DSC for the dissociation of poly(dA)•2poly(dT) into poly(dA)•poly(dT) and poly(dT) at pH 5.5 and 6.8 were 1.67 kcal/mol and 1.59 kcal/mol (Figure 6), suggesting that poly(dA)•2poly(dT) was more stable at lower pH. This observation is consistent with the common belief that the stability of DNA triplex increases with the decrease in pH because of suppression of the electrostatic repulsions between DNA backbones under lower pH conditions. A similar trend was observed for the dissociation of poly(dA)•poly(dT) into poly(dA) and poly(dT), in which the ΔHHS value at pH 5.5 (5.00 kcal/mol) was 150 cal/mol higher than that at pH 6.6 (4.87 cal/mol).
Figure 6.
DSC melting profile of poly(dA)•2poly(dT) in the absence of binding ligand at pH 5.5 (left) and pH 6.8 (right). Experimental conditions: sodium cacodylate (10 mM), EDTA (0.5 mM), KCl (150 mM).
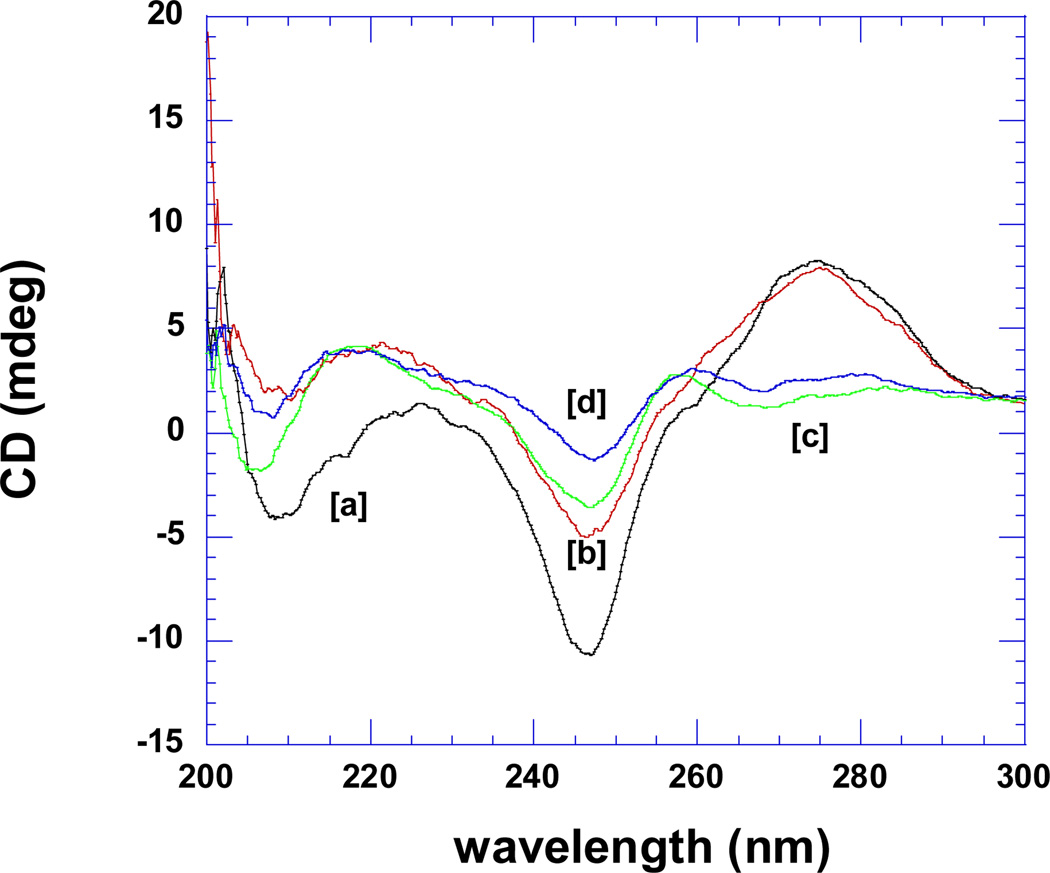
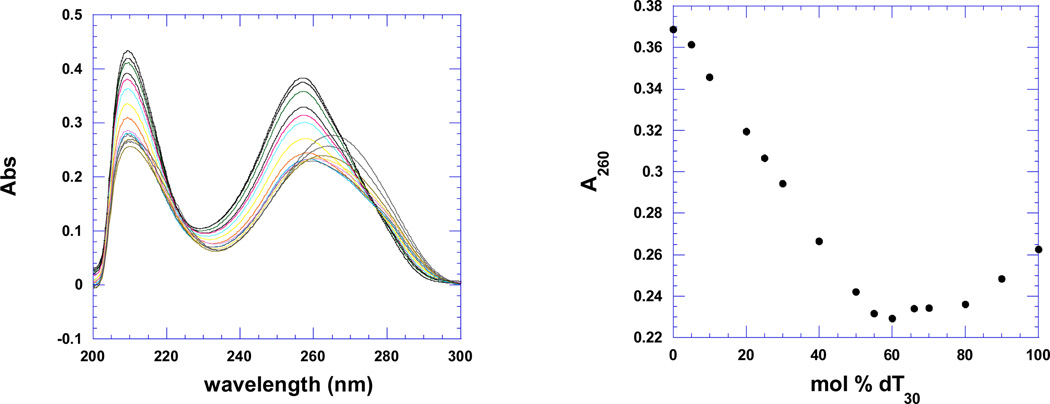
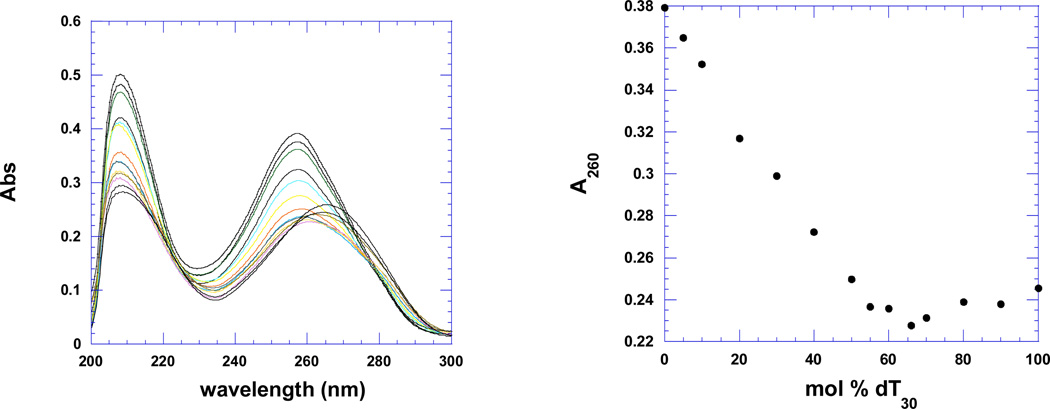
Figure 7a shows the CD spectra of the pre-formed poly(dA)•poly(dT) duplex and poly(dA)•2poly(dT) triplex in the absence (scans a, c) and presence of the tightest binding ligand 2, BQQ-neomycin (scans b, d). The duplex and triplex DNA have signature CD spectra which have been preserved upon addition of the conjugate suggesting that little conformational change takes place when drug is added. Figure 7a shows that upon binding of ligand 2 (BQQ-neomycin) to the duplex or triplex, no red or blue shift is observed in the CD spectrum of the preformed duplex or triplex. Figure 7b shows the continuous variation experiments of poly dA and poly dT in the absence and presence of drug 3. All intercalators and conjugates show a similar minimum at 66% dT, suggesting that the 3-stranded DNA triplex is present in solution in the absence and presence of the drugs.
Figure 7.
CD scans for the poly(dA)•poly(dT) duplex and poly(dA)•2poly(dT) triplex in the absence and presence of 2 (BQQ-neomycin). [a]= poly(dA)•2poly(dT) triplex (no ligand); [b]= poly(dA)•2poly(dT) triplex (0.13 equivalent 2); [c]= poly(dA)•poly(dT) duplex (no ligand); [d]= poly(dA)•poly(dT) duplex (0.13 equivalent 2). Experimental conditions: sodium cacodylate (10 mM, pH 5.5), EDTA (0.5 mM), KCl (150 mM), Temperature = 10°C.
b. Continuous variation scans (left) and A260 plot (right) of dA30 (1 µM/base) and dT30 (1µM/base) in the absence of drug (top) and presence of napthalenediimide-neomycin 3 (rbd =7) (bottom). Experimental conditions: sodium cacodylate (10 mM, pH 5.5), EDTA (0.5 mM), KCl (150 mM), 15°C.
