Abstract
Plant secondary cell walls are deposited mostly in vascular tissues such as xylem vessels, tracheids, and fibers. These cell walls are composed of a complex matrix of compounds including cellulose, hemicellulose, and lignin. Lignin functions primarily to maintain the structural and mechanical integrity of both the transport vessel and the entire plant itself. Since lignin has been identified as a major source of biomass for biofuels, regulation of secondary cell wall biosynthesis has been a topic of much recent investigation. Biosynthesis and patterning of lignin involves many developmental and environmental cues including evolutionarily conserved transcriptional regulatory modules and hormonal signals. Here, we investigate the role of the class I Knotted1-like-homeobox (KNOX) genes and gibberellic acid in the lignin biosynthetic pathway in a representative monocot and a representative eudicot. Knotted1 overexpressing mutant plants showed a reduction in lignin content in both maize and tobacco. Expression of four key lignin biosynthesis genes was analyzed and revealed that KNOX1 genes regulate at least two steps in the lignin biosynthesis pathway. The negative regulation of lignin both in a monocot and a eudicot by the maize Kn1 gene suggests that lignin biosynthesis may be preserved across large phylogenetic distances. The evolutionary implications of regulation of lignification across divergent species are discussed.
Keywords: Knotted1, KNOX, gibberellic acid, lignin, maize, tobacco, tomato
INTRODUCTION
Lignin is a complex polymer and is a fundamental component of cell walls in higher land plants (Gifford and Foster, 1988). Lignin polymers are localized to secondary cell walls found primarily in the xylem (vessel elements and tracheids), phloem fibers, sclerenchyma, and periderm (Esau, 1965). The hardened thickenings of these secondary cell walls, where lignin is covalently attached to carbohydrate components, are necessary to maintain the structural integrity of the plant body. Lignin was a key innovation in the evolution of land plants, allowing for the development of these rigid support structures and robust vessels that are required for the transport of water and minerals throughout the organism. The lignified fibers provide structural support to the vascular system, which allowed for much larger body plans and colonization of a broader range of environments.
The primary driver of carbon-fixation and metabolism on Earth is photosynthesis and a significant majority of all terrestrial photosynthetic activity can be attributed to the lignophytes. The primary chemical forms of reduced carbon that are available from the products of photosynthesis are polysaccharides, with the majority consisting of lignocellulosic cell wall materials. Lignification of cell walls allowed the vascular plants to achieve complex and large forms and dominate the landscape. Unraveling the mechanisms of regulation of lignin content is important for understanding this important structural component of plant cell walls. Understanding the regulation of cell wall development will also be the key to efficiently utilizing biological resources and also help increase utility of plant cell wall materials for biofuel applications on a fuel hungry planet.
Knotted1-like-homeobox (KNOX) genes are transcription factors that may be useful in regulating developmentally and agronomically important plant characteristics to optimize biomass qualities. While many studies have investigated individual genes in the lignin biosynthetic pathway, a gap remains in our understanding of the role of transcription factors in coordinating lignin patterning and deposition during plant development, particularly among taxa. Among the transcription factor families shown to regulate lignin in plants are class I and class II KNOX genes (Mele et al., 2003; Li et al., 2012; Testone et al., 2012), HDZIPIII (Du et al., 2011), NAC (NAM, ATAF1/2, CUC2) transcription factors (Xu et al., 2012), and R2R3 MYB genes (Sablowski et al., 1994; Patzlaff et al., 2003; Omer et al., 2013). The KNOX family of transcription factors is known to regulate and maintain indeterminacy of the cell population in the shoot apical meristem (SAM; Long et al., 1996; Vollbrecht et al., 2000) and vascular cambium (Groover et al., 2006) and plays important and diverse roles during plant development. In Arabidopsis, one particular KNOX gene, BREVIPEDICELLUS (BP)/KNAT1, is a known regulator of plant morphological development as well as lignin biosynthesis (Douglas et al., 2002; Venglat et al., 2002; Mele et al., 2003; Douglas and Riggs, 2005). The loss of function mutation of BP results in an increase in lignin biosynthesis in the inflorescence rachis, while overexpression of this gene results in a suppression of lignin deposition in the plant (Mele et al., 2003). These results suggest that BP might function to repress lignin biosynthetic enzymes thus suppressing lignin deposition (Mele et al., 2003). Therefore, the involvement of KNOX genes, such as BP, in controlling lignin biosynthesis strongly suggests that KNOX genes have the potential to be effective modulators of lignification in plant tissues.
One regulatory mechanism by which KNOXI genes can modulate development and downstream gene expression is through the regulation of small molecule hormones such as gibberellic acid (GA) and cytokinin (Rodriguez et al., 2010). Through this method of regulation, local KNOX gene expression can have cell non-autonomous effects on development. GA is a known regulator of lignin biosynthesis and morphogenesis (Luquita et al., 2009). Increasing the levels of bioactive GA leads to higher levels of lignification in plant tissues (Palatnik et al., 1992), increase in internode growth (Palatnik et al., 1992), as well as a decrease in plastochron index (Palatnik et al., 1990) resulting in increased nodal spacing. The effects on growth and lignification caused by modulating the expression of GA biosynthesis and degradation genes differ depending on the portion of the biosynthetic pathway being affected, with down-regulation of the catabolic enzyme GA2-oxidase showing stronger phenotypic consequences than up-regulation of a GA biosynthetic enzyme GA20-oxidase (Dayan et al., 2010). Direct transcriptional repression of GA20-oxidase and up-regulation of GA2-oxidase genes has been demonstrated in eudicot (Schaposnik et al., 1958; Rodriguez et al., 2010; Spinelli et al., 2011) and monocot (Palatnik et al., 2007; Mateos et al., 2010) model systems, respectively. Gibberellin is a potent target for plant breeding and crop improvement since some agronomic traits such as fiber quality (Larkan et al., 2013), tree height, and growth rate (Yu et al., 2012) can be improved by increasing endogenous GA levels. Other traits such as changes to plant architecture responsible for the green revolution result from decreases in GA levels and/or GA signaling.
In this study, we have used phylogenetically distant model species, maize (monocot) and tobacco (eudicot), to determine how class I KNOX genes are involved in the process of lignification. We also examine the effect of GA on lignification in these species and discuss the evolutionary implications of this regulatory role in the lignin biosynthetic pathway.
MATERIALS AND METHODS
PLANT MATERIAL AND GROWTH CONDITIONS
Maize (Zea mays) lines with overexpression mutations of Kn1 (Knotted1), Gn1 (Gnarley1), and Rs1 (Roughsheath1) were used in this study. Maize plants were grown under field conditions at the University of California, Davis, CA, USA. Tobacco (Nicotiana tobacum cv. Samsun) lines with overexpression constructs of LeT6 (Janssen et al., 1998a, b) and Kn1 (Sinha et al., 1993), the wild-type tomato (Solanum lycopersicum cv. Rutgers), and the mutant plant Mouse ears (Me; Parnis et al., 1997; Cardenas et al., 2012) were also used. Tobacco and tomato seeds were planted directly onto soil and grown in the greenhouse under long day (18 h day: 6 h night) conditions. All plant materials were harvested when they had achieved the onset of reproductive maturity.
HORMONE TREATMENTS
Maize (Z. mays) and tobacco (Nicotiana tobacum) seedlings were grown and treated with external sprays of GA3 (Sigma, MO, USA) and uniconazole (Uni; Sumagic, Valent, CA, USA). Treatments continued twice a week until the earliest evidence of initiation of flowering. GA3 and Uni were diluted into 250 ml water with five drops Tween-20 to aid in leaf surface adhesion. GA treatment was at 100 μm GA3 (Sigma, MO, USA). Uniconazole-P (Sumagic, Valent, CA, USA) treatment was at 50 μm. Control was water with Tween-20 and no additive.
PHYLOGENETIC ANALYSIS
All maize [Liguleless (Lg3) (NM_001112038), (Lg4a) (AF457118), (Lg4b) (AF457119); Knotted1 (Kn1) (NP_001105436); Gnarley1 (Gn1) (AAP76320); Roughsheath1 (Rs1) (NP_001105331)], Arabidopsis [Arabidopsis Knotted1-like (KNAT2) (NM_105719), (KNAT6) (NM_102187); SHOOT MERISTEMLESS (STM) (NM_104916); BREVIPEDICELLUS (BP)/KNAT1 (NM_116884)] class I KNOX genes, as well as the tomato LeT6 (AF000141) gene, were placed into an unrooted cladogram using unambiguously aligned portions of the amino acid sequences and analyzed in Paup (Phylogenetic analysis using parsimony; Sinauer Associates, Inc. Publishing, Sunderland, MA, USA) using neighbor joining with 1000 replications.
