Abstract
Adenosine diphosphate (ADP)-ribosylation is a post-translational protein modification implicated in the regulation of a range of cellular processes. A family of proteins that catalyse ADP-ribosylation reactions are the poly(ADP-ribose) (PAR) polymerases (PARPs). PARPs covalently attach an ADP-ribose nucleotide to target proteins and some PARP family members can subsequently add additional ADP-ribose units to generate a PAR chain. The hydrolysis of PAR chains is catalysed by PAR glycohydrolase (PARG). PARG is unable to cleave the mono(ADP-ribose) unit directly linked to the protein and although the enzymatic activity that catalyses this reaction has been detected in mammalian cell extracts, the protein(s) responsible remain unknown. Here, we report the homozygous mutation of the c6orf130 gene in patients with severe neurodegeneration, and identify C6orf130 as a PARP-interacting protein that removes mono(ADP-ribosyl)ation on glutamate amino acid residues in PARP-modified proteins. X-ray structures and biochemical analysis of C6orf130 suggest a mechanism of catalytic reversal involving a transient C6orf130 lysyl-(ADP-ribose) intermediate. Furthermore, depletion of C6orf130 protein in cells leads to proliferation and DNA repair defects. Collectively, our data suggest that C6orf130 enzymatic activity has a role in the turnover and recycling of protein ADP-ribosylation, and we have implicated the importance of this protein in supporting normal cellular function in humans.
Keywords: ADP-ribose, DNA damage, macrodomain, neurodegeneration, PARP
Introduction
Adenosine diphosphate (ADP)-ribosylation is an evolutionarily conserved reversible post-translational protein modification that regulates a wide range of cellular processes, including DNA repair, transcription, telomere dynamics, cell differentiation and proliferation, the inflammatory and immune responses and apoptosis (Hassa et al, 2006). A large family of proteins that catalyse ADP-ribosylation reactions are the poly(ADP-ribose) (PAR) polymerases (PARPs) (Gibson and Kraus, 2012). PARPs use NAD+ as a substrate and covalently attach an ADP-ribose nucleotide, predominantly to the carboxyl group of glutamate residues on target proteins (D'Amours et al, 1999). To date, 17 members of the PARP family have been identified in humans (Gibson and Kraus, 2012), and some of these proteins (PARP1, PARP2, and tankyrases) can subsequently add additional ADP-ribose units through glycosidic ribose-ribose bonds to generate a PAR chain (D'Amours et al, 1999; Gibson and Kraus, 2012).
ADP-ribosylation is a highly dynamic and reversible process. The specific hydrolysis of ribose-ribose bonds in PAR chains is catalysed by PAR glycohydrolase (PARG) (Lin et al, 1997). However, PARG is unable to cleave the ester bond between the terminal ADP-ribose unit and the ADP-ribosylated glutamate (Slade et al, 2011). The de(ADP-ribosyl)ation of glutamate residues would be of critical importance for cells to complete the reversal of ADP-ribosylation signalling or for the recycling of the covalently linked ADP-ribose. Although the enzymes that catalyse the removal of this type of protein modification have not yet been identified, analogous enzymatic activity has been detected in rat cell extracts (Oka et al, 1984).
Macrodomains are evolutionarily conserved structural protein modules of ∼150 amino acids that bind NAD metabolites, including ADP-ribose/poly(ADP-ribose) and the sirtuin by-product O-acetyl-ADP-ribose (Karras et al, 2005; Ahel et al, 2009; Till and Ladurner, 2009; Timinszky et al, 2009). Some of the macrodomain proteins have been shown to possess catalytic activity on ADP-ribosylated substrates (Chen et al, 2011; Slade et al, 2011; Zaja et al, 2013). In this work, we identify the c6orf130 gene as a defective gene in patients with severe neurodegeneration. We show that the protein product of this gene is a PARP-interacting macrodomain protein with the ability to cleave the mono(ADP-ribose) from PARP-modified proteins. Our X-ray structures of C6orf130 and supporting solution biochemical studies suggest a mechanism of catalytic reversal involving a transient C6orf130 lysyl-(ADP-ribose) intermediate. Finally, we demonstrate that the function of C6orf130 protein is important for normal cellular proliferation and cellular response to DNA damage.
Results
C6orf130 gene is mutated in patients with severe neurodegeneration
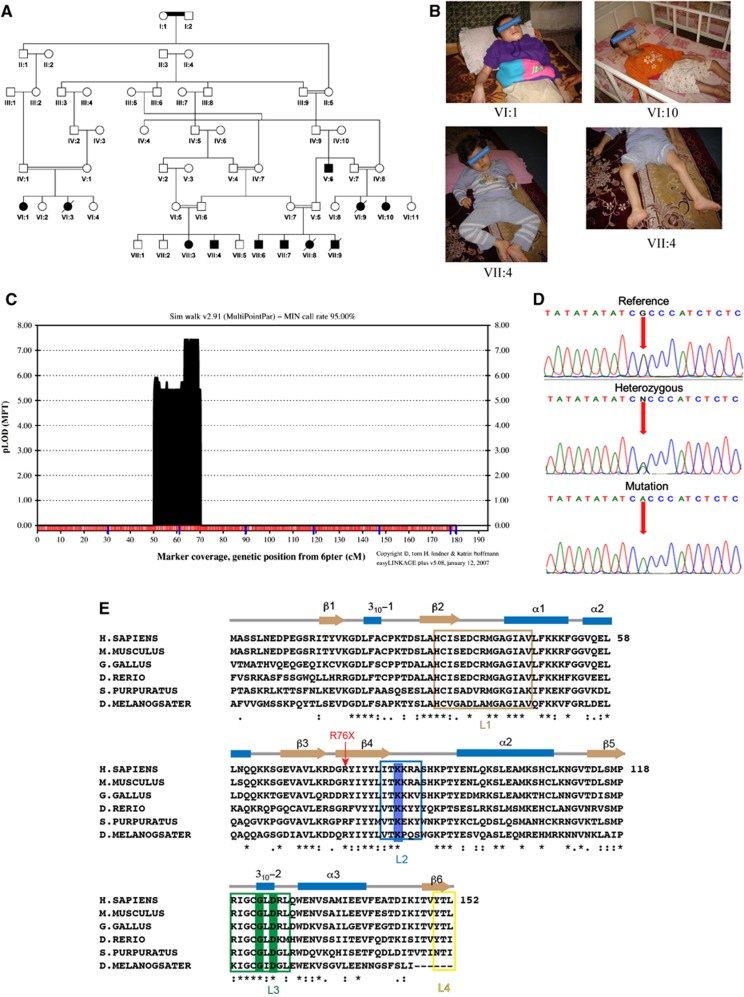
We studied an extended family with an autosomal recessive trait presenting with a severe form of progressive neurodegenerative and seizure disorder without dysmorphic features (Figure 1A and B; Supplementary Table 1). The autozygosity mapping revealed a homozygous region on chromosome 6p21 (20 cM) and linkage analysis produced a significant multipoint LOD score of 7.4 for the mapped region (Figure 1C). Subsequent refinement mapping defined a 6.54-Mb interval flanked by markers D6S1610 and D6S459 and containing a total of 30 labelled genes and 5 open-reading frames (Supplementary Figure 1). To exclude the possible existence of a pathogenic mutation in the linked interval, we performed whole-exome sequencing in one individual from the family and verified three non-sense novel variants within the extended 8.51 Mb linkage region (Supplementary Table 2). In the kindred, we identified a distinct homozygous sequence variant (NC_000006.11:g.41037831G>A; NM_145063.2:c.227C>T) within exon 4 of the c6orf130 gene that segregates with the phenotype and predicts the formation of a truncated C6orf130 protein lacking the C-terminal half of the protein due to a premature stop codon (NP_659500.1:p.R76X) (Figures 1D and E). No likely disease-causing sequence variants were detected in the other genes analysed by direct sequencing (Supplementary Table 3). We did not detect the c.227C>T variant in over 1200 chromosomes assayed from unrelated ethnic matched and European origin control subjects.
Figure 1.
The genetic and clinical data. (A) Pedigree diagram of family. (B) Photographs of individuals VI:1,VI:10, VII:4 affected by neurodegeneration. (C) EasyLinkage Plus v.5.08 output of parametric analysis of chromosome 6 under an autosomal recessive model by Simwalk2.91. (D) Electropherograms showing the identified mutation in c6orf130 gene (NC_000006.11:g.41037831G>A; NM_145063.2:c.227C>T; NP_659500.1:p.R76X). (E) Structure-based sequence alignment of C6orf30 homologues. The conserved residues K84 and G123/D125 are highlighted in blue and green, respectively. The predicted C6orf130 protein truncation in the patients analysed in this study is marked with red arrow.
The c6orf130 gene encodes a macrodomain-containing protein of unknown physiological significance that is ubiquitously expressed in different tissues (Supplementary Figure 2). It was recently demonstrated that the C6orf130 protein can hydrolyse O-acetyl-ADP-ribose in vitro (Peterson et al, 2011). Given the similarity of the chemical bond between the glutamate and ADP-ribose in mono(ADP-ribosyl)ated proteins and the bond in the acetylated ADP-ribose, we postulated that C6orf130 could function as a long-sought protein that reverses the protein mono(ADP-ribosyl)ation synthesized by PARPs. This possibility is further substantiated by another case of severe neurodegeneration that has been described previously (Williams et al, 1984). For this patient, who died after 6 years of progressive neurologic deterioration, it was demonstrated that the primary defect was a genetic abnormality in an unidentified enzyme involved in the cleavage of the bond between glutamate and ADP-ribose.