ITC experiments were carried out for determining the enthalpy changes in the binding of intercalator-neomycin conjugate to poly(dA)•2poly(dT) at pH 5.5 and 6.8 (Figure S13–S20 of the supporting information). The pre-formed DNA triplex solution was incubated in a sample cell at the temperatures which were lower than 20 °C to ensure the presence of DNA triplex (Tm = 34 °C). The enthalpy changes (ΔHT) were recorded as a heat burst curve when small portions of buffered intercalator-neomycin conjugated injected into the DNA solution. The area under each heat burst curve was integrated and recorded as the ΔHT values. For instance, when the pH of the solution was 5.5, the ΔHT values at 10 °C for neomycin, 1, 2, 3, and 4 were −(6.4±0.1), − (3.2±0.1), − (6.8±0.1), −(10.9±0.2), and − (15.9±0.4) kcal/mol, respectively (Table 3). Under this condition, the ΔHT values increased in the order 1 < 3 < 4 < 2, which is consistent with the order of DNA triplex stabilization derived from UV denaturation experiments. A similar trend was also observed when injecting intercalator-neomycin conjugates into the poly(dA)•2poly(dT) solution at different temperatures and pH values (Table 3). The enthalpy change in the binding of neomycin to poly(dA)•2poly(dT) at pH 6.8 was somewhat different from the trend observed in the UV denaturation experiments. Our UV melting studies suggest that 1 has greater stabilization effect on poly(dA)•2poly(dT) than neomycin at pH 6.8. For instance, the melting temperature of poly(dA)•2poly(dT) in the presence of 1 (4 µM) was 24 °C higher than that in the presence of neomycin. However, at pH 6.8, the enthalpy change in the binding of 1 to poly(dA)•2poly(dT) was 3.2 kcal/more less than that in the binding of neomycin to poly(dA)•2poly(dT) even though the fact that 1 has greater DNA triplex stabilization than neomycin. In contrast, at pH 5.5, the enthalpy change in the binding of 1 to poly(dA)•2poly(dT) was 0.9 kcal/mol more than that in the binding of neomycin to poly(dA)•2poly(dT), which was consistent with the order that 1 stabilizes triplex more. In addition, the enthalpy change in the binding of intercalator-neomycin conjugate with poly(dA)•2poly(dT) at pH 6.8 were much higher than that in the corresponding binding at pH 5.5. This observation is attributed to the binding-linked neomycin protonation heat. The amino groups of neomycin, according to their pKa values, are not completely protonated at pH 6.8. The heat produced from such amine protonation at pH 6.8 results in an overestimation of enthalpy changes(57). The enthalpy change measured at pH 5.5 reflects more of an intrinsic interaction between intercalator-neomycin conjugate and poly(dA)•2poly(dT) because neomycin is substantially protonated at this pH value and thus the protonation heat is minimized. However, study such binding event at pH 5.5 can lead to some practical problems. The interactions between these conjugates and poly(dA)•2poly(dT) intend to become more entropy-driven at low pH, thereby, the intensity of ITC signals decreases dramatically necessitating the use of higher concentrations of samples.