HISTOLOGY
Mature stem tissues were hand sectioned with a razor blade; cross sections were stained, and mounted in phloroglucinol–HCl, a stain specific for lignin (Ruzin, 1999). Bright field images were taken on a Zeiss Discovery V8 microscope, digital pictures taken on an AxioCam MRc, and images were saved using an AxioVision Ac camera (Carl Zeiss MicroImaging, Inc., Thornwood, NY, USA). All photographs were processed in Adobe Photoshop CS3 (Adobe Systems Inc., USA).
LIGNIN QUANTIFICATION
Lignin was quantified using a thioglycolic acid protocol as described by Hatfield and Fukushima (2005). Measurements of absorption at 280 nm were made on a NanoDrop Spectrophotometer ND-1000 (NanoDrop Technologies, DE, USA). Tissues used were leaf midrib for maize and stem for tobacco. To ensure consistency among experiments, fresh tissue was placed in 100% methanol with daily changes for 5 days. Contents were dried overnight under vacuum at 37°C to remove the methanol for dry weights. Absorption 280 measurements were made on NANODROP. Comparisons of lignin content were expressed as Absorption 280 divided by dry weight for each sample and relative amounts being proportional to wild-type.
CINNAMYL ALDEHYDE MEASUREMENTS
One to two grams of tobacco and tomato stems were frozen in liquid nitrogen and ground to a fine powder with a mortar and pestle. The powder was added to 50 ml of 100% methanol for 30 min and four changes were made to remove soluble components. Contents were dried under vacuum overnight at 37°C to remove the methanol for dry weight determination. Cinnamyl moiety content in lignin was measured as described by Ralph et al. (1998) and MacKay et al. (1999) for each of the four biological replicates per treatment. Absorption 545 nm readings were taken on a Beckman Du® 640 spectrophotometer (Beckman Coulter, Fullerton, CA, USA) using Acryl Cuvettes 67.740 (Sarstedt, Newton, NC, USA).
DATA COLLECTION AND ANALYSIS
Data for total lignin content in maize and tobacco and cinnamyl aldehyde content in tobacco were assessed using one-way analysis of variance (ANOVA) with a Dunnett comparison test to assess differences between the control (wild-type) versus the other genotypes. Data presented are means of total samples (± standard error, SE; N = 4). Data for LeT6 expression and cinnamyl alcohol dehydrogenase 1 (CAD1) in tomato was assessed using a t-test to compare wild-type to Me. All analyses were performed using SigmaPlot12® software (SPSS Science, IL, USA).
HOMOLOG IDENTIFICATION
Putative tobacco orthologs for lignin biosynthetic genes were obtained by using the NCBI tblastn utility using the Arabidopsis protein sequence query to search translated nucleotide databases: At4CL1 (At1g51680), AtPRXR9GE (At3g21770), AtCAD1 (At4g39330), AtCBR1 (AT1G15950).
QUANTITATIVE REAL-TIME PCR
RNA was isolated from stem tissue of wild-type and overexpression mutant tobacco and tomato plants. Plant tissue was ground with a mortar and pestle in liquid nitrogen and total RNA was isolated using the RNeasy plant mini kit (Qiagen, MD, USA). RNA was treated with RQ1 RNase-Free DNase (Promega, WI, USA). Quantitative real-time PCR (QRT-PCR) was conducted as outlined in Kimura et al. (2008). Beacon software (Bio-Rad Laboratories, Hercules, CA, USA) was used to design the following tobacco primer sequences: for peroxidase (NtPRXR-F 5′-GCTGTGGACCGAT-GCTTCTAC-3′, NtPRXR-R 5′-ACCATTAGTGCCAGTCTTAA-CCTC-3′) (accession AB044153); for CAD1 (NtCAD1-F 5′-ATGGTAACGGACGAGCACTACG-3′, NtCAD1-R 5′-CAAG-ACCACCAAGACCAACAACAC-3′) (accession AM851013); for 4-coumarate:CoA ligase 1 (4CL1) (NtCL1-F 5′-GCTAGACTGGC-TGCTGGTGTTC-3′, NtCL1-R 5′-CTTCAATGGTGGACTTG-TTTAGTTTGTTAC-3′) (accession NTU50845); for cinnamoyl CoA reductase (CCR) (NtCCR-F 5′-TTGGTATTGCTATGGAAA-GATGG-3′, NtCCR-R 5′- AACACTGGCATTCACATTCTG-3′) (accession DV999468). For tomato, the following primer sequences were used: for LeT6 (LeT6-F 5′-ACTACCATCGTCT-CTTGACTGCTTATCTC-3′, LeT6-R 5′-TCTCCAATGATTCCA-CCACCACTACTAC-3′) (accession ES894887); for CAD1 (LeCAD-F 5′- GGAAGCTGCACCATTGTTATG-3′, LeCAD-R 5′- AATGTTCAATCCAGGCTTATCAAG-3′). Data for each gene was normalized to the expression of GAPDPH. Primers used for GAPDH were as follows: for tobacco (NtGAPDH-F4 5′-GGTGCCAAGAAGGTTGTG-3′, NtGAPDH-R4 5′-GCAAGG-CAGTTGGTAGTG-3′) and for tomato (LeGAPDH-F 5′-GGTGCTGACTTCGTTGTTG-3′, LeGAPDH-R 5′-GCTCTGGC-TTGTATTCATTCTC-3′). Reactions were conducted using an iCycler iQ real-time thermal cycler (Bio-Rad Laboratories, Hercules, CA, USA) with iQ SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA, USA). Each experiment contained three technical replicates, utilized two biological replicates per line, and was conducted in duplicate. Data for real-time PCR was assessed using one-way ANOVA with a Tukey multiple comparison test to assess differences between the treatments for all genotypes. Data presented are means of total samples (± SE; N = 3) from Q-RT experiments. These analyses were performed using SigmaPlot12.5® software (SPSS Science, IL, USA).
RESULTS
Kn1, Gn1, AND Rs1 ARE PUTATIVE ORTHOLOGS OF BP IN MAIZE
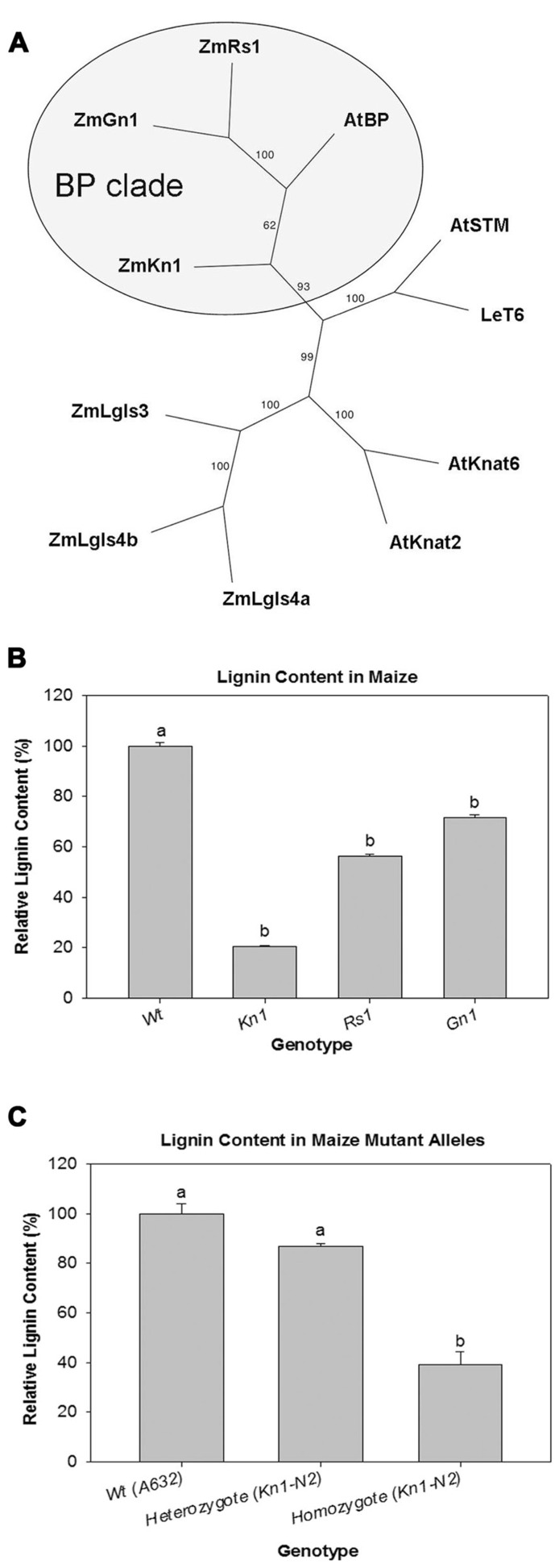
A cladogram generated with maize and Arabidopsis class I KNOX genes was used to determine which genes in maize are orthologs of BP. Amino acid sequences for class I KNOX genes from maize [Liguleless (Lg3), (Lg4a), (Lg4b); Knotted1 (Kn1); Gnarley1 (Gn1); Roughsheath1 (Rs1)], Arabidopsis [Arabidopsis Knotted1-like (KNAT2), (KNAT6); SHOOT MERISTEMLESS (STM); BREVIPEDICELLUS (BP)/KNAT1], and tomato [Lycopersicon esculentum T6 (LeT6)] were aligned. The three maize genes, Kn1, Gn1, and Rs1, formed a clade with Arabidopsis BP with 93% bootstrap support (Figure 1A, circled gray area). Based on their phylogenetic relatedness to BP, the three maize KNOX genes (Kn1, Gn1, and Rs1) were further investigated to determine their effect on lignin deposition.
