Abstract
iNOS localizes to both the cytosol and peroxisomes in hepatocytes in vitro and in vivo. The structural determinants for iNOS localization are not known. One plausible mechanism for iNOS localization to the peroxisome is through the interaction with peroxisomal import proteins PEX5 or PEX7. siRNA knockdown of PEX7 reduced iNOS colocalization with the peroxisomal protein PMP70. Proteomic studies using MALDI-MS identified iNOS association with the 50-kD ezrin binding PDZ protein (EBP50). Confocal microscopy studies and immunoelectron microscopy confirmed iNOS association with EBP50, with greatest colocalization occurring at 8 hours of cytokine exposure. EBP50 associated with peroxisomes in a PEX5 and PEX7-dependent manner. iNOS localization to peroxisomes was contingent on EBP50 expression in LPS-treated mice. Thus, iNOS targeting to peroxisomes in hepatocytes involves interaction with PEX7 and EBP50. The targeting of iNOS protein to the peroxisome may shift the balance of metabolic processes that rely on heme proteins susceptible to modification by radical oxygen and nitrogen radicals.
Keywords: Inflammation, Sepsis, Inducible Nitric Oxide Synthase, Peroxisome, Liver, Subcellular Localization
1.2 Introduction
Three nitric oxide synthases (NOS) have been identified in mammalian cells [1]. The neuronal (Type I) and endothelial (Type III) NOS are constitutively expressed and produce low levels of Nitric Oxide (NO) controlled by calcium transients as well as by other post-translational modifications and molecular interactions. Inducible NOS (Type II or iNOS) is typically absent in unstimulated cells but can be rapidly expressed following exposure to inflammatory stimuli or cell stress such as hypoxia. Once expressed, iNOS can produce sustained and high levels of NO [1]. Distinct subcellular locations have been described for all three NOS enzymes. Targeting NOS enzymes within the cell is a mechanism not only for localizing NO production to a specific site but also for regulating enzyme activity. NOS subcellular localization is thought to be isoform specific and also may be cell-type specific. For example, ~15% of iNOS is targeted to the peroxisome, an organelle particularly abundant in liver cells that function to metabolize long chain fatty acids by β-oxidation. This localization of iNOS to the hepatic peroxisome occurs at higher levels at the earlier time points, as early as 8 hours after inflammatory cytokine stimulation, yet the majority of the iNOS protein is predominantly cytosolic [2–4].
iNOS is readily upregulated in hepatocytes [5], which were the first human cell type in which iNOS was described and cloned [6; 7]. In vivo studies have shown that the consequences of iNOS upregulation in liver depend upon the physiological or pathological circumstances. iNOS generated NO production can be anti-apoptotic during hepatic inflammation. However, NO can also exert pro-apoptotic effects and contribute to hepatocellular damage in ischemia/reperfusion [8]. The expression of iNOS is primarily modulated by transcription and mRNA stability [7; 9], whereas enzymatic function is controlled by substrate availability [10; 11] and dimerization [12; 13]. Because iNOS can have either damaging or protective effects on the liver, we postulated that transport of the iNOS monomer to the peroxisome is one mechanism by which sequestering the enzyme protects the cell from oxidant-induced injury. Here, we sought to define the mechanism of iNOS transport to the peroxisome and focused on protein binding partners for iNOS in hepatocytes. In other cell types iNOS interacts with the GTPases RAC1 and RAC2 [14], NOS-associated protein 110 [15], and the 50-kD ezrin radixin moesin binding protein (EBP50) [16]. EBP50 is important for localizing iNOS in epithelial cells [17]. EBP50, known also as Na+/H+ exchanger regulatory factor-1 (NHERF1), is a modular adaptor protein with two PDZ domains (PDZ1 and PDZ2) and an ERM binding domain at its C-terminus that binds all members of the ezrin-radixin-moesin-merlin family of cytoskeletal proteins [18; 19]. PDZ domain-containing proteins play a role as scaffold proteins for transporters and ion channels, thus controlling their localization, surface stability and function by assembling modulator proteins into multi-molecular complexes [20]. EBP50 binding to the cytoskeleton allows sorting and localization of membrane proteins as well as clustering of proteins to specific cellular domains to facilitate signaling [21].
Peroxisomes, present in all cell types, are distinct in that they are single-membrane organelles that do not require protein unfolding or multimeric complex disruption prior to protein import [22]. In a normal liver, peroxisomes comprise ~ 1% of total cell volume. Peroxisomes, like mitochondria, consume molecular oxygen and metabolize fatty acids, cleaving two carbon atoms per cycle to generate acetyl CoA in a process called β-oxidation. Unlike mitochondria, however, peroxisomal β-oxidation is not coupled to ATP synthesis/energy production. Peroxisomes are rich in antioxidants such as catalase [23]. In the absence of adequate substrate or in its monomeric form, iNOS can generate O2−. Therefore, sequestering monomeric iNOS in peroxisomes could be protective. The peroxisomal targeting sequences PTS1 and PTS2 are consensus sequences that are involved in peroxisomal import and are recognized by the peroxisomal import receptors PEX5 and PEX7 [24]. PTS1 consists of the carboxy-terminal tripeptide (S/A/C)-K/R/H)-L; PTS2 contains an amino-terminal nona-peptide with the consensus sequence (R/K)-(L/V/I)-X5-(H/Q)-(L/A) [24]. There are peroxisomal targeted enzymes in which the peroxisomal targeting sequence of a protein is not 100% identical to the canonical PTS sites. In fact, very large protein oligomers lacking PTS motifs have been shown to piggy-back onto other conventional peroxisomal proteins and gain entry into the peroxisomal matrix in their native three-dimensional configuration [25].
We hypothesized that peroxisomal targeting receptors contribute to iNOS localization in peroxisomes. siRNA approaches revealed a clear role for PEX7. Unexpectedly, we also found that iNOS interacts with EBP50 and that EBP50 localizes to peroxisomes and is required for iNOS localization to peroxisomes. These data identify a novel role for EBP50 in iNOS protein transport into peroxisomes.
1.3 Materials and Methods
Animal Models and Hepatocyte Culture
The Institutional Animal Care and Use Committee of the University of Pittsburgh approved procedures involving animals. Endotoxemia was induced in 6 week old EBP50 wild-type and knockout animals with 5mg/kg lipopolysaccharide (E. coli O111:B4). Livers were perfused in vivo with phosphate-buffered saline (PBS) and a portion of the organ was snap frozen. For imaging purposes, the same animal was fixed with 2% paraformaldehyde in PBS upon removal of a clamp allowing perfusion of remaining tissue. Livers were cryoprotected with 30% sucrose overnight before freezing in liquid nitrogen–cooled isopentane and stored at −80°C until use.
Primary hepatocytes were isolated by collagenase perfusion [12]. After overnight culture, the medium was removed and fresh medium added. Cells were treated with cytokine mixture (CM = 100 U/ml IFN-g, 200 U/ml IL-1, 500 U/ml TNF-a) for 8 hours (CM8). siPEX5 and siPEX7 suppression was accomplished by transiently transfected a combination of siRNA duplexes manufactured by Dharmacon using Lipofectamine™ 2000 (Invitrogen), as per manufacturers protocol (Lafayette, CO). Cultured hepatocytes were treated for 48 hours to suppress endogenous PEX5 and PEX7 protein expression followed by established CM time course. The concentration of siRNA utilized in these experiments was selected based on a dose curve (Figure 1). The efficiency of siPEX suppression was confirmed by measuring membrane thiolase levels, for siPEX7 [26] and catalase levels, a PEX5 dependent peroxisomal imported protein, for PEX5 suppression [27]. Accumulated NO2− was measured by adding equal volume of Griess reagent (1% sulfanilamide/0.1% naphthyethylene diamine dihydrochloride/2% H3PO4) and measuring absorbance at 550 nm along with NaNO2 standards [28].