Table 3.
Thermodynamic profiles of intercalator-neomycin conjugates (1–4) and intercalator 7 with poly(dA)•2poly(dT) at pH 5.5. Experimental condition: sodium cacodylate (10 mM), EDTA (0.5 mM), KCl (150 mM).
| Ligand | [KCl] mM |
ΔHwca (kcal/ mol) |
Tm0a (°C) |
Tma (°C) |
nb | ΔH(10°C)c (kcal/mol) |
ΔH(15°C)c (kcal/mol) |
ΔH(20°C)c (kcal/mol) |
ΔCpd (cal/mol.K) |
KT(20°C)e (M−1) |
|---|---|---|---|---|---|---|---|---|---|---|
| Neomycin | 140 | 1.70 | 31.4 | 34.4 | 6.8 | −0.6±0.1 (12°C) |
- | −1.31±0.02 | −87±4 | (2.4±0.1)×105 |
| 1 | 150 | 1.67 | 33.8 | 49.5 | 7 | −1.5±0.1 | - | −1.64±0.02 | −10±3 | (1.9±0.1)×106 |
| 3 | 150 | 1.67 | 33.8 | 55.4 | 7.5 | −2.4±0.1 | −2.6±0.1 | −2.8±0.1 | −40±20 | (5.5±0.3)×106 |
| 4 | 150 | 1.67 | 33.8 | 64.0 | 7.5 | −5.4±0.2 | −6.1±0.2 | −6.4±0.1 | −110±15 | (3.7±0.1)×107 |
| 2 | 150 | 1.67 | 33.8 | 68.8 | 7.5 | −7.5±0.4 | −8.5±0.4 | −9.7±0.2 | −220±20 | (2.7±0.3)×108 |
| 7 | 150 | 1.67 | 31.0 | 37.2 | 4 | *−1.4±0.1 | −0.38±0.07 | −0.81±0.01 | −131 | (1.89)×105 |
ΔH was calculated at 23°C.
In order to calculate the ΔCp values, the enthalpy changes (ΔHT) were plotted versus temperatures and fitted via linear regression analysis. The slope of linear fitting represents the ΔCp in the binding of intercalator-neomycin conjugate to poly(dA)•2poly(dT) under this experimental condition. In all cases, negative ΔCp values were observed at pH 6.8 and 5.5 and the ΔCp values are in general more negative at pH 6.8 than those at pH 5.5 (Table 3 and 4). Protonation heats at pH 6.8 yields a much higher ΔCp value. In contrast, the relatively low ΔCp values at pH 5.5 indicate a more intrinsic drug-triplex interaction. Additionally, the enthalpy changes in the binding of 3 and 4 with triplex poly(dA)•2poly(dT) were −3.7 kcal/mol and −7.8 kcal/mol more than those with the duplex poly(dA)•poly(dT), respectively, suggesting that interactions between intercalator-neomycin with DNA triplex is more enthalpy-driven than that with DNA duplex.
Table 4.
Thermodynamic profiles of intercalator-neomycin conjugates (1–4) with poly(dA)●2poly(dT) at pH 6.8. Experimental condition: sodium cacodylate (10 mM), EDTA (0.5 mM), KCl (150 mM).