C6orf130 demodifies mono(ADP-ribosyl)ated PARP substrates
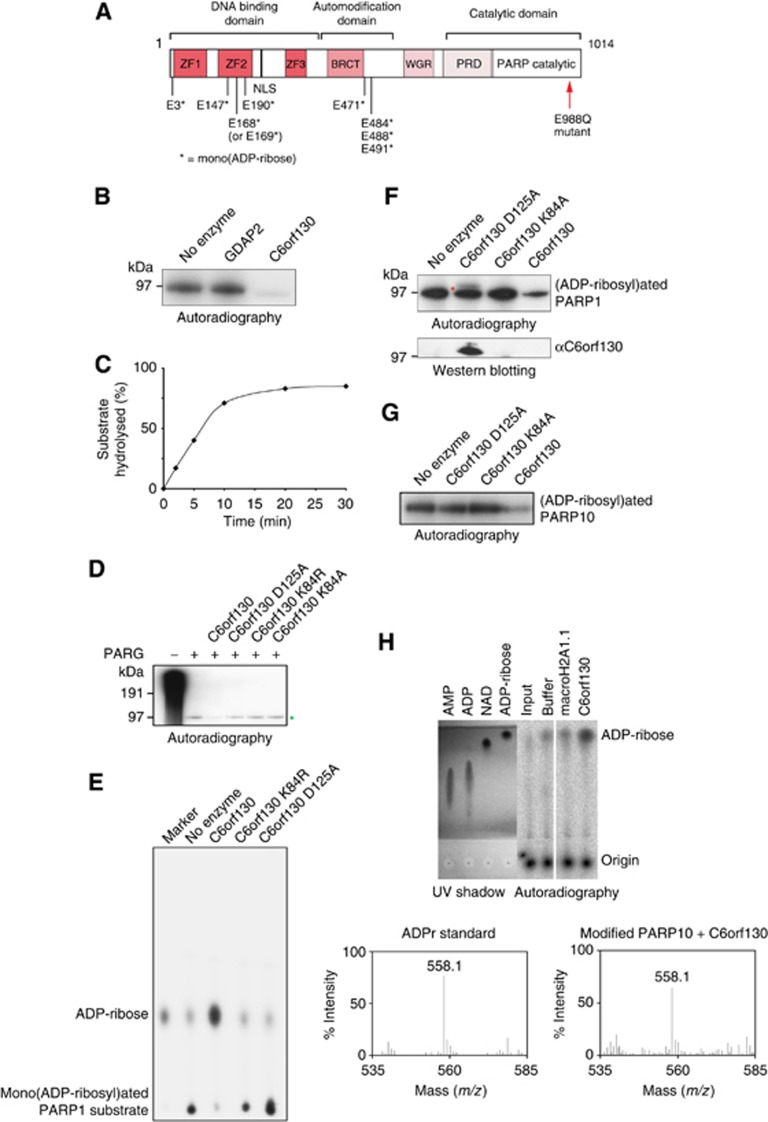
To analyse the ability of C6orf130 to cleave ADP-ribosylated peptides in vitro, we employed an automodified PARP1 E988Q mutant as a substrate. Previous studies demonstrated that the PARP1 E988Q mutant is incapable of poly(ADP-ribosyl)ation activity (Marsischky et al, 1995), but instead mono(ADP-ribosyl)ates itself at two glutamate and one aspartate residues (Tao et al, 2009). We further characterized this substrate and identified several additional modification sites exclusively on glutamate residues clustering in the automodification and the second zinc-finger domains (Figure 2A; Supplementary Dataset 1). Our data showed that the recombinant human C6orf130 protein efficiently cleaved a radioactively labelled PARP1 peptide, whereas another macrodomain protein, GDAP2, was unable to perform the same reaction (Figure 2B, C).
Figure 2.
C6orf130 protein is a mono(ADP-ribosyl) protein hydrolase. (A) Schematic representation of PARP1 E988Q mutant protein. Residues found to be mono(ADP-ribosyl)ated by mass spectrometry analysis of automodified recombinant PARP1 E988Q protein are shown and marked with an asterisk. (B) SDS–PAGE based assay showing that purified recombinant human C6orf130 de(ADP-ribosyl)ates 32P-labelled mono(ADP-ribosyl)ated PARP1 E988Q protein. Macrodomain-containing protein GDAP2 served as a negative control. (C) The time curve showing the activity of C6orf130 on ADP-ribosylated PARP1 E988Q protein. (D) Hydrolytic activity of C6orf130 wild-type and mutated proteins on mono(ADP-ribosyl) PARP1 peptide derived from the activity of PARG protein on [32P]-labelled poly(ADP-ribosyl)ated wt PARP1. Mono(ADP-ribosyl)ated PARP1 species is indicated by green asterisk. (E) Analysis of the product of the C6orf130 reaction on mono(ADP-ribosyl)ated PARP E988Q by thin-layer chromatography (TLC). (F) Catalytic activity of C6orf130 proteins carrying point mutations in the indicated residues (top panel). The band appearing above 97 kDa in the SDS–PAGE in the C6orf130 D125A mutant lane (top panel, red asterisk) contains a crosslinked C6orf130 protein as revealed by western blotting (bottom panel). (G) De(ADP-ribosyl)ation of the PARP10 substrate by C6orf130. (H) Analysis of the reaction products of C6orf130 on ADP-ribosylated PARP10 substrate by TLC (top panel) and mass spectrometry (bottom panel).
We next analysed the ability of C6orf130 to act on mono(ADP-ribosyl)ated substrates that are the products of PARG activity on poly(ADP-ribosyl)ated wild-type PARP1 protein. While PARG is unable to remove the terminal ADP-ribose directly linked to PARP1, the sequential addition of C6orf130 removed most of the remaining peptide ADP-ribosylation (Figure 2D). Analysis of the products of the C6orf130 reaction by thin-layer chromatography (TLC) revealed that C6orf130 released a product with a mobility aligned to the mono(ADP-ribose) marker (Figure 2E), suggesting that C6orf130 acts as an ADP-ribose hydrolase. C6orf130 similarly efficiently de(ADP-ribosyl)ates automodified PARP10, a PARP family member capable of only mono(ADP-ribosyl)ation (Kleine et al, 2008) (Figure 2G and H).
A previously proposed hydrolytic mechanism for the deacylation of O-acetyl-ADP-ribose by C6orf130 suggested a conserved aspartate in C6orf130 (Asp125 in human C6orf130; Figure 1E) participates in general acid base catalysis (Peterson et al, 2011). We therefore analysed the importance of Asp125 for protein de(ADP-ribosyl)ation. The D125A mutation mutant abolished the de(ADP-ribosyl)ation of the automodified PARP1 (Figure 2F). Unexpectedly, however, the D125A mutant protein also formed a stable covalently bound complex with the mono(ADP-ribosyl)ated PARP1 E988Q peptide, as observed by the appearance of a supershifted band above 97 kDa (Figure 2F, top panel) that is recognized by a C6orf130-specific antibody (Figure 2E, bottom panel). Because the D125A mutant was catalytically defective, we hypothesized that this covalent linkage is a reaction intermediate with ADP-ribose. Consistent with this proposal, we also detected covalent adduct formation with the wild-type C6orf130 protein when incubated with free ADP-ribose (instead of ADP-ribosylated peptide) (Supplementary Dataset 2). MS/MS analysis of this adduct showed that the absolutely conserved Lys84 residue reacts with free ADP-ribose, and is conjugated to a dehydrated ADP-ribose adduct. Isothermal titration calorimetry (ITC) demonstrates that the K84A mutation ablates C6orf130 ADP-ribose binding (Supplementary Figure 3), and the mutation of Lys84 to Ala or Met blocked ADP-ribose adduction as monitored by mass spectrometry (Supplementary Dataset 2). Moreover, the K84A mutant ablated de(ADP-ribosyl)ation activity, but unlike the D125A mutant, K84A did not yield a stable covalent adduct with the ADP-ribosylated peptide (Figure 2F). Altogether, these results demonstrate that C6orf130 catalyses the hydrolysis of ADP-ribosylated peptides, and that its catalytic mechanism involves the formation of a transient covalent Lys84-(ADP-ribose) intermediate.
Activity of C6orf130 on poly(ADP-ribosyl)ated PARP substrates
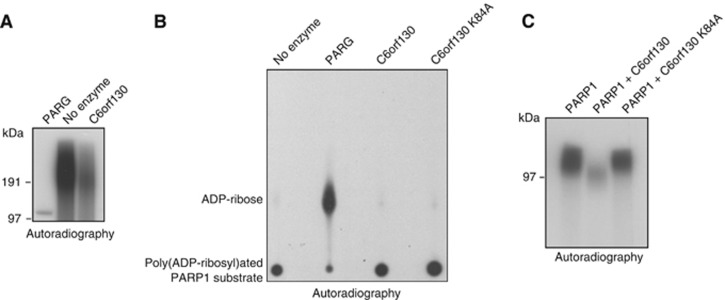
Next, we tested the ability of C6orf130 to directly hydrolyse PAR. While C6orf130 protein exhibited activity in removing PAR chains from poly(ADP-ribosyl)ated PARP1, this activity was lower than the activity observed with PARG protein (Figure 3A). Importantly, we could show that C6orf130 is unable to act on ribose-ribose bonds in PAR chains to release ADP-ribose as a reaction product (Figure 3B), but that this macrodomain protein rather removes the whole PAR chain acting specifically at the glutamate-ADP-ribose ester bonds.