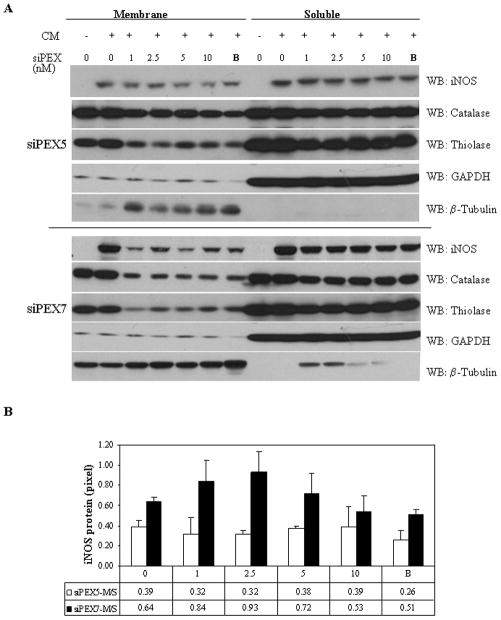
Figure 1. siRNA suppression of PEX5 and PEX7 was effective in the redistribution of PEX dependent Catalase and Thiolase.
Primary rat hepatocytes were treated for 48 hours with both siPEX 5 and siPEX7 over a range of concentrations from 0 to 10nM for 48 hrs followed by 8 hrs of cytokine stimulation to induce iNOS. Cytokine stimulation involves a cytokine mixture (CM) of IL-1, RGI, and TNFα. A. By itself siPEX7 (10 nM) diminished membrane iNOS content. The combination of the two endogenous PEX receptors (B=siPEX5/siPEX7 at concentrations of 5 nM/10 nM, respectively) was found to also further diminish iNOS levels in the membrane fraction (100,000 × g pellet, peroxisomes) when compared to the soluble fraction (100,000 × g supernatant, cytosol). B. Quantitation of iNOS content in membrane and soluble was measured using the analytical software MetaMorph (Molecular Devices, Sunnyvale, CA), with values displayed as a ratio of membrane (pixels)/soluble (pixels) protein content. Results are the mean of 4 separate experiments and are presented −/+ SEM.
Cell lysis and subcellular fractionation
Cell monolayers were washed in cold PBS, and then scraped in lysis buffer (LB:250 mM sucrose, 50 mM Tris-HCl, pH 7.4, 1 mM EDTA, 5 mg/ml pepstatin A, 1 mg/ml chymostatin, 5 mg/ml aprotinin, and 100 mM PMSF). The whole cell lysates were disrupted by 3 freeze thaw cycles, followed by centrifugation at 12,000 ×g for 15 minutes at 4°C to obtain a total cleared fraction (CL). The CL fraction was further centrifuged at 100,000 ×g for 30 minutes at 4°C to obtain the soluble (S) and membrane (M) fractions. The samples were processed for protein determination and immunoblot.
Immunoprecipitation and Immunoblot
Soluble and membrane fractions were incubated in RIPA buffer overnight with mouse anti-iNOS antibody. The immune complexes were then precipitated with protein A/G-agarose beads for 1.5 h and washed extensively with RIPA buffer. Immunoprecipitated proteins (IP) were eluted with 3×SDS loading buffer. Proteins were separated on SDS-PAGE in denaturing conditions and transferred to nitrocellulose membranes. The membranes were hybridized with antibodies for rabbit anti-iNOS (1:1000, BD Transduction), rabbit anti PMP70 (1:2000, Alexis), rabbit anti catalase (1:10,000, Rockland), rabbit anti thiolase (1:2000, Dr. Richard Rachubinski, University of Alberta) and rabbit anti EBP50 (1:1000, Abcam) in Tris Buffered Saline plus Tween-20 (0.01%, TBST) and 1% nonfat milk for 1 hour. The immunoblots were developed with enhanced chemiluminescent (ECL) solution (Pierce) and exposed to x-ray film (Kodak).
2D Gel Electrophoresis and MALDI-MS
IP reactions of the Soluble, Membrane and IgG samples that were prepared as described above were separated on a 2-D gel (Kendrick Laboratory) [29]. In brief, the sample(s) were denatured in SDS buffer and then applied to thin tube gel of 2% pH 3.5–10 Ampholines, with isoelectric focusing was carried out overnight. The tube was sealed after equilibration in SDS buffer. The tube gel was sealed to a stacking gel on top of a slab gel and run at 700 V for 13.5 hr for resolving the sample into the second, molecular weight dimension. The gels were run in duplicate. The first gel was stained for Coomassie and the second gel was probed for iNOS and compared to corresponding Coomassie stained gel. Spot B is the only iNOS positive spot unique to the membrane IP 2-D gel. This sample was excised, trypsin digested, analyzed by MALDI-MS (M. Gawinowicz, Columbia University) and identified as actin. Spots A and C are were found in both 2-D gels and C has been identified as EBP50. The IgG 2-D gel was also probed for iNOS and positive spots were excluded from further characterization.
Microscopy
Primary rat hepatocytes were seeded on collagen-crosslinked coverslips and treated with as indicated in Figure 3. Slides were processed as described [4] either for scanning confocal fluorescence microscopy and viewed on a Fluoview 1000 (Olympus).
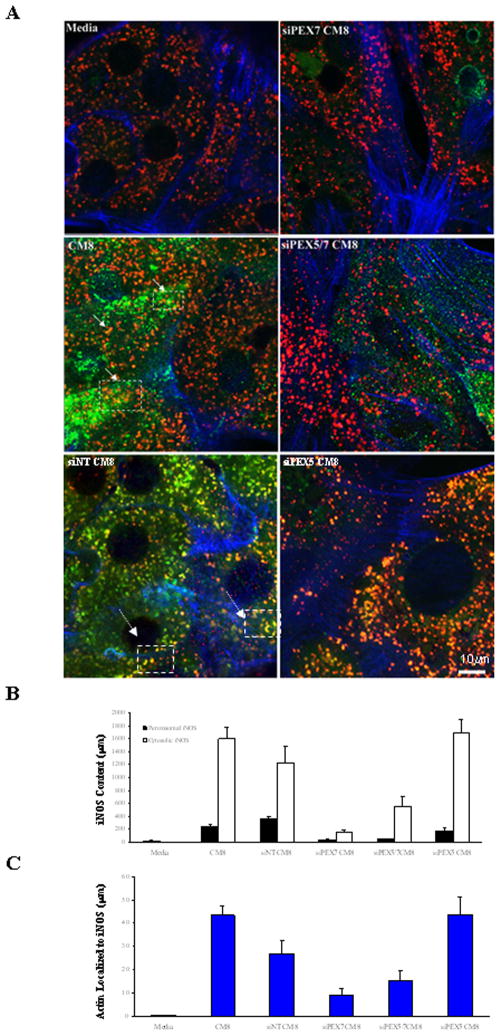
Figure 3. Suppression of PEX5 minimized iNOS co-localization with PMP70 and increases iNOS localized to the cell membrane.