FIGURE 1.
(A) Cladogram depicting the phylogenetic position of LeT6 and Knotted1 among maize and Arabidopsis KNOX1 genes. (B) Relative lignin content in wild-type (Wt) maize and overexpression mutants of Knotted1 (Kn1), Roughsheath1 (Rs1), and Gnarley1 (Gn1). (C) Dosage effect of Kn1-N2 mutant allele. Lignin is more reduced in homozygous Kn1-N2 plants than in Kn1-N2 heterozygous or Wt plants. Error bars in (B) and (C) represent standard error (± SE; N = 4). P < 0.001.
Kn1 IS A MODULATOR OF LIGNIN DEPOSITION IN MAIZE
Relative lignin content was quantified using a thioglycolate acid protocol (Hatfield and Fukushima, 2005) on wild-type and maize mutant lines overexpressing Kn1, Gn1, and Rs1. We found that all mutant lines were significantly different when compared to wild-type. However, in Kn1 mutant plants lignin levels were much lower than wild-type (Figure 1B) showing that Kn1 can strongly affect lignin deposition in the stem. To confirm that Kn1 lignin reduction was not an allele specific effect, a segregating population of Kn1-N2, another Kn1 allele, was selected for analysis. Plants homozygous for Kn1-N2 showed reduced lignin content when compared to segregating wild-types confirming that Kn1 lignin reduction is not allele specific (Figure 1C).
MAIZE Kn1 REDUCES LIGNIN CONTENT IN TOBACCO
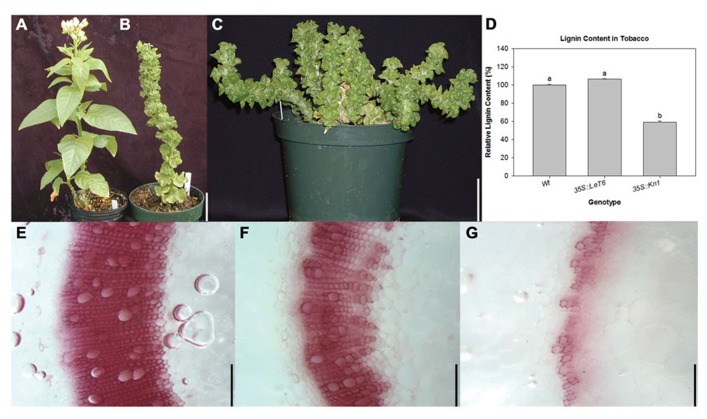
Since lignin content was significantly lower in Kn1 overexpressing maize plants (Figure 1B), transgenic tobacco lines overexpressing Kn1 and LeT6, a non-BP-type KNOX1 gene from tomato orthologous to Arabidopsis STM, were analyzed for lignin content and cellular localization to ascertain whether maize Kn1 can regulate lignin deposition in a eudicot. First, gross morphology of wild-type and the transgenic lines overexpressing Kn1 or LeT6 were observed. In wild-type tobacco, leaves are ovate and arranged in a spiral phyllotaxy (Figure 2A), while leaves of both transgenic tobacco lines are round and curled with short petioles (Figures 2B, C). Notably, only wild-type and 35S:LeT6 showed normal erect stem growth (Figures 2A, B) while 35S:Kn1 plants were less rigid (Figure 2C). To correlate whether the stem weakness observed in 35S::Kn1 plants might due to a decrease in lignin, lignin content was measured in the tobacco lines. Lignin content was not significantly altered in 35S::LeT6 but was significantly lower in 35S::Kn1 plants (Figure 2D). Phloroglucinol stained sections of the tobacco stems confirmed these results showing that maize Kn1 (Figure 2G), but not LeT6 (Figure 2F), reduces lignin content in the eudicot tobacco (Figures 2E–G).
FIGURE 2.
Comparison of (A) wild-type (Wt), (B)35S:LeT6, and (C)35S:Kn1 tobacco plants. (D) Relative lignin content in Wt and transgenic 35S::LeT6 and 35S::Kn1 tobacco plants. Error bars represent standard error (± SE; N = 4) where P < 0.001. Phloroglucinol stained cross sections of (E) Wt, (F) 35S::LeT6, and (G) 35S:Kn1 tobacco stems. Scale bars = 5 cm (A–C); 0.1 mm (E–G).