Figure 3.
Activities of C6orf130 on poly(ADP-ribosyl)ated PARP1 substrates. (A) Hydrolysis of PAR by C6orf130 and PARG analysed by SDS–PAGE and (B) by thin-layer chromatography-based assays. (C) C6orf130 inhibits automodification activity of PARP1 in vitro.
To assess how C6orf130 activities alter PARP1 function in vitro, we performed a PARP1 automodification assay in the presence of wild-type or catalytically inactive C6orf130 proteins. Expectedly, PARP1 activity is greatly suppressed in these conditions, most likely due to the catalytic action of C6orf130 on mono- and poly(ADP-ribosyl)ated PARP1 species as the C6orf130 K84A mutant fails to block PARP1 automodification (Figure 3C).
C6orf130 structure
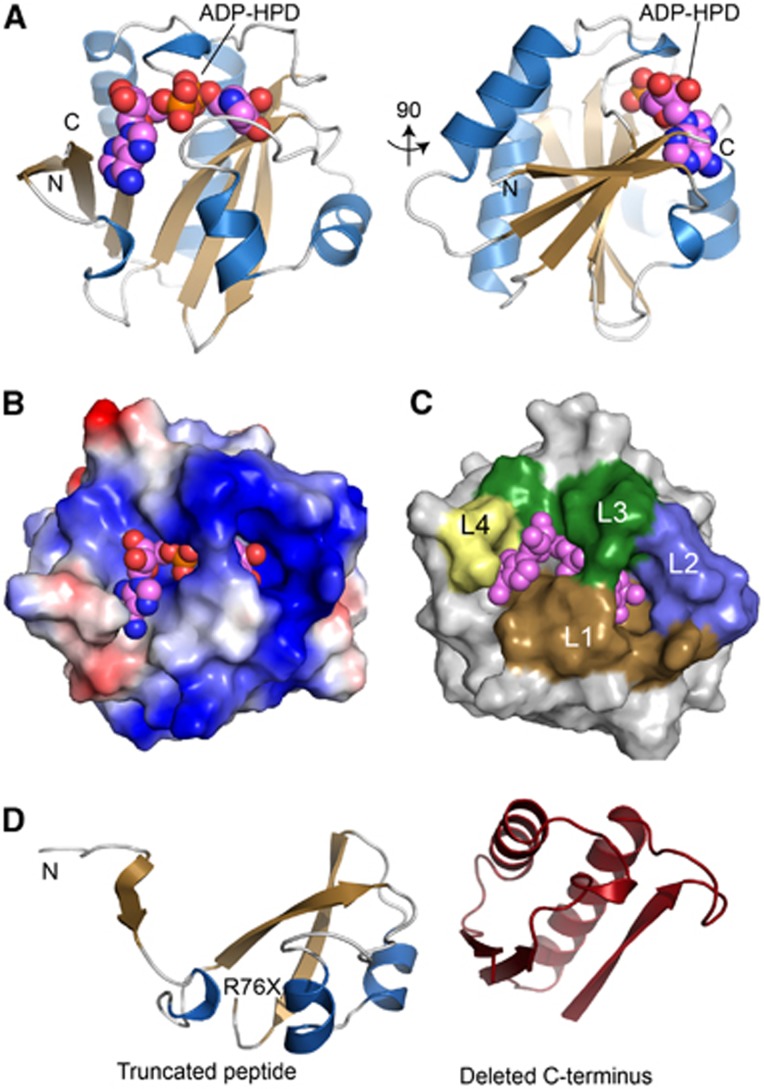
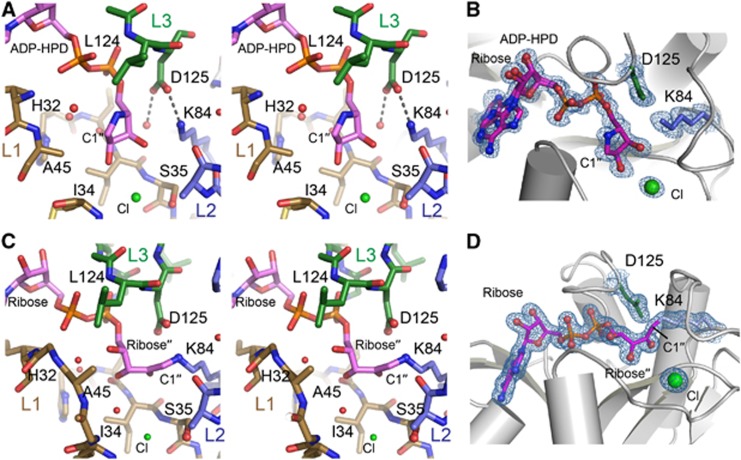
To understand the molecular basis for C6orf130 catalytic activities, we determined high-resolution X-ray crystal structures of C6orf130 in: (1) a ligand-free form at 1.35 Å, (2) bound to the ADP-ribose analogue ADP-HPD at 1.25 Å and (3) as a covalent ADP-ribose complex at 1.55 Å (Figures 4 and 5; Supplementary Table 4). The C6orf130 macrodomain fold adopts a compact 6-stranded β-sheet flanked by four α-helices and two 310-helical elements. Four surface loops coalesce to form the electropositive ligand-binding pocket (L1–L4, Figures 4A–C). The premature truncation of the C6orf130 open reading frame (ORF) in patients (Figure 1) is predicted to generate a non-functional peptide with a stop codon near the β3-β4 junction (Figures 1E and 4D).
Figure 4.
Crystal structure of the C6orf130 ADP-HPD complex. (A) Orthogonal views of C6orf130 (blue and tan) bound to ADP-HPD (spheres). (B) Surface charge representation of C6orf130 (blue, positive; red, negative; grey, neutral or hydrophobic) showing a positively charged binding site for ADP-HPD. (C) ADP-HPD binding site is composed of four sequence motifs, L1–L4. (D) The C6orf130 protein truncation (R76X) in the patients analysed in this study.
Figure 5.
C6orf130 active site architecture and ligand interactions. (A) Stereo view of the active site of C6orf130. A chloride ion (green) interacts with the 2′′- and 3′′-hydroxyls of the ADP-HPD. (B) Final 1.25 Å sigma-A weighted 2Fo-Fc electron density map (contoured at 1.0σ) showing ADP-HPD bound in the C6orf130 active site. Lys84 and Asp125 and a bound chloride ion are in close proximity to the pyrrolidine ring of ADP-HPD. (C) Stereo view of the active site of C6orf130 (ADP-ribose) complex. Molecule D (Supplementary Figure 5) is displayed. (D) Final 1.55 Å sigma-A weighted 2Fo-Fc electron density map (contoured at 1.0σ) showing the covalent lysyl-ADP-ribose adduct of chain D.
Clamping by the L1 and L3 elements grasps the ADP-HPD and ADP-ribose upon binding. Gly123 undergoes a peptide flipping rearrangement, and together with Leu124 secures the pyrophosphate moiety of bound ligands. In the ADP-HPD complex, the pyrophosphate and pyrrolidine groups are well defined, and the adenine base displays two alternate binding conformations in the base interaction groove that is capped by the C6orf130 C-terminal Leu152 (Figures 5A and B). Within the ligand-binding grooves, the structures reveals the active centre comprised of Asp125, Lys84, and Ser35. Notably, although the overall fold of C6orf130 in our X-ray structures is consistent with published C6orf130 NMR structure ensembles (Petersen et al, 2011) the residues forming the active site are found in very different conformations. Specifically, in the NMR ensembles, Lys84 is found excluded from the active site, and the Lys84 epsilon amino group is 10–14 Å away from bound ADP-ribose (Supplementary Figure 4A). By contrast, in the X-ray structures reported here, K84 is closely juxtaposed to D125, is in close proximity to bound ADP-HPD, and is covalently linked to ligand in the ADP-ribose bound structure (Figures 5A–D; Supplementary Figure 4B).
C6orf130 catalytic mechanism
The crystallographic asymmetric unit of the ADP-ribose bound crystal form contains four complexes of the C6orf130 ADP-ribose complex. Consistent with mass spectrometric analysis, the K84-ADP-ribose covalent species is linked to the ribose′′-C1 position for three of the four complexes. Electron density for K84-ADP-ribose covalent adducts is most consistent with an Amadori rearrangement ketoamine product (Cervantes-Laurean et al, 1993) (Supplementary Figure 5), similar to that observed in the structure of the DraG-(ADP-ribose) complex (Berthold et al, 2009) (Figures 5C and D). In addition, other ADP-ribose-derived species arising during the 3-month crystallization period are observed (Supplementary Figure 5).
The C6orf130 Lys84/Asp125 catalytic dyad is reminiscent of the OGG1 DNA glycosylase (Bruner et al, 2000). Analogous to OGG1, we hypothesize that Lys84 acts as a nucleophile to attack the ribose ring (at C1′′), with the release of glutamate (Supplementary Figure 6). Asp125 may promote decomposition of an ensuing Schiff base via the hydrolysis and release of the ADP-ribose product. This scheme would require repositioning of a ribose′′ relative to that observed in the ADP-HPD complex. We suggest that the orientation of the sugar-peptide conjugate is dictated by the binding of the ADP-ribosylated Glu side chain by the L1 loop. Consistent with this hypothesis, Ser42 and the main chain backbone amides from L1 coordinate a bound Cl− ion that marks a possible glutamate binding site (Figures 5A and B).