Primary rat hepatocytes were treated for 48 hours with siPEX5 (10 nM), siNT (10 nM), siPEX7 (5 nM) and siPEX5/PEX7 (10 nM/5 nM) for 48 hrs and then 8 hrs of cytokine treatment to induce iNOS. A. Confocal Immunofluorescent detection of iNOS in hepatocyte culture shows a decrease in iNOS localized to the peroxisome (yellow punctate) in the siPEX5 and siPEX7 samples compared to the siNT. siPEX7 treated cells localize peroxisomal iNOS along the actin tubules. Antibodies against the PMP70, and iNOS proteins are assigned the following colors: Green, iNOS; red, PMP70; Blue, actin; Magenta, co-localized PMP70 and Actin; yellow, co-localized iNOS and PMP70. Experiment is representative of cells isolated from 3 separate rat harvests are presented as the mean −/+ S.E. B. Quantitation of peroxisomal iNOS (black bar) and cytosolic iNOS (white bar) was measured using the analytical software MetaMorph (Molecular Devices, Sunnyvale, CA and normalized to actin within the sample field (METHOD). Values are displayed in units of microns. C. Quantitation of the amount of actin which colocalized with iNOS was measured using the anaytical software MetaMorph within the sample field (Method)
Quantitation of Confocal Immunofluorescent
All confocal immunofluorescent images were uniformly gated for inclusive threshold and iNOS localization to either PMP70 (peroxisomal matrix marker) or actin was analyzed using MetaMorph software (Molecular Devices, Downingtown, PA). Object sizes in all images were calibrated with the “Calibrate Distances” application from the process menu. A scale bar was imaged using the same acquisition configurations as the images of interest. Utilizing the scale bar image, the ratio of pixel size to microns was established. The total area of iNOS was determined by applying a threshold excluding local background fluorescence. The total area of the fluorescent signal specific to iNOS was then measured by using the Show Region Statistics application from the measure menu. The total number of peroxisomes was determined by applying a threshold excluding local background fluorescence and utilizing the Count Cells application from the Apps menu. A standard area was used to determine the standard area of a single peroxisome and ultimately the number of peroxisomes within the sample field. The total area of iNOS localized to a peroxisome was determined by creating a binary mask of all peroxisomes, again excluding local background fluorescence. By using the Arithmetic application in the process menu, specifically the “Logical AND operation” was performed with the iNOS specific channel and the binary mask of peroxisomes yielding an image of iNOS staining only where it overlapped a peroxisome. The same threshold from the total iNOS area measurement was then applied and the Show Region Statistics application was again used to determine the amount of iNOS present. The peroxisomal and cytosolic iNOS were normalized within the sample field in relation to the amount of actin. The total area of actin in the image was measured by applying a threshold, again excluding local background. The Show Region Statistics application was again used to determine the amount of actin present in the image. In order to determine the amount of actin which colocalized to iNOS the “Measure Colocalization” application from the apps menu was used. The same thresholds from previous quantification techniques in this analysis, respectively for iNOS and actin, were used to exclude local background.
For transmission electron microscopy (TEM), liver were fixed with 2.5% glutaraldehyde for 1 hr, followed by 3x PBS wash steps. The liver tissue was sectioned into 2 cm cubes and then postfixed with 1% OsO4 and 1% K3Fe(CN)6 for 1 hr. Samples were washed 3x with PBS and than dehydrated in a graded series of 30% to 100% ethanol before incubation in 3x 1 hr incubations of Polybed 812 expoxy resin. Samples were cured at 37°C for 24hrs followed by 48 hrs at 65°C. Ultrathin sections of 60 nm were collected on 200 mesh grids that were than stained with 2% uranyl acetate in 50% methanol of 10 minutes followed by 1% lead citrate for 7 minutes. Immunoelectron microscopy and observed on a JEOL JEM 1210 electron microscope at 80 kV. Peroxisomes are identifiable by their urate oxidase crystalline core. Samples were stained in two pairings, the first is EBP50 (10 nm gold particle, black arrow) and catalase (5 nm gold particle, white arrow). The second pairing is iNOS (5 nm gold particle, black arrowhead) and EBP50 (10 nm gold, arrowhead) verifying the peroxisomal localization.
Statistical Analysis
Data are presented as means ± S.E. of at least three separate experiments. Experimental results were analyzed for their significance using analysis of variance in Excel.
Ethics
All procedures were approved and performed according to the guidelines of the Council on Animal Care at the University of Pittsburgh and the National Research Council’s Guide for the Care and Use of Laboratory Animals.
1.4 Results
We showed that iNOS localizes to peroxisomes in hepatocytes in vitro and in vivo [2; 30] and is monomeric in this location [3]. To determine if PEX5 and PEX7 receptors target iNOS to the peroxisome we utilized siRNA technology. The efficiency of siPEX7 suppression was confirmed by measuring membrane thiolase levels, for siPEX7 [26]. Catalase subcellular levels was measured as an indicator of siPEX5 suppression efficiency [27]. These two peroxisomal import proteins thereby confirmed protein knockdown as well as functional inhibition (Figure 1). Subcellular fractionation studies showed that the treatment with siRNA to PEX reduced the relative levels of iNOS in the membrane fraction compared to iNOS levels in the soluble fraction. A time course of siRNA suppression at the optimal siRNA concentration was performed to assess the impact of siRNA treatment on iNOS protein levels and NO production (Figure 2). Nitrite levels were reduced in cells treated with siRNA to PEX7 alone or in combination siPEX5 (PEX5/7). Neither non-targeting (NT) siRNA nor siRNA to PEX5 altered protein expression or nitrite levels. Measurement of iNOS protein levels by western blot revealed a decrease with siPEX7 alone or in combination with siPEX5 (Figure 2).
Figure 2. Effect on iNOS protein and Nitrite levels within a time course of suppression of endogenous PEX receptor levels in primary rat hepatocytes.
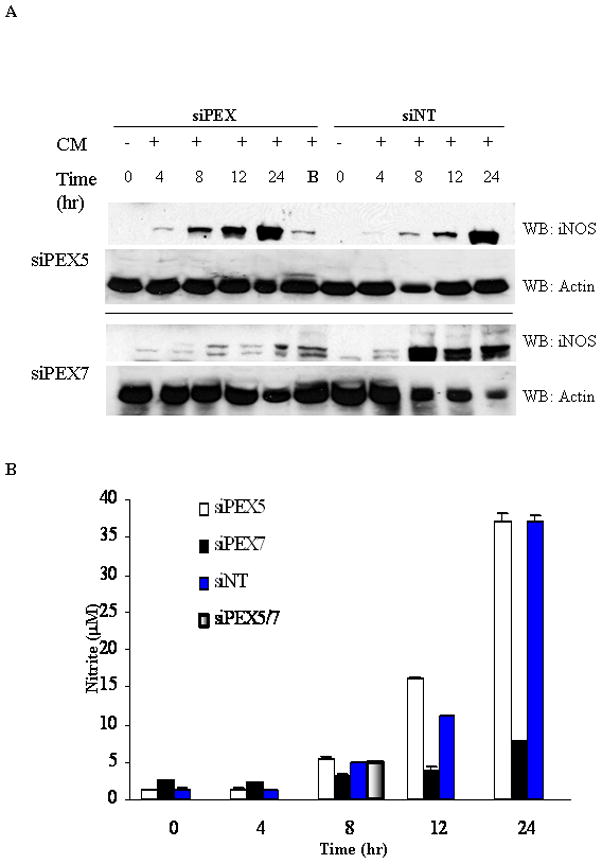
Primary hepatocytes were treated for 48 hrs to diminish endogenous PEX5 and PEX7 followed by 8 hrs of cytokine stimulation to induce iNOS. iNOS content in the cleared lysate (12,000xg) changes over time, as measured by immunoblot, were assessed in comparison to a parallel siNon-Targeting (siNT) time course at levels used for either siPEX5 (5 nM) or siPEX7 (10 nM). Results are representative of 3 separate experiments. Cytokine stimulation involves a cytokine mixture (CM) of IL-1, RGI, and TNFα. The use of B denotes use of both siPEX5 (5nM) in combination with siPEX7 (10nM). Western blot images are representative of 3 separate experiments and the data are presented −/+ SEM. B. NO production in hepatocyte cultures. Cytokine mixture (CM) induced NO production, as measured by accumulation of NO2− in medium over time.