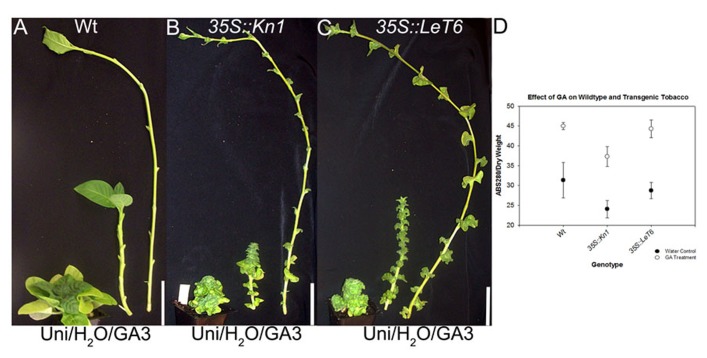
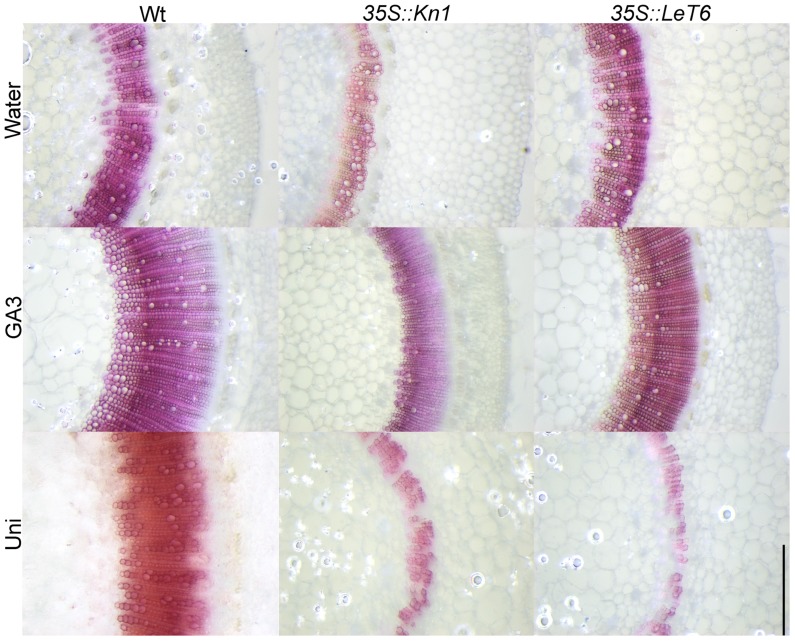
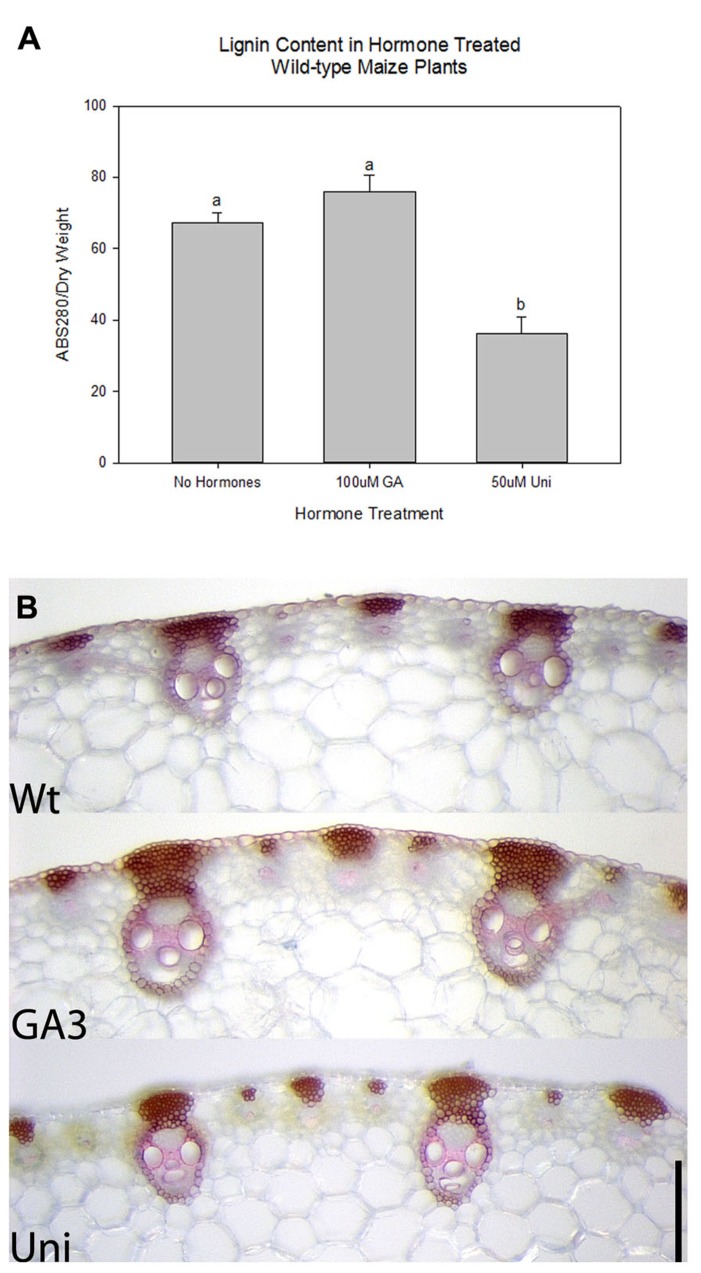
Gibberellic acid is a known regulator of lignification in flowering plants (Wang et al., 2011a, b) while KNOX1 genes are known regulators of GA biosynthesis and degradation (Mun et al., 2010; Parkin et al., 2010; Navabi et al., 2011). To ascertain whether KNOX1 proteins modulate lignin content via attenuation of GA biosynthesis or perception, we treated wild-type and transgenic tobacco with GA3 and Uni, a GA biosynthesis inhibitor. Treatment with GA3 resulted in increased internode lengths in wild-type and transgenic lines (Figures 3A–C) as well as a visible increase in lignin deposition (Figure 4). Measurements of lignin content in stems from wild-type and transgenic lines treated with GA3 or water, each showed a similar increase in lignin upon treatment with GA3 (Figure 3D). While treatment with GA3 caused similar growth stimulation (increased stem elongation) in all tobacco lines (Figures 3A–C), treatment with Uni reduced all treated plants to rosettes yielding insufficient stem material to allow for lignin quantification (Figures 3A–C). Examination of phloroglucinol–HCl stained stem sections showed additional reduction in lignification in Uni treated wild-type and transgenic lines (Figure 4). This reduction in lignification, however, may also be a result of the decreased number of xylem elements that are formed in the Uni treated plants (Figure 4).The additive effects of GA3 or Uni treatments on transgenic lines suggests that lignin regulation by KNOXI genes is not saturated and can be influenced in either direction (increased or decreased lignification) by the addition of GA3 or GA biosynthesis inhibitors (Figures 3 and 4). To establish that GA is a regulator of lignification independent of secondary growth, maize plants were treated with GA3 and uniconizole. Maize was selected because these plants do not undergo secondary growth and reductions in lignin from Kn1 overexpression is a result of reduced levels of lignin in secondary walls of xylem elements and other primary tissues in the vascular bundle. We found that GA3 treated maize tissue did not show a significant increase in lignin, however, Uni treated plants did show a significant decrease in lignin content (P < 0.050; Figures 5A, B).
FIGURE 3.
Effects of gibberellic acid (GA3) and GA biosynthesis inhibitor uniconazole (Uni; left), water (H2O) control (middle), and gibberellic acid3 (GA3; right) on (A) wild-type (Wt), (B)35S::Kn1, and (C)35S::LeT6 tobacco plants. (D) Total lignin content in water control and GA3 treated on Wt and transgenic 35S::LeT6 and 35S::Kn1 tobacco plants. Error bars represent standard error (± SE; N = 4). Scale bars = 5 cm (A–C).
FIGURE 4.
Effects of gibberellic acid3 (GA3) and uniconazole (Uni) treatment on stems of wild-type (Wt) and transgenic 35S::LeT6 and 35S::Kn1 tobacco plants. Stem cross sections were stained with phloroglucinol. Scale bar = 0.5 mm.
FIGURE 5.
Effects of gibberellic acid (GA3) and GA biosynthesis inhibitor uniconazole (Uni) in maize wild-type plants. (A) Relative lignin content in maize plants without hormone treatment and treated with GA3 and Uni. Error bars represent standard error (± SE; N = 4) where P < 0.050. (B) Phloroglucinol stained cross sections of Wt, GA3, and Uni treated maize stems. Scale bars = 0.25 mm (B).