ADP-ribose bound acyl moieties (Sauve et al, 2001; Kasamatsu et al, 2011) can undergo regio-isomerization, such that ADP-ribosylated Glu residues may be linked to C1′′, C2′′, or C3′′ ribose positions in solution (Supplementary Figure 6). A role for Asp125 in promoting hydrolysis of a transient K84-(ADP-ribose) covalent intermediate may thus explain the accumulation of C6orf130-PARP conjugates in a D125A mutant (Figure 2F; Figures 6C and D; Supplementary Figure 6).
Figure 6.
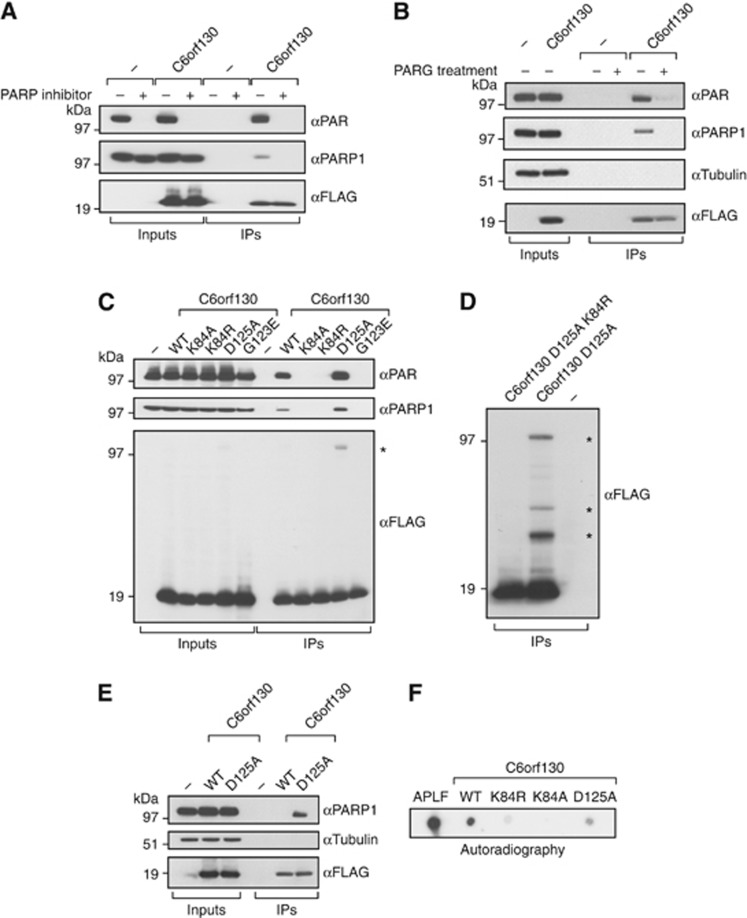
C6orf130 is a PARP1 interacting factor. (A) Interactions of C6orf130 with PARP1 and poly(ADP-ribose) in 293T human cells are diminished by blocking the active poly(ADP-ribose) synthesis using the PARP inhibitor olaparib. (B) Interactions of C6orf130 with PAR and PARP1 are abolished when the complexes are treated with purified, recombinant human PARG protein. (C) Effects of C6orf130 point mutations on its interactions with PARP1 and poly(ADP-ribose). (D) The crosslinking of C6orf130 to proteins is dependent on K84 residue. The crosslinked protein species in C6orf130 Asp125Ala immunoprecipitates in (C, D) are indicated by asterisks. (E) Covalent crosslink between the D125A mutant and PARP1 is retained at high salt concentrations. (F) Binding of C6orf130 and the indicated mutants to free poly(ADP-ribose) in vitro revealed by dot blot assay using radioactively labelled PAR.
C6orf130 interacts with PARP1 and PAR
To examine if C6orf130 associates with proteins involved in PARP-dependent pathways, FLAG-tagged C6orf130 was overexpressed in human 293T cells and its protein complexes were isolated by immunoprecipitation. Although C6orf130 immunoprecipitated endogenous PAR and PARP1, this interaction was abrogated by the pre-incubation of cells with the PARP inhibitor olaparib (Figure 6A). Also, the treatment of the C6orf130 protein complexes with purified human PARG protein diminished the described interactions (Figure 6B). Together, these data suggest that the interaction of C6orf130 with PARP1 largely depends on PAR.
We further analysed the in vivo interactions of C6orf130 mutant proteins containing point mutations in the catalytic residues Lys84 and Asp125, as well as the G123E mutant that is predicted to block the ADP-ribose-binding pocket of C6orf130, as indicated by studies of the conserved residue in other macrodomain proteins (Kustatscher et al, 2005). Analysis revealed that the mutation of Lys84 or Gly123 diminished the interaction between C6orf130 and PARP1/PAR (Figure 6C). In contrast, the mutation of Asp125 to alanine did not negatively affect binding. Furthermore, when the samples were blotted with anti-FLAG antibody to detect the overexpressed C6orf130 protein, additional conjugated polypeptides were observed in the D125A mutant samples (Figures 6C and D, black asterisks), suggesting that this protein is covalently linked to unidentified proteins in the cell extract. Furthermore, when a K84R mutation was introduced in the context of D125A mutation, the crosslinks were abolished (Figure 6D). Notably, when immunoprecipitated samples were subjected to a series of high salt washes prior to elution, the interaction of wild-type C6orf130 but not the D125A mutant with PARP1 was prevented (Figure 6E), suggesting that one of the proteins crosslinked to C6orf130 is PARP1 itself. Altogether, these results mirror the effects observed in the in vitro assay (Figure 2F).
In the X-ray structures, ADP-HPD and ADP-ribose are largely enveloped in the ligand-binding groove. However, the terminal 2′- hydroxyl of the ADP-HPD ribose sugar, and pyrrolidole ring are accessible to solvent, suggesting that C6orf130 also binds ADP-ribose polymers (Figures 4C and D). To test if C6orf130 directly binds to PAR, recombinant C6orf130 variants were analysed by autoradiography using a dot blot assay on nitrocellulose membrane with 32P-labelled-free PAR substrate (Figure 6F). The high affinity PAR-binding protein APLF served as a positive control (Ahel et al, 2008). The assay confirmed that C6orf130 protein directly binds to PAR. In addition, we observed a severely reduced PAR-binding ability for the K84 mutants, supporting the reduced PAR association of these mutants observed in vivo.
C6orf130 is a PAR-regulated DNA damage response factor
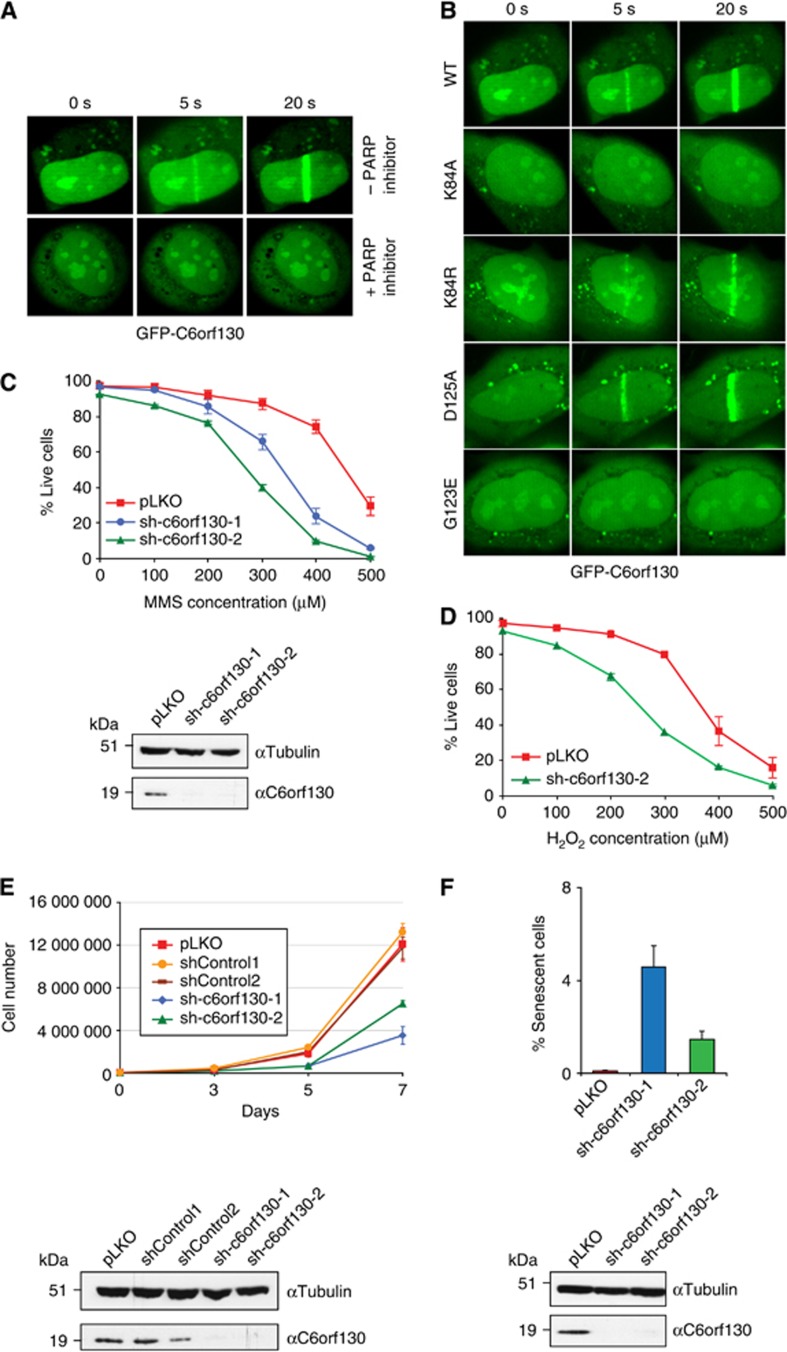
The association of C6orf130 with PARP1 and PAR prompted us to test whether C6orf130 plays a role in the DNA damage response. PARP and PAR-interacting proteins such as XRCC1 and APLF localize to sites of DNA-damaging laser microirradiation (Ahel et al, 2008; Rulten et al, 2008). Similarly, GFP-C6orf130 localizes rapidly to the sites of DNA damage in U2OS cells (Figure 7A). Consistent with a role for C6orf130 in PAR metabolism, this localization was abolished when cells were grown in the presence of PARP inhibitor prior to microirradiation. Also, the recruitment of C6orf130 to the sites of DNA damage was significantly affected by mutating the K84 residue or by blocking the ADP-ribose-binding pocket (G123E mutant) (Figure 7B). We next analysed the importance of C6orf130 in the cellular resistance to the DNA damaging alkylating agent methyl methanosulphonate (MMS) and the oxidizing agent hydrogen peroxide. Short hairpin RNA (shRNA)-induced knockdown of C6orf130 in human 293T cells (estimated 80% efficiency by western blot) (Figure 7C) conferred increased MMS and hydrogen peroxide sensitivity (Figures 7C and D), providing additional evidence for a role of C6orf130 in the DNA damage response.