Immunofluorescent confocal microscopy was performed to assess the distribution of iNOS in cytokine-treated hepatocytes following siRNA treatment. The 8-hour time point was chosen to assess distribution at peak iNOS expression in the membrane fraction. As shown in Figure 3, pre-treatment with siRNA to PEX7 alone or with siRNA to PEX5 resulted in less iNOS (green) within peroxisomes (red) and an increase in cytosolic iNOS. In contrast, iNOS strongly co-localized (punctate yellow staining) with the peroxisomal membrane protein PMP70 in mock or non-targeting siRNA treatments with minimal iNOS being detected outside the peroxisomes. Treatment with siPEX7 also led to an increase in peroxisomal iNOS along actin microfilaments. Thus, PEX7 suppression had the greatest impact on iNOS expression and peroxisomal localization. PEX7 has an internal type 2 Peroxisomal Targeting Sequence (PTS2, see Table 1). Of note, iNOS has a PTS1 at 1151TRL and a PTS2 at 203RIQWSNLQV
Table 1.
Putative Peroxisomal Targeting Sequences
| PTS Type | Species | Protein | Peptide sequence* | Accession Number |
|---|---|---|---|---|
| PTS1 | Mouse/rat | iNOS | 1142TRL | P29477 |
| PTS1 | Human | iNOS | 1151SAL | P35228 |
| PTS2 | Human, mouse/rat | iNOS | 203RIQWSNLQV | P35288528 P29477 AAB26307 |
| PTS2 | Rat | EBP50 | 241VTPSQEHL | Q9JJ119 |
Targeting amino acids are underlines. Peptide sequences were identified using the peroxisome data base (http://www.peroxisomedb.org).
We then carried out studies to identify iNOS binding proteins in the cytosol or peroxisome that could regulate its subcellular localization. The different treatment groups were separated into soluble (S) and membrane (M) fractions and immunoprecipitated (Figure 4A) and the samples were separated on an 8% 2-D gel. A duplicate 2-D gel was transferred to nitrocellulose and probed for iNOS and compared to corresponding Coomassie stained gel (Figure 4B). Subsequent IPs (Figure 4C) with monoclonal EBP50 were positive for EBP50. iNOS was found to interact with EBP50, a PDZ adaptor protein. Both soluble and membrane fractions of immunoprecipitated iNOS were strongly positive for EBP50; IgG controls were negative for both EBP50 and iNOS (Figure 4A).
Figure 4. Isolation of iNOS binding partners unique to the different subcellular populations.
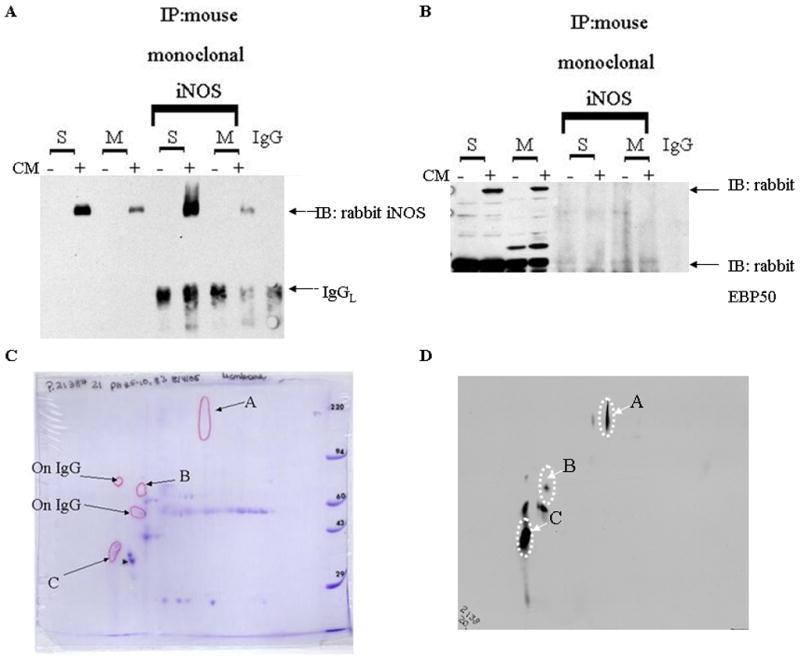
The different treatment groups were separated into subcellular fractions and immunoprecipitated (IP) (METHODS). A. Immunoprecipitation reactions with monoclonal iNOS was WB positive for EBP50. B. Immunoprecipitation reactions with monoclonal EBP50 was iNOS immunonegative. Immunoprecipitation reactions of the Soluble, Membrane and IgG samples were separated on an 8% 2-D gel (Kendrick Laboratory). C. Coomassie stained gel compared to a duplicated gel that was probed for iNOS. D. Spot B is the only iNOS positive spot unique to the membrane IP 2-D gel. This sample was excised, trypsin digested, analyzed by MALDI-MS (M. Gawinowicz, Columbia University) and identified as actin. Spots A and C are found in both 2-D gels and C has been identified as EBP50. The IgG 2-D gel was also probed for iNOS and positive spots were excluded from further characterization. (−)= media; (+)= cytokine mixture (IL-1, RGI, TNFα); S=supernatant of 100,000xg; M=pellet of 100,000xg.
Confocal microscopy and immunoelectron microscopy was performed on CM-treated hepatocytes to seek evidence that iNOS and EBP50 co-localize within cells. As shown in Figure 5A, EBP50 and the peroxisomal protein PMP70 colocalized in the cytosol and on the surface of peroxisomes in cells in which the plasma membrane was selectively permeabilized with digitonin. In cells in which both plasma and peroxisomal membranes were permeabilized, EBP50, PMP70 and iNOS are all strongly colocalized in Triton X-100 treated cells (white, Figure 4A). Immunoelectron microscopy confirmed the presence of EBP50 and iNOS in peroxisomes of CM-treated hepatocytes (Figure 5B).
Figure 5. Scanning laser confocal immunofluorescent detection of the cytosolic iNOS in primary hepatocyte cultures.

Cell monolayers stimulated with cytokines were permeabilized with digitonin (25 μg/ml) or Triton X-100 (0.1%). A. Image shown in grey scale for the purpose of illustrating cell boundaries and location of the nuclei. Hepatocyte boundaries are indicated with a white dashed line and the nuclei are outlined in red. B. Immunofluorescence was performed with antibodies against the PMP70, EBP50 and iNOS proteins. Green, iNOS; red, EBP50; Blue, PMP70; Magenta, co-localized PMP70 and EBP50; Cyan, co-localized iNOS and PMP70; White, all three proteins colocalized. C. Binding partners in the cytosolic (upper panel, permeabilized with digitonin) and total population (lower panel, permeabilized with Triton X-100) is shown in 2 protein combinations. D. Ultrastructural localization of EBP50 and iNOS in hepatocytes treated with cytokines for 8 hrs. Peroxisomes are identifiable by their urate oxidase crystalline core as well as catalase immunolabeled gold beads (white arrow). As available antibodies shared common source species, EBP50 is shown in two pairings. Catalase (white arrow, 5 nm bead) and EBP50 (black arrow, 10 nm bead). The higher magnification insets panels have immunopositive iNOS (black arrowhead, 5 nm bead) and EBP50 (black arrow, 10 nm bead) localized within a common peroxisome. Experiment is representative of a mixture of cytokine stimulated hepatocytes isolated from 2 separate rats.