PEROXIDASE AND CAD TRANSCRIPT LEVELS ARE ALTERED IN TOBACCO
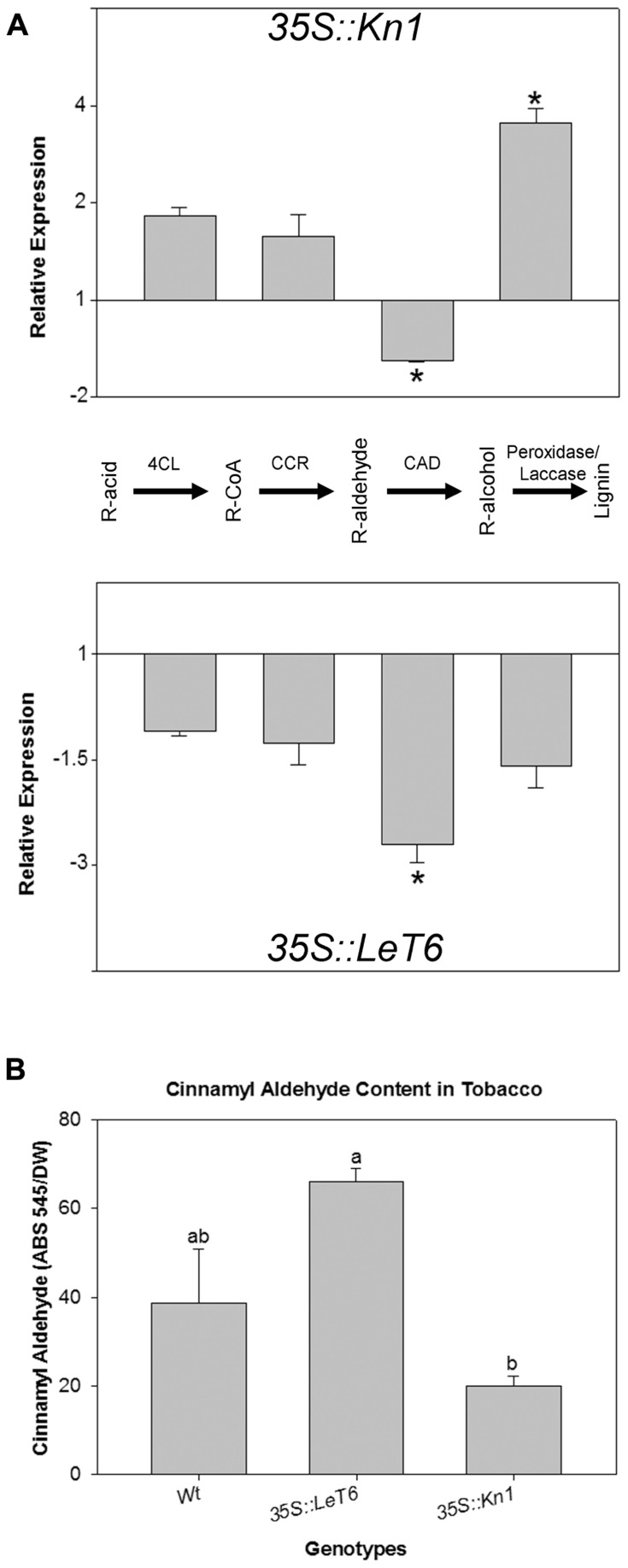
Since our results indicated that lignin content was lower in 35S::Kn1 than in 35S::LeT6 (Figure 2D), transcript levels of key lignin biosynthetic genes were evaluated for possible alterations in these overexpressing plants. Using QRT-PCR, we analyzed the relative expression levels of the four key dedicated lignin biosynthetic genes common to all lignin subunits: 4CL1, CCR, CAD, and PRX (peroxidase; Figure 6A, pathway). We found that in 35S::Kn1 only the PRX gene was significantly increased (P < 0.001) while a significant decrease (P = 0.009) in CAD was observed in both 35S::Kn1 and 35S::LeT6, with respect to wild-type transcript levels (Figure 6A, asterisks) suggesting that KNOX genes regulate at least two crucial steps (CAD and PRX) in the lignin biosynthetic pathway. Although 35S::LeT6 did not show a change in lignin quantity they suggest possible changes to lignin composition, based on these results, cinnamyl aldehyde (the monomeric precursor of lignin; Figure 6A) moiety levels were analyzed to determine if lignin composition was altered in the tobacco lines. We found that cinnamyl aldehyde moieties in 35S::LeT6 was not significantly different (P = 0.064) when compared to wild-type (Figure 6B). Cinnamyl aldehyde moieties in 35S::Kn1 stems, however, were significantly lower (P = 0.023) with respect to wild-type (Figure 6B), likely as a result of the overall reduced lignin content. These results suggest that Kn1 may suppress lignification through changes in levels of biosynthetic steps upstream of 4CL and that CAD levels are not limiting with this reduced flux through the pathway.
FIGURE 6.
(A) Relative expression of four key lignin biosynthetic genes [4CL (4-coumarate:CoA ligase 1), CCR (cinnamoyl CoA reductase), CAD (cinnamyl alcohol dehydrogenase), and PRX (peroxidase) R(lignin monomer functional group)] in 35S::Kn1 and 35S::LeT6 tobacco plants. (B) Cinnamyl aldehyde moiety levels in wild-type (Wt), 35S::LeT6, and 35S::Kn1 tobacco plants. Error bars represent standard error [± SE; N = 3 (A), N = 4 (B)], P = 0.009 (A) and P = 0.005 (B).
NATURAL LeT6 OVEREXPRESSION MUTANTS IN TOMATO SHOW REDUCED LeCAD TRANSCRIPTION
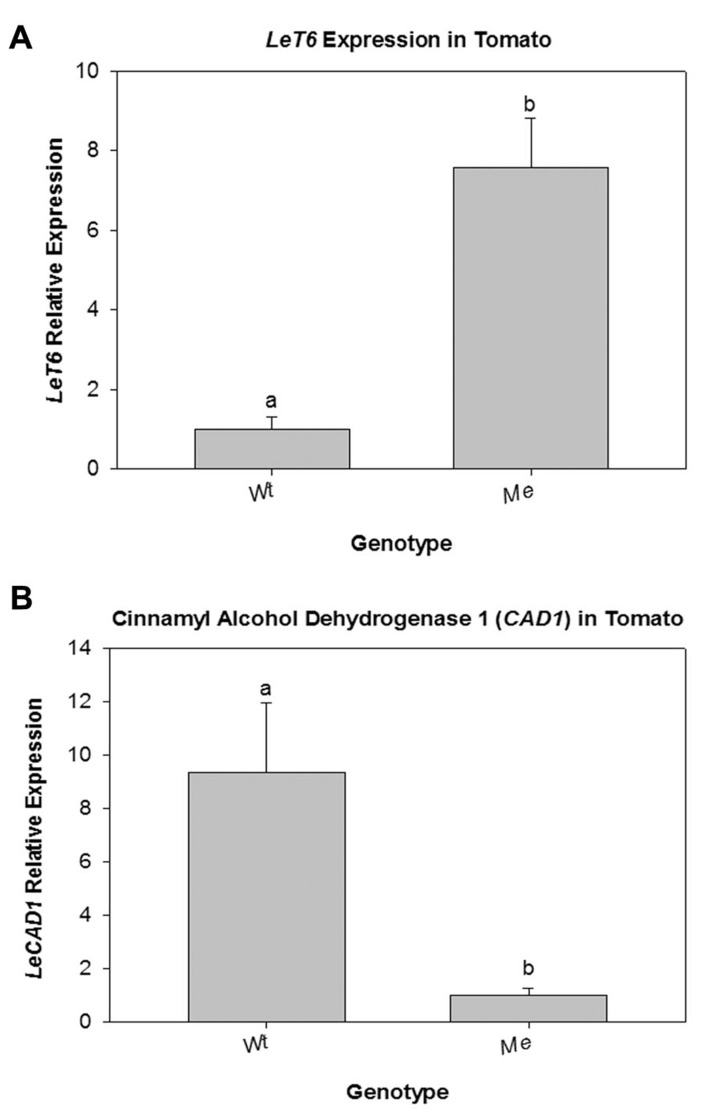
The classical tomato mutant Me is a naturally arising mutant in which the expression of the transcription factor LeT6 is under control of the promoter of the housekeeping gene pyrophosphatase (Chen et al., 1997; Parnis et al., 1997; Janssen et al., 1998a). To determine if CAD is a normal target of LeT6 regulation, stems of the tomato Me mutant were analyzed. First, LeT6 expression levels in wild-type and Me was determined. QRT-PCR showed a 7.5-fold increase in the expression of LeT6 in the stems of Me plants when compared to wild-type (Figure 7A). Transcript levels of LeCAD was then determined for wild-type and Me. We found that transcript levels of LeCAD were significantly lower (P = 0.033) in the Me plants compared to wild-type tomato (Figure 7B) confirming that transcriptional regulation of the CAD gene is conserved between tomato and tobacco.
FIGURE 7.
(A) Relative expression of LeT6 in wild-type (Wt) tomato and in the tomato mutant Mouse ears (Me) plants. (B) Relative expression of LeCAD in Wt and Me tomato plants. Error bars represent standard error [(± SE; N = 3, P = 0.006 (A) and P = 0.033 (B)].