| Ligand | [KCl] mM |
ΔHwca (kcal/mol) |
Tm0a (°C) |
Tma (°C) |
nb | ΔH(10°C)c (kcal/mol) |
ΔH(20°C)c (kcal/mol) |
ΔH(5°C)c (kcal/mol) |
ΔCpd (cal/mol.K) |
KT(20°C)e (M−1) |
|---|---|---|---|---|---|---|---|---|---|---|
| Neomycin | 150 | 1.59 | 34.4 | 49.5 | 6.5 | −6.4±±0.1 | −7.5±0.2 | −8.2±0.04 | −116±4 | - |
| 1 | 150 | 1.59 | 34.4 | 58.0 | 7 | −3.2±0.1 | −3.7±0.1 | - | −45±1 | - |
| 3 | 150 | 1.59 | 34.4 | 65.0 | 7.5 | −6.8±0.1 | −7.8±0.1 | −8.3±0.8 | −104±10 | - |
| 4 | 150 | 1.59 | 34.4 | 68.0 | 7.5 | −1±0.9±0.2 | −12.7±0.2 | −14.5±0.1 | −231±6 | - |
| 2 | 150 | 1.59 | 34.4 | 85.0 | 7.5 | −15.9±0.4 | −19.3±0.4 | −21.1±0.4 | −346±13 | - |
The binding constant of intercalator-neomycin with poly(dA)•2poly(dT) was then derived using all the parameters described above. At pH 5.5, all intercalator-neomycin conjugates seemed to bind poly(dA)•2poly(dT) more tightly than neomycin. The calculated binding constants increased in the order neomycin < 1 < 3 < 4 < 2, with values of (2.7±0.3) ×108 M−1 for 2, (3.7±0.1) ×107 M−1 for 3, (5.5±0.3) ×106 M−1 for 4, (1.9±0.1) ×106 M−1 for 1, and (2.4±0.1) ×105 for neomycin, respectively (Table 3). The binding constant of 2 with poly(dA)•2poly(dT) is (2.7±0.3) ×108 M−1, which is about 1000 fold higher than that of neomycin. This observed high binding constant of 2 is consistent with our previously published result that 2 is a very potent DNA triplex binding ligand. The rationally designed triplex specific binding ligand, BQQ in 2, may play an important role in promoting the significant DNA triplex binding. The binding constant of 2 with poly(dA)•2poly(dT) is about 7.3, 49, and 142 fold higher than that of 4, 3 and 1, respectively. This data, in addition to our FID AC50 values, allows us to quantitate the relative binding strength of the intercalators to poly(dA)•2poly(dT). The larger the surface area of the intercalator, the higher DNA triplex binding affinity of its neomycin conjugate is observed. Pyrene itself does not stabilize DNA triplexes; therefore, its conjugate 1 has the weakest triplex binding ability amongst the four. The slight discrepancy between FID results and calorimetric data, when comparing the affinities, can be attributed to the differences in the nature and sensitivities of the measurements, given that the affinities are quite comparable. FID assay, which is an indirect measure of the ligand affinities, shows a two-four fold higher affinity for 1 over 3, whereas data from Table 3 shows the opposite trend, with 3 showing a two-fold higher affinity. Additionally, the FID measurements were performed at pH 6.8, whereas the affinities in Table 3 are reported at pH 5.5. When the FID results were repeated at pH 5.5, similar trends were observed, with 3 (AC50=408±70 nM) showing a higher affinity than 1 (AC50=240±20 nM). The UV thermal denaturation temperatures at both pH 6.8 and 5.5 (Table 2, Table 3), clearly show a much larger change in Tm3→2 for 3 over 1 (70°C vs. 50°C at pH 6.8 and 55.9°C vs. 49°C at pH 5.5) suggesting that 3 binds tighter than 1 at the two pH values.
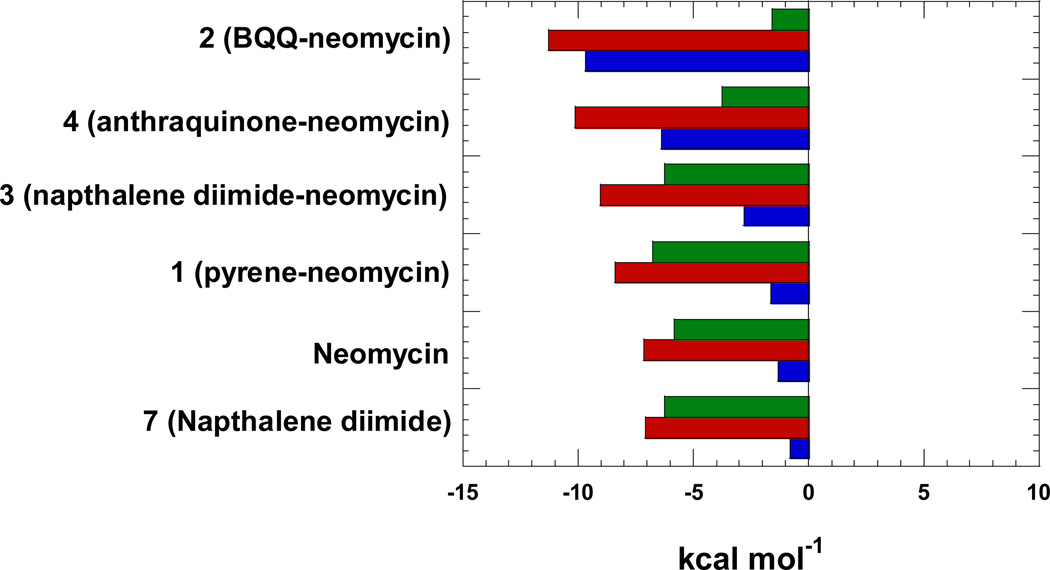
Figure 8 shows the distribution of thermodynamic properties of ligand-triplex interactions as bar graphs. The free energy for each of the ligand has been calculated from binding constant by using Gibbs free energy equation. Free energy change (red bars), enthalpy change (blue bars) and entropic contributions (green bars) help summarize the contribution of the individual ligands as well as the conjugates. Three different classes of ligands are represented in the figure as far as binding mode is concerned. Neomycin is a major groove binder, naphthalene diimide is a well-known intercalator, and neomycin conjugates bind through dual recognition mode (major groove and intercalation). The plot in figure 8 is a good starting point to understand the contribution of thermodynamic properties during the dug-DNA binding interactions. It is clear from the figure that binding of neomycin (groove binder) as well as naphthalene diimide (intercalator) are largely driven by enthalpy contributions. As we go from 1 (pyrene-neomycin) to 2 (BQQ-neomycin), an increase in intercalator surface area leads to an increase in the enthalpy of interaction, which clearly leads to a more negative free energy of binding. However, as seen from the free energies of interaction of neomycin, 7 and 3, the free energy ΔG3-triplex is not a sum of free energy of ΔGneomycin-triplex and ΔG7-triplex leading to a ΔGcoupling of ~5kcal/mol. While the ΔH3-triplex increases substantially, the corresponding increase in the ΔS3-triplex is not observed, suggesting that the conjugate pays an entropic cost of covalently bridging neomycin with the intercalator with the given linker length. As one goes to the higher affinity conjugates 2 and 4, the entropic contribution to the ΔGcomplexation drop substantially. Thermodynamics of interaction of ligands 5, 6 and 8 with the triplex could not be performed due to the poor solubility of these compounds in aqueous solution. Further studies with different linker lengths will be beneficial to maximize the free energies of interaction of the dual binding modes and are currently being explored in our laboratories.