The functional impact of PEX5 and PEX7 on EBP50 localization to peroxisomes was then studied using cells treated with siPEX5 and siPEX7. As before, suppression of PEX7 reduced the level of iNOS localized to the membrane fraction (Figure 6A). Notably, suppression of PEX5 or PEX7 decreased the localization of EBP50 to membranes suggesting that both PEX5 and PEX7 contribute to EBP50 trafficking to the peroxisome (Figure 6B).
Figure 6. siRNA suppression of PEX7 and to a lesser extent both PEX 5 and PEX7 was effective in the suppression of iNOS localization to the membrane fraction and EBP50 levels.
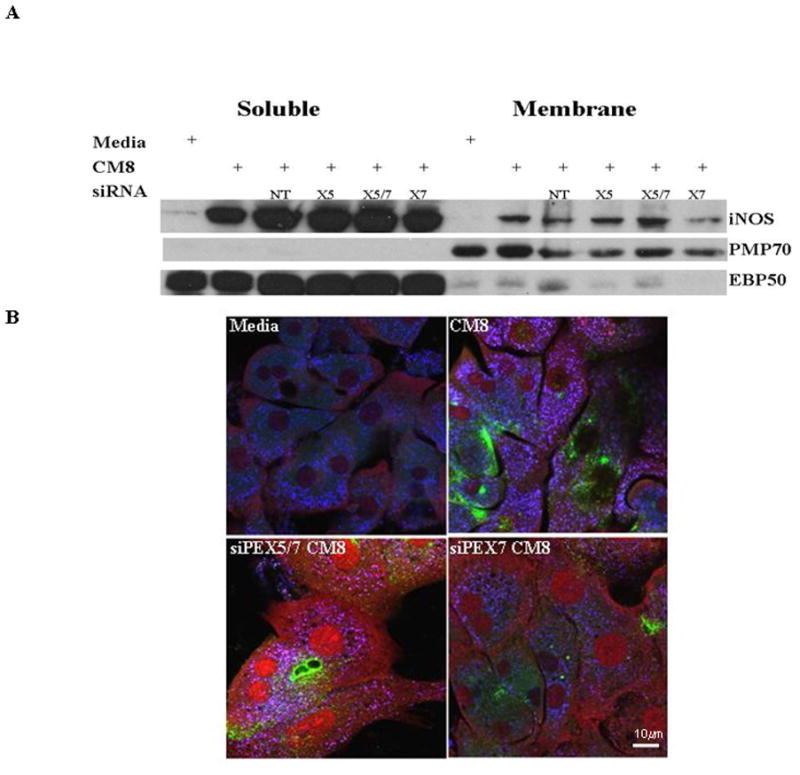
Primary hepatocytes were treated for 48 hrs to diminish endogenous PEX 5 and PEX7. After 48hrs, samples were stimulated with a cytomix to induce iNOS levels. siNon-Targeting (siNT) siRNA was included as an internal control for cellular stress by siRNA transfection. A. iNOS, PMP70 and EBP50 protein levels in the membrane and soluble fractions show variation in response to siPEX treatments. B. Confocal microscopy was performed with these same treatments. There was iNOS colocalized with PMP70 only in the CM8 treatment. siPEX treatments suppressed iNOS trafficking to the peroxisome. Antibodies against the PMP70, EBP50 and iNOS proteins are shown as follows: Green, iNOS; red, EBP50; Blue, PMP70; Magenta co-localized PMP70 and EBP50; Cyan, co-localized iNOS and PMP70; White, all three proteins colocalized. Experiment is representative of cells isolated from 3 separate rat harvests.
The importance of EBP50 in iNOS targeting to the peroxisome was then studied by comparing iNOS co-localization with PMP70 in the liver of LPS-treated wild-type and EBP50−/− mice. As shown in Figure 7B, iNOS co-localized with PMP70 (punctate yellow staining) was easily seen in wild-type livers following LPS treatment. In contrast, no co-localization of iNOS with PMP70 was seen from LPS-treated EBP50−/− mice. The induction of iNOS in the knockout livers was confirmed by the presence of green fluorescence (Figure 7B).
Figure 7. Expression of iNOS and PMP70 in liver of EBP50−/− and EBP50+/+ mice after treatment with lipopolysaccharide.
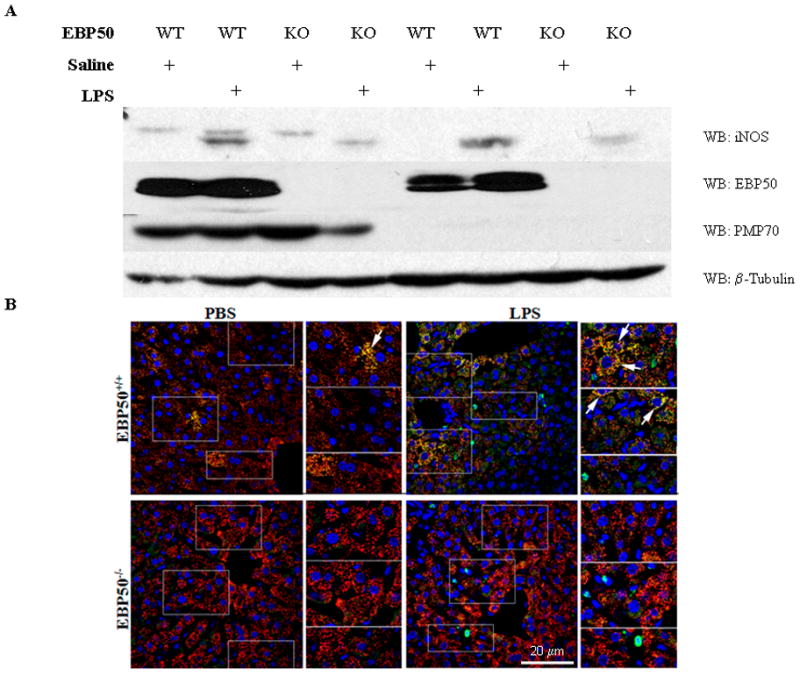
Mice were injected intraperitoneally with 5 mg/kg LPS, and livers were harvested after perfusion fixation 12 hrs after the injection. A. Subcellular fractionation of the liver tissue was separated by SDS-PAGE. Protein levels for iNOS, PMP70 and EBP50 were measured in the membrane and soluble fractions. B. Immunofluorescent levels of iNOS and PMP70 were measured in liver tissue from the same animals as in A. All images are gated uniformly thereby making iNOS localization difference across the sample set obvious. Some selected regions are boxed and magnified in the column to the right to highlight iNOS localization. White arrow heads have been added to bring attention to regions of interest. Experiment is representative N=3. Antibody color assignments are as follows: Green, iNOS; Red, PMP70; Blue, Actin; Yellow, peroxisomal iNOS.