DISCUSSION
We show that the maize class I KNOX gene, Kn1, is a strong regulator of lignification both in a selected monocot and a eudicot. Although three co-orthologs of BP exist in maize, only the Kn1 overexpression mutants seem to negatively regulate lignin, implying sub-functionalization of this role in the maize BP-like KNOX1 genes. This function is retained even when expressed in the heterologous eudicot system, tobacco. Maize Kn1 regulates lignin biosynthetic genes differently than LeT6 and suppresses lignification in maize more efficiently than Gn1 or Rs1, suggesting the possibility of a specialized role for this transcription factor in lignin regulation. Despite the large differences in cell wall composition between monocots and eudicots (Shedletzky et al., 1992), the maize Kn1 gene has the ability to negatively regulate lignin in both of these groups of plants. Additionally, maize Kn1 is able to modulate lignin deposition in vascular tissue during secondary growth. All tissues in grasses are primary but the majority of lignin in the majority of older eudicots is derived from secondary tissues in the stem. This would allow the maize Kn1 gene to be used as a general biotechnological tool in modifying lignin content to improve industrial or forage characteristics in plants. In addition, its efficacy at reducing total lignin across divergent species may allow for many crop species to be optimized for utilization of post-harvest crop residues. We found that LeT6 can regulate lignin composition and based on QRT-PCR results, we further showed that overexpression of LeT6 reduces NtCAD transcript levels by ~2.5-fold. The increase in cinnamoyl aldehyde content in Me and 35S::LeT6 plants is consistent with previous studies where CAD was reduced by antisense expression in tobacco (Hibino et al., 1995; Ralph et al., 1998; Kim et al., 2003). A previous study also found that reducing CAD activity resulted in an accumulation of coniferyl aldehyde, a precursor molecule of the lignin subunit, and incorporation of this molecule into the lignin polymer resulted in lignin with altered chemical composition (MacKay et al., 1999). It has also been shown that CAD down-regulation in several species including maize, Medicago sativa, Pinus taeda, tobacco, and poplar, chemically alters lignin resulting in greater alkaline solubility with unchanged total lignin content and without compromising strength. In the case of poplar, the wood was more amenable to industrial delignification processes including kraft pulping (Hibino et al., 1995; Baucher et al., 1999; MacKay et al., 1999; Vailhe et al., 2000; Marita et al., 2003), while faster growth and higher wood density was observed in loblolly pine (Cavell et al., 1998). Thus, KNOX1 genes may be a useful tool for altering total lignin levels, as well as modulating the lignin chemical makeup to improve forage or processing properties. It remains to be seen, however, by what specific mechanisms these transcription factors regulate genes in the lignin biosynthetic pathway.
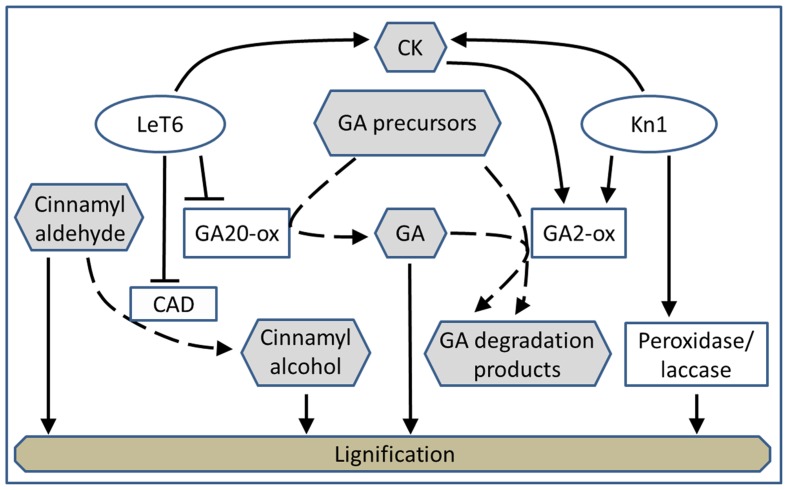
We propose that different KNOX1 genes transgenically expressed in tobacco independently modulate different aspects of lignin biosynthesis and affect GA-related developmental phenotypes by acting on different portions of the GA biosynthetic and catabolic pathways (Figure 8). KNOX1 gene expression is coupled to GA regulation (Parkin and Lydiate, 1997), although the mode of this regulation, via alteration of biosynthesis or degradation of bioactive GAs, can vary depending on the individual KNOX1 gene and the species in which it originates (Vicente et al., 2002; Mayerhofer et al., 2005; Palatnik et al., 2007; Hegedus et al., 2008; Mateos et al., 2010). It has been demonstrated in maize that overexpression of Kn1 results in an increase in GA2-oxidase transcript abundance (Palatnik et al., 2007; Navabi et al., 2010) by a direct interaction of KN1 protein with sequences in a GA2-oxidase intron (Himelblau et al., 2009). Tobacco NTH15, Arabidopsis STM, and potato PotH1 directly reduce transcription of GA20-oxidase in these species (Vicente et al., 2002; Mayerhofer et al., 2005; Hegedus et al., 2008). STM can, via activation of isopentenyl transferases (IPTs; Parkin et al., 2005), lead to GA catabolism by up-regulating GA2-oxidase (Doughty et al., 1998).
FIGURE 8.
Model depicting the proposed regulatory interactions (solid lines) of LeT6 and Kn1 on gibberellic acid biosynthesis and degradation (dashed lines).
We showed that the effects of expression of a maize and tomato gene in tobacco can parallel the effects of these genes in their native species. This supports our hypotheses that expression of specific KNOX genes and their effects on lignin levels and composition are transferable across plant species. Additionally, we found that the large reduction in lignin content in Kn1 overexpressing plants can be explained by reduction of endogenous GA. Since stem elongation in LeT6 overexpressing plants was not as severely affected as in Kn1 overexpressing plants, it is likely that GA concentrations are still high enough for lignification to occur somewhat normally in these plants (Figure 2). The lignin reduction and dwarfing phenotype of the plants overexpressing Kn1 are much more severe than the plants overexpressing LeT6 (Figure 2). Since GA2-oxidase not only leads to deactivation of bioactive GAs, but also acts on precursors of bioactive GAs (Figure 8), regulation of GA by GA2-oxidase expression would be greater than down-regulation of GA20-oxidase. Additionally, inability of GA3 treatment of fully restore lignin content in Kn1 overexpressing plants would be expected since the exogenous GA would be subject to degradation by GA2-oxidase. We found parallel effects of KNOX1 expression and GA regulation of lignification in grass and eudicot species. Here, a monocot species proves a very useful model for the study of secondary cell wall regulation. In dicot plants there can be simultaneous effects of hormonal alterations on both the modulation of secondary cell wall deposition and on secondary growth (xylem development), whereas typical monocot plants lack secondary growth and thus, provides a valuable comparison.
The KNOX1 genes function beyond regulation of cell wall properties. Additional traits under regulation of KNOX1 genes are potentially useful for crop improvement. For instance, alteration of crop architecture and drought resistance could also be targets for crop improvement using KNOX1 genes as regulators of growth processes. Cytokinin biosynthetic genes are up-regulated by expression of KNOX1 and the overexpression phenotypes of IPT and KNOX genes are distinctly similar. Increased levels of cytokinin leads to bud breaking and increased branching (Hegedus et al., 2003; Osborn et al., 2003; Ekuere et al., 2004). These traits (e.g., increased branching resulting in increased leaf production) may be desirable in many circumstances so that light capture can be maximized and inflorescence density increased as is the case in cereal crops such as wheat (Parkin et al., 1995). Expression of Kn1 genes under the control of a senescence-activated promoter in tobacco has been shown to delay onset of senescence in leaves (Lukens et al., 2003). Similarly, increasing levels of IPT in stressed tissues has been demonstrated to lead to significant improvement in survival and recovery from drought in tobacco (Parkin et al., 2003) and in rice (Gao et al., 2003), resulting in significant yield improvement when compared to wild-type plants. KNOXI genes have an indispensable role in compound leaf development (Parkin et al., 2002), with leaf shape being an important determinant of many physiological parameters (Sillito et al., 2000). In tomato interspecies introgression lines, the degree of leaf complexity correlates with brix and sugar levels, suggesting KNOX genes could be also be utilized to alter morphological and nutritional characteristics of crop species (Chitwood and Sinha, unpublished). Use of transcription factors offers the potential to coordinately regulate many genes relating to complex developmental and metabolic processes which in turn could simplify engineering of desirable traits and broaden the range of plants that can be successfully modified when compared to gene specific methods such as RNAi (RNA interference).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by a National Science Foundation grant (0820854) to Neelima R. Sinha.