Figure 8.
Thermodynamics of binding interactions of ligands with poly(dA)•2poly(dT) triplex at pH 5.5.; red bars represent ΔG, blue bars represent ΔH, and green bars represent TΔS. Experimental condition: sodium cacodylate (10 mM), EDTA (0.5 mM), KCl (150 mM).
Conclusions
Decades of work in the recognition of DNA triplexes has led to the design of intercalators that preferentially bind to DNA triplexes, while minimizing the binding to the DNA duplex. Addition of a groove binding DNA triplex selective ligand, such as neomycin has allowed us to achieve DNA triplex affinities that would be difficult to attain with a univalent binding mode without sacrificing the selectivity. Our results here provide a measure of quantification of triplex binding when the ligand surface area is increased in the order pyrene < naphthalenediimide < anthraquinone < BQQ. It is not a surprise that 2 is the most potent DNA triplex stabilizing agent amongst all the intercalator-neomycin conjugates in our study because the BQQ moiety (6) is known as a rationally designed DNA triplex specific binding ligand. All of the other intercalators (pyrene, naphthalenedimide, and anthraquinone) bind to DNA duplex as well as DNA triplex to some extent. The conjugates containing intercalator and neomycin enhance the binding affinity of DNA triplex via a possible cooperative binding mode. Such a “dual binding mode” may be useful in guidance of designing novel DNA triplex binding ligands (and perhaps other nucleic acids). The thermodynamic data also raise the possibility to tune the DNA triplex binding strength of neomycin conjugates, by simply swamping the intercalator moiety. A large data bank of DNA intercalators has already been established; therefore, preparation of such conjugates for screening the nucleic acid binding ligands is practical. Inspection of the ligand-triplex interaction thermodynamics shows a clear additive effect of enthalpies of interaction, and future studies with linker length optimization can lead to even tighter affinity ligands.
Supplementary Material
Figure 2.
A graphical representation of thiazole orange displacement assay. (A) A raw emission data of 1.25 µM thiazole orange upon excitation at 534 nm with buffer only (open circles) and after addition of 0.88 µM/bt poly(dA)•2poly(dT) triplex (black circles). BQQ-neomycin was then titrated from 0.25 µM to 1.06 mM. (B) The decrease in the fluorescence intensity of the complex (DNA-thiazole orange) upon addition of BQQ-neomycin aliquots. (C) Assuming a linear relationship between the changes in fluorescence intensity with the fraction of thiazole orange displaced results in S-shaped binding isotherm. This graph allows the determination of concentration of ligands needed to displace half of the thiazole orange from the DNA triplex.
Buffer conditions: 150 mM KCl, 10 mM SC, 0.5 mM EDTA, pH 6.8. [poly(dA).2poly(dT)] = 0.88 µM/bt. [TO] = 1.25 µM
Acknowledgement
We are grateful for support of this research from the NSF (CHE/MCB-0134972) and NIH (R15CA125724).