Shown in Figure 8 is our proposed model of iNOS-PEX receptor-EBP50 targeting of iNOS to the peroxisomes soluble iNOS in the cytoplasm interacts by its PTS2 with PEX7. The iNOS-PEX7 complex might travel along EBP50, as if the EBP50 was the framework, within the cytoplasm thru the PTS1 sequence of iNOS. The purpose of iNOS localization is believed to be for disposal of monomeric or unstable iNOS that have the potential to generate damaging nitrogen or oxygen radical.
Figure 8. Schematic diagram of our proposed model of PEX7 and EBP50 targeting of iNOS to hepatic peroxisomes.
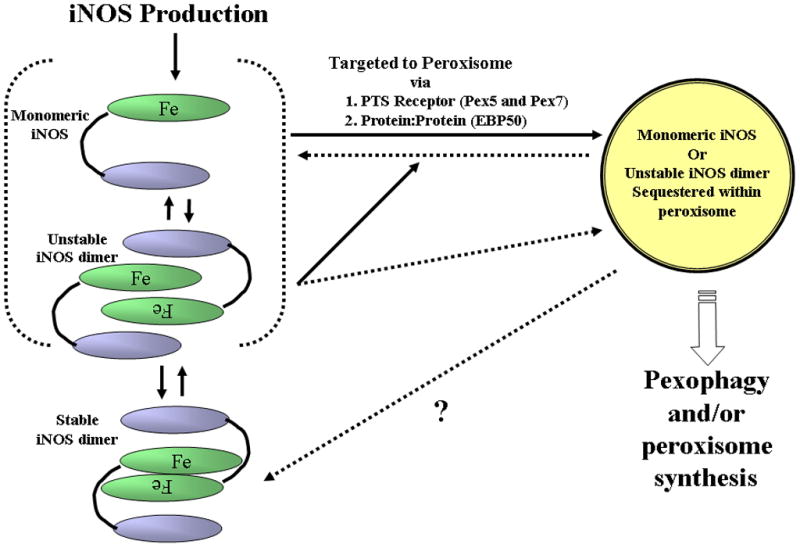
(i) Cytomix or LPS induces iNOS expression predominantly in the cytoplasm. (ii), monomeric or unstable dimeric iNOS binds via the PTS2 sequence to PEX7 (iii) the PTS1 sequence in the carboxy terminus of iNOS interacts with EBP50 that serves as a scaffold and (iv) PEX receptor associated-iNOS travel along the EBP50 into the peroxisome. The purpose of iNOS localization is believed to be for disposal of monomeric or unstable iNOS that have the potential to generate damaging nitrogen or oxygen radical.
1.5 Discussion
This purpose of this study was to identify the mechanism of iNOS targeting to the peroxisome. As previously reported, after hemorrhagic shock peroxisomal iNOS is most abundant at early time points and was independent of NO or cytokine stimulation [2; 30]. In both in vivo and in vitro studies iNOS localization to the peroxisome was heterogeneous and is of interest as we have found reduced peroxisomal catalase expression in the presence of peroxisomal iNOS [4]. Enzyme import into peroxisomes depends upon a variety of factors including cellular environment, protein affinity for peroxisomal targeting receptors (PEX5 and PEX7), and the abundance of available proteins of interest in the cytoplasm. Peroxisomal enzymes are imported posttranslationally by interaction with peroxisomal targeting receptors or protein-protein interactions [4]. The majority of peroxisomal enzymes are imported with a carboxy-terminal tripeptide that is referred to as a PTS1 sequence. Much less frequently, peroxisomal import occurs with an internal or amino terminal sequence called a PTS2. PTS2 is a conserved nonapeptide, RLxxxxx(H/Q)L) that is recognized by the receptor PEX7. Most organisms have the ability to import proteins into the peroxisome by both mechanisms [31]. Upon recognition of the PTS sequence by either PEX5, PEX7 or both, a series of accessory PEX proteins are involved in the trafficking to the peroxisome and recycling of PEX5 and PEX7 out of the peroxisome [32]. There are peroxisomal targeted enzymes in which the peroxisomal targeting sequence of a protein is not 100% identical to the canonical PTS sites. In fact, very large protein oligomers that do not have a PTS motifs have been shown to attach onto other conventional peroxisomal proteins and gain entry into the peroxisomal matrix in their native three-dimensional configuration [25].
With over 50 different enzyme activities occurring in mammalian peroxisomes [33] the cellular needs for import into the peroxisome varies under insults such as inflammation [3; 4] or hypoxia [2; 34]. These clinical models have altered mRNA and protein levels and subsequently affect proteins responsible for oxygen and fatty acid metabolism as they are not imported into the peroxisome [30]. Ghosh and Berg (2010) reported PTS1-containing proteins that had the greatest affinity for the PEX5 receptor, where also the same proteins in the cytoplasm with a lower specific content in the liver. The example these authors provided was the low acyl-CoA oxidase I concentration versus abundant catalase, both of which have a functional PTS1 targeting sequence [35]. To address available protein levels in the cytoplasm, these same investigators compared mRNA levels of peroxisomal proteins in numerous mammalian tissues to the dissociation constant for PTS1-Pex5 receptor. They also reported that it was the lower content proteins in the cytoplasm that had the greatest affinity for their peroxisomal import receptor and calculated the dissociation constant (Kd) of PTS1 containing iNOS to the PEX receptor as being greater than 2 fold higher than the Kd (nM) for catalase [35]. In our study iNOS localization was altered under siPEX7 and protein levels suppressed with siPEX5. The efficacy of these same siRNA treatments conditions effect on PEX7 and PEX5 levels were confirmed by measuring their respective translocation of the PEX7 and PEX5 dependent peroxisomal enzymes, thiolase and catalase, respectively. The PTS1:PEX5 sequence reported necessary for enzymes such as catalase [23] has also been found to be necessary for human PTS2 protein import [36]. This carboxy terminal sequence reported to be necessary for PEX5 receptor recognition has an additional functional role in iNOS as it is necessary for electron flux thru the flavin domain to the heme domain of iNOS [37; 38].
Because complete abrogation of iNOS targeting to the peroxisome was not achieved, we sought additional soluble and membrane fraction binding partners. EBP50 was identified to interact to iNOS by immunoprecipitation and proteomics analysis. This finding was confirmed using immunofluorescence and immunoelectron microscopy. The EBP50 interaction with iNOS is believed to function, as in other models, as a scaffold to facilitate protein-receptor interactions that promote protein complex trafficking [39]. Given that iNOS has both a PTS1 and PTS2 domain (TRL, RIQWSNLQV) we hypothesized a direct interaction between PEX5/7 and iNOS. However, immunoprecipitation and proteomic studies were unable to identify PEX5/7 as a binding partner in hepatocytes. Those studies did reveal that iNOS was bound to EBP50, a PDZ adaptor protein. This would enable soluble iNOS in the cytoplasm to interact via its PTS2 with PEX7. The iNOS-PEX7 complex might travel along EBP50, as if the EBP50 was the framework, within the cytoplasm thru the PTS1 sequence of iNOS. This theory is supported by work by Glynne et al (2002) reported that the mutation of the last three carboxyl terminal amino acids of iNOS (a PTS1) abrogated iNOS interaction with EBP50 and kept iNOS localized at the plasma membrane [16]. Interestingly we also found suppression of endogenous EBP50 following siPEX7 treatment (Fig 5A). Upon further sequence analysis we identified a putative PTS2 sequence within EBP50 that might be responsible for peroxisomal targeting (Table 1).