REFERENCES
- Baucher M., Bernard-Vailhe M. A., Chabbert B., Besle J. M., Opsomer C., Van Montagu M., et al. (1999). Down-regulation of cinnamyl alcohol dehydrogenase in transgenic alfalfa (Medicago sativa L.) and the effect on lignin composition and digestibility. Plant Mol. Biol. 39 437–447 [DOI] [PubMed] [Google Scholar]
- Cardenas P. D., Gajardo H. A., Huebert T., Parkin I. A., Iniguez-Luy F. L., Federico M. L. (2012). Retention of triplicated phytoene synthase (PSY) genes in Brassica napus L. and its diploid progenitors during the evolution of the Brassiceae. Theor. Appl. Genet. 124 1215–1228 [DOI] [PubMed] [Google Scholar]
- Cavell A. C., Lydiate D. J., Parkin I. A., Dean C., Trick M. (1998). Collinearity between a 30-centimorgan segment of Arabidopsis thaliana chromosome 4 and duplicated regions within the Brassica napus genome. Genome 41 62–69 [PubMed] [Google Scholar]
- Chen J. J., Janssen B. J., Williams A., Sinha N. (1997). A gene fusion at a homeobox locus: alterations in leaf shape and implications for morphological evolution. Plant Cell 9 1289–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayan J., Schwarzkopf M., Avni A., Aloni R. (2010). Enhancing plant growth and fiber production by silencing GA 2-oxidase. Plant Biotechnol. J. 8 425–435 [DOI] [PubMed] [Google Scholar]
- Doughty J., Dixon S., Hiscock S. J., Willis A. C., Parkin I. A., Dickinson H. G. (1998). PCP-A1, a defensin-like Brassica pollen coat protein that binds the S locus glycoprotein, is the product of gametophytic gene expression. Plant Cell 10 1333–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas S. J., Chuck G., Dengler R. E., Pelecanda L., Riggs C. D. (2002). KNAT1 and ERECTA regulate inflorescence architecture in Arabidopsis. Plant Cell 14 547–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas S. J., Riggs C. D. (2005). Pedicel development in Arabidopsis thaliana: contribution of vascular positioning and the role of the BREVIPEDICELLUS and ERECTA genes. Dev. Biol. 284 451–463 [DOI] [PubMed] [Google Scholar]
- Du J., Miura E., Robischon M., Martinez C., Groover A. (2011). The Populus Class III HD ZIP transcription factor POPCORONA affects cell differentiation during secondary growth of woody stems. PLoS ONE 6:e17458 10.1371/journal.pone.0017458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekuere U. U., Parkin I. A., Bowman C., Marshall D., Lydiate D. J. (2004). Latent S alleles are widespread in cultivated self-compatible Brassica napus. Genome 47 257–265 [DOI] [PubMed] [Google Scholar]
- Esau K. (1965). Vascular Differentiation in Plants. New York: Holt, Rinehart and Winston [Google Scholar]
- Gao M. J., Schafer U. A., Parkin I. A., Hegedus D. D., Lydiate D. J., Hannoufa A. (2003). A novel protein from Brassica napus has a putative KID domain and responds to low temperature. Plant J. 33 1073–1086 [DOI] [PubMed] [Google Scholar]
- Gifford E. M., Foster A. S. (1988). Morphology and Evolution of Vascular Plants. New York: W. H. Freeman and Co [Google Scholar]
- Groover A. T., Mansfield S. D., DiFazio S. P., Dupper G., Fontana J. R., Millar R., et al. (2006). The Populus homeobox gene ARBORKNOX1 reveals overlapping mechanisms regulating the shoot apical meristem and the vascular cambium. Plant Mol. Biol. 61 917–932 [DOI] [PubMed] [Google Scholar]
- Hatfield R., Fukushima R. (2005). Can lignin be accurately measured? Crop Sci. 45 832–839 [Google Scholar]
- Hegedus D., Yu M., Baldwin D., Gruber M., Sharpe A., Parkin I., et al. (2003). Molecular characterization of Brassica napus NAC domain transcriptional activators induced in response to biotic and abiotic stress. Plant Mol. Biol. 53 383–397 [DOI] [PubMed] [Google Scholar]
- Hegedus D. D., Li R., Buchwaldt L., Parkin I., Whitwill S., Coutu C., et al. (2008). Brassica napus possesses an expanded set of polygalacturonase inhibitor protein genes that are differentially regulated in response to Sclerotinia sclerotiorum infection, wounding and defense hormone treatment. Planta 228 241–253 [DOI] [PubMed] [Google Scholar]
- Hibino T., Takabe K., Shibata D., Higuchi T. (1995). Increase of cinnamaldehyde groups in lignin of transgenic tobacco plants carrying an antisense gene for cinnamyl alcohol dehydrogenase. Biosci. Biotech. Biochem. 59 929–931 [Google Scholar]
- Himelblau E., Gilchrist E. J., Buono K., Bizzell C., Mentzer L., Vogelzang R., et al. (2009). Forward and reverse genetics of rapid-cycling Brassica oleracea. Theor. Appl. Genet. 118 953–961 [DOI] [PubMed] [Google Scholar]
- Janssen B. J., Lund L., Sinha N. (1998a). Overexpression of a homeobox gene, LeT6, reveals indeterminate features in the tomato compound leaf. Plant Physiol. 117 771–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen B. J., Williams A., Chen J. J., Mathern J., Hake S., Sinha N. (1998b). Isolation and characterization of two knotted-like homeobox genes from tomato. Plant Mol. Biol. 36 417–425 [DOI] [PubMed] [Google Scholar]
- Kim H., Ralph J., Lu F., Ralph S. A., Boudet A. M., MacKay J. J., et al. (2003). NMR analysis of lignins in CAD-deficient plants. Part 1. Incorporation of hydroxycinnamaldehydes and hydroxybenzaldehydes into lignins. Org. Biomol. Chem. 1 268–281 [DOI] [PubMed] [Google Scholar]
- Kimura S., Koenig D., Kang J., Yoong F. Y., Sinha N. (2008). Natural variation in leaf morphology results from mutation of a novel KNOX gene. Curr. Biol. 18 672–677 [DOI] [PubMed] [Google Scholar]
- Larkan N. J., Lydiate D. J., Parkin I. A., Nelson M. N., Epp D. J., Cowling W. A., et al. (2013). The Brassica napus blackleg resistance gene LepR3 encodes a receptor-like protein triggered by the Leptosphaeria maculans effector AVRLM1. New Phytol. 197 595–605 [DOI] [PubMed] [Google Scholar]
- Li E., Bhargava A., Qiang W., Friedmann M. C., Forneris N., Savidge R. A., et al. (2012). The Class II KNOX gene KNAT7 negatively regulates secondary wall formation in Arabidopsis and is functionally conserved in Populus. New Phytol. 194 102–115 [DOI] [PubMed] [Google Scholar]
- Long J. A., Moan E. I., Medford J. I., Barton M. K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379 66–69 [DOI] [PubMed] [Google Scholar]
- Lukens L., Zou F., Lydiate D., Parkin I., Osborn T. (2003). Comparison of a Brassica oleracea genetic map with the genome of Arabidopsis thaliana. Genetics 164 359–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luquita A., Urli L., Svetaz M. J., Gennaro A. M., Volpintesta R., Palatnik S., et al. (2009). Erythrocyte aggregation in rheumatoid arthritis: cell and plasma factor’s role. Clin. Hemorheol. Microcirc. 41 49–56 [DOI] [PubMed] [Google Scholar]
- MacKay J., Presnell T., Jameel H., Taneda H., O’Malley D., Sederoff R. (1999). Modified ligin and delignification with a CAD-deficient loblolly pine. Holzforschung 53 403–410 [Google Scholar]
- Marita J. M., Vermerris W., Ralph J., Hatfield R. D. (2003). Variations in the cell wall composition of maize brown midrib mutants. J. Agric. Food Chem. 51 1313–1321 [DOI] [PubMed] [Google Scholar]
- Mateos J. L., Bologna N. G., Chorostecki U., Palatnik J. F. (2010). Identification of microRNA processing determinants by random mutagenesis of Arabidopsis MIR172a precursor. Curr.Biol. 20 49–54 [DOI] [PubMed] [Google Scholar]
- Mayerhofer R., Wilde K., Mayerhofer M., Lydiate D., Bansal V. K., Good A. G., et al. (2005). Complexities of chromosome landing in a highly duplicated genome: toward map-based cloning of a gene controlling blackleg resistance in Brassica napus. Genetics 171 1977–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mele G., Ori N., Sato Y., Hake S. (2003). The knotted1-like homeobox gene BREVIPEDICELLUS regulates cell differentiation by modulating metabolic pathways. Genes Dev. 17 2088–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun J. H., Kwon S. J., Seol Y. J., Kim J. A., Jin M., Kim J. S., et al. (2010). Sequence and structure of Brassica rapa chromosome A3. Genome Biol. 11 R94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navabi Z. K., Parkin I. A., Pires J. C., Xiong Z., Thiagarajah M. R., Good A. G., et al. (2010). Introgression of B-genome chromosomes in a doubled haploid population of Brassica napusxB. carinata. Genome 53 619–629 [DOI] [PubMed] [Google Scholar]