Figure 7.
C6orf130 depletion affects DNA repair, cell proliferation, and senescence. (A) PARP-dependent localization of GFP-tagged C6orf130 to the sites of laser-induced microirradiation. The recruitment is abolished by pre-treatment of cells using the PARP inhibitor olaparib. (B) Recruitment of GFP-tagged C6orf130 point mutants to the sites of laser-induced damage. (C) Sensitivity of C6orf130 knockdown 293T cells to the DNA-damaging agent, methyl methanosulphonate (MMS) and (D) hydrogen peroxide, and the knockdown efficiencies of C6orf130 shRNAs in analysed cells assessed by western blotting (bottom panel). (E) Reduction in cell proliferation observed under stable C6orf130 knockdown in 293T cells and knockdown efficiencies of C6orf130 shRNAs in 293T cells assessed by western blotting (bottom panel). (F) Induction of senescence in c6orf130 knockdown U2OS cells as judged by β-galactosidase staining and knockdown efficiencies of C6orf130 shRNAs in U2OS cells (bottom panel).
C6orf130 depletion affects cell proliferation and senescence
C6orf130 depletion in 293T cells induced cellular phenotypes reminiscent of cellular senescence including large, flat cell morphologies, and significantly reduced proliferation rates (Figure 7E). Similar effects were observed in the human U2OS cell line. To directly address if C6orf130 depletion increased the percentage of senescent cells, we carried out a senescence assay based on the measurement of cellular β-galactosidase activity. Indeed, the C6orf130 knockdown cell lines had a significantly increased proportion of senescent cells compared to control cells (Figure 7F).
Discussion
Our genetic studies of an extended consanguineous family with several affected cases in two generations revealed that defects in the C6orf130 protein cause a form of neurodegenerative disorder with an autosomal recessive inheritance pattern. The predicted C6orf130 truncation presumably leads to a catalytically inactive protein (Figure 4D). Our biochemical data show that wild-type C6orf130 efficiently removes mono(ADP-ribosyl)ated PARP protein substrates, suggesting that these patients’ cells have a defect in PARP-dependent signalling and/or inefficient recycling of ADP-ribosylated peptides that ultimately leads to cell death and neurodegeneration. The physiological importance of ADP-ribose processing activities is corroborated by a report of elevated levels of glutamyl ribose 5-phosphate in an 8-year-old male who died after 6 years of progressive neurologic deterioration and renal failure (Williams et al, 1984). Increased levels of glutamyl ribose 5-phosphate metabolite in this lysosomal storage disease would be consistent with the deficiency of an enzyme(s) able to cleave the protein modification bond between glutamate and ADP-ribose. Here, we propose a cellular role for C6orf130 protein in the removal of the terminal ADP-ribose unit linked to PARP-modified proteins, by directly reversing protein mono(ADP-ribosyl)ation or by completing the reversal of protein poly(ADP-ribosyl)ation following the PARG reaction. Hence, we suggest renaming this protein Terminal ADP-Ribose protein Glycohydrolase (TARG1). The possibility that the ADP-ribosylated peptides are physiological targets of TARG1 protein is further corroborated by the fact that this protein (and in particular the mutated version Asp125Ala) efficiently crosslinks to a number of proteins in human cell extracts.
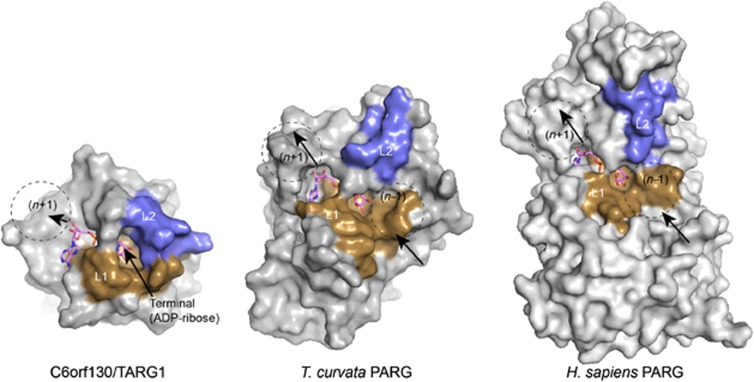
TARG1/C6orf130 catalytic mechanism is very different from any macrodomain proteins described so far, including PARG and MACROD1 (Chen et al, 2011; Slade et al, 2011; Dunstan et al, 2012). Also, the conformation of TARG1 engagement of PAR is such that TARG1 also hydrolyses the ester linkage between glutamate-linked PAR. Consequently, TARG1 is able to release intact PAR chains including the first terminal ADP-ribose unit from proteins. While the activity of TARG1 in removing PAR is low compared with the activity of PARG, it should be noted that TARG1 activity is distinct as it produces free PAR as compared to PARG, which generates principally free mono-ADP-ribose. The differences in PARG and TARG1 substrate specificity are in part accounted for by structural variability in the surface substrate-interaction loops flanking the ADP-ribose binding macrodomain core. The TARG1 L2 loop that bears the active site K84 nucleophile largely envelops the terminal ribose sugar (Figure 8). By contrast, exposed substrate binding surfaces that are anticipated to engage the ribose (n−1) position of a PAR polymer are observed for the topologically equivalent regions of the eubacterial (T. curvata) (Slade et al, 2011) and eukaryotic PARGs (Dunstan et al, 2012; Kim et al, 2012; Tucker et al, 2012) (Figure 8). Differences in the chemistry of the TARG1 catalysed leaving group (glutamate versus ribose (n−1)) also likely contribute to specificity and productive interactions with the TARG1 L1/L2 substrate binding cleft. We hypothesize that TARG1 binding within a PAR polymer, if possible, interacts in a conformation sub-optimal for endoglycosidic cleavage.
Figure 8.
Structural comparisons of TARG1/C6orf130 and PARG enzymes. L1 and L2 substrate binding regions of TARG1 encircle the terminal ADP-ribose. Conversely, exposed surfaces of PARG homologues are well suited for PAR binding and endoglycosidic catalysis.
Protein-free PAR has been identified as a death-signalling molecule that is able to translocate from the nucleus to the mitochondria to trigger apoptosis (Andrabi et al, 2006; Yu et al, 2006). This type of cell death (termed as Parthanatos) has been identified in neuronal cells upon excitotoxicity and in other cell types when exposed to DNA damaging agents. The precise mechanisms through which free PAR is released from the nucleus remains elusive. We hypothesize that C6orf130 may contribute to Parthanatos via its ability to liberate PAR.
It is clear that the ADP-ribosylation metabolism has wide ranging consequences and that the timely modulation of this dynamic post-translational modification is critical. Given the multiplicity of PARPs and the protein/pathways they regulate, it is reasonable to assume that TARG1 might not be the only protein which possesses the ability to remove the mono(ADP-ribosyl)ation synthesized by PARPs. Indeed proteins such as human MACROD1 and MACROD2 were previously shown to possess similar in vitro activities to TARG1 (Chen et al, 2011), suggesting that other cellular enzymes controlling protein mono(ADP-ribosyl)ation might exist in vertebrates.
Here, we provide the first analysis of a macrodomain protein that can fully reverse protein ADP-ribosylation by cleaving the glutamate-linked mono- or poly(ADP-ribose). Further research is required to understand the precise physiological roles of the TARG1/C6orf130 protein and also to uncover and characterize additional proteins modulating protein ADP-ribosylation in living cells.