Our proteomics analysis found an interaction of the membrane iNOS with actin. This fact was not a surprise as our studies using PEX7 treatments localize peroxisomal iNOS along the actin microfilaments and cell membrane (Figure 2). This may result in this multiprotein complex “piggybacking” along cytoskeleton [40]. Work reported by Morales et al (2004) and others found EBP50 mRNA and protein levels to be preferentially expressed at the apical membrane domain ([41] references within; [42]). Furthermore, EBP50 proteins localize to plasma membranes via interactions with F-actin. Specifically, the processing of EBP50 is required for release the ERM binding domain proteins to complex with F-actin. Sites of processing for EBP50 vary by tissue, in models that have not reported interaction with iNOS ([41] references within) and those with an interaction with iNOS [16; 17]. The processing occurs to stabilize the active form that enables stable interaction to F-actin with ERM N-terminal domain interaction with membrane ligands [41]. We propose that the actin-EBP50 interaction scaffolds iNOS until PEX7 receptor shuttling translocates iNOS to the peroxisome (Figure 8). Changes in iNOS localization (cytoplasm, plasma membrane, or peroxisomal) and peroxisome-EBP50 interaction are influenced by length of cytokine exposure. EBP50 expression levels and iNOS targeting were reported to be interrelated as iNOS is located to mycobacterial phagosome [28]. The concentration, duration and cellular environment ultimately determine whether iNOS is protective. We propose that it is these protein interactions may be necessary for achieving iNOS compartmentalization that we defined in this study.
Highlights.
Suppression of PEX5 or PEX7 decreased the localization of EBP50 to membranes
iNOS targeting to peroxisomes in hepatocytes involves interaction with PEX7 and EBP50
Immunoelectron microscopy confirmed the presence of EBP50 and iNOS in peroxisomes
iNOS localization to peroxisomes was required EBP50 expression in LPS-treated mice
Acknowledgments
The authors thank Debra Williams, Danielle Reiser, Carol Meiers, Hong Liao, Mark Ross, Jason Devlin, Morgan Nelson, Callen Wallace, Doris Clay, Mara Sullivan, Jonathan Franks and Ming Sun for their expert technical advice and/or assistance.
1.7 Financial Support
Supported by NIH grant R37-GM044100 (T.R.B.) and DK069998 (P.A.F.)
Abbreviations
- NOS
Nitric Oxide Synthase
- NO
Nitric Oxide
- EBP50
ezrin radixin moesin binding protein
- NHERF1
Na+/H+ exchanger regulatory factor-1
- ERM
ezrin-radixin-moesin-merlin
- PTS
Peroxisomal Targeting Sequence
- LPS
lipopolysaccharide
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
1.9 References
- 1.Loughran PA, Bagci EZ, Zamora R, Vodovotz Y, Billiar TR. The Role of Nitric Oxide in Apoptosis and Autophagy: Biochemical and Computational Studies. In: Ignarro LJ, editor. Nitric Oxide Biology and Pathobiology. Elsevier; Burlington: 2010. pp. 513–537. [Google Scholar]
- 2.Collins JL, Vodovotz Y, Hierholzer C, Villavicencio RT, Liu S, Alber S, Gallo D, Stolz DB, Watkins SC, Godfrey A, Gooding W, Kelly E, Peitzman AB, Billiar TR. Characterization of the expression of inducible nitric oxide synthase in rat and human liver during hemorrhagic shock. Shock. 2003;19:117–122. doi: 10.1097/00024382-200302000-00005. [DOI] [PubMed] [Google Scholar]
- 3.Loughran PA, Stolz DB, Vodovotz Y, Watkins SC, Simmons RL, Billiar TR. Monomeric inducible nitric oxide synthase localizes to peroxisomes in hepatocytes. Proc Natl Acad Sci U S A. 2005;102:13837–13842. doi: 10.1073/pnas.0503926102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stolz DB, Zamora R, Vodovotz Y, Loughran PA, Billiar TR, Kim YM, Simmons RL, Watkins SC. Peroxisomal localization of inducible nitric oxide synthase in hepatocytes. Hepatology. 2002;36:81–93. doi: 10.1053/jhep.2002.33716. [DOI] [PubMed] [Google Scholar]
- 5.Curran RD, Billiar TR, Stuehr DJ, Hoffmann K, Simmons RL. Hepatocytes produce nitrogen oxides from L-arginine in response to inflammatory products of Kupffer cells. J Exp Med. 1989;170:1769–1774. doi: 10.1084/jem.170.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nussler AK, Di Silvio M, Billiar TR, Hoffman RA, Geller DA, Selby R, Madariage J, Simmons RL. Stimulation of the nitric oxide synthase pathway in human hepatocytes by cytokines and endotoxin. J Exp Med. 1992;176:261. doi: 10.1084/jem.176.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geller DA, Lowenstein CJ, Shapiro RA, Nussler AK, Di Silvio M, Wang SC, Nakayama DK, Simmons RL, Snyder SH, Billiar TR. Molecular cloning and expression of inducible nitric oxide synthase from human hepatocytes. Proc Natl Acad Sci USA. 1993;90:3491–3495. doi: 10.1073/pnas.90.8.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor BS, Alarcon LH, Billiar TR. Inducible nitric oxide synthase in the liver: regulation and function. Biochemistry (Mosc ) 1998;63:766–781. [PubMed] [Google Scholar]
- 9.Taylor BS, de Vera ME, Ganster RW, Wang Q, Shapiro RA, Morris SM, Jr, Billiar TR, Geller DA. Multiple NF-kappaB enhancer elements regulate cytokine induction of the human inducible nitric oxide synthase gene. J Biol Chem. 1998;273:15148–15156. doi: 10.1074/jbc.273.24.15148. [DOI] [PubMed] [Google Scholar]
- 10.Hibbs JB, Jr, Vavrin Z, Taintor RR. L-arginine is required for expression of the activated macrophage effector mechanism causing selective metabolic inhibition in target cells. J Immunol. 1987;138:550. [PubMed] [Google Scholar]
- 11.Hibbs JB, Jr, Taintor RR, Vavrin Z. Macrophage cytotoxicity: Role for L-arginine deiminase and imino nitrogen oxidation to nitrite. Science. 1987;235:473–476. doi: 10.1126/science.2432665. [DOI] [PubMed] [Google Scholar]
- 12.Baek KJ, Thiel BA, Lucas S, Stuehr DJ. Macrophage nitric oxide synthase subunits: Purification, characterization, and role of prosthetic groupsand substrate in regulating their association into a dimeric enzyme. J Biol Chem. 1993;268:21120–21129. [PubMed] [Google Scholar]
- 13.Tzeng E, Billiar TR, Robbins PD, Loftus M, Stuehr DJ. Expression of human inducible nitric oxide synthase in a tetrahydrobiopterin (H4B)-deficient cell line: H4B promotes assembly of enzyme subunits into an active dimer. Proc Natl Acad Sci U S A. 1995;92:11771–11775. doi: 10.1073/pnas.92.25.11771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuncewicz T, Balakrishnan P, Snuggs MB, Kone BC. Specific association of nitric oxide synthase-2 with Rac isoforms in activated murine macrophages. Am J Physiol Renal Physiol. 2001;281:F326–F336. doi: 10.1152/ajprenal.2001.281.2.F326. [DOI] [PubMed] [Google Scholar]