- Navabi Z. K., Stead K. E., Pires J. C., Xiong Z., Sharpe A. G., Parkin I. A., et al. (2011). Analysis of B-genome chromosome introgression in interspecific hybrids of Brassica napus x B. carinata. Genetics 187 659–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer S., Kumar S., Khan B. M. (2013). Over-expression of a subgroup 4 R2R3 type MYB transcription factor gene from Leucaena leucocephala reduces lignin content in transgenic tobacco. Plant Cell Rep. 32 161–171 [DOI] [PubMed] [Google Scholar]
- Osborn T. C., Butrulle D. V., Sharpe A. G., Pickering K. J., Parkin I. A., Parker J. S., et al. (2003). Detection and effects of a homeologous reciprocal transposition in Brassica napus. Genetics 165 1569–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik J. F., Wollmann H., Schommer C., Schwab R., Boisbouvier J., Rodriguez R., et al. (2007). Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Dev. Cell 13 115–125 [DOI] [PubMed] [Google Scholar]
- Palatnik M., Simoes M. L., Alves Z. M., Laranjeira N. S. (1990). The 60 and 63 kDa proteolytic peptides of the red cell membrane band-3 protein: their prevalence in human and non-human primates. Hum. Genet. 86 126–130 [DOI] [PubMed] [Google Scholar]
- Palatnik M., Simoes M. L., Guinsburg S. S., Lopes H. (1992). Genetic polymorphism of red cell membrane band 3 in Japanese Brazilians. Gene Geogr. 6 17–20 [PubMed] [Google Scholar]
- Parkin I. A., Clarke W. E., Sidebottom C., Zhang W., Robinson S. J., Links M. G., et al. (2010). Towards unambiguous transcript mapping in the allotetraploid Brassica napus. Genome 53 929–938 [DOI] [PubMed] [Google Scholar]
- Parkin I. A., Gulden S. M., Sharpe A. G., Lukens L., Trick M., Osborn T. C., et al. (2005). Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171 765–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin I. A., Lydiate D. J. (1997). Conserved patterns of chromosome pairing and recombination in Brassica napus crosses. Genome 40 496–504 [DOI] [PubMed] [Google Scholar]
- Parkin I. A., Lydiate D. J., Trick M. (2002). Assessing the level of collinearity between Arabidopsis thaliana and Brassica napus for A. thaliana chromosome 5. Genome 45 356–366 [DOI] [PubMed] [Google Scholar]
- Parkin I. A., Sharpe A. G., Keith D. J., Lydiate D. J. (1995). Identification of the A and C genomes of amphidiploid Brassica napus (oilseed rape). Genome 38 1122–1131 [DOI] [PubMed] [Google Scholar]
- Parkin I. A., Sharpe A. G., Lydiate D. J. (2003). Patterns of genome duplication within the Brassica napus genome. Genome 46 291–303 [DOI] [PubMed] [Google Scholar]
- Parnis A., Cohen O., Gutfinger T., Hareven D., Zamir D., Lifschitz E. (1997). The dominant developmental mutants of tomato, Mouse-ear and Curl, are associated with distinct modes of abnormal transcriptional regulation of a Knotted gene. Plant Cell 9 2143–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzlaff A., McInnis S., Courtenay A., Surman C., Newman L. J., Smith C., et al. (2003). Characterisation of a pine MYB that regulates lignification. Plant J. 36 743–754 [DOI] [PubMed] [Google Scholar]
- Ralph J., Hatfield R. D., Piquemal J., Yahiaoui N., Pean M., Lapierre C., et al. (1998). NMR characterization of altered lignins extracted from tobacco plants down-regulated for lignification enzymes cinnamylalcohol dehydrogenase and cinnamoyl-CoA reductase. Proc. Natl. Acad. Sci. U.S.A. 95 12803–12808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez R. E., Mecchia M. A., Debernardi J. M., Schommer C., Weigel D., Palatnik J. F. (2010). Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137 103–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzin S. E. (1999). Plant Microtechnique and Microscopy. New York: Oxford University Press [Google Scholar]
- Sablowski R. W., Moyano E., Culianez-Macia F. A., Schuch W., Martin C., Bevan M. (1994). A flower-specific Myb protein activates transcription of phenylpropanoid biosynthetic genes. EMBO J. 13 128–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaposnik F., Bottino N. R., Palatnik M., Peluffo R. O., Chalar O. (1958). [Micro cell anemia (sickle-thalassemia)]. Rev. Clin. Esp. 68 284–294 [PubMed] [Google Scholar]
- Shedletzky E., Shmuel M., Trainin T., Kalman S., Delmer D. (1992). Cell wall structure in cells adapted to growth on the cellulose-synthesis inhibitor 2,6-dichlorobenzonitrile: a comparison between two dicotyledonous plants and a graminaceous monocot. Plant Physiol. 100 120–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillito D., Parkin I. A., Mayerhofer R., Lydiate D. J., Good A. G. (2000). Arabidopsis thaliana: a source of candidate disease-resistance genes for Brassica napus. Genome 43 452–460 [DOI] [PubMed] [Google Scholar]
- Sinha N., Williams R. E., Hake S. (1993). Overexpression of the maize homeobox gene, KNOTTED-1, cause a switch from determinate to indeterminate cell fates. Genes Dev. 7 787–795 [DOI] [PubMed] [Google Scholar]
- Spinelli S. V., Martin A. P., Viola I. L., Gonzalez D. H., Palatnik J. F. (2011). A mechanistic link between STM and CUC1 during Arabidopsis development. Plant Physiol. 156 1894–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testone G., Condello E., Verde I., Nicolodi C., Caboni E., Dettori M. T., et al. (2012). The peach (Prunus persica L. Batsch) genome harbours 10 KNOX genes, which are differentially expressed in stem development, and the class 1 KNOPE1 regulates elongation and lignification during primary growth. J. Exp. Bot. 63 5417–5435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vailhe M. A., Provan G. J., Scobbie L., Chesson A., Maillot M. P., Cornu A., et al. (2000). Effect of phenolic structures on the degradability of cell walls isolated from newly extended apical internode of tall fescue (Festuca arundinacea Schreb.). J. Agric. Food Chem. 48 618–623 [DOI] [PubMed] [Google Scholar]
- Venglat S. P., Dumonceaux T., Rozwadowski K., Parnell L., Babic V., Keller W., et al. (2002). The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 99 4730–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente J. G., Taylor J. D., Sharpe A. G., Parkin I. A., Lydiate D. J., King G. J. (2002). Inheritance of race-specific resistance to Xanthomonas campestris pv. campestris in Brassica genomes. Phytopathology 92 1134–1141 [DOI] [PubMed] [Google Scholar]
- Vollbrecht E., Reiser L., Hake S. (2000). Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development 127 3161–3172 [DOI] [PubMed] [Google Scholar]
- Wang J., Lydiate D. J., Parkin I. A., Falentin C., Delourme R., Carion P. W., et al. (2011a). Integration of linkage maps for the Amphidiploid Brassica napus and comparative mapping with Arabidopsis and Brassica rapa. BMC Genomics 12:101 10.1186/1471-2164-12-101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wang H., Wang J., Sun R., Wu J., Liu S., et al. (2011b). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43 1035–1039 [DOI] [PubMed] [Google Scholar]
- Xu B., Sathitsuksanoh N., Tang Y., Udvardi M. K., Zhang J. Y., Shen Z., et al. (2012). Overexpression of AtLOV1 in Switchgrass alters plant architecture, lignin content, and flowering time. PLoS ONE 7:e47399 10.1371/journal.pone.0047399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Gruber M. Y., Khachatourians G. G., Zhou R., Epp D. J., Hegedus D. D., et al. (2012). Arabidopsis cpSRP54 regulates carotenoid accumulation in Arabidopsis and Brassica napus. J. Exp. Bot. 63 5189–5202 [DOI] [PMC free article] [PubMed] [Google Scholar]