Materials and methods
Plasmids and proteins
The human c6orf130 gene was amplified from a HeLa cDNA library and cloned into the pDONR221 entry vector (Invitrogen). C6orf130 and MacroH2A1.1 (aa162–369) were also cloned into pETM-CN vector for expression of 6 × His-TEV-V5 tagged proteins. GST-PARP10 catalytic domain (amino acids 818–1025) was a gift from B Lüscher (Kleine et al, 2008). The QuikChange Lightning site-directed mutagenesis kit (Agilent Technologies) was used to introduce specific mutations into the c6orf130 gene. For expression of FLAG-tagged proteins in mammalian cells for immunoprecipitation, the c6orf130 gene was cloned into the p3X-FLAG-CMV-10 vector (Sigma). For immunofluorescence, the pDEST47 vector (Invitrogen) was used for the expression of C-terminally GFP-tagged proteins. For expression and purification of N-terminally His6-tagged C6orf130 proteins in bacteria for biochemical studies, the pDEST17 vector (Invitrogen) was used. The expression of recombinant proteins was induced with 0.1 mM isopropyl-thiogalactopyranoside (IPTG), and the proteins purified by standard protocols using Ni-NTA beads (Qiagen). The purity of the protein preps was investigated by SDS–PAGE (Supplementary Figure 7). For crystallization studies, an N-terminal truncated variant (aa 11–152) was expressed in E. coli Rosetta2 (DE3) cells (Novagen) from pET15b as an N-terminal 6 × His tag protein with a thrombin cleavage site. Cells were grown in Terrific broth, induced with 0.2 mM IPTG, and expressed at 15°C for 20 h. Following Ni-NTA affinity chromatography, the His tag was removed by overnight incubation with thrombin at 4°C. C6orf130 was further purified by size-exclusion chromatography on a Superdex 75 column (GE Healthcare) and by cation-exchange chromatography. Human PARG was cloned and expressed as described in Dunstan et al (2012).
Antibodies
Rabbit anti-C6orf130 antibody was raised against purified full-length recombinant C6orf130 protein (Eurogentec). Rat anti-tubulin and rabbit anti-PARP1 were obtained from Abcam, rabbit anti-PAR from Trevigen, and anti-FLAG-M2 conjugated to peroxidase from Sigma. Unconjugated anti-FLAG-M2 antibody (Sigma) was used for immunoprecipitation.
Cell lines and cell culture
293T and U2OS cells were maintained as adherent monolayers in DMEM media (Sigma) containing 10% Fetal Bovine Serum, 1% Pen/Strep and 1% L-Glutamine and incubated in a humidified 5% CO2 incubator at 37°C. Cell lines were passaged every 3–4 days, using Trypsin-EDTA (Sigma) to detach cells from the flasks.
C6orf130 activity assays
Both poly(ADP-ribose) and mono(ADP-ribose) substrates were obtained by respectively performing a PARP1, PARP10 or PARP1 E988Q mutant automodification assay using [32P]-NAD+ as described previously (Slade et al, 2011). The reactions were stopped after 20 min incubation at room temperature by the addition of 500 nM of PARP inhibitor olaparib.
In the SDP–PAGE based assays, 0.2–2 μM of recombinant C6orf130 proteins was added to the substrate and the reactions incubated at room temperature for 20 min. In the reactions where poly(ADP-ribosyl)ated PARP1 was treated by PARG, 1 μM of human PARG was used for 20 min at room temperature. All reactions were resolved on 4–12% SDS–PAGE gels. ADP-ribosylated proteins were visualized by autoradiography.
In the TLC-based assay, 1 μM of recombinant C6orf130 protein was added to the mono(ADP-ribosyl)ated PARP1 substrate and the reactions incubated at room temperature for 30 min. The reactions were then spotted onto polyethyleneimine (PEI)-cellulose plate (Macherey-Nagel, Polygram CEL 300 PEI/UV254), which was developed in 0.15 M LiCl and 0.15 M formic acid. The dried plate was exposed to X-ray film and the products of the reactions were visualized by autoradiography. Radiolabelled ADP-ribose marker was prepared as described (Slade et al, 2011).
For the TLC and MS analyses using PARP10 substrates, 30 pmol GST-PARP10 catalytic domain was bound to 10 μl glutathione sepharose beads for 30 min at 4°C. The beads were washed 3 × 1 ml with IP buffer (50 mM HEPES pH 7.2, 150 mM KCl, 4 mM MgCl2, 0.2 mM DTT and 0.1% NP-40), and incubated with 0.625 μM [32P]-NAD+ for 15 min at 37°C. To remove unreacted NAD+, beads were washed 5 × 1 ml IP buffer and incubated with 0.5 μM macrodomains in a total volume of 50 μl for 15 min at 37°C. The reactions were separated on silica gel 60 HPTLC plates (Merck) using 1.5 M LiCl with 20% ethanol as eluent and the plates exposed to imaging plates (Fuji Film).
ADP-ribose and PAR-binding assays
For ITC binding assays either MicroCal VP-ITC or iITC200 Instruments (GE Healthcare) were used. Binding reactions were carried out at 25°C in 25 mM Tris, 100 mM NaCl buffer (pH 7.5), using 20–50 μM macrodomains and ADP-ribose (250–500 μM) as a ligand (Sigma). The ligand in the injection syringe was at 10–15 times higher concentration than the macrodomains. Origin software (OriginLab, USA) was used for data analysis. The PAR binding dot blot assay was performed as previously described (Ahel et al, 2008).
C6orf130 crystallization, structure solution, and refinement
Apo crystals were grown at 4°C by sitting drop vapour diffusion by mixing 0.25 μl of 15 mg/ml C6orf130 in protein solution (300 mM NaCl and 20 mM Bis-Tris, pH 7.0) with 0.25 μl well solution (0.1 M sodium citrate, pH 5.6, 20% (v/v isopropanol, 20% (w/v) PEG 4000. Crystals of C6orf130-ADPHPD and ADP-ribose bound complexes were grown at 4°C by hanging-drop vapour diffusion by mixing 1 μl of 30 mg/ml C6orf130 in protein solution (150 mM NaCl and 20 mM Tris, pH 7.5, 2 mM ADPHPD or 2 mM ADP-ribose) with 1 μl well solution (60–100 mM imidazole, pH 8, 15–20% (w/v) PEG 3350, 200 mM NaCl. Crystals were washed in cryoprotectent (well solution supplemented with 26% ethylene glycol) and flash frozen in liquid nitrogen for data collection. Diffraction data were scaled and reduced with HKL2000. The apo C6orf130 structure was phased by molecular replacement (MOLREP) with RCSB PDB 2JYC. The ADP-HPD and ADP-ribose bound complexes were phased using molecular replacement with the refined X-ray structure of Apo-C6orf130. Following autobuilding using PHENIX Autobuild (Terwilliger, 2000; Adams et al, 2010), molecular models were fit manually in Coot (Emsley et al, 2010) and refined in PHENIX. Final structures display excellent geometry (Supplementary Table 5) and refinement statistics.
Immunoblotting
Cells were harvested and lysed in a lysis buffer containing 50 mM Tris (pH 8), 150 mM NaCl, 1 mM DTT, 1% Triton (Sigma), supplemented with 1 × protease inhibitors (Roche) and 2 U of benzonase nuclease (Sigma). Proteins were resolved on 4–12% gradient Bis-Tris gels (Invitrogen) by SDS–PAGE and transferred onto a nitrocellulose membrane (Amersham Biosciences). The membrane was blocked in a PBS-1% TWEEN solution (Sigma) containing 5% dry milk powder. Membranes were then blotted with the appropriate primary and secondary antibodies as indicated, then developed with ECL Western blotting detection reagent (GE Healthcare).
shRNA constructs design
The following shRNA oligonucleotides were cloned into the pLKO.1-puro plasmid (Addgene) by restriction digest with AgeI and EcoRI (NEB). shControl1 (AGAAGAGTTTAGAGGCAAT) targeted no known human genes, while shControl2 (CAACAAGATGAAGAGCACCAA) was directed against the ORF of c6orf130, but produced no major alteration in c6orf130 expression levels when tested by western blot. shC6orf130-1 (GAGAGATGGGCGATATATA) targeted the ORF of c6orf130 while shC6orf130-2 (GGGCAAACCTACCTTAAAA) targeted the 3′ untranslated region. Stable 293T and U2OS cell lines were generated by lentiviral transduction.
Immunoprecipitation
293T cells were transfected with the construct for the expression of FLAG-tagged proteins using Polyfect reagent, according to manufacturer’s instructions. Where indicated, PARP inhibitor (olaparib—AZD 2281) was added 10–12 h prior to harvesting at a final concentration of 0.5 μM. Cells were washed in PBS and solubilized in lysis buffer (as described for immunoblotting) for 1 h at 4 °C. Whole-cell extracts were clarified by centrifugation and, where indicated, treated with 750 nM purified human PARG protein for 20 min at room temperature. Extracts were then incubated with anti-FLAG M2 antibody (Sigma) pre-bound to magnetic protein A beads (Invitrogen). Following extensive washing with lysis buffer, the immunoprecipitates were eluted with 3 × -FLAG peptide (Sigma) and analysed by western Blotting. Where high salt washes were required, 150 mM NaCl was substituted with 1.5 M NaCl.
Live cell imaging and laser microirradiation
U2OS cells were grown in 24-well glass bottom dishes (Iwaki) transfected with c6orf130 GFP constructs using Polyfect reagent (Qiagen) 48 h prior to damage. Cells were sensitized with 10 μM BrdU in phenol red-free DMEM (Sigma) for 16 h at 37°C. Laser microirradiation was carried out on a spinning-disk confocal microscope (Roper) equipped with an environmental chamber (Solent Scientific) where the CO2/air, temperature of the chamber, and objective lens were maintained at 37°C. To ensure that cells with similar expression levels were assayed, cells showing moderate levels of expression were systematically chosen using identical 488-nm laser settings. To correct for overall bleaching of the signal due to repetitive imaging, fluorescence intensities were normalized against intensities measure in an undamaged nucleus in the same field after background subtraction. Equipment control and image capture were handled by Metamorph (Molecular Devices).