Abbreviations used
- FID
Fluoroscent Intercalator Displacement
- TFO
triplex forming oligonucleotide
- BQQ
benzoquinoquinoxaline
Footnotes
Supporting information available NMR spectra of 3 and 4, spectra for ITC experiments of compound 1–4 titrating into large excess of poly(dA)•2poly(dT) at pH 5.5 and pH 6.8, and ΔCp plots. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Felsenfeld G, Davies D, Rich A. Formation of a Three Stranded Polynucleotide Molecule. J. Am. Chem. Soc. 1957;79:2023–2024. [Google Scholar]
- 2.Radhakrishnan I, Patel DJ. Solution Structure of a Pyrimidine.Purine.Pyrimidine DNA Triplex Containing T.AT, C+.GC and G.TA Triples. Structure. 1994;2:17–32. doi: 10.1016/s0969-2126(00)00005-8. [DOI] [PubMed] [Google Scholar]
- 3.Praseuth D, Guieysse AL, Helene C. Triple Helix Formation and the Antigene Strategy for Sequence-Specific Control of Gene Expression. Biochim. Biophys. Acta. 1999;1489:181–206. doi: 10.1016/s0167-4781(99)00149-9. [DOI] [PubMed] [Google Scholar]
- 4.Beal PA, Dervan PB. Second Structural Motif for Recognition of DNA by Oligonucleotide-Directed Triple-Helix Formation. Science. 1991;251:1360–1363. doi: 10.1126/science.2003222. [DOI] [PubMed] [Google Scholar]
- 5.Chen FM. Intramolecular Triplex Formation of the Purine.Purine.Pyrimidine Type. Biochemistry (N.Y.) 1991;30:4472–4479. doi: 10.1021/bi00232a014. [DOI] [PubMed] [Google Scholar]
- 6.Radhakrishnan I, Patel DJ. Solution Structure of a Purine.Purine.Pyrimidine DNA Triplex Containing G.GC and T.AT Triplexes. 1993;1:135–152. doi: 10.1016/0969-2126(93)90028-f. [DOI] [PubMed] [Google Scholar]
- 7.Michel D, Chatelain G, Herault Y, Brun G. The Long Repetitive Polypurine Polypyrimidine Sequence (Ttccc)48 Forms DNA Triplex with Pu-Pu-Py Base Triplets Invivo. Nucleic Acids Res. 1992;20:439–443. doi: 10.1093/nar/20.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agazie YM, Lee JS, Burkholder GD. Characterization of a New Monoclonal Antibody to Triplex DNA and Immunofluorescent Staining of Mammalian Chromosomes. Journal of Biological Chemistry. 1994;269:7019–7023. [PubMed] [Google Scholar]
- 9.Kopel V, Pozner A, Baran N, Manor H. Unwinding of the Third Strand of a DNA Triple Helix, a Novel Activity of the SV40 Large T-Antigen Helicase. Nucl. Acids Res. 1996;24:330–335. doi: 10.1093/nar/24.2.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spitzner JR, Chung IK, Muller MT. Determination of 5’ and 3’ DNA Triplex Interference Boundaries Reveals the Core DNA Binding Sequence for Topoisomerase II. Journal of Biological Chemistry. 1995;270:5932–5943. doi: 10.1074/jbc.270.11.5932. [DOI] [PubMed] [Google Scholar]
- 11.Cooney M, Czernuszewicz G, Postel EH, Flint SJ, Hogan ME. Site-Specific Oligonucleotide Binding Represses Transcription of the Human c-Myc Gene in Vitro. Science. 1988;241:456–459. doi: 10.1126/science.3293213. [DOI] [PubMed] [Google Scholar]
- 12.Postel EH, Flint SJ, Kessler DJ, Hogan ME. Evidence that a Triplex-Forming Oligodeoxyribonucleotide Binds to the c-Myc Promoter in HeLa Cells, Thereby Reducing c-Myc mRNA Levels. Proc. Natl. Acad. Sci. USA. 1991;88:8227–8231. doi: 10.1073/pnas.88.18.8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grigoriev M, Praseuth D, Guieysse AL, Robin P, Thuong NT, Helene C, Harel-Bellan A. Inhibition of Gene Expression by Triple Helix-Directed DNA Cross-Linking at Specific Sites. Proc. Natl. Acad. Sci. USA. 1993;90:3501–3505. doi: 10.1073/pnas.90.8.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grigoriev M, Praseuth D, Robin P, Hemar A, Saison-Behmoaras T, Dautry-Varsat A, Thuong NT, Helene C, Harel-Bellan A. A Triple Helix-Forming Oligonucleotide-Intercalator Conjugate Acts as a Transcriptional Repressor Via Inhibition of NF kB Binding to Interleukin-2 Receptor a-Regulatory Sequence. J. Biol. Chem. 1992;267:3389–3395. [PubMed] [Google Scholar]
- 15.Murray JAH. Antisense RNA and DNA. New york: Wiley-Liss (New York); 1992. [Google Scholar]
- 16.Aviño A, Grimau MG, Frieden M, Eritja R. Synthesis and Triple-Helix-Stabilization Properties of Branched Oligonucleotides Carrying 8-Aminoadenine Moieties. Helv. Chim. Acta. 2004;87:303–316. [Google Scholar]
- 17.Sollogoub M, Dominguez B, Brown T, Fox KR. Synthesis of a Novel Bis-Amino-Modified Thymidine Monomer for use in DNA Triplex Stabilisation. Chem. Commun. 2000;23:2315–2316. [Google Scholar]