- 15.Ratovitski EA, Bao C, Quick RA, McMillan A, Kozlovsky C, Lowenstein CJ. An inducible nitric-oxide synthase (NOS)-associated protein inhibits NOS dimerization and activity. J Biol Chem. 1999;274:30250–30257. doi: 10.1074/jbc.274.42.30250. [DOI] [PubMed] [Google Scholar]
- 16.Glynne PA, Darling KE, Picot J, Evans TJ. Epithelial inducible nitric-oxide synthase is an apical EBP50-binding protein that directs vectorial nitric oxide output. J Biol Chem. 2002;277:33132–33138. doi: 10.1074/jbc.M205764200. [DOI] [PubMed] [Google Scholar]
- 17.Zhang W, Kuncewicz T, Yu ZY, Zou L, Xu X, Kone BC. Protein-protein interactions involving inducible nitric oxide synthase. Acta Physiol Scand. 2003;179:137–142. doi: 10.1046/j.1365-201X.2003.01119.x. [DOI] [PubMed] [Google Scholar]
- 18.Scott JD, Pawson T. Cell signaling in space and time: where proteins come together and when they’re apart. Science. 2009;326:1220–1224. doi: 10.1126/science.1175668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fehon RG, McClatchey AI, Bretscher A. Organizing the cell cortex: the role of ERM proteins 1. Nat Rev Mol Cell Biol. 2010;11:276–287. doi: 10.1038/nrm2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fanning AS, Anderson JM. PDZ domains: fundamental building blocks in the organization of protein complexes at the plasma membrane. J Clin Invest. 1999;103:767–772. doi: 10.1172/JCI6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ardura JA, Friedman PA. Regulation of G protein-coupled receptor function by Na+/H+ exchange regulatory factors. Pharmacol Rev. 2011;63:882–900. doi: 10.1124/pr.110.004176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subramani S. Components involved in peroxisome import, biogenesis, proliferation, turnover, and movement. Physiol Rev. 1998;78:171–188. doi: 10.1152/physrev.1998.78.1.171. [DOI] [PubMed] [Google Scholar]
- 23.Purdue PE, Lazarow PB. Targeting of human catalase to peroxisomes is dependent upon a novel COOH-terminal peroxisomal targeting sequence. J Cell Biol. 1996;134:849–862. doi: 10.1083/jcb.134.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heiland I, Erdmann R. Biogenesis of peroxisomes. Topogenesis of the peroxisomal membrane and matrix proteins. FEBS J. 2005;272:2362–2372. doi: 10.1111/j.1742-4658.2005.04690.x. [DOI] [PubMed] [Google Scholar]
- 25.McNew JA, Goodman JM. An oligomeric protein is imported into peroxisomes in vivo. J Cell Biol. 1994;127:1245–1257. doi: 10.1083/jcb.127.5.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Braverman N, Steel G, Obie C, Moser A, Moser H, Gould SJ, Valle D. Human PEX7 encodes the peroxisomal PTS2 receptor and is responsible for rhizomelic chondrodysplasia punctata. Nat Genet. 1997;15:369–376. doi: 10.1038/ng0497-369. [DOI] [PubMed] [Google Scholar]
- 27.Gould SJ, Keller GA, Schneider M, Howell SH, Garrard LJ, Goodman JM, Distel B, Tabak H, Subramani S. Peroxisomal protein import is conserved between yeast, plants, insects and mammals. EMBO J. 1990;9:85–90. doi: 10.1002/j.1460-2075.1990.tb08083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis AS, Vergne I, Master SS, Kyei GB, Chua J, Deretic V. Mechanism of inducible nitric oxide synthase exclusion from mycobacterial phagosomes. PLoS Pathog. 2007;3:e186. doi: 10.1371/journal.ppat.0030186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burgess-Cassler A, Johansen JJ, Santek DA, Ide JR, Kendrick NC. Computerized quantitative analysis of coomassie-blue-stained serum proteins separated by two-dimensional electrophoresis 1. Clin Chem. 1989;35:2297–2304. [PubMed] [Google Scholar]
- 30.Schrader M, Fahimi HD. Peroxisomes and oxidative stress. Biochim Biophys Acta. 2006;1763:1755–1766. doi: 10.1016/j.bbamcr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 31.Lazarow PB. The import receptor Pex7p and the PTS2 targeting sequence. Biochim Biophys Acta. 2006;1763:1599–1604. doi: 10.1016/j.bbamcr.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 32.Lanyon-Hogg T, Warriner SL, Baker A. Getting a camel through the eye of a needle: the import of folded proteins by peroxisomes. Biol Cell. 2010;102:245–263. doi: 10.1042/BC20090159. [DOI] [PubMed] [Google Scholar]
- 33.Wanders RJ, Waterham HR. Biochemistry of mammalian peroxisomes revisited. Annu Rev Biochem. 2006;75:295–332. doi: 10.1146/annurev.biochem.74.082803.133329. [DOI] [PubMed] [Google Scholar]
- 34.Khan Z, Michalopoulos GK, Stolz DB. Peroxisomal localization of hypoxia-inducible factors and hypoxia-inducible factor regulatory hydroxylases in primary rat hepatocytes exposed to hypoxia-reoxygenation. Am J Pathol. 2006;169:1251–1269. doi: 10.2353/ajpath.2006.060360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghosh D, Berg JM. A proteome-wide perspective on peroxisome targeting signal 1(PTS1)-Pex5p affinities. J Am Chem Soc. 2010;132:3973–3979. doi: 10.1021/ja9109049. [DOI] [PubMed] [Google Scholar]
- 36.Braverman N, Dodt G, Gould SJ, Valle D. An isoform of pex5p, the human PTS1 receptor, is required for the import of PTS2 proteins into peroxisomes. Hum Mol Genet. 1998;7:1195–1205. doi: 10.1093/hmg/7.8.1195. [DOI] [PubMed] [Google Scholar]
- 37.McMillan K, Bredt DS, Hirsch DJ, Snyder SH, Clark JE, Masters BS. Cloned, expressed rat cerebellar nitric oxide synthase contains stoichiometric amounts of heme, which binds carbon monoxide. Proc Natl Acad Sci U S A. 1992;89:11141–11145. doi: 10.1073/pnas.89.23.11141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seglen PO. Preparation of isolated rat liver cells. Methods Cell Biol. 1976;13:29–83. doi: 10.1016/s0091-679x(08)61797-5. [DOI] [PubMed] [Google Scholar]
- 39.Bretscher A, Chambers D, Nguyen R, Reczek D. ERM-Merlin and EBP50 protein families in plasma membrane organization and function. Annu Rev Cell Dev Biol. 2000;16:113–143. doi: 10.1146/annurev.cellbio.16.1.113. [DOI] [PubMed] [Google Scholar]
- 40.Wheeler D, Sneddon WB, Wang B, Friedman PA, Romero G. NHERF-1 and the cytoskeleton regulate the traffic and membrane dynamics of G protein-coupled receptors. J Biol Chem. 2007;282:25076–25087. doi: 10.1074/jbc.M701544200. [DOI] [PubMed] [Google Scholar]
- 41.Morales FC, Takahashi Y, Kreimann EL, Georgescu MM. Ezrin-radixin-moesin (ERM)-binding phosphoprotein 50 organizes ERM proteins at the apical membrane of polarized epithelia. Proc Natl Acad Sci U S A. 2004;101:17705–17710. doi: 10.1073/pnas.0407974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fouassier L, Duan CY, Feranchak AP, Yun CH, Sutherland E, Simon F, Fitz JG, Doctor RB. Ezrin-radixin-moesin-binding phosphoprotein 50 is expressed at the apical membrane of rat liver epithelia. Hepatology. 2001;33:166–176. doi: 10.1053/jhep.2001.21143. [DOI] [PubMed] [Google Scholar]