DNA-damage sensitivity assays
C6orf130-shRNA and control cells were seeded in triplicate into 6-well plates in media containing the indicated concentrations of MMS (Sigma-Aldrich, UK) or hydrogen peroxide (Sigma-Aldrich). After 4 days, cells and the overlaying media were collected, washed, and stained with the LIVE/DEAD Fixable Far Red Dead Cell Stain Kit (Invitrogen) following the manufacturer’s instructions. Proportions of live and dead cells were quantified by FACS (Callibur Cytometer) and survival curves were generated using FlowJo software. Error bars were calculated using standard deviations from triplicate repeats.
Proliferation assay
C6orf130-shRNA and control cells were seeded in triplicate into 6-well plates (day 3), 6-cm plates (day 5), and 10-cm plates (day 7). Cells were harvested and counted at the indicated times using a Neubauer Cell Counter (Hawksley).
Senescence assays
Cells were seeded in duplicate onto coverslips in 24-well plates. After 24–48 h, cells were fixed and stained using the Senescent Cells Histochemical Staining Kit (Invitrogen), as per manufacturer’s instructions. After staining overnight, the wells were washed with PBS and the coverslips mounted onto microscope slides using ProLong Gold antifade reagent with DAPI (Invitrogen). Cells were observed under a light microscope (Zeiss). Total cells in a field of view were counted using DAPI visualization (405 nm), while β-galactosidase-positive cells were observed as blue-stained cells under white light.
Analysis of ADP-ribosylation sites by mass spectrometry
Mono(ADP-ribosyl)ated PARP1 mutant E988Q, treated with PDE or left untreated, was digested with either Lys-C or trypsin according to the FASP method (Wisniewski et al, 2009). Briefly, the protein mixture was solubilized in 8 M urea and loaded onto Vivacon 500 μl ultrafiltration spin columns with nominal cutoff of 50 kDa (Sartorius Stedim Biotech, Epsom, UK) to remove unreacted NAD+. After removal of urea, the sample was digested overnight at 37°C with trypsin (Promega, Madison, USA) or endoproteinase Lys-C (Wako Biochemicals, Osaka, Japan) in water. In a separate experiment, before the digestion the sample was treated on the filter with phosphodiesterase (PDE; Sigma-Aldrich; buffer 50 mM TRIS pH 8 MgCl2), which cleaves the ADP-ribose and converts it into ribose phosphate (Hengel and Goodlett, 2012).
Mass spectrometric analysis of peptides was performed by nanoflow LC-MS/MS using Q Exactive, a recently introduced quadrupole Orbitrap mass spectrometer capable of fast high-resolution HCD MS/MS spectra. Following separation on a Thermo Scientific Acclaim PepMap RSLC C18 capillary column (50 μm i.d. × 15 cm, 2 μm) using an EASY-nLC 1000 HPLC system (Thermo Scientific, Odense, Denmark), peptides were directly injected into the mass spectrometer via a Thermo Scientific nanoelectrospray interface. Data were acquired in the data-dependent mode at a resolution of 70 000 at m/z 200 for survey scans and 17 500 at m/z 200 for MS/MS spectra.
Raw data were analysed with the MaxQuant proteomics framework (version 1.3.0.5) (Cox and Mann, 2008). The search was performed against a database containing the sequence of PARP1 mutant as described. Enzyme specificity was set to either trypsin or Lys-C allowing for up to four missed cleavages. Methionine oxidation, protein N-acetylation, ADP-ribosylation (mass shift of 541.0611) or phosphate ribosylation (sample treated with PDE; mass shift of 212.0086) on glutamate, aspartate and lysine were set as variable modifications. MaxQuant was set up to automatically search for the following diagnostic ions in MS/MS spectra matched to ADP-ribosylated peptides: adenine (mass 135.0545), adenosine –H2O (mass 249.0862), adenosine monophosphate (AMP; mass 347.0631), adenosine diphosphate (ADP). All reported fragmentation spectra were manually validated using stringent criteria (Matic et al, 2012).
Analysis of C6orf130-ADP-ribose crosslinks
Approximately 100 pmol of the C6orf130 (wild type or mutants with or without ADP-ribose) was desalted using C18 ZipTips (Millipore) using essentially the manufacturer’s recommendations. The protein samples were eluted in 10 μl using 30:70 (v:v) of 0.1% formic acid:0.1% formic acid in acetonitrile. MS spectra were acquired on a Micromass Ultima hybrid quadrupole-ToF mass spectrometer that had been calibrated using the multiply-charged envelope of horse heart cytochrome C. Samples were individually introduced at ∼500 nl/min via flow injection from a pressurized bomb. Instrument settings included a capillary voltage of 3.8 kV, a cone voltage of 40 V, cone gas flow of 50 l/h, and RF1 lense setting of 140 V. To obtain the mass of the protein, the multiply-charged envelope was then deconvoluted using the MaxEnt1 algorithm of the MassLynx software package.
For peptide analysis, 10 pmol of wild-type C6orf130 were digested for 16 h at 37°C with trypsin (Promega) and spotted (0.3 μl) onto a stainless steel target. A 33% saturated solution of cyano-hydroxycinnamic acid in 50:50 (v:v) 0.1% formic acid:0.1% formic acid in acetonitrile was added (0.3 μl) to the peptide digest and mixed on target. MALDI-ToF and MALDI-ToF/ToF experiments were performed on an Applied Biosystems 4800 Plus MALDI TOF/TOF Analyzer in either positive ion reflector or negative ion reflector modes. The MS was internally calibrated using autolytic tryptic peptides and the MS/MS calibrated externally using the fragment ions of the angiotensis I M+H ion (m/z 1298.68). A focus mass of m/z 2000 was used for the MS acquisitions. For MS/MS 1000, kV was used for the collision energy.
Single-nucleotide polymorphism chip arrays and linkage analysis
This research has been approved by the local Ethical Research Review Board (Ahwaz Shahid Chamran University) and all participants were consented according to approved protocols. EDTA blood samples were taken from all consenting participants from the family and genomic DNA was extracted according to the FlexiGene DNA kit handbook (Qiagen, UK). Single-nucleotide polymorphism (SNP) analysis was performed for six of the affected patients and samples were run on the Illumina HumanCytoSNP-12 (Illumina v2.1 array), Bead Station 500X version (CA, USA) containing 301 232 markers. The statistical package EasyLinkage Plus v.5.08 (Hoffmann and Lindner, 2005) designed to perform automated linkage analyses using large-scale SNP data was used for all analyses. All SNPs showing inconsistency in transmission were removed from further analyses. Simwalk 2.91 software (Sobel and Lange, 1996) was used to perform haplotype and parametric linkage analysis. LOD scores were obtained using a recessive model of inheritance, with full penetrance and a disease allele frequency of 1:10 000. Allele frequencies of genotyped SNPs were set to codominant. Map order and genetic inter-SNPs distances were taken from the Illumina website. Since closely spaced SNP markers were used for the linkage analysis, the genome analyses were performed with a predefined spacing of 0.2 cM, in blocks of 125 SNPs.
Direct sequencing
Primers were designed using Primer 3 software. PCR was performed using 5 pmol of primer (Sigma-Aldrich) and purified following the ExoSAP protocol for sequencing reactions. Sequencing was performed using the BigDye Terminator Chemistry (Sequenase v3.1 Cycle Sequencing Kit; Applied Biosystems, CA, USA). A 3130xl automated DNA sequencer (Applied Biosystems) was used to run the samples. Sequencing results were obtained using the CLC Bio Sequence Viewer 6.5.2 (CLC Bio, Denmark) and Finch TV (GeoSpiza, Seattle, WA). Wild-type sequencing was established from the Ensembl Genome Browser (Wellcome Trust Sanger Institute, Cambridge, UK).
Exome sequencing
Exome sequencing was undertaken in an affected patient using the SureSelect All Exon 50 Mb Target Enrichment System (Agilent) (Coffey et al, 2011) and sequenced on a Genome Analyser IIx (Illumina) with 76 bp paired end reads. Sequence reads were aligned to the reference genome (hg19) with Novoalign (Novocraft Technologies Sdn Bhd). Duplicate reads, resulting from PCR clonality or optical duplicates, and reads mapping to multiple locations were excluded from downstream analysis. Depth and breadth of sequence coverage was calculated with custom scripts and the BedTools package (Quinlan and Hall, 2010). Single-nucleotide substitutions and small insertion deletions were identified and quality filtered within the SamTools software package and in-house software tools. Variants were annotated with respect to genes and transcripts with the Variant Classifier tool (Li and Stockwell, 2010). Filtering of variants for novelty was performed by comparison to dbSNP132 and 1000 Genomes SNP calls (December 2010) and variants identified in 150 control exomes sequenced and analysed by the same method described above.
Coordinates for the TARG1/C6orf130 apo X-ray structure (4J5Q), ADP-HPD complex (4J5R), and ADP-ribose complex (4J5S) have been deposited in the RCSB protein data bank.