- 18.Kawai K, Saito I, Sugiyama H. Stabilization of Hoogsteen Base Pairing by Introduction of NH2 Group at the C8 Position of Adenine. Tetrahedron Lett. 1998;39:5221–5224. [Google Scholar]
- 19.Gianolio DA, Segismundo JM, McLaughlin LW. Tethered Naphthalene Diimide-Based Intercalators for DNA Triplex Stabilization. Nucl. Acids Res. 2000;28:2128–2134. doi: 10.1093/nar/28.10.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun J, Carestier T, Hélène C. Oligonucleotide Directed Triple Helix Formation. Curr. Opin. Struct. Biol. 1996;6:327–333. doi: 10.1016/s0959-440x(96)80051-0. [DOI] [PubMed] [Google Scholar]
- 21.Baudoin O, Marchand C, Teulade-Fichou M, Vigneron J, Sun JS, Garestier T, Helene C, Lehn JM. Stabilization of DNA Triple Helices by Crescent-Shaped Dibenzophenanthrolines. Chemistry - A European Journal. 1998;4:1504–1508. [Google Scholar]
- 22.Robles J, McLaughlin LW. DNA Triplex Stabilization using a Tethered Minor Groove Binding Hoechst 33258 Analogue. J. Am. Chem. Soc. 1997;119:6014–6021. [Google Scholar]
- 23.Scaria PV, Shafer RH. Binding of Ethidium Bromide to a DNA Triple Helix. J. Biol. Chem. 1991;266:5417–5423. [PubMed] [Google Scholar]
- 24.Pilch DS, Martin M, Nguyen CH, Sun J, Bisagni E, Garestier T, Helene C. Self-Association and DNA-Binding Properties of Two Triple Helix-Specific Ligands: Comparison of a Benzo[e]Pyridoindole and a Benzo[g]Pyridoindole. J. Am. Chem. Soc. 1993;115:9942–9951. [Google Scholar]
- 25.Escude C, Nguyen CH, Mergny J, Sun J, Bisagni E, Garestier T, Helene C. Selective Stabilization of DNA Triple Helices by Benzopyridoindole Derivatives. J. Am. Chem. Soc. 1995;117:10212–10219. [Google Scholar]
- 26.Arya DP, Jr, RLC, Willis B, Abramovitch AI. Aminoglycoside-Nucleic Acid Interactions: Remarkable Stabilization of DNA and RNA Triple Helices by Neomycin. J. Am. Chem. Soc. 2001;123:5385–5395. doi: 10.1021/ja003052x. [DOI] [PubMed] [Google Scholar]
- 27.Arya DP, Micovic L, Charles I, Jr, RLC, Willis B, Xue L. Neomycin Binding to Watson-Hoogsteen (W-H) DNA Triplex Groove: A Model. J. Am. Chem. Soc. 2003;125:3733–3744. doi: 10.1021/ja027765m. [DOI] [PubMed] [Google Scholar]
- 28.Willis B, Arya DP. Triple Recognition of B-DNA. Bioorg. Med. Chem. Lett. 2009;19:4974–4979. doi: 10.1016/j.bmcl.2009.07.079. [DOI] [PubMed] [Google Scholar]
- 29.Willis B, Arya DP. Major Groove Recognition of DNA by Carbohydrates. Current Organic Chemistry. 2006;10:663–673. [Google Scholar]
- 30.Willis B, Arya DP. An Expanding View of Aminoglycoside-Nucleic Acid Recognition. Adv. Carbohydr. Chem. Biochem. 2006;60:251–302. doi: 10.1016/S0065-2318(06)60006-1. [DOI] [PubMed] [Google Scholar]
- 31.Arya DP, Willis B. Reaching into the Major Groove of B-DNA: Synthesis and Nucleic Acid Binding of a Neomycin–Hoechst 33258 Conjugate Doi:10.1021/ja036742k. J. Am. Chem. Soc. 2003;125:12398–12399. doi: 10.1021/ja036742k. [DOI] [PubMed] [Google Scholar]
- 32.Shaw NN, Arya DP. Recognition of the Unique Structure of DNA:RNA Hybrids. Biochimie. 2008;90:1026–1039. doi: 10.1016/j.biochi.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 33.Shaw NN, Xi H, Arya DP. Molecular Recognition of a DNA:RNA Hybrid: Sub-Nanomolar Binding by a Neomycin-Methidium Conjugate. Bioorg. Med. Chem. Lett. 2008;18:4142–4145. doi: 10.1016/j.bmcl.2008.05.090. [DOI] [PubMed] [Google Scholar]
- 34.Arya DP. Aminoglycoside-Nucleic Acid Interactions: The Case for Neomycin. Top. Curr. Chem. 2005;253:149–178. [Google Scholar]
- 35.Arya DP, Coffee RL, Jr, Charles I. Neomycin-Induced Hybrid Triplex Formation. J. Am. Chem. Soc. 2001;123:11093–11094. doi: 10.1021/ja016481j. [DOI] [PubMed] [Google Scholar]
- 36.Charles I, Xi H, Arya DP. Sequence-Specific Targeting of RNA with an Oligonucleotide-Neomycin Conjugate. Bioconjug. Chem. 2007;18:160–169. doi: 10.1021/bc060249r. [DOI] [PubMed] [Google Scholar]
- 37.Charles I, Xue L, Arya DP. Synthesis of Aminoglycoside-DNA Conjugates. Bioorg. Med. Chem. Lett. 2002;12:1259–1262. doi: 10.1016/s0960-894x(02)00157-9. [DOI] [PubMed] [Google Scholar]