Supplementary Material
Acknowledgments
RSW is supported by the intramural research program of the US National Institutes of Health (NIH), National Institute of Environmental Health Sciences (NIEHS) grant 1Z01ES102765. We thank L Pedersen of the NIEHS collaborative crystallography group, and the Advanced Photon Source (APS) Southeast Regional Collaborative Access Team (SER-CAT) staff for assistance with crystallographic data collection. IA is supported by Cancer Research UK and the European Research Council. IM is a Sir Henry Wellcome Postdoctoral Fellow. AGL is supported by CIPSM, SyNergy, HFSP, the LMU and the EU ITN network ‘Nucleosome4D’, which partly funded the PhD research of GJ. GJ is a member of the International Max Planck Research School for Molecular and Cellular Life Sciences (IMPRS). GT is supported by the LMU. RS is supported by the grants awarded from MRC, UK, and SGUL. Research at KCL was supported by the UK Department of Health via the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre award to Guy’s and St Thomas’ National Health Service (NHS) Foundation Trust in partnership with King’s College London and King’s College Hospital NHS Foundation Trust. We thank the patients and families participated in this study. We also thank the Blood Transfusion Organisation-IRAN-Khouzestan.
Author Contributions: AGL and RS initiated the basic science-clinical collaboration. RM and GJ performed biochemical experiments; CDA, MJS, JK, and RSW performed crystallization and structural studies; RS performed patients clinical evaluation; MAS and RCT performed exome sequencing experiments; BC, EO, HG, AW, AHC, and RS performed the genetic studies; IM, RH, and JGW performed mass spectrometry analyses; MT, BG, DA, RW, and MNR performed cell biology experiments, GT, RSW, CDA, RS, and IA wrote the manuscript, designed experiments, and analysed data.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahel D, Horejsí Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, Owen-Hughes T, Boulton SJ (2009) Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science 325: 1240–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, West SC (2008) Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature 451: 81–85 [DOI] [PubMed] [Google Scholar]
- Andrabi SA, Kim NS, Yu SW, Wang H, Koh DW, Sasaki M, Klaus JA, Otsuka T, Zhang Z, Koehler RC, Hurn PD, Poirier GG, Dawson VL, Dawson TM (2006) Poly(ADP-ribose) (PAR) polymer is a death signal. Proc Natl Acad Sci USA 103: 18308–18313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthold CL, Wang H, Nordlund S, Hogbom M (2009) Mechanism of ADP-ribosylation removal revealed by the structure and ligand complexes of the dimanganese mono-ADP-ribosylhydrolase DraG. Proc Natl Acad USA 106: 14247–14252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruner SD, Norman DP, Verdine GL (2000) Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature 403: 859–866 [DOI] [PubMed] [Google Scholar]
- Cervantes-Laurean D, Minter DE, Jacobson EL, Jacobson MK (1993) Protein glycation by ADP-ribose: studies of model conjugates. Biochemistry 32: 1528–1534 [DOI] [PubMed] [Google Scholar]
- Chen D, Vollmar M, Rossi MN, Phillips C, Kraehenbuehl R, Slade D, Mehrotra PV, von Delft F, Crosthwaite SK, Gileadi O, Denu JM, Ahel I (2011) Identification of macrodomain proteins as novel O-acetyl-ADP-ribose deacetylases. J Biol Chem 286: 13261–13271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey AJ, Kokocinski F, Calafato MS, Scott CE, Palta P, Drury E, Joyce CJ, Leproust EM, Harrow J, Hunt S, Lehesjoki AE, Turner DJ, Hubbard TJ, Palotie A (2011) The GENCODE exome: sequencing the complete human exome. Eur J Hum Genet 19: 827–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- D'Amours D, Desnoyers S, D'Silva I, Poirier GG (1999) Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J 342(Pt 2): 249–268 [PMC free article] [PubMed] [Google Scholar]
- Dunstan MS, Barkauskaite E, Lafite P, Knezevic CE, Brassington A, Ahel M, Hergenrother PJ, Leys D, Ahel I (2012) Structure and mechanism of a canonical poly(ADP-ribose) glycohydrolase. Nat Commun 3: 878. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson BA, Kraus WL (2012) New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol 13: 411–424 [DOI] [PubMed] [Google Scholar]
- Hassa PO, Haenni SS, Elser M, Hottiger MO (2006) Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol Mol Biol Rev 70: 789–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengel SM, Goodlett DR (2012) A review of tandem mass spectrometry characterization of adenosine diphosphate-ribosylated peptides. Int J Mass Spectrom 312: 114–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann K, Lindner TH (2005) easyLINKAGE-Plus--automated linkage analyses using large-scale SNP data. Bioinformatics 21: 3565–3567 [DOI] [PubMed] [Google Scholar]
- Karras GI, Kustatscher G, Buhecha HR, Allen MD, Pugieux C, Sait F, Bycroft M, Ladurner AG (2005) The macro domain is an ADP-ribose binding module. EMBO J 24: 1911–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasamatsu A, Nakao M, Smith BC, Comstock LR, Ono T, Kato J, Denu JM, Moss J (2011) Hydrolysis of O-acetyl-ADP-ribose isomers by ADP-ribosylhydrolase 3. J Biol Chem 286: 21110–21117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IK, Kiefer JR, Ho CM, Stegeman RA, Classen S, Tainer JA, Ellenberger T (2012) Structure of mammalian poly(ADP-ribose) glycohydrolase reveals a flexible tyrosine clasp as a substrate-binding element. Nat Struct Mol Biol 19: 653–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine H, Poreba E, Lesniewicz K, Hassa PO, Hottiger MO, Litchfield DW, Shilton BH, Lüscher B (2008) Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol Cell 32: 57–69 [DOI] [PubMed] [Google Scholar]
- Kustatscher G, Hothorn M, Pugieux C, Scheffzek K, Ladurner AG (2005) Splicing regulates NAD metabolite binding to histone macroH2A. Nat Struct Mol Biol 12: 624–625 [DOI] [PubMed] [Google Scholar]
- Li K, Stockwell TB (2010) VariantClassifier: a hierarchical variant classifier for annotated genomes. BMC Res Notes 3: 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Ame JC, Aboul-Ela N, Jacobson EL, Jacobson MK (1997) Isolation and characterization of the cDNA encoding bovine poly(ADP-ribose) glycohydrolase. J Biol Chem 272: 11895–11901 [DOI] [PubMed] [Google Scholar]
- Marsischky GT, Wilson BA, Collier RJ (1995) Role of glutamic acid 988 of human poly-ADP-ribose polymerase in polymer formation. Evidence for active site similarities to the ADP-ribosylating toxins. J Biol Chem 270: 3247–3254 [DOI] [PubMed] [Google Scholar]
- Matic I, Ahel I, Hay RT (2012) Reanalysis of phosphoproteomics data uncovers ADP-ribosylation sites. Nat Methods 9: 771–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka J, Ueda K, Hayaishi O, Komura H, Nakanishi K (1984) ADP-ribosyl protein lyase. Purification, properties, and identification of the product. J Biol Chem 259: 986–995 [PubMed] [Google Scholar]
- Peterson FC, Chen D, Lytle BL, Rossi MN, Ahel I, Denu JM, Volkman BF (2011) Orphan macrodomain protein (human C6orf130) is an O-acyl-ADP-ribose deacylase: solution structure and catalytic properties. J Biol Chem 286: 35955–35965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulten SL, Cortes-Ledesma F, Guo L, Iles NJ, Caldecott KW (2008) APLF (C2orf13) is a novel component of poly(ADP-ribose) signaling in mammalian cells. Mol Cell Biol 28: 4620–4628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauve AA, Celic I, Avalos J, Deng H, Boeke JD, Schramm VL (2001) Chemistry of gene silencing: the mechanism of NAD+-dependent deacetylation reactions. Biochemistry 40: 15456–15463 [DOI] [PubMed] [Google Scholar]
- Slade D, Dunstan MS, Barkauskaite E, Weston R, Lafite P, Dixon N, Ahel M, Leys D, Ahel I (2011) The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature 477: 616–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel E, Lange K (1996) Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet 58: 1323–1337 [PMC free article] [PubMed] [Google Scholar]
- Tao Z, Gao P, Liu HW (2009) Identification of the ADP-ribosylation sites in the PARP-1 automodification domain: analysis and implications. J Am Chem Soc 131: 14258–14260 [DOI] [PubMed] [Google Scholar]
- Terwilliger TC (2000) Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr 56: 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till S, Ladurner AG (2009) Sensing NAD metabolites through macro domains. Front Biosci 14: 3246–3258 [DOI] [PubMed] [Google Scholar]
- Timinszky G, Till S, Hassa PO, Hothorn M, Kustatscher G, Nijmeijer B, Colombelli J, Altmeyer M, Stelzer EH, Scheffzek K, Hottiger MO, Ladurner AG (2009) A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat Struct Mol Biol 16: 923–929 [DOI] [PubMed] [Google Scholar]
- Tucker JA, Bennett N, Brassington C, Durant ST, Hassall G, Holdgate G, McAlister M, Nissink JW, Truman C, Watson M (2012) Structures of the human poly (ADP-Ribose) glycohydrolase catalytic domain confirm catalytic mechanism and explain inhibition by ADP-HPD derivatives. PLoS ONE 7: e50889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JC, Chambers JP, Liehr JG (1984) Glutamyl ribose 5-phosphate storage disease. A hereditary defect in the degradation of poly(ADP-ribosylated) proteins. J Biol Chem 259: 1037–1042 [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Nagaraj N, Mann M (2009) Universal sample preparation method for proteome analysis. Nat Methods 6: 359–362 [DOI] [PubMed] [Google Scholar]
- Yu SW, Andrabi SA, Wang H, Kim NS, Poirier GG, Dawson TM, Dawson VL (2006) Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proc Natl Acad Sci USA 103: 18314–18319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaja R, Mikoc A, Barkauskaite E, Ahel I (2013) Molecular insights into poly(ADP-ribose) recognition and processing. Biomolecules 3: 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.