- 38.Xi H, Gray D, Kumar S, Arya DP. Molecular Recognition of Single-Stranded RNA: Neomycin Binding to Poly(A) FEBS Lett. 2009;583:2269–2275. doi: 10.1016/j.febslet.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 39.Arya DP, Coffee RLJ, Xue L. From Triplex to B-Form Duplex Stabilization: Reversal of Target Selectivity by Aminoglycoside Dimers. Bioorg. Med. Chem. Lett. 2004;14:4643–4646. doi: 10.1016/j.bmcl.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 40.Arya DP, Coffee RL, Jr, Willis A, I II. Triple Helix Stabilization by Aminoglycoside Antibiotics. Abstr.Pap.- Am.Chem.Soc. 2001;221st doi: 10.1016/s0960-894x(00)00372-3. CARB-088. [DOI] [PubMed] [Google Scholar]
- 41.Arya DP, Coffee RL., Jr DNA Triple Helix Stabilization by Aminoglycoside Antibiotics. Bioorganic and Medicinal Chemistry Letters. 2000;10:1897–1899. doi: 10.1016/s0960-894x(00)00372-3. [DOI] [PubMed] [Google Scholar]
- 42.Arya DP, Shaw N, Xi H. Novel Targets for Aminoglycosides. In: Arya DP, editor. Aminoglycoside Antibiotics: From chemical biology to drug discovery. New York: Wiley; 2007. pp. 289–314. 1 plate. [Google Scholar]
- 43.Xi H, Arya DP. Recognition of Triple Helical Nucleic Acids by Aminoglycosides. Curr Med Chem Anticancer Agents. 2005;5:327–338. doi: 10.2174/1568011054222328. [DOI] [PubMed] [Google Scholar]
- 44.Arya DP, Xue L, Tennant P. Combining the Best in Triplex Recognition: Synthesis and Nucleic Acid Binding of a BQQ-Neomycin Conjugate. J. Am. Chem. Soc. 2003;125:8070–8071. doi: 10.1021/ja034241t. [DOI] [PubMed] [Google Scholar]
- 45.Xue L, Charles I, Arya DP. Pyrene-Neomycin Conjugate: Dual Recognition of a DNA Triple Helix. Chem. Commun. 2002;1:70–71. doi: 10.1039/b108171c. [DOI] [PubMed] [Google Scholar]
- 46.Doyle ML, Brigham-Burke M, Blackburn MN, Brooks IS, Smith TM, Newman R, Reff M, Stafford WF, Sweet RW, Truneh A, Hensley P, O'Shannessy DJ. Measurement of Protein Interaction Bioenergetics: Application to Structural Variants of Anti-sCD4 Antibody. Methods Enzymol. 2000;323:207–230. doi: 10.1016/s0076-6879(00)23368-5. [DOI] [PubMed] [Google Scholar]
- 47.Ren J, Chaires JB. Rapid Screening of Structurally Selective Ligand Binding to Nucleic Acids. Methods Enzymol. 2001;340:99–108. doi: 10.1016/s0076-6879(01)40419-8. [DOI] [PubMed] [Google Scholar]
- 48.Kirk SR, Luedtke NW, Tor Y. Neomycin-Acridine Conjugate: A Potent Inhibitor of Rev-RRE Binding. J. Am. Chem. Soc. 2000;122:980–981. [Google Scholar]
- 49.Arya DP, Xue L, Willis B. Aminoglycoside (Neomycin) Preference is for A-Form Nucleic Acids, Not just RNA: Results from a Competition Dialysis Study. J. Am. Chem. Soc. 2003;125:10148–10149. doi: 10.1021/ja035117c. [DOI] [PubMed] [Google Scholar]
- 50.Wang H, Tor Y. Electrostatic Interactions in RNA Aminoglycosides Binding. J. Am. Chem. Soc. 1997;119:8734–8735. [Google Scholar]
- 51.Charles I, Arya DP. Synthesis of Neomycin-DNA/peptide Nucleic Acid Conjugates. J. Carbohydr. Chem. 2005;24:145–160. [Google Scholar]
- 52.Boger DL, Fink BE, Brunette SR, Tse WC, Hedrick MP. A Simple, High- Resolution Method for Establishing DNA Binding Affinity and Sequence Selectivity. J. Am. Chem. Soc. 2001;123:5878–5891. doi: 10.1021/ja010041a. [DOI] [PubMed] [Google Scholar]
- 53.Boger DL, Tse WC. Thiazole Orange as the Fluorescent Intercalator in a High Resolution Fid Assay for Determining DNA Binding Affinity and Sequence Selectivity of Small Molecules. Bioorg. Med. Chem. 2001;9:2511–2518. doi: 10.1016/s0968-0896(01)00243-7. [DOI] [PubMed] [Google Scholar]
- 54.Escude C, Nguyen CH, Kukreti S, Janin Y, Sun J, Bisagni E, Garestier T, Helene C. Rational Design of a Triple Helix-Specific Intercalating Ligand. Proc. Natl. Acad. Sci. U. S. A. 1998;95:3591–3596. doi: 10.1073/pnas.95.7.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ren J, Chaires JB. Sequence and Structural Selectivity of Nucleic Acid Binding Ligands. Biochemistry. 1999;38:16067–16075. doi: 10.1021/bi992070s. [DOI] [PubMed] [Google Scholar]
- 56.McGhee JD. Theoretical Calculations of the Helix-Coil Transition of DNA in the Presence of Large, Cooperatively Binding Ligands. Biopolymers. 1976;15:1345–1375. doi: 10.1002/bip.1976.360150710. [DOI] [PubMed] [Google Scholar]
- 57.Barbieri CM, Pilch DS. Complete Thermodynamic Characterization of the Multiple Protonation Equilibria of the Aminoglycoside Antibiotic Paromomycin: A Calorimetric and Natural Abundance 15N NMR Study. Biophys. J. 2006;90:1338–1349. doi: 10.1529/biophysj.105.075028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.